Abstract
The genetic diversity of pathogens presents a challenge to the development of broadly effective vaccines. In this issue, Scarselli et al. (2011) combine atomic-level structural information with genomics and classical vaccinology to design a single immunogen that elicits protective immunity against more than 300 natural variants of the bacterial pathogen, meningococcus B. This accomplishment provides a glimpse of the power of structure-based vaccine design to create immunogens capable of eliciting protective responses against genetically diverse pathogens.
Single Sentence Summary:
Structure-based vaccine design produces an immunogen capable of eliciting protective responses against diverse strains of meningococcus B, a primary cause of sepsis and bacterial meningitis.
MENINGOCOCCAL DISEASE AND VACCINES
The bacterial pathogen Neisseria meningitidis can overcome host defenses to invade the bloodstream and brain leading to sepsis or meningitis, respectively. Mortality rates are 5–15% even when antibiotics are used due to the fast onset of disease; indeed, within a few hours the condition of an infected individual can change from healthy to critically ill (1). Because many people harbor the bacterium without symptoms, the small percentage that progress to illness has a significant public health impact, with up to 100 cases per 100,000 people during epidemics. N. meningitidis shows significant genetic diversity with thirteen serogroups each of which consists of multiple serotypes (2).
Five serogroups, A, B, C, W135, and Y, cause epidemics (3). Fortunately vaccines have been licensed for all of these serogroups, except group B (meningococcus B) (4). These vaccines teach the immune system to recognize distinct polysaccharides in the bacterial outer coat. For serotype B, however, the coat polysaccharide unit is sialic acid, which is also a component of human tissue and thus it is not a useful immunogen. In the search for an effective meningococcus B vaccine, the N. meningitidis genome has been mined for suitable immunogens, with candidate protein antigens identified from open reading frames and assessed for their ability to induce bactericidal antibodies (5–6). Indeed, four of these proteins identified by genomics comprise the meningococcus B vaccine (4CMenB) currently in phase III clinical trials (7). One of these proteins, the factor H binding protein (fHBP), enables meningococcus B to evade complement-mediated lysis by the host immune system, and this immunogen has been proposed as the basis for a single-component vaccine.
The human complement system provides a rapid innate response to bacterial infection. Complement factor H, a serum protein present at high concentrations (~1mg/ml), plays a critical role in controlling complement activation (8). Meningococcal fHBP, a lipid-anchored component of the bacterial outer membrane, binds to human factor H blocking complement-mediated lysis and thus is essential for bacterial pathogenicity (9).
Sequencing of the fhbp gene in a large number of meningococcal isolates revealed the presence of three main sequence groups termed variants 1, 2 and 3, which comprise 65%, 25%, and 10%, respectively, of the variant sequences of meningococcus B (Figure 1). Immune sera raised using immunogens from each of the three variant groups are cross-reactive between variants, but bactericidal activity is limited to the variant group of the immunogen (Figure 1). Currently, a vaccine composed of two immunogens from variant groups 1 and 2 and predicted to cover 90% of meningococcus B variants is in phase II clinical trials (10).
Figure 1. Rational design of an immunogen.
(Top) The more than 300 sequence variants of factor H binding protein (fHBP) of meningococcus B can be classified into three groups, variants 1, 2 and 3 (red, blue and green, respectively). The immunodominant surfaces of variants 1, 2, and 3 (shown as solid colors in the schematic representations of sequence and protein structure) elicit bactericidal antibodies in mice, but these antibodies are only effective at neutralizing meningococcal strains within each group. The left half of the bilobed fHBP structural schematic represents the C-terminal β-barrel region, which carries key amino acids that elicit bactericidal antibodies. (Bottom) In a new study, Scarselli et al. use structure-based design to engineer a single chimeric immunogen that is capable of eliciting antibodies, which neutralize meningococcal variants from all three groups. Manipulation of immunogens at the sub-molecular level may enable the development of vaccines against genetically diverse pathogens.
FROM 3 TO 1
Immunogens comprised of fHBP variants could, in principle, elicit protection against 100% of meningococcus B infections. One approach is to combine separate immunogens in a single multivalent vaccine. This approach has been successfully used for vaccines to protect against polio (three weakened polio virus strains, one from each serotype) (11) and cervical cancer (four recombinant variants of the major capsid protein of human papillomavirus types 6, 11, 16 and 18) (12). Although such vaccines can be highly effective, each additional vaccine component introduces additional complications and costs. An alternative approach would be to create a single chimeric immunogen that contains the relevant immunological structures (epitopes) to allow for broad recognition of all of the pathogenic variants.
