ABSTRACT
Porcine deltacoronavirus (PDCoV) is a recently discovered coronavirus that poses a potential threat to the global swine industry. Although we know that aminopeptidase N (APN) is important for PDCoV replication, it is unclear whether it is the primary functional receptor, and the mechanism by which it promotes viral replication is not fully understood. Here, we systematically investigated the roles of porcine APN (pAPN) during PDCoV infection of nonsusceptible cells, including in viral attachment and internalization. Using a viral entry assay, we found that PDCoV can enter nonsusceptible cells but then fails to initiate efficient replication. pAPN and PDCoV virions clearly colocalized with the endocytotic markers RAB5, RAB7, and LAMP1, suggesting that pAPN mediates PDCoV entry by an endocytotic pathway. Most importantly, our study shows that regardless of which receptor PDCoV engages, only entry by an endocytotic route ultimately leads to efficient viral replication. This knowledge should contribute to the development of efficient antiviral treatments, which are especially useful in preventing cross-species transmission.
IMPORTANCE PDCoV is a pathogen with the potential for transmission across diverse species, although the mechanism of such host-switching events (from swine to other species) is poorly understood. Here, we show that PDCoV enters nonsusceptible cells but without efficient replication. We also investigated the key role played by aminopeptidase N in mediating PDCoV entry via an endocytotic pathway. Our results demonstrate that viral entry via endocytosis is a major determinant of efficient PDCoV replication. This knowledge provides a basis for future studies of the cross-species transmissibility of PDCoV and the development of appropriate antiviral drugs.
KEYWORDS: PDCoV, aminopeptidase N, entry, endocytosis, cross-species
INTRODUCTION
Porcine deltacoronavirus (PDCoV) (genus Deltacoronavirus; family Coronaviridae) is a recently discovered enteropathogenic swine coronavirus (CoV) that poses a threat to sows and nursing piglets, often with lethal outcomes (1–6). There are currently PDCoV outbreaks in several countries, resulting in economic losses to the swine industry (7). It is believed to originate from sparrow CoV HKU17 because the two viruses are closely related (8). CoVs have a remarkable capacity to change tropism and may frequently cross the host species barrier. In fact, over the past 30 years, several cross-species transmission events, as well as changes in viral tropism, have resulted in significant outbreaks of animal and human diseases (9, 10). Many important pathogens, such as the endemic severe acute respiratory syndrome coronavirus, human coronavirus HKU1 (HCoV-HKU1), HCoV-229E, HCoV-NL63, HCoV-OC43, and Middle East respiratory syndrome coronavirus, have a zoonotic origin (11–17). Notably, PDCoV is reported to successfully infect chickens and calves (18, 19). Therefore, studying the entry mechanism of PDCoV may provide insight into the likelihood of future host-switching events from swine to other species.
Aminopeptidase N (APN) is a ubiquitous transmembrane ectoenzyme that is reportedly involved in a number of processes, including cell survival, cell migration, angiogenesis, blood pressure regulation, and virus uptake (20, 21). Porcine aminopeptidase N (pAPN) is reported to act as a functional receptor for PDCoV (22, 23), and it has been shown that PDCoV can use APN from many different species (including humans) as a receptor for entry into nonsusceptible cells (23). However, Zhu et al. provided some evidence that pAPN contributes to viral entry but does not act as the primary receptor for PDCoV (24). Although APN is a functional receptor of PDCoV, the mechanism by which APN mediates PDCoV entry remains unclear.
To address these issues, we systematically evaluated the role of pAPN during PDCoV infection. We established stable pAPN expression in nonsusceptible cells to investigate the impact of pAPN on viral entry. Our findings indicate that pAPN is critical for PDCoV replication in nonsusceptible cells. We also demonstrate that whereas PDCoV can attach to and enter nonsusceptible cells, it fails to replicate efficiently in them. Finally, we demonstrate that APN is an entry cofactor for PDCoV and that only pAPN-mediated PDCoV internalization via an endocytotic pathway initiates efficient PDCoV replication.
RESULTS
Exogenous pAPN expression in nonsusceptible HEK293 and HEK293T cells permits PDCoV infection.
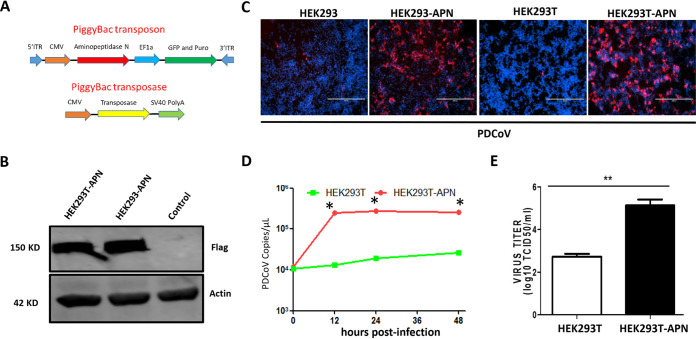
To investigate the role of APN in PDCoV replication, we first constructed cell lines stably expressing pAPN, using the PiggyBac transposon system, in otherwise nonsusceptible cell lines (Fig. 1A). Cell surface labeling with APN indicated that APN was expressed at the cell surface (data not shown). Stable pAPN expression in the resulting HEK293T-APN and HEK293-APN cell lines was further confirmed by Western blotting (Fig. 1B). To examine the role of APN in these cell lines, we infected HEK293-APN, HEK293T-APN, and both parental cell lines with PDCoV at a multiplicity of infection (MOI) of 0.1 and tested for viral replication at 24 h postinfection (hpi) by immunofluorescence. We found that only very few of the parental HEK293 and HEK293T cells were infected by PDCoV, whereas those expressing pAPN were highly susceptible to PDCoV infection (Fig. 1C). We also evaluated APN’s role in nonsusceptible BHK-21 cells, with similar results (data not shown). The viral RNA levels and viral titers in the pAPN-expressing cell lines were also higher than those in the parental cell lines (Fig. 1D and E). All these data suggest that pAPN expression permits PDCoV entry and replication in nonsusceptible cell lines.
