Abstract
Organoids, or miniaturized organs formed in vitro, hold potential to revolutionize how researchers approach and answer fundamental biological and pathological questions. In the context of cardiac biology, development of a bona fide cardiac organoid enables study of heart development, function, and pathogenesis in a dish, providing insight into the nature of congenital heart disease and offering the opportunity for high-throughput probing of adult heart disease and drug discovery. Recently, multiple groups have reported novel methods for generating in vitro models of the heart; however, there are substantial conceptual and methodological differences. In this review we will evaluate recent cardiac organoid studies through the lens of the core principles of organoid technology: patterned self-organization of multiple cell types resembling the in vivo organ. Based on this, we will classify systems into the following related types of tissues: developmental cardiac organoids, chamber cardiac organoids, microtissues, and engineered heart tissues. Furthermore, we highlight the interventions which allow for organoid formation, such as modulation of highly conserved cardiogenic signaling pathways mediated by developmental morphogens. We expect that consolidation and categorization of existing organoid models will help eliminate confusion in the field and facilitate progress towards creation of an ideal cardiac organoid.
Keywords: Cardiac organoid, Engineered heart tissue, Self-organization, Gastruloid
Introduction:
Over the past decade, organoid generation has emerged as a new discipline in the field of developmental biology and regenerative medicine [1–3]. Organoids, or in vitro 3D miniaturized organ-like structures, have been broadly defined as being composed of multiple self-organizing cell types, patterned similarly to their in vivo equivalent, and undergoing in vivo-like developmental trajectories [1, 2, 4], though there has been debate over minor nuances [5]. Organoids of multiple organ systems have been previously described, including brain [4, 6–10], intestine [11–15], and optic cup [16], each with varying levels of resemblance to in vivo development. Such systems and their applications represent the potential for organoids to answer crucial biological questions related to human health. While multitudes of organoids in a variety of systems have been developed, progress in the field of cardiac organoids conspicuously lagged, in part due to the complexity of early heart morphogenesis. Recently, a flurry of preprints and publications detailing cardiac organoids have generated hope and excitement in the field, although each system looks highly dissimilar and undergoes vastly different developmental processes. Such dissonance has exposed a fundamental need for understanding a) what exactly is a cardiac organoid and b) why does making such a distinction matter?
Heart development can conceptually be split into two broad stages – early cardiogenesis, during which cells of the embryonic heart are specified, differentiated, and organized into a complex structure which supplies the embryo [17–24] (Fig 1A–D), and maturation, during which the heart undergoes adaptational changes required for adult function [25–30] (Fig 1E). In vitro models of the heart have largely sought to recapitulate one of these two processes. For example, developmental biologists have aimed to construct systems that recapitulate key developmental milestones and embryonic structures of the heart. Likewise, various tissue engineering approaches have been used to model the adult cardiac state, though the extent to which these approaches are biomimetic is somewhat unknown. In this review, we will attempt to group existing in vitro models of the heart into four categories: developmental cardiac organoids, chamber cardiac organoids, microtissues, and engineered heart tissues. We will outline the methods that are utilized for generation of each system to consolidate the modulations of biological signaling pathways and tissue engineering that are necessary for optimal formation (Table 1). We will then evaluate the potential for each organoid model to add to our understanding of heart development and function – based on how these in vitro models are similar to in vivo, and, perhaps equally as important, how they are unable to recapitulate in vivo development. Finally, we will project how such technologies may evolve in the future. By exploring these differences, we hope to outline important properties that can guide the development of the next generation of cardiac organoid and tissue models.
Figure 1:
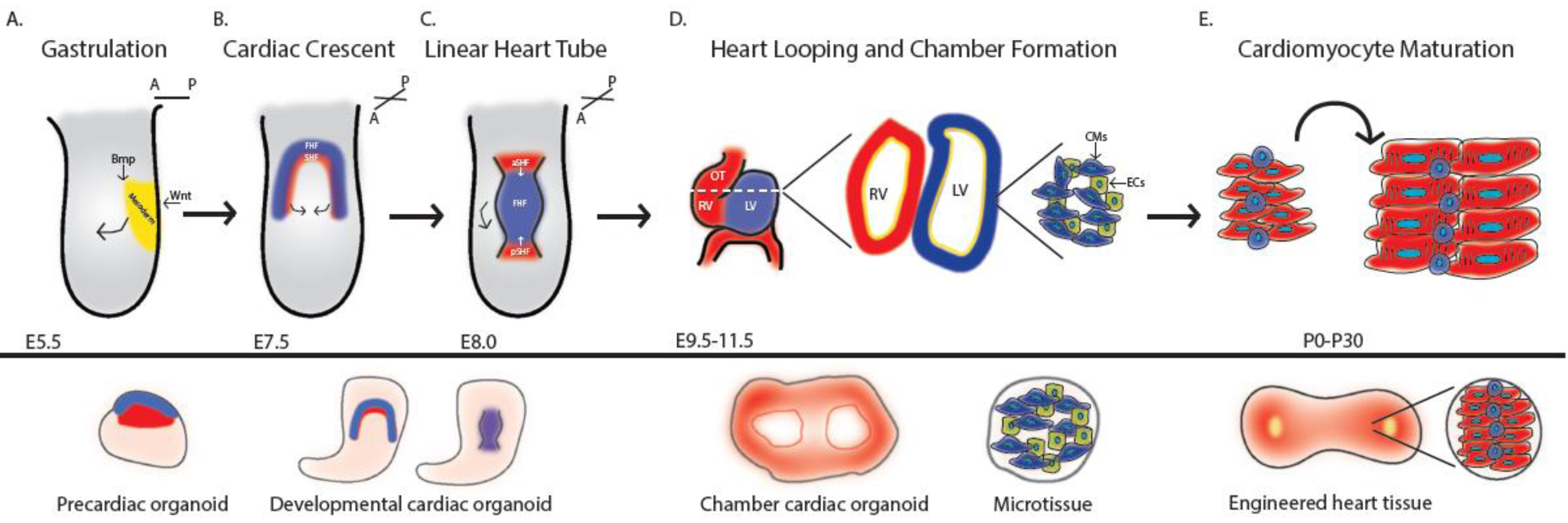
Staging in vitro models of the heart to in vivo heart development. A. Embryo undergoing gastrulation. Precardiac organoid models attempt to recapitulate this stage of development. B.-C. Cardiac crescent formation and merging to form the linear heart tube. Developmental cardiac organoids model this stage of development. D. Embryo undergoing rightward looping and chamber formation. Chamber cardiac organoids recapitulate this stage of development and microtissues offer insight into the cell type-cell type interactions between cardiomyocytes and stromal cells. E. Maturation of cardiomyocytes to promote adult heart function. Engineered heart tissues attempt to promote a more adult and in vivo-like state of cardiac tissues.
