ABSTRACT
Dengue virus (DENV) constitutes one of the most important arboviral pathogens affecting humans. The high prevalence of DENV infections, which cause more than 20,000 deaths annually, and the lack of effective vaccines or direct-acting antiviral drugs make it a global health concern. DENV genome replication occurs in close association with the host endomembrane system, which is remodeled to form the viral replication organelle that originates from endoplasmic reticulum (ER) membranes. To date, the viral and cellular determinants responsible for the biogenesis of DENV replication organelles are still poorly defined. The viral nonstructural protein 4A (NS4A) can remodel membranes and has been shown to associate with numerous host factors in DENV-replicating cells. In the present study, we used reverse and forward genetic screens and identified sites within NS4A required for DENV replication. We also mapped the determinants in NS4A required for interactions with other viral proteins. Moreover, taking advantage of our recently developed polyprotein expression system, we evaluated the role of NS4A in the formation of DENV replication organelles. Together, we report a detailed map of determinants within NS4A required for RNA replication, interaction with other viral proteins, and replication organelle formation. Our results suggest that NS4A might be an attractive target for antiviral therapy.
IMPORTANCE DENV is the most prevalent mosquito-borne virus, causing around 390 million infections each year. There are no approved therapies to treat DENV infection, and the only available vaccine shows limited efficacy. The viral nonstructural proteins have emerged as attractive drug targets due to their pivotal role in RNA replication and establishment of virus-induced membranous compartments, designated replication organelles (ROs). The transmembrane protein NS4A, generated by cleavage of the NS4A-2K-4B precursor, contributes to DENV replication by unknown mechanisms. Here, we report a detailed genetic interaction map of NS4A and identify residues required for RNA replication and interaction between NS4A-2K-4B and NS2B-3 as well as NS1. Importantly, by means of an expression-based system, we demonstrate the essential role of NS4A in RO biogenesis and identify determinants in NS4A required for this process. Our data suggest that NS4A is an attractive target for antiviral therapy.
KEYWORDS: dengue virus, flavivirus, genetic mapping, membrane remodeling, nonstructural protein 4A, replication organelles
INTRODUCTION
Dengue virus (DENV) is an enveloped positive-strand RNA virus belonging to the Flavivirus genus within the Flaviviridae family (1). DENV is an arthropod-borne virus that is transmitted to humans by the bite of infected mosquitos. Infections are usually either asymptomatic or lead to mild symptoms resembling febrile illness-like diseases. However, in a minority of cases, DENV infections can manifest severe symptoms such as hemorrhagic fever and shock syndrome that can lead to death. Although the estimated percentage of severe diseases associated with DENV infections is low (∼1:1,000 infections), the high worldwide incidence of DENV infections (∼390 million each year) makes these severe diseases a significant health concern (2). Moreover, dynamic global climate changes, together with modern population migration and deforestation, increase the global spread of mosquito populations and virus infection, thus putting millions of people at risk of infection (3). At the moment, no antiviral drugs are available, and the only approved prophylactic vaccine has limited efficacy, especially in immunologically naive individuals, and its administration is not recommended for those with higher risk of serious disease, which are young children or elderly people (4).
The DENV genome is composed of a single-strand RNA of positive polarity containing a 5′ cap but no 3′ poly(A) tail (1). Following receptor-mediated entry into the cell and release into the cytosol, the genome is translated at the rough endoplasmic reticulum (ER), giving rise to a single polyprotein. This polyprotein is co- and posttranslationally processed into three structural proteins (capsid, precursor membrane protein, and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). While the structural proteins, together with the RNA genome, are assembled to form the virion, the nonstructural proteins (NS) are responsible for the replication of the viral genome within virus-induced membrane compartments called the viral “replication organelle” (RO). For DENV and all the related flaviviruses studied thus far, the NS proteins, together with host cell factors, induce the formation of ROs within ER membranes. These ROs are composed of ordered arrays of vesicles, referred to as vesicle packets (VPs) that form as result of invaginations of the ER membranes. The viral RNA is presumably replicated within the vesicles’ interior, and newly synthesized genomes exit the VPs through a pore-like opening that also allows for exchange of metabolites. Electron tomography evaluation of these VPs has provided structural and morphological information, but the molecular constituents and processes of VP biogenesis are still unknown.
Evidence suggests prominent roles for NS1, NS2A, NS4A, and NS4B in RO biogenesis. NS1, the only luminal-resident NS protein, has been shown to interact with and remodel lipid membranes in vitro and to be required for VPs formation (5, 6). NS2A was recently shown to interact with the viral genome and was proposed to shuttle the viral RNA between the replication and the virion assembly sites. NS4A and NS4B possess several transmembrane spanning regions, as well as an amphipathic α-helix, that can act as a wedge, possibly inducing the negative-membrane curvature required for ER invagination. NS4A is composed of three α-helices and is linked to NS4B by a 2-kDa-long transmembrane segment termed the 2K peptide. This peptide acts as signal peptide for NS4B and is cleaved at the N and C termini by both the viral NS2B/3 protease and the cellular signal peptidase, respectively (7). For DENV, it was shown that the sole expression of mature NS4A, i.e., NS4A lacking the 2K peptide, is able to promote the formation of convoluted membrane structures that have previously been associated with DENV infection (8). Conversely, West Nile virus (WNV) NS4A was able to induce convoluted membranes more efficiently when the 2K peptide was retained (9). Membrane remodeling by NS4A is supported by direct interaction with host cell proteins, most notably the reticulon 3.1a protein, an ER-resident membrane-bending protein able to stabilize highly curved ER membrane tubules (10, 11). In addition, both NS4A and NS4B form homo- and hetero-oligomers (12–14), and genetic interactions between NS4A and either NS1 or NS4B were reported for yellow fever virus (YFV) and WNV, two closely related flaviviruses (15). It has been reported that the N-terminal region of NS4A binds highly curved membranes and contributes to protein oligomerization and mutations introduced into this region affect NS4A folding and stability, at least in the case of WNV (13, 14, 16).
Recently, several studies have provided evidence that the uncleaved NS4A-2K-4B intermediate has a prominent role in DENV replication. This protein precursor was shown to specifically interact with NS1 in infected cells, and mutations in NS1 abrogating this interaction also reduced viral replication (5). Thus, NS4A might have essential roles during DENV replication, not only as mature protein but also as part of polyprotein cleavage intermediates.
In this study, we combined genetic, biochemical, and imaging approaches to elucidate the molecular determinants of NS4A involved in DENV replication. Using mutagenesis analysis, we identified residues within NS4A essential for viral replication. Selection for compensatory mutations allowed for the definition of a genetic NS4A interaction map. Biochemical characterization of NS4A mutations that abrogate viral replication revealed specific residues within NS4A responsible for modulating interactions with NS1 and NS3. Finally, taking advantage of our recently developed polyprotein expression system (17, 18) that supports VP formation in a replication-independent manner, we characterized the contribution of NS4A to DENV RO biogenesis and identified residues within NS4A required for this process.
RESULTS
Identification of residues within NS4A essential for DENV replication.
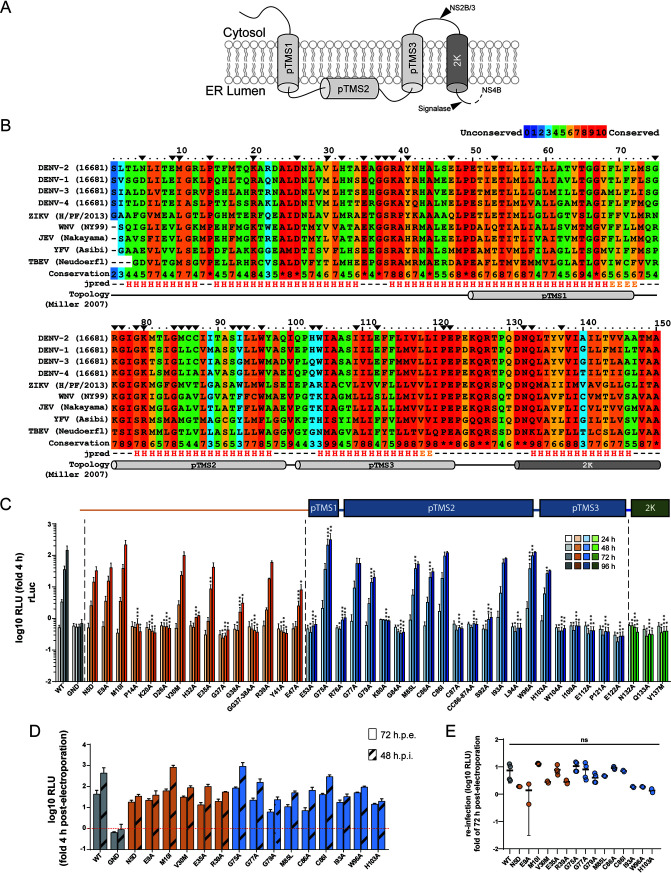
NS4A is an integral membrane protein (Fig. 1A) that can induce membrane rearrangements and contributes to viral replication in a poorly understood manner. In order to identify determinants within NS4A required for viral RNA replication, we performed a mutational analysis through reverse genetic engineering of DENV NS4A mutants. Multiple-sequence alignments of the NS4A-2K-coding region of the different DENV serotypes, as well as other members of the Flavivirus genus, identified 32 highly conserved residues, which were selected as targets for site-directed mutagenesis along with 5 residues located in highly variable regions (Fig. 1B). Moreover, for positions with two identical consecutive amino acid residues, we decided to target, in addition, both residues at the same time by introducing double mutations. Each of the NS4A mutations was introduced into the DENV luciferase reporter virus genome DVs-R2A, and in vitro transcripts generated therefrom were transfected into Huh7 cells. Luciferase activity was evaluated at different time points after transfection as surrogate for viral replication (Fig. 1C). Wild-type (WT) reporter virus and a replication-deficient NS5 mutant (GND) carrying a mutation that abrogates RNA-dependent RNA polymerase activity were used as positive and negative controls, respectively. Replication kinetics for 15 of the mutants showed only minor differences from the WT, whereas 25 of the mutations severely impaired (>90% reduction compared to WT) (Table 1) or completely abrogated viral replication. While several mutations in the N-terminal cytoplasmic region and within putative transmembrane segment 2 (pTMS2) (8) were tolerated, save for H103A, all mutations in pTMS3 and the 2K peptide-coding region completely blocked viral replication, arguing for a critical role, especially of these regions for genome amplification.