FIG 1.
pAPN expression allows PDCoV replication in nonsusceptible HEK293 and HEK293T cells. (A) Scheme of the PiggyBac transposon system. ITR, internal transcribed spacer region; CMV, cytomegalovirus; SV40, simian virus 40. (B) HEK293-APN or HEK293T-APN cells were plated into six-well plates, and after 24 h, stable pAPN expression was confirmed by Western blotting using an anti-Flag antibody, with actin as the loading control. (C) HEK293, HEK293-APN, HEK293T, and HEK293T-APN cells were infected with PDCoV at an MOI of 0.1 in the presence of 200 ng/ml trypsin. PDCoV N protein was analyzed by immunofluorescence at 48 h postinfection (hpi). (D and E) Viral RNA in HEK293T and HEK293T-APN cells was collected at the indicated times (hours postinfection) and quantified by qPCR (D), and viral titers were quantified at 72 h (E). Each experiment was repeated at least three times. Error bars represent standard deviations (SD). *, P < 0.05; **, P < 0.01. TCID50, 50% tissue culture infective dose.
pAPN promotes PDCoV attachment and entry in a viral-dose-dependent manner in nonsusceptible cells.
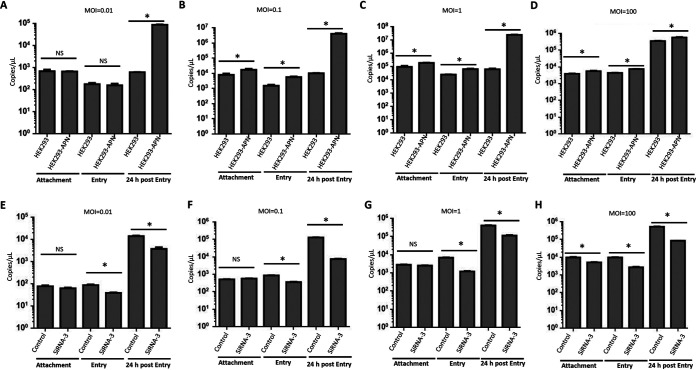
We next investigated the step in the viral replication cycle that is affected by pAPN, starting with attachment and entry. To evaluate pAPN’s role in viral attachment, we first inoculated HEK293 and HEK293-APN cells with PDCoV at the same MOI (0.01), allowed the virus to attach to the cells for 2 h at 4°C, and then washed the cells three times with cold phosphate-buffered saline (PBS). The viral RNA was extracted and measured by real-time PCR. To our surprise, there was no significant difference in the attachment of PDCoV to HEK293 and HEK293-APN cells at a low MOI of 0.01 (Fig. 2A). However, when the infection dose was increased to an MOI of 0.1, 1, or 100, PDCoV attachment was significantly increased in the HEK293-APN cells compared with that in the parental control cells (Fig. 2B to D).
FIG 2.
pAPN promotes PDCoV attachment and entry in a viral-dose-dependent manner. (A to D) HEK293 and HEK293-APN cells were plated into six-well plates, and at 90% confluence, the cells were infected at an MOI of 0.01 (A), an MOI of 0.1 (B), an MOI of 1 (C), or an MOI of 100 (D). The cells were then subjected to attachment and entry assays. At 24 h postinfection, the viral RNA copies were confirmed as the positive infection control. (E to H) IPI-21 cells were plated into 12-well plates, and at 90% confluence, the cells were transfected with siRNA-3 or the scrambled siRNA control. After 24 h, the cells were infected at an MOI of 0.01 (E), an MOI of 0.1 (F), an MOI of 1 (G), or an MOI of 100 (H). The cells were then subjected to attachment and entry assays. At 24 h postinfection, the viral RNA copies were confirmed as the positive infection control. Each experiment was repeated at least three times. Error bars represent SD. *, P < 0.05; NS, not significantly different.
We next evaluated whether pAPN is involved in PDCoV entry after PDCoV inoculation. We first inoculated HEK293 and HEK293-APN cells with PDCoV at the same MOI (0.01), allowed the virus to enter the cells for 2 h at 37°C, and then washed the cells three times with cold PBS. The viral RNA was extracted and measured by real-time PCR, which detected no significant difference in the entry of PDCoV into the parental HEK293 and HEK293-APN cells (Fig. 2A). Similar to the attachment results, PDCoV entry was significantly higher in the pAPN-expressing cells at the higher infection doses (MOI = 0.1, 1, or 100) (Fig. 2B to D). The viral RNAs were also quantified at 24 h postinfection as the positive control. This indicated that pAPN significantly promotes PDCoV replication (Fig. 2A to D). To confirm the role of pAPN, we knocked down endogenous pAPN expression with a specific small interfering RNA (siRNA) (siRNA-3) in porcine epithelioid intestinal (IPI-21) cells. The knockdown efficacy is illustrated in Fig. 3C. We then performed attachment and entry assays in IPI-21 cells. The results indicated that, unlike HEK293 cells, pAPN did not influence the attachment of PDCoV to IPI-21 cells, except when the MOI was 100 (Fig. 2E to H). However, PDCoV entry and replication were significantly reduced by pAPN knockdown in IPI-21 cells (Fig. 2E to H). These results demonstrate that nonsusceptible cells lacking pAPN expression still allow PDCoV attachment and entry.