In the case of meningococcus B, the three variant groups of fHBP immunogens elicit bactericidal antibodies directed against different fHBP surfaces that vary between the three groups. A single chimeric fHBP immunogen would thus need to account for immunodominance (the ability of a particular surface to generate a substantial immune response) and antigenic variation (the manner by which genetic variation affects recognition by antibodies). Antibodies that are effective against fHBP variant 1 are elicited by a unique, mostly contiguous surface. By contrast, the immunodominant surfaces of variants 2 and 3 are not unique and overlap with each other (Fig. 1). This overlap complicates the design of a single, broadly effective immunogen.
In this issue of Science Translational Medicine, Rappuoli and colleagues (Scarselli et al., 2011) tackle this problem by transplanting elements of fHBP variants 2 and 3 into the fHBP parent variant 1. They modified the surface of their variant 1 fHBP immunogen (part of the vaccine currently in phase III trials), by importing patches of surface-exposed amino acids from variants 2 and 3. They focused their design efforts on the C-terminal β-barrel domain of fHBP. This domain contains the majority of the amino acids important for recognition by antibodies that have bactericidal activity. The investigators then divided the surface of this domain into 11 partially overlapping areas of sufficient size to accommodate the binding of an antibody. Within each of these 11 surface regions, groups of point mutations were introduced to reproduce co-occurring variation observed in natural variant 2 and 3 bacterial subpopulations. Fifty-four fHBP immunogen candidates were created and then were used to immunize mice. Mouse sera were then tested for the breadth of bactericidal activity of antibodies elicited by the immunogens against a panel of diverse meningococcus B strains. The best 8 immunogen candidates were subjected to extensive biophysical characterization to ensure their quality as vaccine candidates. These 8 candidates were then mixed with adjuvant and tested in further immunization experiments against a larger and more diverse panel of meningococcus B variants. The best of the immunogen candidates (G1), which contained 22 point mutations relative to the parental variant-1 fHBP, was further characterized by X-ray crystallography and engineered into a vaccine format in which two copies of the immunogen were fused to GNA2091, a meningococcal protein that enhances the immunogenicity of fHBP immunogens (5, 13). Vaccination with this immunogen elicited antibodies in mice which, in the presence of human complement, showed bacterialytic activity against ten different strains of meningococcus B, including members of all 3 serogroups. This immunogen is predicted to elicit protective immunity against all 300 natural variants of meningococcus B.
VACCINE DESIGN IMPLICATIONS
In principle, the information required to design a 3-into-1 fHBP immunogen is present in the genomic sequences of meningococcus B. The potential number of combinatorial variants, however, is too large to be used for identification of an immunogen by functional testing. Even with only two choices for each of the 22 altered amino acid residues selected in the final immunogen, more than a million candidate immunogens would need to be tested. The procedure used by Scarselli et al. reduces the complexity of the screening problem by focusing on immunodominant surfaces likely to be relevant and by screening with combinations of surface subsets, each of which contained multiple simultaneous alterations.
The Scarselli et al. strategy is essentially a combinatorial experiment with functional readout. In this case, the combinatorial experiment involves constructing a library of immunogens, each with mutations designed to represent a spatial subset of the most important immunodominant surfaces. Then, within each spatial subset, mutations are clustered to reflect natural pathogen diversity. This design process captures much of the diversity of fHBP with only 54 candidate immunogens. Despite this greatly reduced number, the ability of several candidate immunogens (G1, G2, F3, and H9; more than 5% of the candidate set) to achieve broad bactericidal effects indicates that the design procedure has sufficient redundancy to robustly identify effective immunogens.
It is unclear whether the current procedure could be used to reduce the valency of other vaccines. Could a single immunogen replace the three immunogens of the trivalent polio vaccine? Scarselli et al. show that such design would require intimate knowledge of (1) immunogen atomic-level structure, (2) antigenic variation of the pathogen, (3) epitopes of neutralizing antibody, and (4) relative immunogenicity of these epitopes, especially related to immunodominance. Because of the difficulty presented by overlapping epitopes, success is not assured. Nonetheless, this new study provides proof of principle that even in this circumstance the Scarselli et al. approach can meet with success.