Table 1:
Summary of methods for generation of precardiac and cardiac organoids
| Study | Organoid type | Method of organoid generation | Culture conditions | Extent of morphogenesis | Uses | Limitations | Species | Reference |
|---|---|---|---|---|---|---|---|---|
| Domian | Precardiac | Spontaneous (Hanging Drop) | Suspension | Not studied | Isolation of ventricular progenitors | No patterning or structure formation | Mouse | 93 |
| Andersen | Precardiac | Bmp and Activin A (D2-D4) | Suspension | FHF forms exteriorly to SHF (verified by heart field fluorescent reporters) | Study of heart field specification and development | Limited patterning and structure formation | Mouse | 23 |
| Silva | Develop mental | Wnt activation (D0-D1), Wnt inactivation (D3-D5), Ascorbic Acid (D7-end) | Monolayer and reaggregation in pyramidal agarose wells. Then, maintenance in rotary suspension culture. | Cardiac core, gut-like tube, epicardial lining | Study of co-development of mesoderm and endoderm, enhanced maturation of CMs | Limited patterning and in vivo-like structure formation | Human | 100 |
| Rossi | Develop mental | Wnt activation (D2-D3), bFgf (D4-D6), Vegf (D4-D6), Ascorbic Acid (D4-D6) | Individual gastruloids formed and maintained in ultra-low 96 U-well plates. Transferred to 24-well ultra-low on rotary shaker. | Cardiac crescent-like, Linear heart tube-like, gut-like tube, endocardial-like structures | Study of early heart development in the context of the early embryo | Does not persist past linear heart tube stage | Mouse | 103 |
| Drakhlis | Develop mental | Wnt activation (D0-D1), Wnt inactivation (D3-D5), embed in Matrigel | Aggregates preformed in ultra-low 96 U-well plates. Embedded in Matrigel on D0 | Layered 3D structure with 3 layers of endodermal-like, endocardial-like, and cardiomyocyte cells | Study of co-development of foregut endoderm and heart lineages | Limited morphology and in vivo-like structure formation. Limited contribution of FHF lineage | Human | 104 |
| Israeli | Chamber | Wnt activation (D0-D1), Bmp (D0-D1), Activin A (D0-D1), Wnt inactivation (D2-D4), Wnt activation (D7 to D7+1hr) | Ultra-low 96 U-well plates | Microcham ber formation | Study of human heart development with enhance d in vivo-like transcriptomes | Chambers are small and variable | Human | 105 |
| Hofbauer | Chamber | Fgf2 (D0-D7.5), Bmp (D0-D7.5), Activin A (D0-D1.5), Wnt activation (D0-D1.5), Wnt inactivation (D1.5-D5.5), Retinoic Acid (D1.5-D5.5) | Non-adherent 3D culture | Chamber formation | Modeling congenital heart defects and studying early chamber formation | Unclear to the extent which organoids develop and mature | Human | 106 |
| Lee | Chamber | ECM, Fgf4(D4-end), Bmp (D9-end), Wnt activation (BIO)(D9-end), LIF (D9-end) | Form EBs in suspension, transfer individual EBS to LN/ET culture wells | Four chamber-like structure | Modeling congenital heart defects and studying early chamber formation | Further characterization of structures and identity | Mouse | 107 |
Precardiac organoids exhibit primitive patterning and lack morphogenesis:
Organoids in multiple systems have been utilized to study the dynamics and mechanisms underlying their respective organ’s development. The study of development, especially in humans, is inherently complicated by the limited access to and control of embryonic tissue, in addition to a myriad of ethical concerns. Thus, it is preferable that a developmental organoid not only resembles the organ in structure, composition, and spatial organization, but undergoes a similar developmental trajectory. Specifically, in the context of heart development, it is not sufficient for the organoid to merely form a structure resembling the early heart, but it should also recapitulate similar developmental steps. Through this, the organoid will allow for interrogation of the processes governing heart development. The potential for such research in cardiac organoid systems will be pivotal for developing treatments and tools for diagnosing congenital heart disease. To appropriately evaluate each organoid, it is crucial to compare their methods of induction and structures formed to the developmental trajectories and structures of the in vivo heart.
Long before the first heart structure is observed during development, the embryo has already been meticulously coordinating the differentiation and migration of cells that will eventually make up the heart. Importantly, during the period of early development, the embryo is patterned along multiple axes, instructing the embryo on how to proceed with development [31–34]. After mesoderm is formed during gastrulation [35] (Fig 1A), cardiac progenitor cells undergo a complex morphogenetic process of specification, differentiation, and migration to eventually form an embryonic, four chambered heart. Briefly, the heart is formed from two heart fields [19] (Fig 1B), and dysregulation of heart field development can lead to a variety of congenital heart defects [36]. The first heart field (FHF) forms the left ventricle (LV) and parts of the atria while the second heart field (SHF) forms the right ventricle (RV), outflow tract (OT) and parts of the atria [18]. The FHF and SHF are specified near the onset of gastrulation [24, 37–39] and are regulated by multiple signaling pathways such as Wnt [22, 40–48], Bmp [22, 49–51], Notch [52–58] and Numb [59–63]. The first observable cardiac structure is composed of FHF cells and is known as the cardiac crescent (Fig 1B), which is transiently present before merging to become a linear heart tube [64] (Fig 1C). SHF-derived cells continue to add to the arterial and venous poles of the heart tube [65–67] (Fig 1C), initiating rightward looping [68] and chamber formation [69–75] (Fig 1D). In addition to the cardiac muscle cells that make up the myocardium, the heart is encapsulated by a layer of epicardial cells [76–84], supplied by networks of vasculature [85], and supported by endocardial cells [86–88], lineages that should be included in the ideal cardiac organoid.
To study these early developmental processes, the field of cardiac organoids naturally began as an extension of the widely used embryoid body (EB) 3D differentiation cultures. Multiple studies differentiated cardiomyocytes (CMs) in 3D EB culture; however, such studies were not focused on investigating self-organization or patterning of EBs, but rather focused primarily on directed differentiation of CMs [89–92]. In an important advance in cardiac EB differentiations, Domian et al., developed a method to visualize dynamics of differentiation within an EB through creation of a fluorescent reporter EB marking each heart field [93] (Table 1). However, similar to other approaches, the study was focused on isolation of ventricular CMs, and not dynamics of the EB system itself. Furthermore, this study utilized the then popular, hanging drop method of differentiation, in which cells of interest are generated spontaneously in aggregated EBs [94]. Thus the level of organization or patterning in these reporter EBs is unclear [93].
Building on this work, we utilized an updated differentiation protocol utilizing Bmp and Activin A (activating pathways necessary for mesodermal and cardiac differentiation[95–96]), to develop an EB system using fluorescent reporters of the FHF and SHF (Hcn4 and Tbx1, respectively) [23, 97] (Table 1). Interestingly, in agreement with in vivo development, each heart field was formed from spatially distinct regions, composed of multiple cell types present in the early developing heart, and exhibited in vivo-like FHF and SHF transcriptomes. However, there was a notable lack of polarity (in vivo, the FHF forms anterior to the SHF) and morphogenesis (no cardiac crescent or linear heart tube-like structures formed), suggesting that the Bmp/Activin A differentiation protocol in spheroids may not be sufficient for early patterning of the body axis and initiating cardiac morphogenesis. We therefore coined this as a precardiac organoid system, as it was able to self-organize and pattern development prior to the cardiac crescent stage, but not beyond.
Both Domian et al. [93] and Andersen et al. [23] utilize fluorescent reporters to visualize the FHF and SHF, however with varying differentiation protocols. As previously mentioned, there was limited investigation into the developmental aspects of EB differentiation in Domian et al.; however, in the available fluorescent images, there appeared to be minimal organization and patterning. In Andersen et al., there was clear segregation of the FHF and SHF regions during differentiation, though not further morphogenesis. The probable explanation for this discrepancy is the method for differentiation, where Domian et al. utilized spontaneous differentiation of hanging drops (extremely low efficiency) and Andersen et al. utilized BMP/Activin A cardiac directed differentiation (high mesodermal/cardiac differentiation efficiency). Taken together, these results suggest exogenous Bmp and Activin A are needed to induce sufficient cardiac mesoderm to initiate primitive heart field self-organization programs in the EB. However, further organization and morphogenesis would require additional signaling modulation and intervention.
Developmental cardiac organoids utilize signals from non-cardiac cell types and exogenous cardiogenic factors to promote in vivo-like development:
A critical shift that has aided the development of cardiac organoid models is the use of the small molecule Chir99021, a Gsk3β inhibitor that activates Wnt signaling. Wnt signaling is necessary to form mesoderm, and has been frequently used to induce cardiogenesis in stem cell systems [98]. Furthermore, a burst of Chir-induced Wnt signaling has been shown to be sufficient to induce symmetry breaking and self-organization [99], pointing to a pivotal role of Wnt signaling in inducing germ-layer specification. In this section, we will highlight two recent studies which utilize Chir to initiate developmental programs. These two studies are unique, in that both emphasize the importance of coordinated co-development with other non-cardiac cell types for organoid generation, leading us to categorize these models as developmental cardiac organoids.
In a recent study using hiPSCs, Silva et al. reported that with the addition of the cardiogenic factor ascorbic acid, Chir treatment promotes the formation of a multi-lineage cardiac organoid that consists of cardiac and endoderm domains [100] (Table 1). The domains contain a cardiac core and gut-like region composed of Paneth and Goblet-like cells that exhibit peristalsis-like contraction. A Tbx18+ epicardial-like layer forms in these organoids; however, this layer surrounds the entirety of the organoid, and not just the cardiac region, as it does in vivo. The degree of morphogenesis in these organoids is thus difficult to assess. While there are cardiac and endodermal regions present, the cardiac region does not resemble its in vivo counterpart. Instead, it forms a cardiac core, which is dissimilar to the cardiac crescent (present in early development), linear heart tube/looping heart tube, and the chambered heart. This could be a result of an early dissociation step in their organoid generation protocol, where cells are pre-differentiated in 2D culture before being dissociated and reaggregated in 3D culture on differentiation day 5. Nonetheless, the ability of these organoids to reorganize after differentiation is biologically interesting. Finally, the authors suggest that cardiac co-development with endoderm enhances the maturation of the CMs through various functional metrics. Further comparison to in vivo CMs, which can be done with single cell RNA sequencing or functional analysis would be useful for more precise staging [101].