FIG 1.
Residues of DENV NS4A critical for viral replication identified by alanine-scanning mutagenesis. (A) Schematic representation of NS4A membrane topology according to reference 8. pTMS, putative transmembrane segment; 2K, 2 kDa peptide. (B) Multiple-sequence alignment of NS4A from several flaviviruses. Color grades indicate the degree of conservation according to the 0-to-10 coloring scheme given on the top right. The secondary structure prediction of each residue in context is indicated on the bottom (H, helix; E, extended). The putative transmembrane segments and 2K peptide are indicated by cylinders below the corresponding amino acid residues. Black arrowheads indicate residues selected for alanine-scanning mutagenesis. (C) Effects of mutations in NS4A on DENV replication. Mutations specified on the bottom were inserted into a DENV-2 (strain 16681) full-length reporter genome (DVs-R2A). In vitro transcripts were electroporated into Huh7 cells that were collected and lysed at different time points to measure renilla luciferase activity, which reflects RNA replication. Replication efficiency is reported as relative light units (RLU) normalized to the 4-h value (fold 4 h) that reflects transfection efficiency. Values represent the mean and SD from three independent experiments. Significance has been calculated with one-way ANOVA with Dunnett’s posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Supernatants from cells electroporated as in panel C were collected 72 h postelectroporation and used to infect Huh7 cells. Cells were lysed after 48 h, and renilla luciferase activity was measured (48 h postinfection [h.p.i.). Replication efficiency is reported as relative light units (RLU) normalized to the value determined 4 h postelectroporation. For comparison, replication levels of each construct at 72 h postelectroporation are shown (72 h.p.e.). Values represent the mean and SD from at least two independent experiments. (E) Reinfection values from panel D normalized to the respective values determined 72 h postelectroporation. Each dot represents one experiment. Significance has been calculated with one-way ANOVA with Dunnett’s posttest. ns, not significant.
TABLE 1.
Effects of mutations in NS4A on DENV reporter virus replication
| Mutation | DVs-R2A replication at 96 ha |
|---|---|
| WT | 145.1 ± 118.5 |
| GND | 0.7 ± 0.5 |
| N5D | 32.8 ± 18.3 |
| E9A | 41.1 ± 27.6 |
| M10I | 216.8 ± 142.8 |
| P14A | 0.4 ± 0.2 |
| K20A | 0.4 ± 0.2 |
| D26A | 0.5 ± 0.3 |
| V30M | 100.3 ± 48.9 |
| H32A | 1.2 ± 0.1 |
| E35A | 42.3 ± 27.1 |
| G37A | 0.4 ± 0.2 |
| G38A | 3.1 ± 2.4 |
| GG37-38AA | 0.4 ± 0.3 |
| R39A | 60.4 ± 11.7 |
| Y41A | 0.3 ± 0.2 |
| E47A | 8.1 ± 6.3 |
| E53A | 0.7 ± 0.5 |
| G75A | 301.3 ± 244.3 |
| R76A | 1.1 ± 0.4 |
| G77A | 73.3 ± 37.7 |
| G79A | 22.7 ± 9.3 |
| K80A | 0.9 ± 0 |
| G84A | 0.4 ± 0.2 |
| M85L | 53.3 ± 16.2 |
| C86A | 29.7 ± 12.1 |
| C86I | 122.4 ± 17.3 |
| C87A | 0.5 ± 0.1 |
| CC86-87AA | 0.7 ± 0.2 |
| S92A | 1.1 ± 0.8 |
| I93A | 79.9 ± 7.1 |
| L94A | 0.5 ± 0.3 |
| W96A | 126.8 ± 10.6 |
| H103A | 33 ± 2.8 |
| W104A | 0.5 ± 0.2 |
| I109A | 0.6 ± 0.3 |
| E112A | 0.4 ± 0.2 |
| P121A | 0.4 ± 0.2 |
| E122A | 0.3 ± 0.2 |
| N132A | 0.5 ± 0.2 |
| Q133A | 0.3 ± 0.2 |
| V137M | 0.4 ± 0.3 |
To correct for transfection efficiency, values were normalized to those measured at 4 h postelectroporation.
To test if any of the NS4A mutations might have an effect on virus production, we collected the supernatants from cells 72 h postelectroporation and used them to infect Huh7 cells. After 48 h, cells were lysed, and luciferase activity was evaluated (Fig. 1D and E). For this assay, we excluded the replication-incompetent NS4A mutants because these cannot produce virus a priori. None of the tested mutants showed significantly altered virus production (Fig. 1E), indicating that those NS4A mutations do not affect virion assembly or release.
Identification of compensatory second-site mutations.
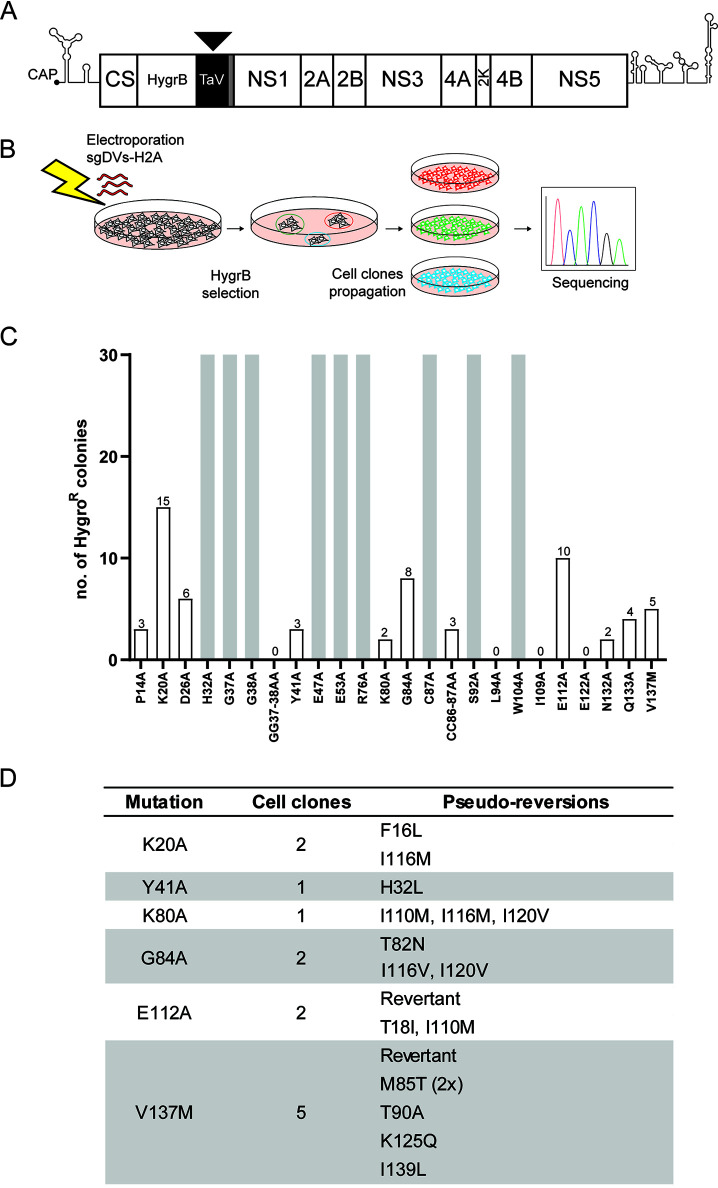
To establish a genetic interaction map of NS4A, we performed a forward genetic screen to select for second-site mutations that could compensate for the replication defects caused by the primary NS4A mutations. Toward this aim, we inserted 24 replication-suppressing mutations (reduction of replication > 90%) (Fig. 1C and Table 1) into a selectable subgenomic reporter system (sgDV-H2A), which encodes a hygromycin resistance gene (Fig. 2A). In vitro-transcribed RNA was prepared and electroporated into Vero E6 cells. Twenty-four hours postelectroporation, hygromycin B was added to the medium to select for cells containing a stably replicating replicon (Fig. 2B). For 9 of the 24 primary mutants tested, confluent cell monolayers were obtained (Fig. 2C). These included mutants H32A, G38A, E47A, and S92A, which retained low-level replication in Huh7 cells (Fig. 1C and Table 1) that was probably sufficient to exert antibiotic resistance even without the need for second-site mutation. For the other mutants (G37A, E53A, R76A, C87A, and W104A), replication might be due to either decreased replication suppression by these mutations in Vero E6 cells compared to Huh7 cells used for the transient replication assay or to rapid reversion to WT. In any case, this outcome prevented further evaluation of these 9 mutants. For mutants L94A, I109A, E122A, and GG37-38AA, no cell colonies were obtained after 3 weeks of selection (Fig. 2C). Conversely, for the other 11 mutants, 2 to 15 individual cell colonies were obtained.