ATOMIC LEVEL DESIGN
Atomic-level information can be used in many different ways. In the design of toxoid vaccines against diphtheria toxin, for example, knowledge of the diphtheria toxin structure (14) allows for design of inactive variants useful as immunogens. The atomic-level structure of polio virus (15), meanwhile, allows antigenic variation to be mapped spatially on the physical structure of the virus. And in the current work with fHBP, atomic-level details determined by NMR were used to validate candidate immunogens. However, inactive diphtheria toxins can be identified genetically, structural details of polio antigenic variation were not used in the design of the polio vaccine, and with fHBP the NMR data merely served to confirm protein integrity. Thus, none of these examples use atomic-level information in the vaccine design process in a way that cannot be replicated with other experimental information.
For design of the 3-into-1 fHBP immunogen, the critical structural data flows from the crystal structure of fHBP (16). The crystal structure enabled the mapping of the molecular surfaces of fHBP relative to the elicitation of bactericidal antibodies as well as the direct visualization of antigenic variation and the localization of residues responsible for immunodominance on the physical structure. This knowledge permitted surfaces roughly the size of antibody epitopes to be swapped between variants, and individually optimized. Because atomic-level structure played a critical role in this design process, we consider this to be one of the first successful uses of atomic-level structure in vaccine design. Other notable examples of atomic-level design involve the conformational stabilization of the HIV-1 gp120 envelope (17) to elicit antibodies against the co-receptor-binding site (18) and the use of the structure of foot-and-mouth disease virus (19) to increase the stability of the vaccine (20).
Despite the advances made by Scarselli et al., it is fair to ask whether the goal of obtaining a single immunogen effective against diverse meningoccocus B variants could have been achieved by other methods. For example, an immunogen composed of three fHBP molecules linked in tandem, one from each of the variant groups, could in principle elicit broad protection. Also, the final GNA2091-G1-G1 immunogen produced by Scarselli et al. has only been tested in mice. It is not clear that human immunogenicity will mirror that of the mouse, especially in qualities such as immunodominance, which may depend upon species-specific immunological subtleties. Lastly, the connection between recognizing antibodies and epitopes is generalized from polyclonal or mouse antibody responses, and the link between immunogen, recognizing antibody, and development of that recognizing antibody is tenuous.
Nonetheless, because of the significant degree of information inherent in atomic-level structure, with implicit links to both genetic sequences and chemical functionality, it is likely that such information – spurred by the success here – will play an increasing role in the design of future vaccine immunogens.
STRUCTURE-BASED VACCINE DESIGN
A number of approaches are currently being used that require atomic-level information in vaccine design. The Scarselli et al. approach falls within a general class of design involving epitope transplantation, that is, the relocation of antibody-recognized surfaces to an acceptor protein that preserves its antigenicity. In the current case, epitopes from one group of fHBP variants were transplanted to a different fHBP variant. In other cases, structurally-defined epitopes have been transplanted between non-homologous proteins that nonetheless preserve antigenicity. For example, epitopes for the broadly neutralizing antibodies 2F5 and 4E10, which recognize the membrane-proximal external region of HIV-1 gp41, have been transplanted to acceptor scaffolds from diverse protein families (21–22). Antibodies elicited by these “epitope scaffolds” recognize gp41 and induce a nearly identical epitope conformation as found in the template antibody-antigen complex (22). Despite the faithful recapitulation of antibody-epitope recognition, the anti-gp41 antibodies elicited by the epitope scaffold are not effective at neutralization, likely because of additional constraints related to recognition of this part of gp41 (23–24). Similar results have recently been achieved with epitopes transplanted from respiratory syncytial virus (25).
Structure-based vaccine design provides a number of methodologies to optimize the vaccine potential of target epitopes (26). With truncation, the immunogen can be minimized to remove surfaces that are deleterious to the immunogenicity of the target epitope (27–28). With glycan masking, such deleterious regions can be covered with N-linked glycosylation, to make them immunologically silent (29). With stabilization, conformationally diverse immunogens can be fixed into immunologically relevant states, and alterations such as the introduction of disulfide linkages or cavity-filling mutations may also assist with the physical stability of the target epitope (18, 20). And with resurfacing, antigenic surfaces other than the target epitope are replaced, altering their antigenicity and allowing for the creation of epitope-specific probes (30).
Overall, these strategies and methodologies often seek to replicate the elicitation of desired antibodies. A general strategy of this type has been articulated by Burton and colleagues (31–32). With this retrovaccinology approach, monoclonal antibodies with useful characteristics are identified, the recognized epitope is characterized, and immunogens, based on the recognized epitope, are designed and presented to the immune system to re-elicit the original template antibody. A modification of retrovaccinology has been suggested with a “site of vulnerability” approach, where knowledge of biological function is used to define the target surface (17). For example, the CD4-binding site of the HIV-1 gp120 envelope glycoprotein is critical for viral entry, and numerous antibodies (both neutralizing and non-neutralizing) target this site (33). It is by understanding the interplay between function, antibody recognition, and antigen conservation that an appropriate target for vaccine design emerges (17, 34).