Contrasting with the dissociation step, which affects spatial information and cell-cell interactions in Silva et al., the gastruloid system allows for preserved cell type-cell type interactions throughout in vitro development. Gastruloids leverage the innate ability of stem cells to organize and undergo in vivo-like development to form unprecedented, multiaxial embryo-like structures of cell types from multiple germ layers [99, 102]. Thus, utilizing gastruloids to study heart development would offer the ability to answer questions of how cross-talk between cell types are important in development in an unprecedented way. A recent study sought to apply the principles of gastruloids to create a developmental cardiac organoid. Rossi et al. aimed to utilize the cell-cell interactions and signaling of the early embryo to create cardiac structures that underwent in vivo-like developmental processes [103]. A burst of Wnt signaling from differentiation day 2–3, orbital shaking, and the addition of cardiogenic factors ascorbic acid, bFgf, and Vegf on differentiation day 4 (Fig 2), was able to enhance the cardiogenic capability of gastruloids. This resulted in a developmental cardiac organoid that was patterned along multiple axes and consisted of cell types from multiple germ layers (Table 1). Furthermore, these organoids reproducibly formed structures that resembled the early heart – the cardiac crescent and linear heart tube. Importantly, correct spatial and temporal development of multiple events was shown to be conserved in this in vitro system, such as vascularization, FHF and SHF development, and endocardial and gut tube-like structure formation. Thus, these developmental cardiac organoids are faithful to early development – recapitulating aspects of self-organization, patterning, and structure formation of heart development. One limitation of this system is that these cardiac organoids do not persist long after the linear heart tube stage, limiting their use for study of later-occurring heart morphogenic defects. Nonetheless, to date, this gastruloid system presents the most faithful recapitulation of early heart development, at structural, functional, and transcriptomic levels.
Figure 2:
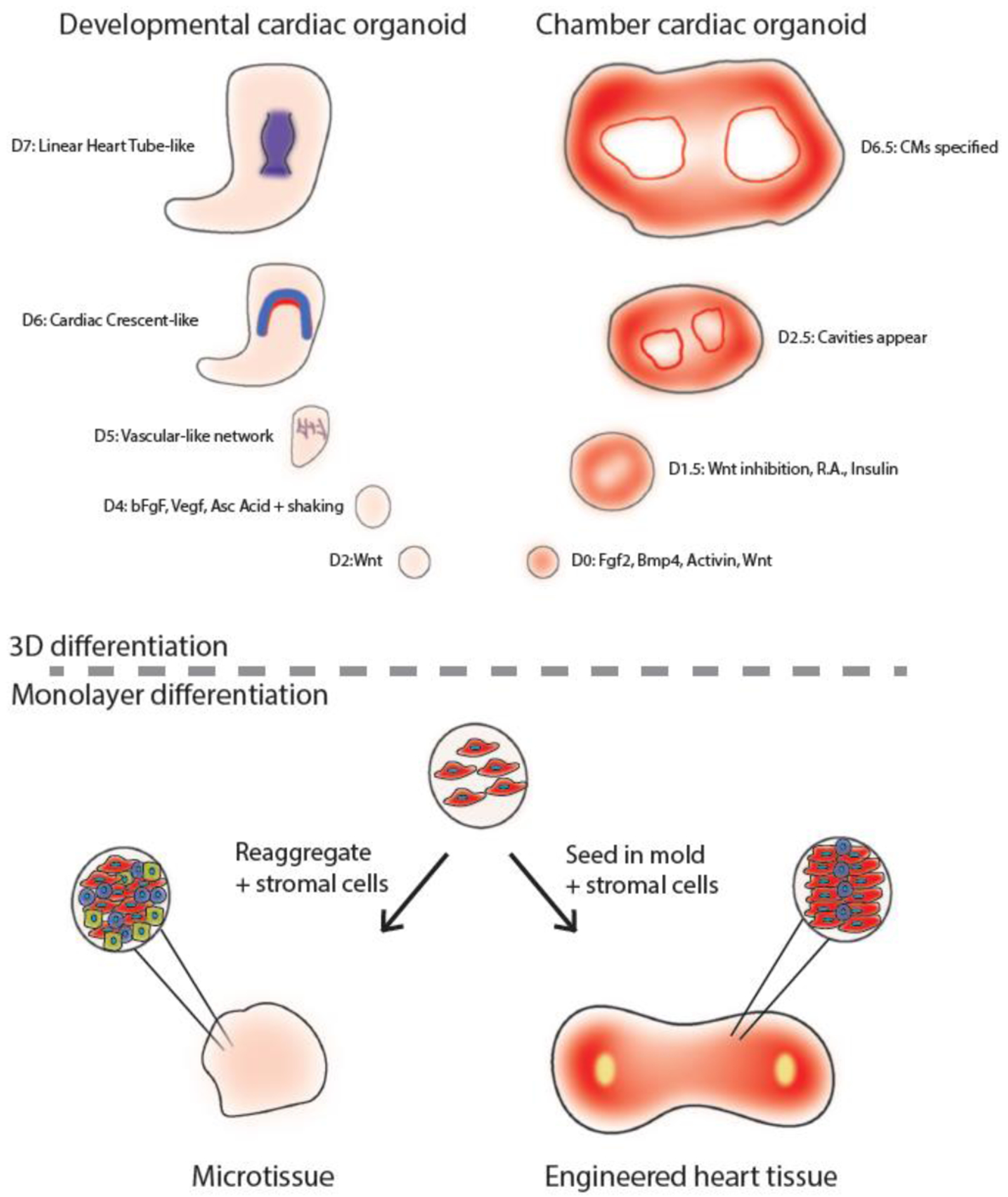
Summary of noteworthy models of in vitro models of the heart. Developmental cardiac organoids [103] form vascular networks, cardiac crescent-like, and linear heart tube-like structures with the addition of pro-cardiogenic factors bFgf, Vegf, and ascorbic acid. Chamber cardiac organoids [106] form patterned chambers regulated by a Wnt-Bmp signaling axis. Microtissues are aggregates of CMs and stromal cell types which allow for study of cell type-cell type interactions. Engineered heart tissues are models of a piece of the heart, and enable enhanced CM maturation for drug discovery and disease modeling.
When comparing Silva et al. [100] and Rossi et al. [103], there are two systems that emphasize the co-development of the heart and other cell types to create a cardiac organoid. However, the approaches for generating these organoids have some differences which may underlie the discrepancies in the resulting structure. Three notable areas of difference in the protocols are: species of PSC lines, early dissociation in Silva et al., and the additional cardiogenic factors (bFgf and Vegf) in Rossi et al. Firstly, Silva et al. used human iPSCs and Rossi et al. utilizes a variety of mouse PSC lines. Early gastruloid protocols have noted that there is an increased difficulty in generation of hiPSC gastruloids [99]. How the intrinsic species differences affect organoid formation has yet to be fully understood, and this is likely driving some of the differences observed between these two studies – a very important consideration when assessing all organoid protocols. Next, the early dissociation step in Silva et al. is in opposition to one of the main tenets of gastruloid technology. In the gastruloid system, there is a significant emphasis on the cell-cell interactions from the earliest stages of differentiation that help instruct self-organization and patterning in the resulting gastruloid, similar to how development occurs in vivo. However, after dissociation, these interactions are likely reset, and the mechanisms underlying the reorganization are unclear. This early dissociation step provides insight into the level of organization progenitor cells are able to achieve, which will provide another interesting set of questions to probe further. Finally, since exogenous bFgf and Vegf enhance generation of in vivo-like structures in Rossi et al., it would be interesting to see how addition of these factors would affect morphogenesis in the organoids of Silva et al.