FIG 2.
Selection for pseudoreversions rescuing replication deficiency of primary mutations in NS4A. (A) Schematic representation of the selectable subgenomic DENV-2 (strain 16681) replicon (sgDVs-H2A) used to select for pseudoreversions. The hygromycin phosphotransferase gene (HygrB) is preceded by the cis-acting cyclization sequence residing in the capsid-coding region (CS). The sequence coding for the 2A-like peptide from the Thosea asigna virus (TaV) ensures cotranslational processing of the polyprotein to release properly cleaved NS1. The sequence coding for the last transmembrane helix of the envelope protein (gray box) and acting as signal sequence is retained upstream of the NS1-coding sequence to allow for ER membrane passage of NS1. (B) To select for second-site mutations rescuing viral replication, Vero E6 cells were electroporated with in vitro transcripts of sgDVs-H2A containing given primary mutations in NS4A. Electroporated cells were cultured in the presence of hygromycin B, and single-cell clones were selected and expanded. Total RNA was extracted, and coding regions of the DENV replicons were amplified by reverse transcriptase PCR (RT-PCR). Amplicons were subjected to nucleotide sequence analysis in the NS4A-coding region. (C) Number of hygromycin B-resistant cell colonies obtained after selection for each of the given constructs. Gray bars indicate confluent cell monolayers that were obtained with cells transfected with given replicon mutants. (D) List of pseudoreversions in NS4A identified in replicons after hygromycin B selection. The numbering of amino acid residues refers to strain 16681. For each cell clone, the pseudoreversion identified in replicon RNA of said clone is given. Revertant indicates a nucleotide substitution that restores the original amino acid residue.
To determine if second-site mutations in NS4A of replicon RNAs in these cell clones were compensating for the primary mutations, cell colonies were expanded, total RNA extracted, and amplicons were generated that were subjected to nucleotide sequence analysis using primers covering the NS4A-coding region. Expansion of cell colonies harboring the P14A, D26A, CC86-87AA, N132A, and Q133A replicon mutants failed; thus, viral RNA could not be amplified. For two cell clones, nucleotide substitutions that reverted the mutated residues back to the WT NS4A sequence were observed. In all the other cases, second-site mutations affecting other NS4A residues were found in addition to the primary mutations. Interestingly, four different primary mutations yielded similar second-site mutations. Primary mutation K80A led to 3 S site mutations, I110M, I116M, and I120V, each located in close proximity of the pTMS3 region. Different combinations of these second-site mutations were also found for the K20A, G84A, or E112A primary mutants.
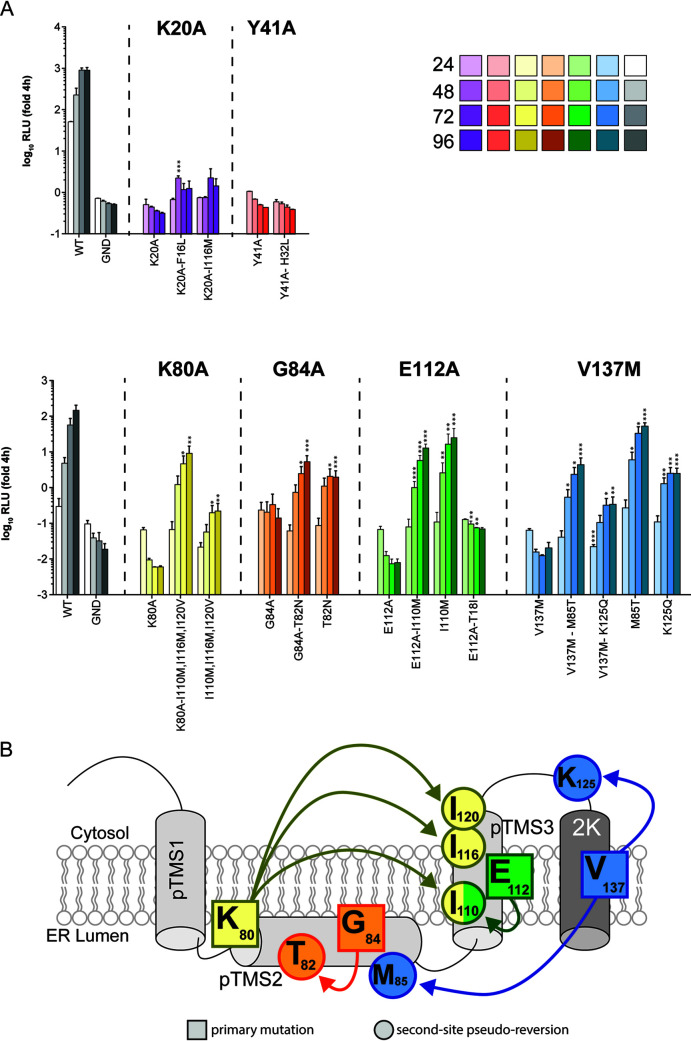
To confirm that the second-site mutations were able to rescue viral replication, pseudoreversions were inserted together with their respective primary mutations into the DENV luciferase reporter virus genome DVs-R2A, and in vitro transcripts were transfected into Vero E6 cells. Importantly, none of the tested pseudoreversions on their own conferred replication levels higher than the WT construct (Fig. 3A), suggesting that these compensatory mutations did not increase replication fitness per se. The second-site mutations identified for K20A or Y41A, as well as the T18I second-site mutation, coselected with the primary E112A mutation, did not restore viral replication (Fig. 3A). Possibly for these 3 mutants, second-site mutations might have arisen outside the NS4A-coding region, which we would have missed in our study. For all of the other identified second-site mutations, we observed a significant rescue of viral replication of the respective primary mutant (Fig. 3A). Although in all cases, replication was lower than the one of the WT, this reduction is not unexpected since replication levels far below the WT are sufficient to confer antibiotic resistance to the cell, and therefore, such attenuated mutants can be enriched during the selection process.
FIG 3.
Effect of second-site mutations on replication of wild-type and NS4A mutants. (A) Vero E6 cells were electroporated with the reporter virus DVs-R2A containing the mutations specified on the bottom of each panel. Cell lysates were collected 24, 48, 72, and 96 h after transfection, and renilla luciferase activity was measured as surrogate for viral replication. Values were normalized to the 4-h time point, reflecting transfection efficiency. Data represent the mean and SD of three independent experiments. Significance has been calculated by Student’s t test using, for comparison, values from the same time point obtained with the primary mutant. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) NS4A genetic interaction map as determined through reverse and forward genetic screens. Arrows connect primary mutations (squares) and the corresponding second-site compensatory mutations (circles).
Analysis of obtained replication data allowed to establish a genetic interaction map of NS4A (Fig. 3B). For residues G84A, E112A, and V137M, pseudoreversions resided in close proximity of the primary mutations. Interestingly, the V137M mutation, located within the 2K peptide, was also rescued by the M85T mutation located within the pTMS2, while mutation K80A in the pTMS2 was rescued by residues in pTMS3, arguing for genetic interactions between the different pTMSs of NS4A.
Determinants in NS4A required for interaction between the NS4A-2K-4B precursor and NS3.
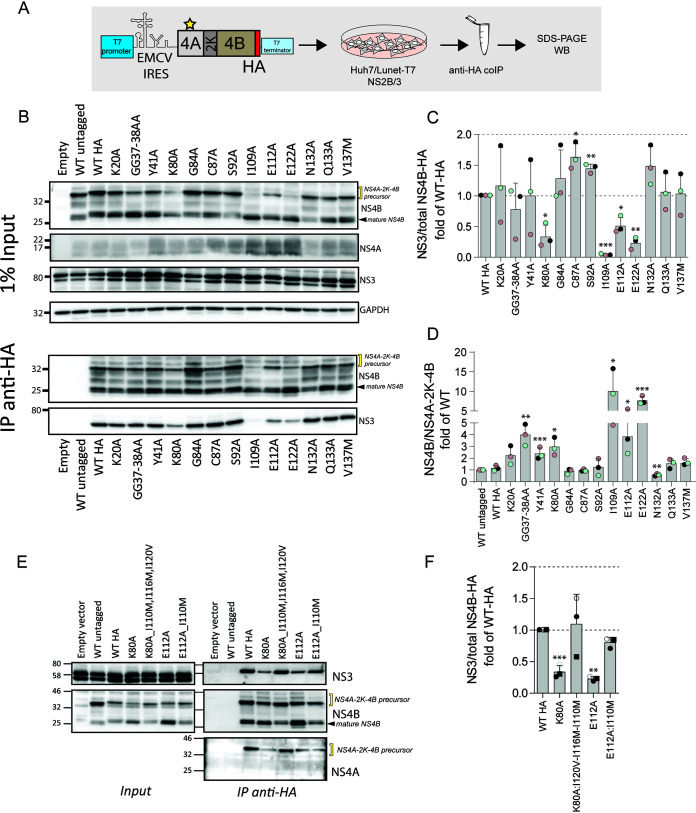
Previously, we have shown that the interaction between NS3 and NS4B is essential for viral replication (19). Moreover, we recently demonstrated that the NS4A-2K-4B precursor produced during DENV infection interacts with NS1, indicating an important role for NS4A-2K-4B in virus replication (5). Thus, mutations in NS4A might influence cleavage of this precursor or affect the interaction of this precursor, and indirectly of NS4B, with the other viral proteins. To investigate the effect of NS4A mutations on the interaction between NS4A and NS4B or NS3, we selected 13 mutations from those that severely reduced viral replication and evaluated changes in protein interactions. Each mutation was inserted into an NS4A-2K-4B expression vector encoding a C-terminally hemagglutinin (HA) epitope-tagged NS4B (Fig. 4A). The coding sequences were placed under the control of the T7 RNA polymerase promoter to allow cytoplasmic transcription in Huh7 cells stably expressing both T7 RNA polymerase and the DENV NS2B-3, the latter mediating NS4A-2K-4B cleavage. Cell extracts were prepared, and protein interactions were evaluated by coimmunoprecipitation. Consistent with previous studies, NS3 was efficiently coimmunoprecipitated with NS4B (Fig. 4B). Although the majority of NS4A mutations did not alter interactions between NS4B and NS3, a significant reduction in this association was observed for specific mutations, including I109A, E112A, and E122A (Fig. 4B; summarized in Fig. 4C). Interestingly, these mutations cluster within pTMS3 of NS4A (Fig. 1B), consistent with the notion that pTMS3 plays a particularly important role in viral replication.