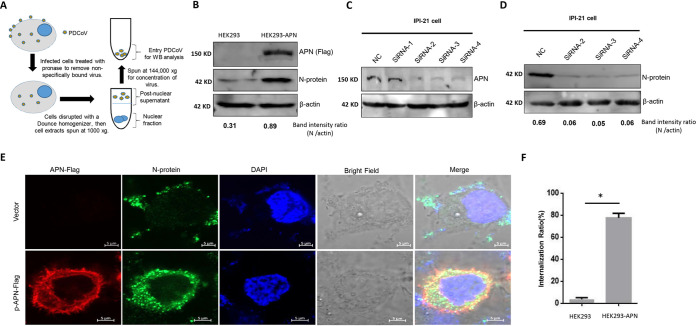
FIG 3.
pAPN promotes PDCoV internalization. (A) Scheme of the PDCoV entry assay. The same quantities of HEK293 and HEK293-APN cells were plated into 10-cm2 dishes, and after the cells had adhered, they were infected with PDCoV at an MOI of 100 for 2 h at 37°C. The cells were then harvested, treated with pronase (7 mg/ml), washed three times with ice-cold PBS, and centrifuged at 500 × g for 7 min at 4°C. The cell pellet was treated with hypotonic lysis buffer, lysed with a Dounce homogenizer, and centrifuged at 1,000 × g for 4 min at 4°C to remove the nuclear pellet. The supernatant fraction was centrifuged at 144,000 × g for 2 h at 4°C in a 50% sucrose solution in PBS. (B) The pellet fraction was analyzed by Western blotting (WB). The protein bands were then quantified by densitometry relative to the levels of N protein. This experiment was performed three times. A representative result is shown. (C) Knockdown of pAPN in IPI-21 cells. IPI-21 cells were transfected with either the indicated pAPN-specific siRNAs (100 nM siRNA-1 to −4) or 100 nM scrambled siRNA control (nontargeting control [NC]). At 24 h posttransfection (hpt), pAPN was quantified by Western blotting. This experiment was performed three times, and a representative result is shown. (D) Knockdown of pAPN reduced PDCoV replication in IPI-21 cells. IPI-21 cells were transfected with the indicated pAPN-specific siRNAs (100 nM siRNA-1 to -4) or 100 nM scrambled siRNA control (siScr) and then infected with PDCoV at an MOI of 100 for 2 h at 37°C, as described above. The protein bands then were quantified by densitometry relative to the levels of N protein. This experiment was performed three times. A representative result is shown. (E) Upon reaching 80% confluence, HEK293 cells were cotransfected with 1 μg of the pAPN-Flag plasmid (or the control vector). At 24 hpt, the cells were infected with PDCoV at an MOI of 100 at 37°C. After 2 h, we detected the internalized virions by immunofluorescence specific for the viral N protein. Each experiment was repeated at least three times. (F) The internalization ratios were analyzed as the number of cells with internalized virions/total cell numbers. Error bars represent the SD. *, P < 0.05.
pAPN promotes PDCoV internalization.
We next investigated how pAPN affects the PDCoV life cycle. Because a previous study indicated that pAPN contributes to the early steps of PDCoV replication (24), we first tested whether PDCoV is internalized into cells, which we detected at the protein level. We first performed this experiment at a lower MOI, but it was very difficult to detect the viral N protein at that MOI (data not shown). Therefore, we performed this experiment at a higher MOI of 100. HEK293 and HEK293-APN cells were infected at the high MOI (100) at 37°C, and after 2 h, the internalized virions were quantified by Western blotting. The procedure is shown in Fig. 3A. The infected cells were treated with pronase before the internalized virions were purified to exclude any virions on the cell surface. PDCoV successfully entered the cells regardless of their pAPN expression (Fig. 3B), although the quantity of internalized virions was significantly smaller when pAPN was absent (Fig. 3B).
We confirmed this finding in IPI-21 cells. We knocked down endogenous pAPN with specific siRNAs (siRNA-1, siRNA-2, siRNA-3, or siRNA-4) or a nonspecific scrambled control siRNA (siScr) in IPI-21 cells and found that siRNA-2, siRNA-3, and siRNA-4 significantly knocked down pAPN expression (Fig. 3C). Cells transfected with these specific siRNAs or the nonspecific scrambled siRNA were then inoculated with PDCoV at 24 h posttransfection (hpt), and the level of PDCoV replication was measured 48 h later. As shown in Fig. 3D, the knockdown of pAPN significantly suppressed the production of PDCoV compared with that in the siScr-transfected control cells.