In the most recent version of a developmental cardiac organoid, Drakhlis et al. [104], presented a method to form hiPSC-derived developmental cardiac organoids which consisted of both cardiac and endodermal lineages (similar to Silva et al. [100]). To do this, they activated and subsequently blocked Wnt signaling in Matrigel-embedded, preformed aggregates to produce an organoid with a layered structure. Interestingly, this formation required Matrigel, and such structure could not be replicated with commonly utilized alternatives. Furthermore, the resulting organoids formed a unique, layered, 3D structure that consisted of an inner endodermal core, surrounded by endocardial-like cells, a dense cardiomyocyte layer, and cardiomyocytes interspersed with liver anlagen and septum transversum-like cells. The morphogenesis of these organoids was very thought provoking, however, did not match the organization of in vivo biology. The authors also noted that the cardiomyocytes seem to be of SHF organism (primarily ventricular-like with a smaller portion of atrial-like cells), without much contribution from the FHF. Thus, as seen in these three iterations of developmental cardiac organoids, we are able to appreciate aspects of the elegance of in vivo development in the dish, while also witnessing where in vitro heart development falls short. With further optimization, innovation, and collaboration, we will hopefully move towards more developmental cardiac organoids which faithfully recapitulate human heart development to form more in vivo-like cardiac structures.
Chamber cardiac organoids form chamber-like structures in the absence of endodermal cells:
The developmental cardiac organoids developed by Silva et al. and Rossi et al. represent a key improvement in recapitulating early cardiogenesis in vitro. However, neither are composed specifically of cardiac cell types nor do they recapitulate an important development step unique to the heart – chamber formation. Recently, multiple groups have attempted to generate cardiac chambers in vitro [105–107]. We define chamber cardiac organoids as 3D in vitro models of the heart that reliably generate standardizable chambers, display patterning, and are composed of multiple cardiac cell types.
Israeli et al. describes a method for formation of multiple microchambers within each organoid [105]. After formation and stabilization of EBs in pluripotent stem cell culture for 2 days and an initial activation of Wnt signaling, Bmp, Activin A, with subsequent Wnt inactivation, multiple “microchambers” were formed within each cardiac organoid (Table 1). Furthermore, epicardial-like cells formed after a second burst of Wnt signaling. However, in this system, the microchambers were notably small, and did not form with a standardizable pattern. The presence of microchambers alone does not indicate resemblance to the embryonic heart, given that such chambers can be formed in other, non-in vivo-like contexts using tissue engineering approaches [108] as well. Nonetheless, transcriptomic analysis comparing these cardiac organoids and conventional monolayer differentiation to fetal heart tissue suggest that these organoids are more similar to fetal heart tissue than monolayers. Further validation of these microchambers, and whether they are controlled by processes related to in vivo chamber formation, will be necessary.
As an improved method of chamber formation, the cardioid system [106] offers a protocol to generate more consistent and robust chamber formation. By utilizing a multitude of canonical cardiogenic factors such as Bmp, Fgf, Wnt activation and inhibition, and retinoic acid (Fig 2), self-organized chamber formation of cardiac tissue was enabled in the absence of endodermal cells (Table 1). While this system is quite reproducible, it potentially differs from in vivo development, specifically in that cavity formation initiates very shortly after mesoderm induction (around differentiation day 2.5). Furthermore, the authors note that smaller chambers at earlier stages tended to eventually coalesce into larger chambers as differentiation continued. However, interestingly, in agreement with in vivo development [109–110], Hand1, was shown to be necessary for the formation of the organoid chambers, and was regulated by a Wnt-Bmp signaling axis. The authors noted that the formation of cavities is not necessarily linked to cardiac differentiation – high Wnt concentrations lead to large chamber formations at the expense of CM development – suggesting that cavity formation and cardiac specification are not necessarily linked processes. In an elegant conclusion to this paper, the authors added exogenous epicardial cells to the cardioids to form pro-epicardial organ-like structures, or an area of epicardial cell accumulation on the periphery of the cardioid. It would be interesting to see whether these pro-epicardial organ-like accumulations are directed to a particular region of the cardioid, indicating self-organization based on patterning of each cardioid, or adhere non-specifically. This system exhibits the self-organization, primitive patterning, and some resemblance of chamber-like formation, suggestive of a chamber cardiac organoid. Further study into why this system does not appear to undergo the early morphogenic stages of the heart (cardiac crescent and linear heart tube), while still forming chamber-like structures that are regulated by in vivo-like processes, would provide interesting insight into what is required to recapitulate each specific aspect of heart development. As the cardioid model lacks endodermal cells, such experiments would allow for comparison to developmental cardiac organoids, which are composed of multiple non-cardiac cell types. This would likely facilitate elucidation of the aspects of morphogenesis which are regulated by cell type-cell type interactions and which can be intrinsically regulated by cardiac mesoderm.
In one of the most exciting recent chamber cardiac organoid systems, Lee et al. describes a method to form four-chambered heart organoids through modulation of ECM and addition of Fgf4 [107] (Table 1). The authors report the formation of RA, LA, RV, and LV-like chambers. While the four-chamber structure was verified by combinatorial immunofluorescence, there appears to be some level of variability in the morphology of such organoids. Furthermore, the authors claim that the organoids form cardiac crescent-like structures in the process of chamber formation, and suggest this crescent is composed of FHF-like cells with adjacent SHF-like cells based on Tbx5 (to suggest FHF-like) and Nkx2–5 (to suggest SHF-like) immunofluorescence. However, this will likely need to be verified using more encompassing approaches incorporating additional markers of each heart field, as Nkx2–5 also marks the FHF contributions to the cardiac crescent (in addition to the SHF) [111], and utilization of 3D light sheet microscopy [103] (or similar techniques) to better appreciate the spatial patterning and resulting structures.
When comparing such chamber organoids, it is again important to consider the species of PSC line when evaluating the resulting structure formation. Both Israeli et al. [105] and Hofbauer et al. [106] utilize human PSCs while Lee et al. [107] uses mouse PSCs. Similar to the comparison between developmental organoids, it appears that mouse PSCs are better able to recapitulate the morphogenic events of in vivo development. The culture conditions or mechanisms allowing for ease of mouse cardiac organoid generation compared to human cardiac organoid remain unclear and will be interesting to probe further. Additionally, there are multiple differences in protocols for organoid generation between studies that may underlie differences between the two human cardiac organoid protocols [105–106], such as the addition of Fgf, retinoic acid, and prolonged Bmp exposure. Further probing into the role of these developmental signaling pathways will be crucial in optimizing human chamber cardiac organoid protocols in the future.
The field of cardiac organoid development is gaining momentum, as researchers continue to optimize conditions to marry recapitulation of early development (i.e., heart field and linear heart tube formation) with subsequent aspects of development (chamber formation). Such optimizations will likely involve stage-specific modulation of the canonical cardiogenic pathways utilized in the aforementioned studies (Bmp, Nodal, Wnt, Fgf, and Retinoic acid), and leverage the self-organizing capabilities of stem and progenitor cells throughout the process. Honest evaluation of each model – where the organoid faithfully recapitulates in vivo development and where it falls short – will be essential for such technologies to mature and develop to their full potential.
Microtissues provide insight into interactions between multiple cardiac cell types:
Another interesting approach that cardiac and stem cell biologists have taken to study interactions between multiple cell types is through co-aggregation of multiple cell types in 3D culture [113–117]. These models incorporate already differentiated CMs with supporting cell types such as cardiac fibroblasts [113, 115–117] and/or endothelial cells [113–117] in 3D aggregates (Fig 2). We define microtissues as 3D in vitro models of the heart which incorporate cardiac cell types but display limited self-organization, patterning, and structure formation. Nonetheless, these models allow for probing of CM-stromal cell interactions and enhanced functionality of CMs.
Interestingly, co-culture with either cardiac fibroblasts or endothelial cells appears to have benefits on the cardiomyocytes within each aggregate [113, 115]; however, the biological relevance of such interactions remains to be determined. Richards et al. described and optimized a system for aggregation of hiPSC derived CMs with exogenous human cardiac ventricular fibroblasts and human umbilical vein endothelial cells (HUVECs). In this system, after co-aggregation, a putative vasculature formed within the spheroid with a primitive lumen-like structure. This model is amenable to modeling infarct, a condition intimately involving non-CM cell types [114], and exhibits the pathological fibrosis associated with infarct. However, while some level of organization occurs within the vasculature of the spheroid, these lack elements of patterning or chamber formation of the heart. Giacomelli et al. combined iPSC-derived CMs and endothelial cells. After formation of the spheroids, immunofluorescence showed diffuse expression of endothelial markers throughout the microtissue, although the level of vasculature organization is unknown. However, compared to microtissues composed of CMs alone, the endothelial microtissues showed more adult-like gene expression [113]. When hiPSC cardiac fibroblasts were added to their microtissues, maturation of CMs was further improved, pointing to an important role of non-CMs in maturation [115]. Again, these spheroids do not exhibit notable organization or patterning. However, the functional improvements to the CMs suggests that these cellular interactions are crucial in enhancing CM maturation and organoid models in the future. Curiously, in Hookway et al., the authors suggest that such functional benefits diminish over time [116]. This suggests that the cellular crosstalk is not sufficient to maintain or follow the developmental program of CM maturation.