From a historical context, vaccine design appears to incorporate ever finer levels of detail (Figure 2). This trend is likely to continue as greater detail engenders increased information. Such information can be used to define critical differences between pathogen and self. The increased resolution also allows more precise manipulation. The current work incorporates some aspects of atomic-level design, but does not present a general atomic-level solution to the vaccine design problem. A general solution would integrate a chemical understanding of antibody recognition and maturation into the immunogen design process. It also would likely require new tools for structural bioinformatics to predict such recognition and deep sequencing to follow maturation of antibody lineages in B cells. The new work by Scarselli et al. provides a glimpse of the potential power of atomic-level design, as well as a foretaste of the daunting complexities of measuring, intervening, and guiding the immune response that will be required to exploit this potential.
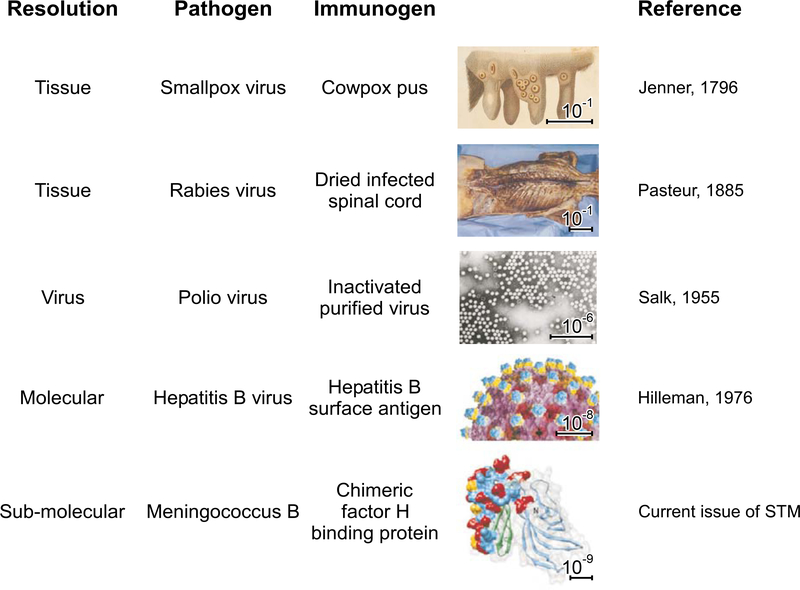
Figure 2. Vaccine design at progressively increasing resolution.
Vaccines teach the immune system to recognize pathogens so that appropriate host defenses are primed for protection. Because of the exquisite precision of antibody recognition, a single amino acid change can be sufficient to permit self versus non-self distinction. In principle, an effective vaccine may only need to elicit a robust immune response against a single conserved pathogen surface that is susceptible to antibody-mediated neutralization. Starting from the semi-pure tissue samples of early vaccinology (exemplified by cowpox pus and dried infected rabbit spinal cord used to prepare the first vaccines against smallpox and rabies, respectively), numerous advances have been made. The tools of tissue culture enabled preparation of purified virus (allowing for the polio vaccine); the tools of molecular biology enabled expression of individual recombinant molecules (leading to subunit vaccines against, for example, hepatitis B); and most recently, the tools of structure-based design enabled the construction of chimeric molecular immunogens. This is exemplified here by the chimeric fHBP molecule that induces bactericidal antibodies against diverse strains of meningococcus B (Scarselli et al., 2011). The scale bars indicate this progression in the ability to understand and to manipulate pathogens at ever greater resolution (from 10−1 m to 10−9 m, a 100-million-fold increase in resolution), allowing effective vaccines to be developed against more and more difficult pathogens.
Acknowledgments.
We thank J. Stuckey for assistance with figures and G.J. Nabel and members of the structural biology and structural bioinformatics core sections of the Vaccine Research Institute for helpful discussions.