To identify the step that is influenced by pAPN, we visualized the internalization status of the virus by immunofluorescence. HEK293 cells were transfected with 1 μg of the pAPN-Flag plasmid (or the control vector), and 24 h later, the cells were infected with PDCoV at an MOI of 100 at 37°C. After 2 h, we detected the PDCoV N protein by immunofluorescence. To our surprise, without pAPN expression, the majority of PDCoV virions localized to the cell surface, and a few virions were detected in the cytoplasm (Fig. 3E and F). In contrast, in the pAPN-expressing cells, most of the virions were found within the cytoplasm (Fig. 3E and F). This finding demonstrates that pAPN significantly facilitates the internalization of PDCoV virions.
pAPN may mediate PDCoV internalization via an endocytotic pathway.
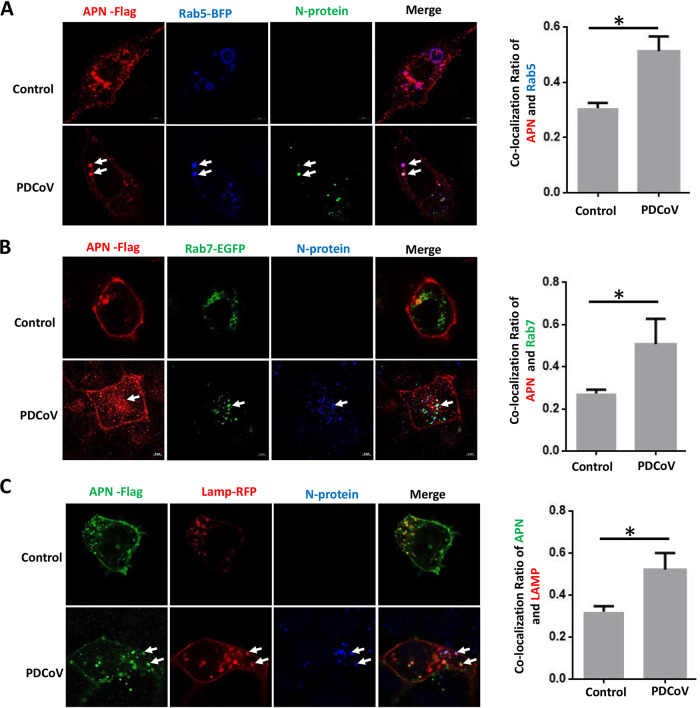
Many CoVs are known to enter host cells predominantly via an endocytotic pathway, so we hypothesized that PDCoV internalization is related to endocytosis. We first evaluated whether the location of pAPN correlates with the endocytotic route. Because RAB5, RAB7, and LAMP1 are cellular proteins commonly involved in viral trafficking through the early endosome, late endosome, and lysosome, respectively, we investigated whether pAPN is trafficked via these compartments. We cotransfected HEK293 cells with plasmids encoding pAPN and fluorescently labeled RAB5, RAB7, or LAMP1 and found significant colocalization 2 h after viral inoculation (Fig. 4A to C, left). Based on these results, we speculated that pAPN mediates PDCoV internalization through an endocytotic pathway. To determine whether pAPN is essential for the endocytotic internalization of PDCoV, we first coexpressed pAPN and fluorescently labeled RAB5, RAB7, or LAMP1 in HEK293 cells, and 24 h later, we infected the cells with PDCoV at an MOI of 100. After 2 h, we detected the colocalization of pAPN and PDCoV with RAB5, RAB7, and LAMP1 (Fig. 4A to C, left). Most importantly, PDCoV inoculation significantly increased the colocalization of pAPN with RAB5, RAB7, and LAMP1 (Fig. 4A to C, right). This indicated that pAPN may mediate PDCoV internalization via an endocytotic pathway.
FIG 4.
pAPN may mediate PDCoV internalization via an endocytotic pathway. HEK293 cells were plated into confocal dishes, and upon reaching 80% confluence, they were cotransfected with 1 μg of the pAPN-Flag plasmid (or the control vector) and either 1 μg of Rab5-BFP (blue) (A), 1 μg of Rab7-EGFP (green) (B), or 1 μg of LAMP1-RFP (red) (C). The cells were then infected with PDCoV (MOI = 100) for 2 h at 37°C, and the N protein was visualized by an immunofluorescence assay. The colocalization ratios in the right panel were analyzed with ImageJ. Error bars represent the SD. *, P < 0.05.
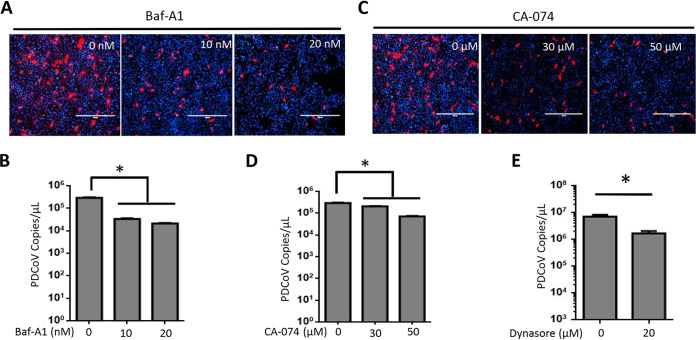
Inhibition of endosomal acidification and lysosomal proteases significantly reduced PDCoV replication in susceptible and less susceptible cells.