Interestingly, in microtissues, dissociation and reaggregation does not lead to significant levels of reorganization or patterning. This is in contrast with the developmental organoid system of Silva et al., which displayed patterning and organization after dissociation. The fundamental difference between these two dissociation approaches is the extent to which cells are pre-differentiated before dissociation and reaggregation. In microtissues, terminally differentiated CMs and stromal cells are dissociated and reaggregated, while in Silva et al., mesendodermal progenitors, but not terminally differentiated CMs, are dissociated and reaggregated in 3D culture. Taken together, these findings suggest that cardiac organoids are unable to be generated by reaggregation of differentiated cardiac and related cell types, but are better generated by aggregation of progenitor cells to allow for sharing of developmental trajectories to achieve more in vivo-like patterning.
Engineered heart tissues allow for advances in high-throughput drug discovery and disease modeling:
Generating adult-like heart tissues from human pluripotent stem cells has emerged as one of the most exciting areas of cardiac regenerative medicine. Using techniques established by tissue engineering, research groups have created tissues that can serve as unique systems for modeling human disease and drug discovery. Specifically, such models are proposed to help PSC-CMs overcome the developmental arrest which has historically limited their applicability as a system. These tissues utilize various interventions such as mechanical stretch, electrical stimulation, co-culture with stromal cells, and bioprinting (Fig 2). We believe that such tissues, coined by others as engineered heart tissues, are distinct from cardiac organoids based on the limited patterning and tissue-level, instead of organ level, scale. Furthermore, they are distinct from microtissues, as they are forced into their structure with some level of biomimetic-inspired engineering (stretch, electrical stimulation, etc.).
Many of these exciting new cardiac engineered heart tissue models utilize seeding cells to form strips of cardiac tissue-like structures [117–123]. The “Biowire” platform requires seeding of CMs with fibroblasts into a microwell between two flexible wires at either end of the well [117, 122]. After seeding, the cells undergo a process of compaction where they form a tube-like structure that is suspended in the well and attached to wires on both sides, and are subjected to electrical stimulation. In a triumph of cardiac tissue engineering, the authors then created an atrioventricular Biowire, by which each piece of tissue has distinct atria-like and ventricle-like regions that can be studied in the same sample. Again, highlighting a key difference between engineered heart tissues and cardiac organoids, these atrioventricular Biowires are formed by adding atrial and ventricular CMs to opposite ends of the microwell. Such forced aggregation is in contrast with self-organized patterning and structure formation in cardiac organoids. However, as is the case with many of these engineered heart tissue systems, the goal of such tissues may not be to recapitulate development on the organ-scale, but rather provide a system that allows for efficient disease modeling and drug discovery in vitro.
Similarly, multiple other studies have reported on methods involving seeding cells in molds surrounding posts, which can then be used to either mechanically stretch the tissues [121] or hold tissues in place as CMs contract [118–121, 123]. Interestingly, most of these methods require differentiation in monolayer to generate cardiac and stromal cell types before seeding into various molds. The resulting tissues are composed of multiple cell types present in the heart (CMs, vasculature, epicardial-like cells) and provide a compelling system for modeling disease and drug discovery as the resulting tissue displays properties of more adult-like heart tissue. In these models, CMs are aligned, tubular endothelial structures form, and epicardial cells are located towards the periphery of the tissue. The aligning and stretching of the CMs may promote the maturation of their tissues, potentially modeling an interesting developmental process essential to the function of the heart. These tissues do not display appreciable patterning, highlighting a key difference with cardiac organoids. However, these tissues will offer incredible value to the fields of regenerative medicine and developmental biology, by facilitating high-throughput study of adult-like cardiac tissue in vitro.
Conclusions:
In this review we highlight recent progress and challenges in the cardiac organoid and tissue engineering fields. By applying the common organoid principles of self-organization, patterning, and structure formation, we discriminate between studies to help add clarity to the rapidly evolving field. Based on such principles, marrying concepts of developmental cardiac organoids and chamber cardiac organoids with principles of bioengineering seems to be a promising approach. As discussed in [124], this will likely involve harmonization of technologies such as microfluidics, engineering of cell surfaces, and high-throughput scaling with the innate ability of progenitor cells to self-organize and pattern. Additionally, as mentioned, other non-organoid in vitro models of the heart such as microtissues and engineered heart tissues are providing unique tools to approach generating adult-like cardiac tissue in vitro for disease modeling and drug discovery. Overall, the cardiac organoid/tissue engineering fields are rapidly evolving and progressing, and it will be exciting to see how these technologies continue to develop and mature.
Acknowledgments:
We thank Dr. Giuliana Rossi and Dr. Michael Miyamoto for providing insightful comments and feedback in preparing this manuscript. This work was supported by grants from NIH, AHA, MSCRF, and DOD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Rossi G, Manfrin A, and Lutolf MP, “Progress and potential in organoid research,” Nature Reviews Genetics, vol. 19, no. 11. Nature Publishing Group, pp. 671–687, 01-November-2018, doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- [2].Clevers H, “Modeling Development and Disease with Organoids,” Cell, vol. 165, no. 7. Cell Press, pp. 1586–1597, 16-June-2016, doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- [3].Fu J, Warmflash A, and Lutolf MP, “Stem-cell-based embryo models for fundamental research and translation,” Nature Materials. Nature Research, 2020, doi: 10.1038/s41563-020-00829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Qian X, Song H, and Ming GL, “Brain organoids: Advances, applications and challenges,” Development (Cambridge), vol. 146, no. 8. Company of Biologists Ltd, 15-April-2019, doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Simian M and Bissell MJ, “Organoids: A historical perspective of thinking in three dimensions,” J. Cell Biol, vol. 216, no. 1, pp. 31–40, 2017, doi: 10.1083/jcb.201610056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jacob F et al. , “A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity,” Cell, vol. 180, no. 1, pp. 188–204.e22, January. 2020, doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mariani J et al. , “FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders,” Cell, vol. 162, no. 2, pp. 375–390, July. 2015, doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lancaster MA et al. , “Cerebral organoids model human brain development and microcephaly,” Nature, vol. 501, no. 7467, pp. 373–379, September. 2013, doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Qian X et al. , “Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure,” Cell, vol. 165, no. 5, pp. 1238–1254, May 2016, doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jo J et al. , “Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons,” Cell Stem Cell, vol. 19, no. 2, pp. 248–257, August. 2016, doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sato T et al. , “Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche,” Nature, vol. 459, no. 7244, pp. 262–265, May 2009, doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- [12].Spence JR et al. , “Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro,” Nature, vol. 470, no. 7332, pp. 105–110, February. 2011, doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nikolaev M et al. , “Homeostatic mini-intestines through scaffold-guided organoid morphogenesis,” Nature, vol. 585, no. 7826, September. 2020, doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- [14].Brassard JA, Nikolaev M, Hübscher T, Hofer M, and Lutolf MP, “Recapitulating macro-scale tissue self-organization through organoid bioprinting,” Nat. Mater, 2020, doi: 10.1038/s41563-020-00803-5. [DOI] [PubMed] [Google Scholar]