FIG 4.
Impact of mutations in NS4A on the interaction between the NS4A-2K-4B precursor and the NS2B-3 complex. (A) Experimental approach. Plasmids encoding the HA-tagged NS4A-2K-4B precursor into which NS4A mutations had been inserted were transfected into Huh7/Lunet-T7 cells stably expressing DENV NS2B-3. Cell extracts were prepared 16 h posttransfection and subjected to HA-specific immunoprecipitation. Captured immunocomplexes were analyzed by SDS-PAGE and Western blotting. (B) Western blot analysis of captured protein complexes. A representative experiment of three independent experiments is shown. GAPDH served as loading control of the input. (C) Quantification of coimmunoprecipitation experiments representing the ratio of NS3 and HA-reactive proteins, i.e., uncleaved NS4A-2K-4B-HA precursor and processed NS4B-HA. Values were normalized against the HA-tagged wild-type sample that was set to 1. Data are the mean and SD from three independent experiments. (D) Quantification of the ratio between mature NS4B and NS4A-2K-4B precursor signals. Data are the mean and SD from three independent experiments normalized against the wild-type untagged sample. (E) Restoration of the interaction between NS2B-3- and NS4B-containing proteins (NS4A-2K-4B-HA and NS4B-HA) by second-site compensatory mutations in NS4A. Plasmids expressing the HA-tagged NS4A-2K-4B precursor containing primary and the respective compensatory mutations in NS4A were transfected into Huh7/Lunet-T7 cells stably expressing DENV NS2B-3. Immunoprecipitations were performed as described above. Abundance of proteins specified on the right of each panel was determined by Western blot analysis. A representative experiment of three independent experiments is shown. (F) Quantification of panel E. Values representing the ratio between NS3 and NS4B reactive proteins (uncleaved NS4A-2K-4B-HA precursor and processed NS4B-HA) were normalized against the wild-type sample that was arbitrarily set to 1. Data are the mean and SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by two-tailed t test.
Trans-cleavage of the NS4A-2K-4B precursor is inefficient, with less than 40% of mature NS4B being released from the precursor at the steady state (Fig. 4B, WT). Interestingly, we observed an inverse correlation between NS3 binding to NS4B and NS4A-2K-4B polyprotein cleavage, especially for the mutants clustering within the pTMS3. There, the I109A mutation leads to the lowest NS3-NS4B association and the largest amounts of mature NS4B, relative to the NS4A-2K-4B precursor (Fig. 4B and D), indicating that the NS3-NS4B interaction occurs primarily via the precursor.
The relevance of NS3-NS4B association for viral replication was corroborated when we tested the capability of the pseudoreversions to restore this association. These pseudoreversions were available for primary mutations K80A and E122A. Their combination with the respective pseudoreversions restored the ratio of uncleaved NS4A-2K-4B to NS4B and rescued the NS4B-NS3 interaction (Fig. 4E and F) as well as viral replication (Fig. 3A). These results reinforce the notion that a strict correlation exists between viral replication and interaction between NS3 and NS4B. Moreover, our results suggest that regulated cleavage of the NS4A-2K-4B precursor is important for NS3 interaction with NS4B and viral replication.
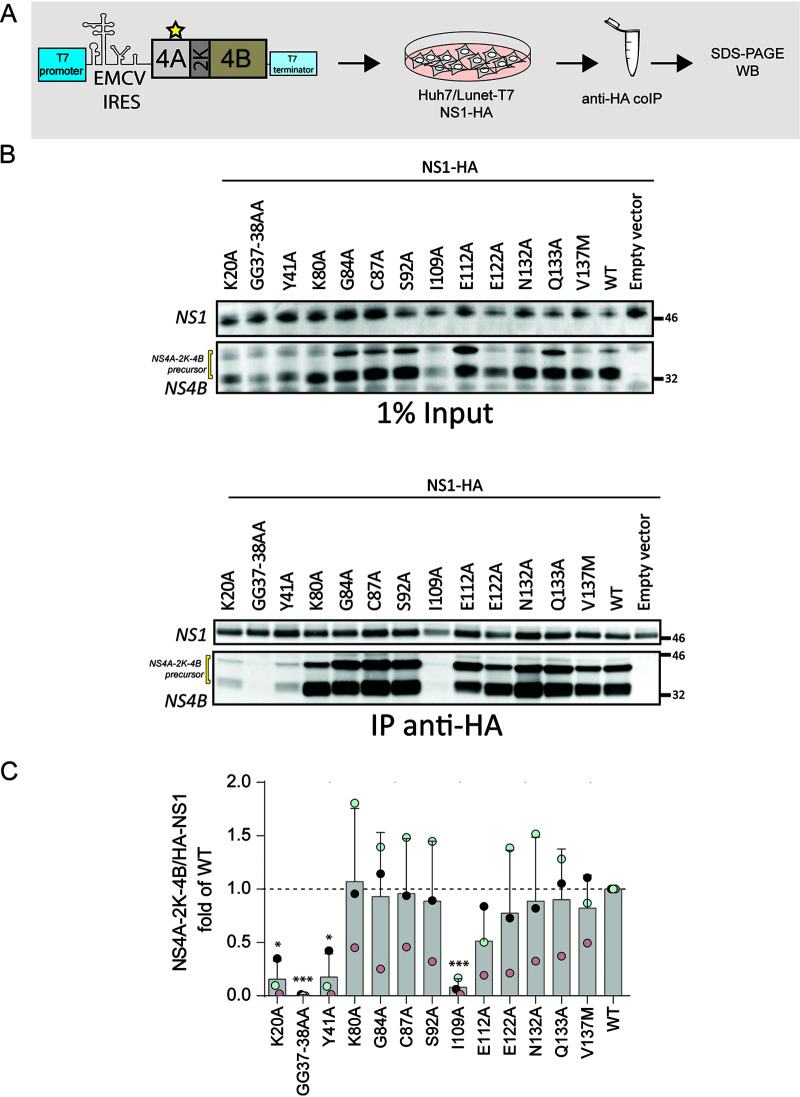
Determinants in NS4A required for interaction between the NS4A-2K-4B precursor and NS1.
Several studies have demonstrated genetic interactions between NS4A or NS4B and NS1 for different flaviviruses (5, 15), but direct interaction between NS4A or NS4B and NS1 has not yet been described. We reported earlier the interaction of the NS4A-2K-4B precursor with NS1, both in infected cells and in transient expression systems (5). To determine if mutations in NS4A alter the association between the NS4A-2K-4B cleavage intermediate and NS1, constructs containing specific NS4A point mutations were transiently expressed in NS1-HA-expressing Huh7 cells using the cytoplasmic T7 system as described above (Fig. 5A). Interactions were evaluated by HA-specific immunoprecipitation and Western blotting (Fig. 5B). For the majority of mutants, interaction between NS1 and the NS4A-2K-4B precursor was comparable to the WT. In the case of mutants K20A, GG37-38AA, Y41A, and I109A, association with NS1 was significantly reduced, which might be due in part to reduced protein abundance, best visible in the case of mutants GG37-38AA and I109A (Fig. 5B and C). Except for the latter, these NS4A mutations reside within the cytosolic N-terminal region of NS4A, highlighting a role for this region in precursor cleavage/stability and interaction with NS1.
FIG 5.
Mutations in NS4A affecting the interaction between the NS4A-2K-4B precursor and NS1. (A) Schematic representation of the experimental approach. Plasmids encoding the NS4A-2K-4B precursor and containing mutations in NS4A were transfected into Huh7/Lunet-T7 cells stably expressing HA-tagged DENV NS1. Cell extracts were prepared 16 h posttransfection and used for HA-specific immunoprecipitation. Captured immunocomplexes were analyzed by SDS-PAGE and Western blotting. (B) Western blot analysis of captured immunocomplexes. A representative experiment of three independent experiments is shown. (C) Quantification of coimmunoprecipitation experiments showing the ratio between HA-NS1 and the NS4A-2K-4B precursor. Values were normalized against the wild-type (WT) sample that was set to 1. Data are the mean and SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by two-tailed t test.