Because most CoVs require endosomal acidification and lysosomal proteases for their efficient replication, we tested the effect of the specific inhibition of endosomal acidification and lysosomal proteases on PDCoV replication in a susceptible cell line, LLC-PK. The endosomal acidification inhibitor bafilomycin A1 (Baf-A1) and the lysosomal protease inhibitor CA-074 were tested for cell toxicity by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (data not shown). The results indicated that both Baf-A1 and CA-074 significantly reduced the replication of PDCoV (Fig. 5A to D). These data demonstrate that PDCoV also uses the endocytotic pathway to initiate efficient viral replication in susceptible LLC-PK cells. To confirm that pAPN mediates its entry via endocytosis, we then pretreated HEK293-APN cells with dynasore, an endocytosis inhibitor. Dynasore significantly blocked the entry of PDCoV into HEK293-APN cells (Fig. 5E).
FIG 5.
Specific inhibitors of endosomal acidification and lysosomal proteases significantly inhibited PDCoV replication in susceptible cells. LLC-PK cells were treated with the indicated concentrations of bafilomycin A1 (Baf-A1), CA-074 methyl ester, or the same volume of dimethyl sulfoxide (DMSO) (control) for 2 h at 37°C and then infected with PDCoV at an MOI of 0.1. (A and C) Immunofluorescence assay for detecting the viral N protein at 24 h postinfection (hpi). (B and D) Viral RNA was collected at 24 hpi and quantified by qPCR. (E) HEK293-APN cells were treated with 20 μM dynasore or the same volume of DMSO (control) for 2 h at 37°C and then infected with PDCoV at an MOI of 1. Viral RNA was collected at 24 hpi and quantified by qPCR. Experiments were repeated at least three times. Error bars represent the SD. *, P < 0.05.
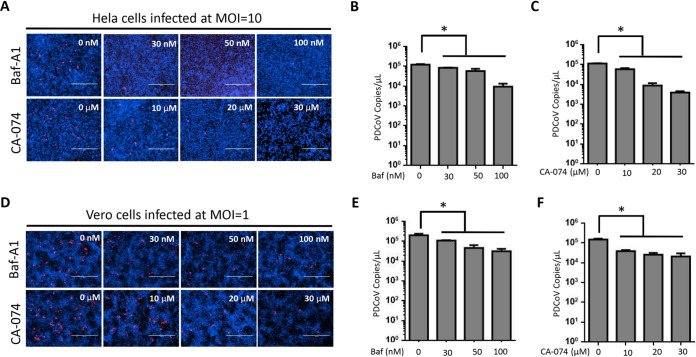
Although HeLa and Vero cells do not express APN, they support PDCoV replication to some extent. Therefore, we investigated whether an endocytotic pathway is a determinant for efficient viral replication. We treated both these cell lines with Baf-A1 or CA-074 before PDCoV infection and found that both inhibitors significantly reduced PDCoV replication (Fig. 6A to F). These data suggest that regardless of receptor usage, only entry via an endocytotic pathway leads to efficient PDCoV replication.
FIG 6.
Inhibition of endosomal acidification and lysosomal proteases significantly reduced PDCoV replication in HeLa and Vero cells. (A and D) HeLa (A) and Vero (D) cells were treated with the indicated concentrations of CA-074 methyl ester, bafilomycin A1 (Baf-A1), or the same volume of DMSO (control) for 2 h at 37°C and then infected with PDCoV at an MOI of 10 or 1, respectively, for 24 h. Viral N protein was visualized by an immunofluorescence assay. (B, C, E, and F) Viral RNA was collected at 24 h postinfection and quantified by qPCR. Experiments were repeated at least three times. Error bars represent SD. *, P < 0.05.
DISCUSSION
PDCoV is an emerging enteropathogenic swine virus, and its potential for cross-species transmission (23) makes understanding its entry mechanism very important. APN is the major receptor for most alphacoronaviruses, including porcine, feline, and human CoVs (25–27). To resolve a recent dispute about whether APN is a critical functional receptor for PDCoV (22–24), we demonstrate here that the stable expression of pAPN in otherwise nonsusceptible HEK293 and HEK293T cells rendered those cells susceptible to PDCoV infection (Fig. 1). This conclusion is also supported by previous studies that showed that pAPN expression renders other nonsusceptible cells, such as BHK, HeLa, and Vero cells, susceptible to PDCoV infection (22–24). Furthermore, APN from other species can also facilitate PDCoV infection in nonpermissive cells (23). All these data support the notion that APN is an important cofactor for PDCoV entry. However, our data also support the existence of an alternative receptor(s) because PDCoV attachment and entry were not influenced by pAPN at a low infectious dose (MOI = 0.01). Furthermore, pAPN knockout in IPI-21 and swine testis (ST) cells only partly reduced PDCoV infection, whereas transmissible gastroenteritis virus (TGEV) infection was absolutely blocked (23, 24). Recent studies have also demonstrated that pAPN knockout pigs were susceptible to PDCoV infection, indicating the presence of another receptor for PDCoV entry (28, 29).