- [15].Brandenberg N et al. , “High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays,” Nat. Biomed. Eng, vol. 4, no. 9, September. 2020, doi: 10.1038/s41551-020-0565-2. [DOI] [PubMed] [Google Scholar]
- [16].Eiraku M et al. , “Self-organizing optic-cup morphogenesis in three-dimensional culture,” Nature, vol. 472, no. 7341, pp. 51–58, April. 2011, doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- [17].Bruneau BG, “The developmental genetics of congenital heart disease,” Nature, vol. 451, no. 7181, pp. 943–948, February. 2008, doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- [18].Meilhac SM and Buckingham ME, “The deployment of cell lineages that form the mammalian heart,” Nature Reviews Cardiology, vol. 15, no. 11. Nature Publishing Group, pp. 705–724, 01-November-2018, doi: 10.1038/s41569-018-0086-9. [DOI] [PubMed] [Google Scholar]
- [19].Buckingham M, Meilhac S, and Zaffran S, “Building the mammalian heart from two sources of myocardial cells,” Nat. Rev. Genet, vol. 6, no. 11, pp. 826–835, November. 2005, doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- [20].Kathiriya IS, Nora EP, and Bruneau BG, “Investigating the transcriptional control of cardiovascular development,” Circulation Research, vol. 116, no. 4. Lippincott Williams and Wilkins, pp. 700–714, 13-February-2015, doi: 10.1161/CIRCRESAHA.116.302832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Galdos FX, Guo Y, Paige SL, Vandusen NJ, Wu SM, and Pu WT, “Cardiac Regeneration: Lessons from Development,” Circulation Research, vol. 120, no. 6. Lippincott Williams and Wilkins, pp. 941–959, 17-March-2017, doi: 10.1161/CIRCRESAHA.116.309040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Klaus A, Saga Y, Taketo MM, Tzahor E, and Birchmeier W, “Distinct roles of Wnt/-catenin and Bmp signaling during early cardiogenesis,” 2007. [DOI] [PMC free article] [PubMed]
- [23].Andersen P et al. , “Precardiac organoids form two heart fields via Bmp/Wnt signaling,” Nat. Commun, vol. 9, no. 1, p. 3140, December. 2018, doi: 10.1038/s41467-018-05604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ivanovitch K et al. , “Ventricular, atrial and outflow tract heart progenitors arise from spatially and molecularly distinct regions of the primitive streak,” bioRxiv, p. 2020.07.12.198994, July. 2020, doi: 10.1101/2020.07.12.198994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kannan S and Kwon C, “Regulation of cardiomyocyte maturation during critical perinatal window,” J. Physiol, vol. 598, no. 14, pp. 2941–2956, July. 2020, doi: 10.1113/JP276754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Karbassi E et al. , “Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine,” Nature Reviews Cardiology, vol. 17, no. 6. Nature Research, pp. 341–359, 01-June-2020, doi: 10.1038/s41569-019-0331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Marchianò S, Bertero A, and Murry CE, “Learn from Your Elders: Developmental Biology Lessons to Guide Maturation of Stem Cell-Derived Cardiomyocytes,” Pediatr. Cardiol, vol. 40, no. 7, pp. 1367–1387, October. 2019, doi: 10.1007/s00246-019-02165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Uosaki H et al. , “Transcriptional Landscape of Cardiomyocyte Maturation,” Cell Rep, vol. 13, no. 8, 2015, doi: 10.1016/j.celrep.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Guo Y and Pu WT, “Cardiomyocyte maturation: New phase in development,” Circulation Research, vol. 126, no. 8. Lippincott Williams and Wilkins, pp. 1086–1106, 2020, doi: 10.1161/CIRCRESAHA.119.315862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].DeLaughter DM et al. , “Single-Cell Resolution of Temporal Gene Expression during Heart Development,” Dev. Cell, vol. 39, no. 4, pp. 480–490, November. 2016, doi: 10.1016/j.devcel.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RSP, and Robertson EJ, “Nodal signalling in the epiblast patterns the early mouse embryo,” Nature, vol. 411, no. 6840, pp. 965–969, June. 2001, doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- [32].Perea-Gomez A, Shawlot W, Sasaki H, Behringer RR, and Ang S, “HNF3beta and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior-posterior polarity in the mouse embryo,” Development, vol. 126, no. 20, 1999. [DOI] [PubMed] [Google Scholar]
- [33].Norris DP and Robertson EJ, “Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements,” Genes Dev, vol. 13, no. 12, pp. 1575–1588, June. 1999, doi: 10.1101/gad.13.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chazaud C and Rossant J, “Disruption of early proximodistal patterning and AVE formation in Apc mutants,” Development, vol. 133, no. 17, pp. 3379–3387, September. 2006, doi: 10.1242/dev.02523. [DOI] [PubMed] [Google Scholar]
- [35].Arnold SJ and Robertson EJ, “Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo,” Nature Reviews Molecular Cell Biology, vol. 10, no. 2. Nature Publishing Group, pp. 91–103, 08-February-2009, doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- [36].Miyamoto M, Gangrade H, and Tampakakis E, “Understanding Heart Field Progenitor Cells for Modeling Congenital Heart Diseases,” Curr. Cardiol. Rep, vol. 23, no. 5, p. 38, May 2021, doi: 10.1007/s11886-021-01468-5. [DOI] [PubMed] [Google Scholar]
- [37].Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, and Bruneau BG, “Early patterning and specification of cardiac progenitors in gastrulating mesoderm,” Elife, vol. 3, October. 2014, doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lescroart F et al. , “Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development,” Nat. Cell Biol, vol. 16, no. 9, pp. 829–840, September. 2014, doi: 10.1038/ncb3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lescroart F et al. , “Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq,” Science (80-. ), vol. 359, no. 6380, pp. 1177–1181, March. 2018, doi: 10.1126/science.aao4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kwon C, Cordes KR, and Srivastava D, “Wnt/β-catenin signaling acts at multiple developmental stages to promote mammalian cardiogenesis,” Cell Cycle, vol. 7, no. 24. Taylor and Francis Inc., pp. 3815–3818, 15-December-2008, doi: 10.4161/cc.7.24.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ai D et al. , “Canonical Wnt signaling functions in second heart field to promote right ventricular growth,” Proc. Natl. Acad. Sci. U. S. A, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cohen ED et al. , “Wnt/β-catenin signaling promotes expansion of Isl-1–positive cardiac progenitor cells through regulation of FGF signaling,” J. Clin. Invest, vol. 117, no. 7, pp. 1794–1804, July. 2007, doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qyang Y et al. , “The Renewal and Differentiation of Isl1 + Cardiovascular Progenitors Are Controlled by a Wnt/b-Catenin Pathway,” Cell Stem Cell, 2007, doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- [44].Abdul-Ghani M, Dufort D, Stiles R, De Repentigny Y, Kothary R, and Megeney LA, “Wnt11 Promotes Cardiomyocyte Development by Caspase-Mediated Suppression of Canonical Wnt Signals †,” Mol. Cell. Biol, vol. 31, no. 1, pp. 163–178, 2011, doi: 10.1128/MCB.01539-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bisson JA, Mills B, Helt JCP, Zwaka TP, and Cohen ED, “Wnt5a and Wnt11 inhibit the canonical Wnt pathway and promote cardiac progenitor development via the Caspase-dependent degradation of AKT,” Dev. Biol, vol. 398, no. 1, pp. 80–96, 2015, doi: 10.1016/j.ydbio.2014.11.015. [DOI] [PubMed] [Google Scholar]
- [46].Cohen ED, Miller MF, Wang Z, Moon RT, and Morrisey EE, “Wnt5a and wnt11 are essential for second heart field progenitor development,” Development, vol. 139, no. 11, pp. 1931–1940, June. 2012, doi: 10.1242/dev.069377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Miyamoto M et al. , “Cardiac progenitors auto-regulate second heart field cell fate via Wnt secretion,” bioRxiv, p. 2021.01.31.428968, January. 2021, doi: 10.1101/2021.01.31.428968. [DOI] [Google Scholar]
- [48].Klaus A, Muller M, Schulz H, Saga Y, Martin JF, and Birchmeier W, “Wnt/ - catenin and Bmp signals control distinct sets of transcription factors in cardiac progenitor cells,” Proc. Natl. Acad. Sci, vol. 109, no. 27, pp. 10921–10926, July. 2012, doi: 10.1073/pnas.1121236109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Prall OWJ et al. , “An Nkx2–5/Bmp2/Smad1 Negative Feedback Loop Controls Heart Progenitor Specification and Proliferation,” Cell, vol. 128, no. 5, pp. 947–959, March. 2007, doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jain R et al. , “Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts,” Science (80-. ), vol. 348, no. 6242, June. 2015, doi: 10.1126/science.aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].McCulley DJ, Kang JO, Martin JF, and Black BL, “BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development,” Dev. Dyn, vol. 237, no. 11, pp. 3200–3209, November. 2008, doi: 10.1002/dvdy.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].MacGrogan D, Münch J, and de la Pompa JL, “Notch and interacting signalling pathways in cardiac development, disease, and regeneration,” Nature Reviews Cardiology, vol. 15, no. 11. Nature Publishing Group, pp. 685–704, 01-November-2018, doi: 10.1038/s41569-018-0100-2. [DOI] [PubMed] [Google Scholar]