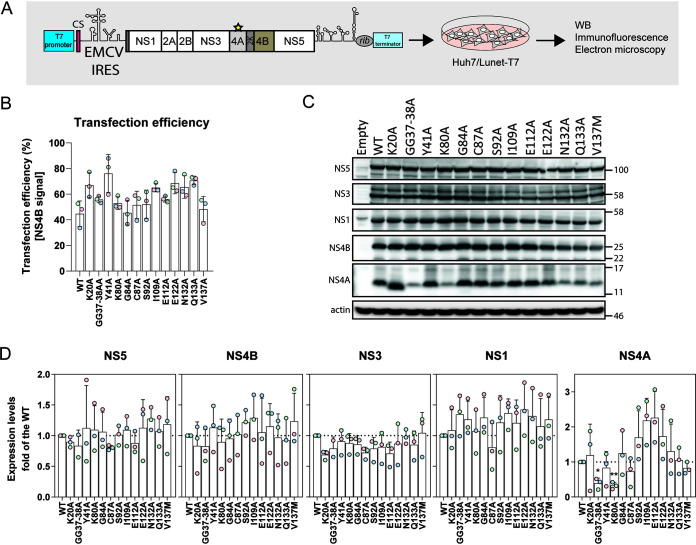
Effect of NS4A mutations on polyprotein cleavage.
Our results show that several NS4A mutations affect NS4A-2K-4B precursor maturation in the context of the trans-cleavage system, where NS4A-2K-4B and NS2B-3 are expressed separately. We next tested if these mutations also altered viral protein stability or processing in the context of a polyprotein comprising all NS proteins. To this end, we employed the recently described pIRO-D system that allows expression of the DENV-2 NS1-5 polyprotein (Fig. 6A) (17). While this system is not suitable for analysis of NS4A-2K-4B processing because of very fast cleavage kinetics (5), it allows evaluation of polyprotein processing for all the mature DENV NS proteins. In addition, this system supports RO biogenesis in the absence of RNA replication, allowing for the evaluation of replication-abolishing mutations for their impact on RO formation. To this end, we inserted selected NS4A mutations individually into the pIRO-D plasmid and evaluated effects on abundance of mature, i.e., fully processed, NS proteins (Fig. 6B and C). None of the mutations significantly affected production of NS1, NS3, NS4B, and NS5 at steady state (Fig. 6C and D). In the case of NS4A, both the double-glycine mutation GG37-38AA and the K80A mutation located in the N-terminal cytosolic region and at the start of pTMS2, respectively, reduced abundance of NS4A, arguing that these residues might be involved in the stability of NS4A.
FIG 6.
Effect of NS4A mutations on DENV polyprotein processing. (A) Schematic representation of the DENV expression construct (pIRO-D) (17) used in this study. It comprises the T7 RNA polymerase promoter, part of the 5′ nontranslated region (NTR) and the cyclization sequence in the DENV capsid-coding region (CS), the encephalomyocarditis virus internal ribosome entry site (EMCV IRES), the coding region from the signal sequence of NS1 in the 3′ terminal region of E up to the stop codon of the polyprotein, the complete 3′ NTR, the ribozyme of the hepatitis D virus, and the T7 terminator. Mutations specified in the individual panels were inserted into NS4A, and plasmids were transfected into Huh7/Lunet-T7 cells. After 16 h, cells were harvested for immunofluorescence, Western blot analysis, and electron microscopy. (B) Quantification of the transfection efficiency obtained with the different pIRO-D constructs as determined by immunofluorescence using NS4B-specific antibody. Mean and SD of three independent experiments are shown. (C) Polyprotein cleavage and abundance of cleavage products was determined by Western blot analysis using antibodies with specificity given on the left of each panel. Actin was used as loading control. A representative experiment of three independent experiments is shown. (D) Quantification of three experiments, including the one in panel C. Values are the mean and SD of three independent experiments. *, P < 0.05; **, P < 0.01, as determined by two-tailed t test.
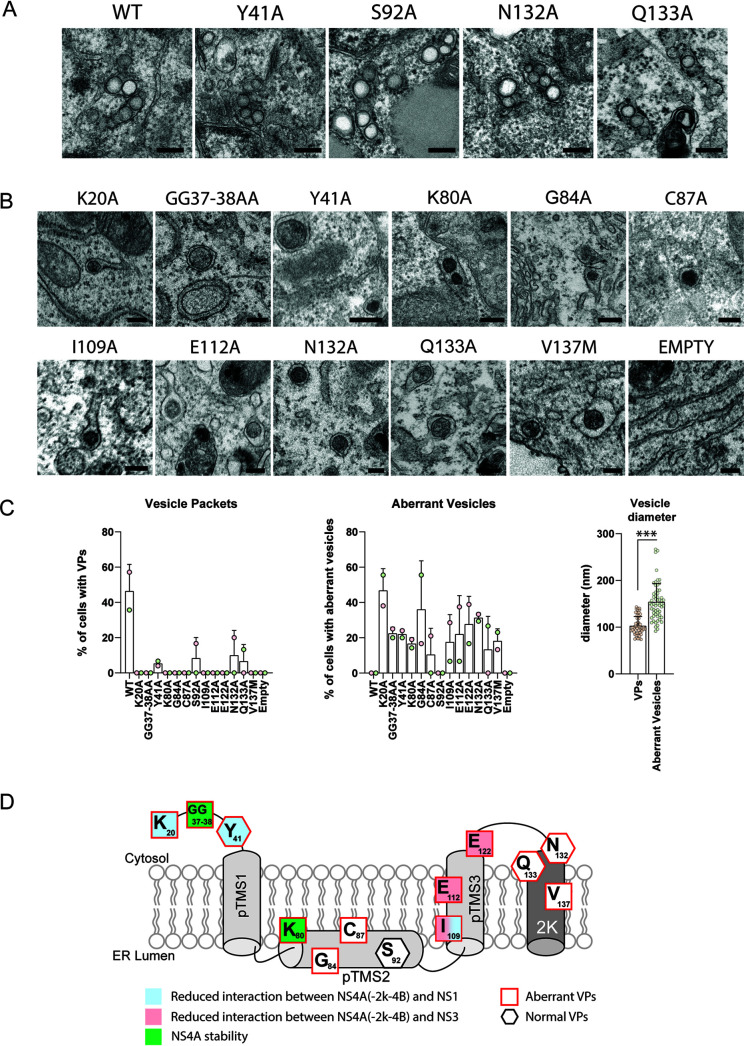
Effect of NS4A mutations on replication organelle biogenesis.
It has been proposed that NS4A, together with other NS proteins such as NS4B and NS2A, might contribute to the formation of the viral ROs that appear in transmission electron microscopy as clusters of vesicles designated VPs. Indeed, several studies support a role for NS4A in membrane remodeling (reviewed in reference 1), but its contribution to VP formation is poorly defined. To test if NS4A is important for VP biogenesis, we performed electron microscopy analysis of cells transfected with pIRO-D constructs harboring various NS4A mutations. Consistent with our recent report (17), expression of the DENV WT polyprotein through the pIRO-D system induces vesicular ER invaginations that are structurally identical to those observed during infection (Fig. 7A). Notably, all of the NS4A mutants were severely impaired in VP formation. In the case of NS4A mutants Y41A, S92A, N132A, and Q133A, efficiency of VP formation was drastically reduced, and those VPs being detectable had a morphology either comparable to VPs induced by the WT polyprotein or of aberrant morphology, with the exception of mutant S92A (Fig. 7A and C). All other NS4A mutants either failed to induce VPs or induced VPs with an aberrant morphology, notably the accumulation of electron-dense material in the vesicle lumen and a larger diameter with respect to WT VPs (Fig. 7B and C).
FIG 7.
Effect of NS4A mutations on the biogenesis of vesicle packets. (A and B) Electron microscopy analysis of Huh7/Lunet-T7 cells transfected with polyprotein constructs specified in Fig. 6A. Transfected cells were fixed and embedded into Epon resin, and thin sections were examined by transmission electron microscopy. For each sample, at least 15 cells were analyzed. Representative examples of vesicle packets or aberrant vesicles are shown in panels A and B, respectively. The experiment has been repeated twice. Scale bar, 200 nm. (C) Left and middle panels show the percentage of cells presenting VPs and aberrant vesicles, respectively. Each dot represents the mean of at least 15 cells analyzed. Two independent experiments were performed. The right panel shows the diameter of aberrant vesicles (nm) as found in cells transfected with the mutants specified in the middle panel. Each dot represents one vesicle. Means and SD are shown. ***, P < 0.001, as determined by two-tailed t test. (D) Graphical summary of the NS4A mutation analysis. Residues in NS4A and their contributions to interaction with NS1 and NS3, as well as their role in protein stability and VP formation, are displayed. Note that mutations affecting residues Y41, N132, and Q133 induced both regular VPs and VPs of aberrant morphology.
In summary, these results demonstrate that DENV NS4A is required for RO formation. Moreover, our data suggest that impaired replication of the NS4A mutants is due to impaired ability to form viral ROs. We speculate that, for some mutants, this might be the result of altered cleavage kinetics of the NS4A-2K-4B precursor, changes in NS4A stability, or altered interaction of NS4A, either as such or as part of the precursor, with other viral proteins (Fig. 7D).