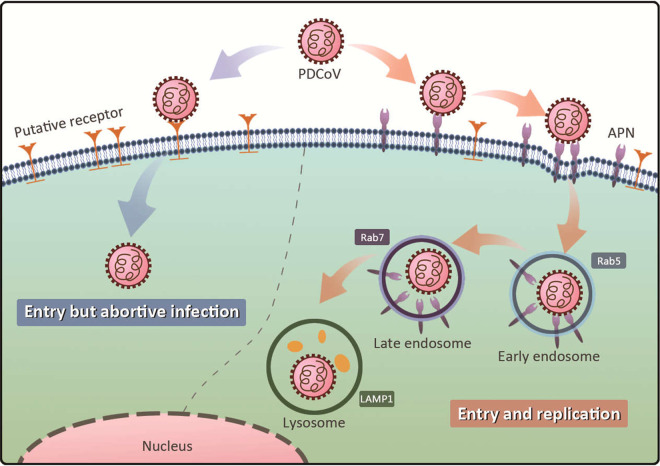
In the present study, although the nonsusceptible cell lines supported PDCoV entry, the virus failed to replicate efficiently in those cells. We consider that the putative receptor(s) utilized in nonsusceptible cells is incapable of initiating effective virus-cell fusion, which is critically dependent upon the interaction between the viral spike (S) protein and the cell receptor (Fig. 7). CoVs are known to depend upon the acidification of the endosome (30–32) and the proteolytic activation of the S protein by proteases along the endocytotic route (33, 34). In a recent study, Zhang et al. demonstrated that PDCoV initiates protease-mediated entry at the cell surface and that cathepsins facilitate its entry into the endosome (35). We believe that PDCoV entry mainly occurs via the endosomal pathway because PDCoV entry is significantly inhibited by these endosome inhibitors. In a recent study, we used a pseudovirus entry assay to demonstrate that PDCoV entry is not trypsin dependent (36). The inhibition of intracellular proteases has also been shown to block viral entry and virus-cell fusion (10, 33–35, 37). Therefore, we conclude that pAPN mediates PDCoV entry into its target cells predominantly via an endocytotic pathway rather than via membrane fusion at the cell surface (Fig. 7).
FIG 7.
Illustration of APN’s role in PDCoV replication. PDCoV attaches to and enters nonsusceptible cells but fails to initiate efficient replication because pAPN mediates PDCoV internalization via the endocytotic pathway, and only this pathway initiates efficient viral replication.
CoV entry includes two principal steps, receptor binding and membrane fusion, and the latter step requires the activation of the viral spike protein by host proteases, particularly lysosomal proteases (38). Lysosomal proteases have been shown to play a key role in CoV infection. We agree that IPI-21 and ST cells may have another functional receptor(s) (23, 24), which may also mediate PDCoV entry by an endocytotic pathway and may use lysosomal proteases to activate the S protein to facilitate viral membrane fusion. In nonsusceptible cells, although lysosomal proteases can activate the S protein and facilitate viral membrane fusion, without a guiding receptor (such as APN) to bring the virus into an endocytotic pathway, the initiation of efficient replication is difficult.
In summary, we have demonstrated that APN is a cofactor for PDCoV entry and that although PDCoV can attach to and enter cells lacking APN, it fails to initiate efficient replication in them. This is probably because pAPN mediates PDCoV internalization via an endocytotic pathway, and only this pathway can initiate efficient viral replication.
MATERIALS AND METHODS
Cells, virus, reagents, and plasmids.
The LLC-PK cell line (porcine kidney; ATCC CL-101); human embryonic kidney HEK293T and HEK293 cells; and BHK, IPI-21 (a porcine intestinal cell line that is always used as a cell model for PDCoV), HeLa, and Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (HyClone, USA). The HEK293T-APN and HEK293-APN cell lines (stably expressing pAPN) were generated with the PiggyBac transposon system, as previously described (36). We chose the HEK293T and HEK293 cell lines for the experiment because these cells are nonpermissive for PDCoV infection, and the transfection efficiency of HEK293T and HEK293 cell lines is high, allowing us to investigate the role of APN in PDCoV replication.
PDCoV attachment assay.
HEK293 and HEK293-APN cells were seeded into six-well plates, and when they reached 90% confluence, they were infected with PDCoV at different MOIs (0.01, 0.1, 1, or 100) for 2 h at 4°C. The cells were then washed three times with cold PBS, and the RNA was extracted and quantified by quantitative PCR (qPCR), as described previously (22).
PDCoV entry assays.
HEK293 and HEK293-APN cells were seeded into six-well plates, and upon reaching 90% confluence, they were infected with PDCoV at different MOIs (0.01, 0.1, 1, or 100) for 2 h at 4°C. The cells were then washed three times with cold PBS and cultured at 37°C under 5% CO2 for another 1 h to allow virus internalization. Viral entry was quantified by qPCR, as described previously (22).
Virus entry was also detected by a Western blot assay. Briefly, equal numbers of HEK293 and HEK293-APN cells were plated into 10-cm2 dishes, and after the cells had adhered, they were infected with PDCoV (MOI = 100) for 2 h at 37°C. The cells were harvested by scraping, treated with 1 ml of DMEM containing pronase (7 mg/ml), and incubated on ice for 5 min. They were then washed three times with ice-cold PBS (with centrifugation at 500 × g for 7 min at 4°C). After washing, 2 ml of hypotonic lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM KCl, 1 mM EDTA, and a protease inhibitor tablet) was added, and the samples were incubated on ice for 15 min. The cells were lysed with 15 strokes of pestle B in a Dounce homogenizer. The lysates were centrifuged at 1,000 × g for 4 min at 4°C to separate the nuclear pellet from the cytosolic fraction. An aliquot (100 μl) of the supernatant was collected for the actin input, and the remainder was centrifuged (Beckman SW41 rotor at 144,000 × g for 2 h at 4°C) on 50% sucrose in PBS. After the careful removal of the sucrose, the pellet was resuspended in 40 μl of 4× loading buffer and analyzed by Western blotting. The band intensity ratios were analyzed using ImageJ software.