- [53].Miazga CM and McLaughlin KA, “Coordinating the timing of cardiac precursor development during gastrulation: A new role for Notch signaling,” Dev. Biol, vol. 333, no. 2, pp. 285–296, September. 2009, doi: 10.1016/j.ydbio.2009.06.040. [DOI] [PubMed] [Google Scholar]
- [54].Kwon C, Qian L, Cheng P, Nigam V, Arnold J, and Srivastava D, “A regulatory pathway involving Notch1/β-catenin/Isl1 determines cardiac progenitor cell fate,” Nat. Cell Biol, vol. 11, no. 8, pp. 951–957, 2009, doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kwon C et al. , “Notch post-translationally regulates β-catenin protein in stem and progenitor cells,” Nat. Cell Biol, vol. 13, no. 10, pp. 1244–1251, October. 2011, doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rones MS, McLaughlin KA, Raffin M, and Mercola M, “Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis,” Development, vol. 127, no. 17, 2000. [DOI] [PubMed] [Google Scholar]
- [57].Schroeder T et al. , “Recombination signal sequence-binding protein Jκ alters mesodermal cell fate decisions by suppressing cardiomyogenesis,” Proc. Natl. Acad. Sci. U. S. A, vol. 100, no. 7, pp. 4018–4023, April. 2003, doi: 10.1073/pnas.0438008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].De Zoysa P et al. , “Delta-like ligand 4-mediated Notch signaling controls proliferation of second heart field progenitor cells by regulating Fgf8 expression,” Development, vol. 147, no. 17, September. 2020, doi: 10.1242/dev.185249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shenje LT et al. , “Precardiac deletion of numb and numblike reveals renewal of cardiac progenitors,” Elife, vol. 2014, no. 3, p. e02164, April. 2014, doi: 10.7554/eLife.02164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhao C et al. , “Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation,” Dev, vol. 141, no. 2, pp. 281–295, January. 2014, doi: 10.1242/dev.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wu M and Li J, “Numb family proteins: Novel players in cardiac morphogenesis and cardiac progenitor cell differentiation,” Biomolecular Concepts, vol. 6, no. 2. De Gruyter Mouton, pp. 137–148, 01-April-2015, doi: 10.1515/bmc-2015-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Miao L et al. , “Cardiomyocyte orientation modulated by the Numb family proteins-N-cadherin axis is essential for ventricular wall morphogenesis,” Proc. Natl. Acad. Sci. U. S. A, vol. 116, no. 31, pp. 15560–15569, July. 2019, doi: 10.1073/pnas.1904684116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gibbs BC, Shenje L, Andersen P, Miyamoto M, and Kwon C, “β1-integrin is a cell-autonomous factor mediating the Numb pathway for cardiac progenitor maintenance,” Biochem. Biophys. Res. Commun, 2018, doi: 10.1016/j.bbrc.2018.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ivanovitch K, Temiño S, and Torres M, “Live imaging of heart tube development in mouse reveals alternating phases of cardiac differentiation and morphogenesis,” Elife, vol. 6, 2017, doi: 10.7554/eLife.30668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rochais F, Mesbah K, and Kelly RG, “Signaling pathways controlling second heart field development,” Circulation Research, vol. 104, no. 8. Lippincott Williams & Wilkins, pp. 933–942, 24-April-2009, doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- [66].Laugwitz KL, Moretti A, Caron L, Nakano A, and Chien KR, “Islet1 cardiovascular progenitors: A single source for heart lineages?,” Development, vol. 135, no. 2. The Company of Biologists Ltd, pp. 193–205, 15-January-2008, doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- [67].Laugwitz KL et al. , “Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages,” Nature, vol. 433, no. 7026, pp. 647–653, February. 2005, doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Desgrange A, Le Garrec JF, and Meilhac SM, “Left-right asymmetry in heart development and disease: Forming the right loop,” Development (Cambridge), vol. 145, no. 22. Company of Biologists Ltd, 01-November-2018, doi: 10.1242/dev.162776. [DOI] [PubMed] [Google Scholar]
- [69].Gittenberger-De Groot AC, Bartelings MM, Deruiter MC, and Poelmann RE, “Basics of cardiac development for the understanding of congenital heart malformations,” Pediatric Research, vol. 57, no. 2. Pediatr Res, pp. 169–176, February-2005, doi: 10.1203/01.PDR.0000148710.69159.61. [DOI] [PubMed] [Google Scholar]
- [70].Kula-Alwar D, Marber MS, Hughes SM, and Hinits Y, “Mef2c factors are required for early but not late addition of cardiomyocytes to the ventricle,” Dev. Biol, vol. 470, pp. 95–107, February. 2021, doi: 10.1016/j.ydbio.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Boogerd CJJ, Moorman AFM, and Barnett P, “Protein interactions at the heart of cardiac chamber formation,” Annals of Anatomy, vol. 191, no. 6. Ann Anat, pp. 505–517, 20-November-2009, doi: 10.1016/j.aanat.2009.06.004. [DOI] [PubMed] [Google Scholar]
- [72].Kelly RG, Buckingham ME, and Moorman AF, “Heart fields and cardiac morphogenesis,” Cold Spring Harb. Perspect. Med, vol. 4, no. 10, 2014, doi: 10.1101/cshperspect.a015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nakajima Y, “Retinoic acid signaling in heart development,” Genesis, vol. 57, no. 7. John Wiley and Sons Inc., 2019, doi: 10.1002/dvg.23300. [DOI] [PubMed] [Google Scholar]
- [74].Moorman AFM and Christoffels VM, “Cardiac chamber formation: Development, genes, and evolution,” Physiological Reviews, vol. 83, no. 4. American Physiological Society, pp. 1223–1267, 2003, doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- [75].del Monte-Nieto G et al. , “Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation,” Nature, vol. 557, no. 7705, pp. 439–471, 2018, doi: 10.1038/s41586-018-0110-6. [DOI] [PubMed] [Google Scholar]
- [76].Cao J and Poss KD, “The epicardium as a hub for heart regeneration,” Nat. Rev. Cardiol, vol. 15, no. 10, pp. 631–647, October. 2018, doi: 10.1038/s41569-018-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Quijada P, Trembley MA, and Small EM, “The Role of the Epicardium during Heart Development and Repair,” Circ. Res, vol. 126, no. 3, pp. 377–394, 2020, doi: 10.1161/CIRCRESAHA.119.315857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Carmona R et al. , “The embryonic epicardium: An essential element of cardiac development,” J. Cell. Mol. Med, vol. 14, no. 8, pp. 2066–2072, August. 2010, doi: 10.1111/j.1582-4934.2010.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Cao Y and Cao J, “Covering and re-covering the heart: Development and regeneration of the epicardium,” J. Cardiovasc. Dev. Dis, vol. 6, no. 1, pp. 1–12, March. 2019, doi: 10.3390/jcdd6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lupu IE, Redpath AN, and Smart N, “Spatiotemporal Analysis Reveals Overlap of Key Proepicardial Markers in the Developing Murine Heart,” Stem Cell Reports, vol. 14, no. 5, pp. 770–787, May 2020, doi: 10.1016/j.stemcr.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM, and Conlon FL, “Tcf21 regulates the specification and maturation of proepicardial cells,” Dev, vol. 140, no. 11, pp. 2409–2421, June. 2012, doi: 10.1242/dev.093385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cao Y, Duca S, and Cao J, “Epicardium in heart development,” Cold Spring Harb. Perspect. Biol, vol. 12, no. 2, February. 2020, doi: 10.1101/cshperspect.a037192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hatcher CJ et al. , “A role for Tbx5 in proepicardial cell migration during cardiogenesis,” Physiol. Genomics, vol. 18, no. 2, pp. 129–140, October. 2004, doi: 10.1152/physiolgenomics.00060.2004. [DOI] [PubMed] [Google Scholar]
- [84].Zhou B et al. , “Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart,” Nature, vol. 454, no. 7200, pp. 109–113, July. 2008, doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Brutsaert DL, “Cardiac endothelial-myocardial signaling: Its role in cardiac growth, contractile performance, and rhythmicity,” Physiological Reviews, vol. 83, no. 1. American Physiological Society, pp. 59–115, 2003, doi: 10.1152/physrev.00017.2002. [DOI] [PubMed] [Google Scholar]