DISCUSSION
In order to replicate their genome and produce virus progeny, positive-strand RNA viruses reorganize cellular membranes to create a microenvironment conducive to virus-driven processes. For DENV, the morphology of these membrane alterations has been well documented, but the underlying mechanisms of biogenesis and function are still poorly understood. The DENV genome encodes several trans-membrane proteins, including NS4A and NS4B, that can reshape cellular membranes and have been predicted to serve essential functions in RO formation. In order to gain a better understanding of the role of these viral membrane proteins in the DENV replication cycle, we have established a genetic map of NS4A, identifying amino acid residues that are crucial for the various functions of this protein, i.e., stability, interaction with NS1 and NS4B, induction of ROs, and RNA replication (Fig. 7D).
Our initial mutation analysis targeted 37 amino acid residues in NS4A and identified multiple positions of relevance for viral replication. Among those, mutations affecting 3 amino acid residues highly conserved between flaviviruses (L94, I109, and E122) did not select for pseudoreversions, arguing for complete loss of viral replication and indicating a vital role of these residues in maintaining NS4A structure, membrane association, or associations with other viral proteins or host cell factors. Conversely, we identified 4 mutations that could be rescued by second-site mutations within NS4A. Save for K125Q, these pseudoreversions resided in NS4A pTMS2 and pTMS3, indicating genetic flexibility within these domains that allows for the selection of compensatory mutations restoring protein structure or binding affinity disturbed by primary mutations. For example, the impairment in NS3 binding conferred by the E112A mutation could be completely rescued by an additional mutation in pTMS3 (I110M). Interestingly, a previous study showed that the NS4A pTMS3 mutations I110M and I116M could both compensate for a K8A substitution introduced into NS4B, arguing that NS4A also associates with NS4B (19).
Nonstructural proteins can have a dual function, being involved in viral genome replication and virus particle assembly or release, as exemplified with DENV NS2A (20). Therefore, we also determined whether mutations in NS4A with little or no impact on viral replication might affect the production of infectious DENV particles. However, we did not observe such a phenotype, indicating that NS4A is not involved in assembly or release of infectious DENV particles, although we cannot exclude that residues in NS4A not studied here might contribute to this process.
The location of primary and second-site mutations, in combination with protein interaction studies, provides insight into the structure and function of specific NS4A regions (Fig. 7D). For instance, we observed mutations in pTMS3 that severely limit virus replication and disrupt interactions with the NS2B-3 complex. In addition, the pTMS2 K80A mutation reduced abundance of NS4A without affecting the levels of the other nonstructural proteins. This defect was rescued by the combination of 3 mutations in pTMS3. Of note, this K80A mutation, as well as mutations I109A and E122A, profoundly reduces the ratio between uncleaved and cleaved NS4A-2K-4B, suggesting that these mutations affect polyprotein processing kinetics or decrease precursor stability or increase stability of the cleavage products.
Pseudoreversions for replication-impairing NS4B mutations inserted into the very N-terminal region of this protein have been identified in NS4A pTMS3, indicating associations between this domain and NS4B (19). Although biochemical analysis mapped the interaction domain of NS4A with NS4B to the N-terminal region of NS4A (12), these studies were done with individually expressed proteins and did not consider the NS4A-2K-4B precursor. We note that each of the pTMS3-linked mutations limits virus replication and viral RO formation, arguing that associations between pTMS3 and the NS4B N-terminal region are critically involved in these processes.
Mutations K20A and Y41A residing in the NS4A N-terminal region did not alter associations with NS3 but led to a decreased interaction with NS1 (Fig. 7D). The N-terminal region of NS4A has a predicted α-helix structure, associates with highly curved membranes, and is important for NS4A oligomerization (13, 21). Recent studies suggest that NS1 interacts with the NS4A-2K-4B precursor, but not with fully processed NS4A or NS4B, and this interaction is required for VP formation (5). Here, we found that the N-terminal NS4A region plays a critical role in this interaction, as mutations inserted into this region reduced interaction with NS1 and impaired VP formation. The GG37-38AA mutation residing in this region appears to reduce stability of fully processed NS4A since decreased NS4A levels were observed when these substitutions were analyzed in the context of the polyprotein. Since abundance of the other viral proteins was unaltered, these NS4A mutations did not affect general polyprotein processing but, rather, reduced the stability of NS4A.
Our genetic analysis combined with interaction studies identified several residues in NS4A involved in the interaction between NS4A-2k-4B and NS1 and NS3. Interestingly, the NS4A N-terminal region is in the cytosol, while NS1 resides in the ER lumen; thus, a direct interaction between these two proteins mediated by the N-terminal NS4A residues is not conceivable. Similarly, NS4A residues modulating the interaction with NS3 are either embedded in the ER membrane or located in the ER lumen and, as such, cannot mediate a direct interaction with NS3. One possible explanation for the loss of these interactions could be that these residues influence the topology, or protein stability, or subcellular distribution of NS4A or the NS4A-2k-4B precursor. For example, mutations E112A or E122A might affect membrane insertion and, thus, membrane topology of NS4A. The same could apply to the mutations in the N-terminal region of NS4A that might affect NS4A structure in a way that association with NS1 is reduced. Alternatively, the N-terminal region of NS4A was shown to bind highly curved membranes and to contribute to protein oligomerization (13, 14). Therefore, this segment might contribute to localizing NS4A or its precursor to specific membrane regions favorable to interacting with NS1 (perhaps involving I109 located in pTMS2 facing the ER lumen). However, in the absence of information about the in-membrane structure of NS4A, the precise impact of individual mutations on NS4A structure and membrane topology remains unknown.
We also identified several replication-inactivating mutations within the 2K peptide-coding region, one of which (V137M) being rescued by a reversion in NS4A, arguing for cross talk between the 2K peptide and NS4A. In line with that, we reported earlier that fully cleaved NS4A, expressed on its own, induced membrane alterations, whereas NS4A retaining the 2K peptide did not, indicating a suppressive function of 2K on NS4A-induced membrane remodeling (8). Whether this is mediated by direct protein-protein interaction remains to be determined.
By using our recently established pIRO-D polyprotein expression system (17), we dissected the specific step of the viral replication cycle requiring NS4A. We found that each NS4A mutation limiting virus replication also impaired the formation of viral ROs. In fact, of all the 13 tested point mutations, only Y41A, S92A, N132A, and Q133A were still able to induce VPs, but with very limited efficiency. Interestingly, for 10 out of 15 mutants, including the VPs-forming Y41A, N132A, and Q133A mutants, we observed formation of morphologically aberrant vesicles within the ER lumen. While resembling invaginated vesicles, these structures were often larger and presented an electron-dense interior, clearly dissimilar to the electron-lucent VPs’ interior as found in infected cells or cells expressing the WT polyprotein. These findings confirm a role for NS4A in virus-driven membrane-shaping events. We envisage at least two ways how NS4A could contribute to RO formation. First, NS4A could drive RO formation as a structural component by forming oligomeric complexes and inserting protein wedges, such as pTMS2, into one layer of the membrane bilayer, thus causing asymmetric membrane surface expansion and curvature. Consistently, NS4A has been reported to induce membrane alterations when expressed in cells (8, 9). Proteins of similar function have been reported for other positive-strand RNA viruses. For instance, Flock House virus replicates its genome in membrane invaginations derived from the outer mitochondrial membrane (22). These invaginations are driven by the viral protein A that has two membrane-association domains and requires RNA replication catalyzed by the RNA-dependent RNA polymerase residing within protein A (23). In the case of hepatitis C virus (HCV), the integral membrane protein NS4B that is located at viral ROs is critically involved in shaping cellular membranes (24).
A second possibility of how NS4A might drive RO formation is by coordination of a sequence of interactions between NS1, NS2B-3, and the NS4A-2K-4B precursor. These interactions might be linked to cleavage kinetics, giving rise to processing intermediates such as 2K-4B and a balanced ratio of fully processed proteins (7). Further experiments will be required to determine the exact order and kinetics of these interactions and their precise contribution to RO formation as well as determining the role of host cell factors in this process.
In conclusion, our study has shed light on the genetic landscape of NS4A and identified regions within NS4A contributing to interactions with NS1 and NS2B-3, RO formation, and viral replication. Additionally, our findings point toward a complex series of interactions, including polyprotein-processing intermediates, which are required for virus-driven membrane alterations. Several studies have shown that viral proteins lacking enzymatic activity, such as DENV NS4B and HCV NS5A, are well-suitable drug targets for highly efficacious antiviral therapy (25–28). These proteins have in common with DENV NS4A their contribution to different processes in the viral replication cycle and their mode of action, which is mainly mediated by serving as a hub for interaction with several viral and cellular factors. In the light of these considerations, DENV NS4A might serve as a target for novel therapeutic strategies.
MATERIAL AND METHODS
Cells, viruses, and antibodies.
Human hepatoma (Huh7), Vero E6, and BHK-21 cells were grown in Dulbecco’s modified Eagle medium (DMEM; Thermo Scientific) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum. Huh7_T7, Huh7/Lunet_T7, and Huh7_T7_NS2B-3 cells were described before (19). For Huh7_T7 and Huh7/Lunet_T7 cells, zeocin was added to the culture medium at a final concentration of 5 μg/ml. Huh7_T7_NS2B-3 and the Huh7_T7_NS1-HA cells were generated by transduction of Huh7_T7 cells with lentiviruses encoding either DENV2 16681 NS2B-3 or NS1-HA and were cultured in the presence of 5 μg/ml and 1 μg/ml of puromycin, respectively. Generation of rabbit polyclonal antisera against DENV NS1, NS3, NS4A, NS4B, and NS5 has been described previously (29). Infectious DENV2 (strain 16681) and DENV2-derived reporter viruses used for infection studies were produced as described elsewhere (30).