Immunofluorescence analysis of PDCoV entry.
HEK293 cells were plated into laser confocal dishes, and upon reaching 80% confluence, they were cotransfected with 1 μg of the pAPN-Flag plasmid (or the control vector). After 24 h, the cell monolayers were infected with PDCoV (MOI = 100) for 2 h at 37°C. The cells were fixed in 4% paraformaldehyde for 1 h and then treated with 0.1 M glycine for 10 min to neutralize the paraformaldehyde. They were washed three times with PBS, permeabilized with 0.2% Triton X-100 for 1 h, washed three times again, and blocked with 1% bovine serum albumin for 2 h. After the cells were blocked, they were incubated with a mouse anti-N antibody or a rabbit anti-Flag antibody for 1 h at room temperature and then with an Alexa Fluor 488-conjugated goat anti-mouse IgG secondary antibody (Sigma) or an Alexa Fluor 568-conjugated goat anti-rabbit IgG secondary antibody (Sigma), respectively. To visualize the cell nuclei, the cells were treated with 4′,6-diamidino-2-phenylindole (DAPI; Sigma).
Knockdown of pAPN expression in IPI-21 cells with siRNA.
Four pAPN-specific siRNAs (siRNA-1 [sense, 5′-GCGAGUUGUCACUGUGAUUTT-3′; antisense, 5′-AAUCACAGUGACAACUCGCTT-3′], siRNA-2 [sense, 5′-GCGAGAUGGUUUGACUCCAUTT-3′; antisense, 5′-AUGGAGUCAAACAUCUCGCTT-3′], siRNA-3 [sense, 5′-GGUCCACCAUCUACUGCAATT-3′; antisense, 5′-UUGCAGUAGAUGGUGGACCTT-3′], and siRNA-4 [described in a previous study {22}]) and a scrambled negative siRNA control (siScr) were synthesized by RiboBio, China. IPI-21 cells were seeded into 12-well plates and transfected with either a pAPN-specific or a negative-control siRNA that had been diluted to 100 nM in serum-free medium. The knockdown efficacy of pAPN was verified by Western blotting. The anti-APN antibody was purchased from ABclonal (China).
Subcellular localization of pAPN.
HEK293 cells were plated into confocal dishes, and upon reaching 80% confluence, they were cotransfected with 1 μg of the pAPN-Flag plasmid and either 1 μg of Rab5-blue fluorescent protein (BFP) (blue), 1 μg Rab7-enhanced green fluorescent protein (EGFP) (green), or 1 μg LAMP1-red fluorescent protein (RFP) (red). The cells were then infected with PDCoV (MOI = 100) for 2 h at 37°C, and an immunofluorescence assay was performed, as described above.
Treatment with specific inhibitors.
LLC-PK, HeLa, Vero, and HEK293-APN cells were seeded into 24-well plates, and upon reaching 90% confluence, they were treated with the indicated concentrations of CA-074 methyl ester (Selleck), Baf-A1 (MedChemExpress [MCE]), or dynasore (MCE) for 2 h at 37°C. The cells were then infected with the indicated dose of PDCoV for 24 h at 37°C, and an immunofluorescence assay was performed, as described above.
Statistical analysis.
Origin GraphPad Prism 8.0 software was used for all graphical representations. Statistical significance was analyzed by one-way analysis of variance (ANOVA) and Tukey’s multiple-comparison test or the independent-sample Student t test. All P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
Y.-D.T., F.M., and L.F. designed the experiments. Y.-L.Y. and the other authors performed the experiments. All the authors analyzed the data. Y.-D.T. and Y.-L.Y. wrote the manuscript.
This work was funded by the National Key Research and Development Program of China (grant 2016YFD0500100).
Contributor Information
Yan-Dong Tang, Email: tangyandong2008@163.com.