- [86].Feng W, Chen L, Nguyen PK, Wu SM, and Li G, “Single Cell Analysis of Endothelial Cells Identified Organ-Specific Molecular Signatures and Heart-Specific Cell Populations and Molecular Features,” Front. Cardiovasc. Med, vol. 6, November. 2019, doi: 10.3389/fcvm.2019.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Harris IS and Black BL, “Development of the endocardium,” in Pediatric Cardiology, 2010, vol. 31, no. 3, pp. 391–399, doi: 10.1007/s00246-010-9642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Luxán G, D’Amato G, MacGrogan D, and De La Pompa JL, “Endocardial Notch Signaling in Cardiac Development and Disease,” Circulation Research, vol. 118, no. 1. Lippincott Williams and Wilkins, pp. e1–e18, 08-January-2016, doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- [89].Correia C et al. , “3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes,” Biotechnol. Bioeng, vol. 115, no. 3, pp. 630–644, March. 2018, doi: 10.1002/bit.26504. [DOI] [PubMed] [Google Scholar]
- [90].Chen VC et al. , “Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells,” Stem Cell Res, vol. 15, no. 2, pp. 365–375, September. 2015, doi: 10.1016/j.scr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kensah G et al. , “Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro,” Eur. Heart J, vol. 34, no. 15, pp. 1134–1146, April. 2013, doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- [92].Kempf H, Kropp C, Olmer R, Martin U, and Zweigerdt R, “Cardiac differentiation of human pluripotent stem cells in scalable suspension culture,” Nat. Protoc, vol. 10, no. 9, pp. 1345–1361, October. 2015, doi: 10.1038/nprot.2015.089. [DOI] [PubMed] [Google Scholar]
- [93].Domian IJ et al. , “Generation of functional ventricular heart muscle from mouse ventricular progenitor cells,” Science (80-. ), vol. 326, no. 5951, pp. 426–429, October. 2009, doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wobus AM, Wallukat G, and Hescheler J, “Pluripotent mouse embryonic stem cells are able to differentiate into cardiomyocytes expressing chronotropic responses to adrenergic and cholinergic agents and Ca2+ channel blockers,” Differentiation, vol. 48, no. 3, pp. 173–182, December. 1991, doi: 10.1111/j.1432-0436.1991.tb00255.x. [DOI] [PubMed] [Google Scholar]
- [95].Kattman SJ et al. , “Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines,” Cell Stem Cell, vol. 8, no. 2, pp. 228–240, February. 2011, doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- [96].Yang L et al. , “Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population,” Nature, vol. 453, no. 7194, pp. 524–528, May 2008, doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- [97].Tampakakis E, Miyamoto M, and Kwon C, “In vitro generation of heart field-specific cardiac progenitor cells,” J. Vis. Exp, vol. 2019, no. 149, p. e59826, July. 2019, doi: 10.3791/59826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lian X et al. , “Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions,” Nat. Protoc, vol. 8, no. 1, pp. 162–175, December. 2012, doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Beccari L et al. , “Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids,” Nature, vol. 562, no. 7726, pp. 272–276, October. 2018, doi: 10.1038/s41586-018-0578-0. [DOI] [PubMed] [Google Scholar]
- [100].Silva AC et al. , “Developmental co-emergence of cardiac and gut tissues modeled by human iPSC-derived organoids,” bioRxiv. bioRxiv, p. 2020.04.30.071472, 01-May-2020, doi: 10.1101/2020.04.30.071472. [DOI] [Google Scholar]
- [101].Kannan S, Farid M, Lin B, Miyamoto M, and Kwon C, “Transcriptomic entropy benchmarks stem cell-derived cardiomyocyte maturation against endogenous tissue at single cell level,” bioRxiv, p. 2020.04.02.022632, September. 2020, doi: 10.1101/2020.04.02.022632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Moris N et al. , “An in vitro model of early anteroposterior organization during human development,” Nature, vol. 582, no. 7812, pp. 410–415, June. 2020, doi: 10.1038/s41586-020-2383-9. [DOI] [PubMed] [Google Scholar]
- [103].Rossi G et al. , “Capturing Cardiogenesis in Gastruloids,” Cell Stem Cell, vol. 0, no. 0, November. 2020, doi: 10.1016/j.stem.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Drakhlis L et al. , “Human heart-forming organoids recapitulate early heart and foregut development,” Nat. Biotechnol, February. 2021, doi: 10.1038/s41587-021-00815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Israeli Y et al. , “Generation of Heart Organoids Modeling Early Human Cardiac Development Under Defined Conditions,” bioRxiv, p. 2020.06.25.171611, June. 2020, doi: 10.1101/2020.06.25.171611. [DOI] [Google Scholar]
- [106].Hofbauer P et al. , “Cardioids reveal self-organizing principles of human cardiogenesis,” bioRxiv, p. 2020.07.06.189431, July. 2020, doi: 10.1101/2020.07.06.189431. [DOI] [PubMed] [Google Scholar]
- [107].Lee J et al. , “In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix,” Nat. Commun, vol. 11, no. 1, December. 2020, doi: 10.1038/s41467-020-18031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Ma Z et al. , “Self-organizing human cardiac microchambers mediated by geometric confinement,” Nat. Commun, vol. 6, no. 1, pp. 1–10, July. 2015, doi: 10.1038/ncomms8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Firulli AB, McFadden DG, Lin Q, Srivastava D, and Olson EN, “Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1,” Nat. Genet, vol. 18, no. 3, pp. 266–270, 1998, doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- [110].McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, and Olson EN, “The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner,” Development, vol. 132, no. 1, pp. 189–201, January. 2005, doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- [111].Zhang L et al. , “Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development,” Dev. Biol, vol. 390, no. 1, pp. 68–79, June. 2014, doi: 10.1016/j.ydbio.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Richards DJ et al. , “Inspiration from heart development: Biomimetic development of functional human cardiac organoids,” Biomaterials, vol. 142, pp. 112–123, October. 2017, doi: 10.1016/j.biomaterials.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Giacomelli E et al. , “Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells,” Dev, vol. 144, no. 6, pp. 1008–1017, March. 2017, doi: 10.1242/dev.143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Richards DJ et al. , “Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity,” Nat. Biomed. Eng, vol. 4, no. 4, pp. 446–462, April. 2020, doi: 10.1038/s41551-020-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Giacomelli E et al. , “Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease,” Cell Stem Cell, vol. 26, no. 6, pp. 862–879.e11, June. 2020, doi: 10.1016/j.stem.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Hookway T, Matthys O, Joy D, Sepulveda J, Thomas R, and McDevitt T, “Bi-directional Impacts of Heterotypic Interactions in Engineered 3D Human Cardiac Microtissues Revealed by Single-Cell RNA-Sequencing and Functional Analysis,” bioRxiv, p. 2020.07.06.190504, July. 2020, doi: 10.1101/2020.07.06.190504. [DOI] [Google Scholar]
- [117].Nunes SS et al. , “Biowire: A platform for maturation of human pluripotent stem cell-derived cardiomyocytes,” Nat. Methods, vol. 10, no. 8, pp. 781–787, August. 2013, doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tiburcy M et al. , “Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair,” Circulation, vol. 135, no. 19, pp. 1832–1847, May 2017, doi: 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Mills RJ et al. , “Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest,” Proc. Natl. Acad. Sci. U. S. A, vol. 114, no. 40, pp. E8372–E8381, October. 2017, doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Voges HK, Mills RJ, Elliott DA, Parton RG, Porrello ER, and Hudson JE, “Development of a human cardiac organoid injury model reveals innate regenerative potential,” Dev, vol. 144, no. 6, pp. 1118–1127, March. 2017, doi: 10.1242/dev.143966. [DOI] [PubMed] [Google Scholar]
- [121].Ronaldson-Bouchard K et al. , “Advanced maturation of human cardiac tissue grown from pluripotent stem cells,” Nature, vol. 556, no. 7700, pp. 239–243, April. 2018, doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhao Y et al. , “A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling,” Cell, vol. 176, no. 4, pp. 913–927.e18, February. 2019, doi: 10.1016/j.cell.2018.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Mills RJ et al. , “Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway,” Cell Stem Cell, vol. 24, no. 6, pp. 895–907.e6, June. 2019, doi: 10.1016/j.stem.2019.03.009. [DOI] [PubMed] [Google Scholar]
- [124].Hofer M and Lutolf MP, “Engineering organoids,” Nature Reviews Materials. Nature Research, pp. 1–19, 19-February-2021, doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