Plasmids.
The plasmid pFK-DVs containing the full-length 16681 DENV2 strain and its derivates, the reporter plasmids pFK-DVs-R2A, encoding the Renilla reniformis luciferase (Rluc) gene, as well as the subgenomic replicon plasmids pFK-sgDVs-R2A and pFK-sgDVs-H2A encoding the Rluc or the hygromycin B resistance gene, respectively, were described elsewhere (30, 31). Primary NS4A mutations and pseudoreversions were inserted into the DENV2 strain 16681 by site-directed mutagenesis using an overlap PCR-based approach. The full list of primers is available on request. A silent mutation was inserted to destroy the BstBI restriction site present in the Rluc gene in the pFK-DVs-R2A plasmid to generate the pFK-DVs-R2A_ΔBstBI. Amplicons containing the mutated NS4A gene were inserted into the NheI/BstBI, or the BstBI/NruI or the NheI/NruI cassette of the pFK-DVs-R2A_ΔBstBI plasmid. In the case of selectable replicons, NS4A mutations were inserted into the BstBI/NruI cassette of the pFK-DVs-H2A plasmid. The pTM-based plasmids encoding NS4A-2K-4B, NS4A, or 2K-NS4B with or without C-terminal HA tag (HAct) were described elsewhere (19). The NS4A mutations were inserted into the pTM-NS4A-2K-4B and in the pTM-NS4A-2K-4B-HAct plasmids using an overlap PCR-based approach. Plasmids for polyprotein expression experiments were generated by restricting the pFK-DVs-R2A_ΔBstBI plasmids containing the NS4A mutations with NruI and XhoI and insertion of the DNA fragments into the pIRO-D plasmid (17, 18).
Transient transfection.
Cells were transiently transfected using the liposomal-based TransIt-LT1 (Mirus, Madison, WI, USA) reagent according to the manufacturer’s instructions. Cell culture medium was changed before and 4 h after transfection, and cells were collected for analysis 16 h posttransfection.
In vitro transcription and cell electroporation.
For in vitro transcription (IVT), pFK-based plasmids containing DENV genomes were linearized by restriction digest with XbaI, and linearized DNA was purified using a column-based purification kit (Macherey-Nagel). A total of 10 μg of linearized DNA was used for each IVT reaction. The reaction was performed in a total volume of 100 μl containing 20 μl of SP6 buffer (400 mM HEPES [pH 7.5], 80 mM MgCl2, 10 mM spermidine, and 200 mM dithiothreitol [DTT]), 12.5 μl of NTP mix (50 mM ATP, UTP, and CTP and 25 mM GTP), 20 μl of a 5 mM solution of 7mG(ppp)G RNA cap (NEB), 2.5 μl of RNAsin (40 U/μl; Promega), and 4 μl of SP6 RNA polymerase (20 U/μl; NEB). The reaction mixture was incubated at 40°C for 5 h, and then 20 μl of RQ1 RNase-free DNase (1 U/μl; Promega) was added to the sample, and incubation was continued at 37°C for 30 min. In vitro-transcribed RNA was purified by phenol-chloroform extraction and precipitation with isopropanol. Pellet was resuspended in RNase-free water, and integrity was evaluated by agarose gel electrophoresis.
For RNA electroporation, BHK-21, Huh7, or Vero E6 cells were trypsinized, centrifuged for 5 min at 200 × g, washed once in phosphate-buffered saline (PBS), and resuspended in cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM potassium phosphate buffer, 25 mM HEPES [pH 7.6], 2 mM EGTA, and 5 mM MgCl2) to a concentration of 1 × 107 cells/ml for Huh7 and Vero E6 cells and 1.5 × 107 cells/ml for BHK-21 cells. For each electroporation, 400 μl of cell suspension was mixed with 10 μg of in vitro transcript and transferred to a 0.4-cm gap width electroporation cuvette (Bio-Rad). Electroporation was performed by pulsing the cells once with 975 μF and 270 V (Gene Pulser II, Bio-Rad), and cells were transferred to prewarmed complete DMEM and seeded into culture dishes.
Viral replication assay.
The activity of DENV-encoded Rluc was monitored as a surrogate measurement of intracellular viral replication. Cells infected with DV-R2A reporter virus were lysed in Rluc lysis buffer (1% Triton X-100, 25 mM glycine-glycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, and 1 mM DTT) and stored at −20°C for 1 day. Rluc activity was measured using a Lumat LB 9507 luminometer (Berthold Technologies) by diluting 20 μl of cell lysate with 100 μl of Rluc assay buffer (15 mM potassium phosphate [pH 7.8], 25 mM glycine-glycine, 15 mM MgSO4, 4 mM EGTA, and 1.43 μM freshly added coelenterazine).
Selection for pseudorevertants.
The selection for pseudorevertants was carried out as previously described (19) with slight modifications. Vero E6 cells were electroporated with in vitro-transcribed RNA of the selectable DV-H2A DENV2 full-length virus encoding the hygromycin B resistance gene. The medium was exchanged after 24 h and supplemented with 500 μg/ml of hygromycin B. Medium supplemented with antibiotic was regularly exchanged to maintain a constant selection pressure. Mock-electroporated cells treated with the same amount of antibiotic were used to monitor the efficiency of antibiotic-induced cell death. After 4 weeks of selection, cell colonies were isolated, transferred to a 96-well plate, and cultured in medium containing hygromycin B. Cell clones were expanded to larger cell culture formats before being harvested to extract total RNA. Approximately 106 cells were used for RNA extraction for which the NucleoSpin RNA II kit was used according to the manufacturer’s protocol (Macherey-Nagel). One microgram of total RNA was used for reverse transcription (SuperScript III reverse transcriptase; Thermo Scientific) using a specific antisense primer (5′-CGACCTGACTTCTAGCCTTGTTTC-3′). The cDNA was used to amplify a portion of the coding region of DENV2 (nucleotides 2,422 to 10,248) using the Expand long template PCR system with forward primer 5′-ATTAGAGCTCGATAGTGGTTGCGTTGTGAGCT-3′ and reverse primer 5′-ATAATCTAGACCACAGAACTCCTGCTTCTTCC-3′. Amplified fragments were purified by agarose gel electrophoresis, and the sequence of the NS4A-coding region was determined.
Immunoprecipitation assay.
Immunoprecipitation assays were performed as previously described with slight modifications (19, 32). Briefly, cells were washed in PBS twice before being lysed in lysis buffer containing 0.5% dodecyl-β-d-maltoside, 100 mM NaCl, 20 mM Tris-HCl (pH 7.5), and EDTA-free protease inhibitors (Roche) for 30 min on ice. Samples were centrifuged for 30 min at 4°C, supernatants were transferred to a new tube, and protein concentration was calculated using Bradford protein assay (Bio-Rad). Lysates were incubated with anti-HA-conjugated magnetic beads (Dynabeads; Thermo Scientific) and equilibrated in lysis buffer for 3 h at 4°C (approximatively 20 μl of beads for 500 μg of total lysate). Samples were collected, washed 3 times with 20× beads volume of lysis buffer, transferred to a new tube, and washed twice with 10× beads volume of 50 mM Tris-HCl (pH 7.5) and 150 mM NaCl. Immunocomplexes were eluted by sequential incubation with 3% SDS in PBS, followed by a second elution with PBS only. The two fractions were pooled, diluted in loading buffer, and subjected to SDS-PAGE and Western blot analysis. Images were acquired with an Intas ECL Chemocam Imager (Intas), and relative protein amounts were quantified using the LabImage 1D software package (Kapelan).
Immunofluorescence analysis.
Cells seeded on glass coverslips were fixed in 4% paraformaldehyde (PFA) in PBS for 30 min at room temperature. Samples were washed 3 times in PBS, permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature, rinsed in PBS, and incubated in blocking buffer (0.5% milk in PBS). Primary antibodies were incubated for 1 h at room temperature in blocking buffer, followed by 3 washes in PBS containing 0.01% Tween 20. Incubation with Alexa Fluor secondary antibody (Thermo Scientific) was performed in PBS in the dark for 1 h. Samples were washed again as before and mounted on a glass slide using Fluoromount-G medium containing DAPI (4′,6-diamidino-2-phenylindole). Images were acquired with a Nikon Eclipse widefield microscope (Nikon).
Electron microscopy.
All electron microscopy (EM) buffers were prepared with high-performance liquid chromatography (HPLC)-grade water, and EM was performed as previously described (33, 34). In brief, cells grown on glass coverslips were fixed 16 h posttransfection with EM fixative containing 2.5% glutaraldehyde in sodium cacodylate buffer (50 mM sodium cacodylate, 50 mM KCl, 2.6 mM CaCl2, 2.6 mM MgCl2, and 2% sucrose) for 1 h at room temperature. Samples were washed 5 times in 50 mM cacodylate buffer and then incubated with 2% OsO4 in 50 mM sodium cacodylate on ice for 40 min. After 3 washes in water, samples were incubated with 0.5% uranyl acetate in water for 30 min in the dark, washed 3 times with water, and dehydrated with increasing concentrations of ethanol before being embedded in a mixture of Epon-based resin. Polymerization was promoted by incubation at 60°C for 72 h. Ultrathin sections of 70 nm were generated by sectioning with a Leica EM UC6 Ultramicrotome and a diamond knife. Sections were collected on EM grids and counterstained with lead citrate and uranyl acetate. Grids were examined with either a Zeiss EM 10C (Carl Zeiss) or a Jeol transmission electron microscope operating at 80 kV.