References
- 1.Andersen BM, Mortality in meningococcal infections. Scand J Infect Dis 10, 277 (1978). [DOI] [PubMed] [Google Scholar]
- 2.Caugant DA, Maiden MC, Meningococcal carriage and disease--population biology and evolution. Vaccine 27 Suppl 2, B64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotschlich EC, Liu TY, Artenstein MS, Human immunity to the meningococcus. 3. Preparation and immunochemical properties of the group A, group B, and group C meningococcal polysaccharides. J Exp Med 129, 1349 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rappuoli R, Conjugates and reverse vaccinology to eliminate bacterial meningitis. Vaccine 19, 2319 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, Cartocci E, Ciucchi L, Di Marcello F, Ferlicca F, Galli B, Luzzi E, Masignani V, Serruto D, Veggi D, Contorni M, Morandi M, Bartalesi A, Cinotti V, Mannucci D, Titta F, Ovidi E, Welsch JA, Granoff D, Rappuoli R, Pizza M, A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A 103, 10834 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappuoli R, Reverse vaccinology. Curr Opin Microbiol 3, 445 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R, Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Ferreira VP, Pangburn MK, Cortes C, Complement control protein factor H: the good, the bad, and the inadequate. Mol Immunol 47, 2187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, Brunelli B, Pieri A, Santini L, Savino S, Serruto D, Litt D, Kroll S, Welsch JA, Granoff DM, Rappuoli R, Pizza M, Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med 197, 789 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granoff DM, Review of meningococcal group B vaccines. Clin Infect Dis 50 Suppl 2, S54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearce JM, Salk and Sabin: poliomyelitis immunisation. J Neurol Neurosurg Psychiatry 75, 1552 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampl M, Prevention of human papilloma virus-induced preneoplasia and cancer by prophylactic HPV vaccines. Minerva Med 98, 121 (2007). [PubMed] [Google Scholar]
- 13.Seib KL, Oriente F, Adu-Bobie J, Montanari P, Ferlicca F, Giuliani MM, Rappuoli R, Pizza M, Delany I, Influence of serogroup B meningococcal vaccine antigens on growth and survival of the meningococcus in vitro and in ex vivo and in vivo models of infection. Vaccine 28, 2416 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Choe S, Bennett MJ, Fujii G, Curmi PM, Kantardjieff KA, Collier RJ, Eisenberg D, The crystal structure of diphtheria toxin. Nature 357, 216 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Hogle JM, Chow M, Filman DJ, Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229, 1358 (1985). [DOI] [PubMed] [Google Scholar]
- 16.Cendron L, Veggi D, Girardi E, Zanotti G, Structure of the uncomplexed Neisseria meningitidis factor H-binding protein fHbp (rLP2086). Acta Crystallogr Sect F Struct Biol Cryst Commun 67, 531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD, Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT, Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog 5, e1000445 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F, The three-dimensional structure of foot-and-mouth disease virus at 2.9 A resolution. Nature 337, 709 (1989). [DOI] [PubMed] [Google Scholar]
- 20.Mateo R, Luna E, Rincon V, Mateu MG, Engineering viable foot-and-mouth disease viruses with increased thermostability as a step in the development of improved vaccines. J Virol 82, 12232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correia BE, Ban YE, Holmes MA, Xu H, Ellingson K, Kraft Z, Carrico C, Boni E, Sather DN, Zenobia C, Burke KY, Bradley-Hewitt T, Bruhn-Johannsen JF, Kalyuzhniy O, Baker D, Strong RK, Stamatatos L, Schief WR, Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. J. Mol. Biol 18, 1116 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD, Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A 107, 17880 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD, Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 78, 10724 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ofek G, McKee K, Yang Y, Yang ZY, Skinner J, Guenaga FJ, Wyatt R, Zwick MB, Nabel GJ, Mascola JR, Kwong PD, Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol 84, 2955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLellan JS, Correia BE, Chen M, Yang Y, Graham BS, Schief WR, Kwong PD, Design and Characterization of Epitope-Scaffold Immunogens That Present the Motavizumab Epitope from Respiratory Syncytial Virus. J Mol Biol, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwong PD, Nabel GJ, Acharya P, Boyington JC, Hood C, Chen L, Kim A, Kong L, Kwon YD, Majeed S, McLellan J, Ofek G, Pancera M, Sastry M, Shah AC, Stuckey J, Zhou T, Eds., Structural biology and the design of effective vaccines against HIV-1 and other viruses., vol. III (Humana Press, Totwa, New Jersey, 2011), vol. III. [Google Scholar]
- 27.Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J, Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol 78, 12975 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L, Zhou T, Yang ZY, Svehla K, O’Dell S, Louder MK, Xu L, Mascola JR, Burton DR, Hoxie JA, Doms RW, Kwong PD, Nabel GJ, Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol 83, 5077 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pantophlet R, Burton DR, GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol 24, 739 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR, Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burton DR, Antibodies, viruses and vaccines. Nat Rev Immunol 2, 706 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT, HIV vaccine design and the neutralizing antibody problem. Nat Immunol 5, 233 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD, Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326, 1123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD, Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]