Li Feng, Email: fengli@caas.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y, Xia L, Zhou J, Zhen W, Feng Y, Guo H, Zhang H, Tu C. 2014. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J Virol 88:7070–7082. 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Q, Li G, Stasko J, Thomas JT, Stensland WR, Pillatzki AE, Gauger PC, Schwartz KJ, Madson D, Yoon KJ, Stevenson GW, Burrough ER, Harmon KM, Main RG, Zhang J. 2014. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol 52:234–243. 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau SKP, Woo PCY, Yip CCY, Fan RYY, Huang Y, Wang M, Guo R, Lam CSF, Tsang AKL, Lai KKY, Chan K-H, Che X-Y, Zheng B-J, Yuen K-Y. 2012. Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol 86:5481–5496. 10.1128/JVI.06927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung K, Hu H, Saif LJ. 2016. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res 226:50–59. 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. 2015. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 6:e00064-15. 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Vlasova AN, Kenney SP, Saif LJ. 2019. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol 34:39–49. 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niederwerder MC, Hesse RA. 2018. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound Emerg Dis 65:660–675. 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, Bai R, Teng JLL, Tsang CCC, Wang M, Zheng B-J, Chan K-H, Yuen K-Y. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J Virol 86:3995–4008. 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham RL, Baric RS. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol 84:3134–3146. 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulswit RJ, de Haan CA, Bosch BJ. 2016. Coronavirus spike protein and tropism changes. Adv Virus Res 96:29–57. 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijgen L, Keyaerts E, Lemey P, Maes P, Van Reeth K, Nauwynck H, Pensaert M, Van Ranst M. 2006. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol 80:7270–7274. 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman VM, Eckerle I, Memish ZA, Liljander AM, Dijkman R, Jonsdottir H, Juma Ngeiywa KJ, Kamau E, Younan M, Al Masri M, Assiri A, Gluecks I, Musa BE, Meyer B, Muller MA, Hilali M, Bornstein S, Wernery U, Thiel V, Jores J, Drexler JF, Drosten C. 2016. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci USA 113:9864–9869. 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu-Sarkodie Y, Vallo P, da Silva Filho LV, Leroy EM, Thiel V, van der Hoek L, Poon LL, Tschapka M, Drosten C, Drexler JF. 2015. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol 89:11858–11870. 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S. 2017. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol 91:e01953-16. 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau SKP, Woo PCY, Li KSM, Tsang AKL, Fan RYY, Luk HKH, Cai J-P, Chan K-H, Zheng B-J, Wang M, Yuen K-Y. 2015. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J Virol 89:3076–3092. 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Wong SK, Li F, Kuhn JH, Huang IC, Choe H, Farzan M. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol 80:4211–4219. 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD, de Wit E, Munster VJ, Hensley LE, Zalmout IS, Kapoor A, Epstein JH, Karesh WB, Daszak P, Mohammed OB, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5:e00884-14. 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Q, Zhang H, Li B, Ding Q, Wang Y, Gao W, Guo D, Wei Z, Hu H. 2019. Susceptibility of chickens to porcine deltacoronavirus infection. Viruses 11:573. 10.3390/v11060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung K, Hu H, Saif LJ. 2017. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol 162:2357–2362. 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mina-Osorio P. 2008. The moonlighting enzyme CD13: old and new functions to target. Trends Mol Med 14:361–371. 10.1016/j.molmed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amin SA, Adhikari N, Jha T. 2018. Design of aminopeptidase N inhibitors as anti-cancer agents. J Med Chem 61:6468–6490. 10.1021/acs.jmedchem.7b00782. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Liu Y, Ji C-M, Yang Y-L, Liang Q-Z, Zhao P, Xu L-D, Lei X-M, Luo W-T, Qin P, Zhou J, Huang Y-W. 2018. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J Virol 92:e00318-18. 10.1128/JVI.00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA 115:E5135–E5143. 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Liu S, Wang X, Luo Z, Shi Y, Wang D, Peng G, Chen H, Fang L, Xiao S. 2018. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg Microbes Infect 7:65. 10.1038/s41426-018-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delmas B, Gelfi J, L’Haridon R, Vogel LK, Sjostrom H, Noren O, Laude H. 1992. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357:417–420. 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422. 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tresnan DB, Levis R, Holmes KV. 1996. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J Virol 70:8669–8674. 10.1128/JVI.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoian A, Rowland RRR, Petrovan V, Sheahan M, Samuel MS, Whitworth KM, Wells KD, Zhang J, Beaton B, Cigan M, Prather RS. 2020. The use of cells from ANPEP knockout pigs to evaluate the role of aminopeptidase N (APN) as a receptor for porcine deltacoronavirus (PDCoV). Virology 541:136–140. 10.1016/j.virol.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu K, Zhou Y, Mu Y, Liu Z, Hou S, Xiong Y, Fang L, Ge C, Wei Y, Zhang X, Xu C, Che J, Fan Z, Xiang G, Guo J, Shang H, Li H, Xiao S, Li J, Li K. 2020. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance. Elife 9:e57132. 10.7554/eLife.57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eifart P, Ludwig K, Bottcher C, de Haan CA, Rottier PJ, Korte T, Herrmann A. 2007. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J Virol 81:10758–10768. 10.1128/JVI.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama S, Taguchi F. 2009. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J Virol 83:11133–11141. 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zelus BD, Schickli JH, Blau DM, Weiss SR, Holmes KV. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J Virol 77:830–840. 10.1128/jvi.77.2.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJ, Bosch BJ, de Haan CA. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102:11876–11881. 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Chen J, Shi D, Shi H, Zhang X, Liu J, Cao L, Zhu X, Liu Y, Wang X, Ji Z, Feng L. 2019. Porcine deltacoronavirus enters cells via two pathways: a protease-mediated one at the cell surface and another facilitated by cathepsins in the endosome. J Biol Chem 294:9830–9843. 10.1074/jbc.RA119.007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang YL, Meng F, Qin P, Herrler G, Huang YW, Tang YD. 2020. Trypsin promotes porcine deltacoronavirus mediating cell-to-cell fusion in a cell type-dependent manner. Emerg Microbes Infect 9:457–468. 10.1080/22221751.2020.1730245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frana MF, Behnke JN, Sturman LS, Holmes KV. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J Virol 56:912–920. 10.1128/JVI.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Y, Shang J, Yang Y, Liu C, Wan Y, Geng Q, Wang M, Baric R, Li F. 2018. Lysosomal proteases are a determinant of coronavirus tropism. J Virol 92:e01504-18. 10.1128/JVI.01504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]