Multiple-sequence alignments.
Protein sequences of NS4A of the 4 DENV serotypes (DENV1 strain Hawaii, GenBank accession no. KM204119.1; DENV2 strain 16681, GenBank accession no. U87411.1; DENV3 strain H87, GenBank accession no. M93130.1; and DENV4 strain B5, GenBank accession no. AF289029.1) and prototype strains of different flaviviruses (Zika virus strain H/PF/2013, GenBank accession no. KJ776791.2; West Nile virus strain NY99, GenBank accession no. DQ211652.1; yellow fever virus strain Asibi, GenBank accession no. KF769016.1; Japanese encephalitis virus strain Nakayama, GenBank accession no. EF571853.1; and tick-borne encephalitis virus strain Neudoerfl, GenBank accession no. U27495.1) were aligned using PRALINE (35). The JPred4 server was used to predict the protein secondary structure (36).
Statistical analysis.
All statistical information related to individual experiments is reported in the figure legends. Significance was calculated with either Student’s test or with one-way analysis of variance (ANOVA) with Dunnett’s post hoc test.
ACKNOWLEDGMENTS
This project was supported by the Deutsche Forschungsgemeinschaft (SFB1129, TP11, and BA1505/8-1, both to R.B.). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Ralf Bartenschlager, Email: ralf.bartenschlager@med.uni-heidelberg.de.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16:125–142. 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaaijk P, Luytjes W. 2018. Are we prepared for emerging flaviviruses in Europe? Challenges for vaccination. Hum Vaccin Immunother 14:337–344. 10.1080/21645515.2017.1389363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortes M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M, CYD-TDV Dengue Vaccine Working Group. 2015. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med 373:1195–1206. 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- 5.Płaszczyca A, Scaturro P, Neufeldt CJ, Cortese M, Cerikan B, Ferla S, Brancale A, Pichlmair A, Bartenschlager R. 2019. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog 15:e1007736. 10.1371/journal.ppat.1007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akey DL, Brown WC, Dutta S, Konwerski J, Jose J, Jurkiw TJ, DelProposto J, Ogata CM, Skiniotis G, Kuhn RJ, Smith JL. 2014. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 343:881–885. 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C, Amberg SM, Chambers TJ, Rice CM. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol 67:2327–2335. 10.1128/JVI.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282:8873–8882. 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 9.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol 80:4623–4632. 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aktepe TE, Liebscher S, Prier JE, Simmons CP, Mackenzie JM. 2017. The host protein reticulon 3.1A is utilized by flaviviruses to facilitate membrane remodelling. Cell Rep 21:1639–1654. 10.1016/j.celrep.2017.10.055. [DOI] [PubMed] [Google Scholar]
- 11.Aktepe TE, Mackenzie JM. 2018. Shaping the flavivirus replication complex: it is curvaceous! Cell Microbiol 20:e12884. 10.1111/cmi.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou J, Xie X, Wang QY, Dong H, Lee MY, Kang C, Yuan Z, Shi PY. 2015. Characterization of dengue virus NS4A and NS4B protein interaction. J Virol 89:3455–3470. 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stern O, Hung YF, Valdau O, Yaffe Y, Harris E, Hoffmann S, Willbold D, Sklan EH. 2013. An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J Virol 87:4080–4085. 10.1128/JVI.01900-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CM, Xie X, Zou J, Li SH, Lee MY, Dong H, Qin CF, Kang C, Shi PY. 2015. Determinants of dengue virus NS4A protein oligomerization. J Virol 89:6171–6183. 10.1128/JVI.00546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindenbach BD, Rice CM. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol 73:4611–4621. 10.1128/JVI.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrose RL, Mackenzie JM. 2015. Conserved amino acids within the N-terminus of the West Nile virus NS4A protein contribute to virus replication, protein stability and membrane proliferation. Virology 481:95–106. 10.1016/j.virol.2015.02.045. [DOI] [PubMed] [Google Scholar]
- 17.Cerikan B, Goellner S, Neufeldt CJ, Haselmann U, Mulder K, Chatel-Chaix L, Cortese M, Bartenschlager R. 2020. A non-replicative role of the 3' terminal sequence of the dengue virus genome in membranous replication organelle formation. Cell Rep 32:107859. 10.1016/j.celrep.2020.107859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goellner S, Cerikan B, Cortese M, Neufeldt CJ, Haselmann U, Bartenschlager R. 2020. Replication-independent generation and morphological analysis of flavivirus replication organelles. STAR Protoc 1:100173. 10.1016/j.xpro.2020.100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatel-Chaix L, Fischl W, Scaturro P, Cortese M, Kallis S, Bartenschlager M, Fischer B, Bartenschlager R. 2015. A combined genetic-proteomic approach identifies residues within dengue virus NS4B critical for interaction with NS3 and viral replication. J Virol 89:7170–7186. 10.1128/JVI.00867-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X, Zou J, Zhang X, Zhou Y, Routh AL, Kang C, Popov VL, Chen X, Wang QY, Dong H, Shi PY. 2019. Dengue NS2A protein orchestrates virus assembly. Cell Host Microbe 26:606–622.e8. 10.1016/j.chom.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Hung YF, Schwarten M, Schunke S, Thiagarajan-Rosenkranz P, Hoffmann S, Sklan EH, Willbold D, Koenig BW. 2015. Dengue virus NS4A cytoplasmic domain binding to liposomes is sensitive to membrane curvature. Biochim Biophys Acta 1848:1119–1126. 10.1016/j.bbamem.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Miller DJ, Schwartz MD, Ahlquist P. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol 75:11664–11676. 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopek BG, Settles EW, Friesen PD, Ahlquist P. 2010. Nodavirus-induced membrane rearrangement in replication complex assembly requires replicase protein A, RNA templates, and polymerase activity. J Virol 84:12492–12503. 10.1128/JVI.01495-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. 2011. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J Virol 85:6963–6976. 10.1128/JVI.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moquin SA, Simon O, Karuna R, Lakshminarayana SB, Yokokawa F, Wang F, Saravanan C, Zhang J, Day CW, Chan K, Wang QY, Lu S, Dong H, Wan KF, Lim SP, Liu W, Seh CC, Chen YL, Xu H, Barkan DT, Kounde CS, Sim WLS, Wang G, Yeo HQ, Zou B, Chan WL, Ding M, Song JG, Li M, Osborne C, Blasco F, Sarko C, Beer D, Bonamy GMC, Sasseville VG, Shi PY, Diagana TT, Yeung BKS, Gu F. 2021. NITD-688, a pan-serotype inhibitor of the dengue virus NS4B protein, shows favorable pharmacokinetics and efficacy in preclinical animal models. Sci Transl Med 13:eabb2181. 10.1126/scitranslmed.abb2181. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Xie X, Ye N, Zou J, Chen H, White MA, Shi PY, Zhou J. 2019. Design, synthesis, and biological evaluation of substituted 4,6-dihydrospiro[[1,2,3]triazolo[4,5-b]pyridine-7,3'-indoline]-2',5(3H)-dione analogues as potent NS4B inhibitors for the treatment of dengue virus infection. J Med Chem 62:7941–7960. 10.1021/acs.jmedchem.9b00698. [DOI] [PubMed] [Google Scholar]
- 27.van Cleef KW, Overheul GJ, Thomassen MC, Kaptein SJ, Davidson AD, Jacobs M, Neyts J, van Kuppeveld FJ, van Rij RP. 2013. Identification of a new dengue virus inhibitor that targets the viral NS4B protein and restricts genomic RNA replication. Antiviral Res 99:165–171. 10.1016/j.antiviral.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O'Boyle DR, II, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100. 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischl W, Bartenschlager R. 2013. High-throughput screening using dengue virus reporter genomes. Methods Mol Biol 1030:205–219. 10.1007/978-1-62703-484-5_17. [DOI] [PubMed] [Google Scholar]
- 31.Chatel-Chaix L, Cortese M, Romero-Brey I, Bender S, Neufeldt CJ, Fischl W, Scaturro P, Schieber N, Schwab Y, Fischer B, Ruggieri A, Bartenschlager R. 2016. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe 20:342–356. 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaturro P, Cortese M, Chatel-Chaix L, Fischl W, Bartenschlager R. 2015. Dengue virus non-structural protein 1 modulates infectious particle production via interaction with the structural proteins. PLoS Pathog 11:e1005277. 10.1371/journal.ppat.1005277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, Schorb M, Pruunsild P, Schwab Y, Chatel-Chaix L, Ruggieri A, Bartenschlager R. 2017. Ultrastructural characterization of Zika virus replication factories. Cell Rep 18:2113–2123. 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortese M, Lee JY, Cerikan B, Neufeldt CJ, Oorschot VMJ, Kohrer S, Hennies J, Schieber NL, Ronchi P, Mizzon G, Romero-Brey I, Santarella-Mellwig R, Schorb M, Boermel M, Mocaer K, Beckwith MS, Templin RM, Gross V, Pape C, Tischer C, Frankish J, Horvat NK, Laketa V, Stanifer M, Boulant S, Ruggieri A, Chatel-Chaix L, Schwab Y, Bartenschlager R. 2020. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe 28:853–866.e5. 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simossis VA, Heringa J. 2005. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33:W289–W294. 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]