ABSTRACT
Long noncoding RNAs (lncRNAs) are involved in numerous cellular processes. Increasing evidence suggests that some lncRNAs function in immunity through various complex mechanisms. However, implication of a large fraction of lncRNAs in antiviral innate immunity remains uncharacterized. Here, we identified an lncRNA called lncRNA IFITM4P that was transcribed from interferon-induced transmembrane protein 4 pseudogene (IFITM4P), a pseudogene belonging to the interferon-induced transmembrane protein (IFITM) family. We found that expression of lncRNA IFITM4P was significantly induced by infection with several viruses, including influenza A virus (IAV). Importantly, lncRNA IFITM4P acted as a positive regulator of innate antiviral immunity. Ectopic expression of lncRNA IFITM4P significantly suppressed IAV replication in vitro, whereas IFITM4P deficiency promoted viral production. We further observed that expression of lncRNA IFITM4P was upregulated by interferon (IFN) signaling during viral infection, and altering the expression of this lncRNA had significant effects on the mRNA levels of several IFITM family members, including IFITM1, IFITM2, and IFITM3. Moreover, lncRNA IFITM4P was identified as a target of the microRNA miR-24-3p, which represses mRNA of IFITM1, IFITM2, and IFITM3. The experiments demonstrated that lncRNA IFITM4P was able to cross-regulate the expression of IFITM family members as a competing endogenous RNA (ceRNA), leading to increased stability of these IFITM mRNAs. Together, our results reveal that lncRNA IFITM4P, as a ceRNA, is involved in innate immunity against viral infection through the lncRNA IFITM4P–miR-24-3p–IFITM1/2/3 regulatory network.
IMPORTANCE lncRNAs play important roles in various biological processes, but their involvement in host antiviral responses remains largely unknown. In this study, we revealed that the pseudogene IFITM4P belonging to the IFITM family can transcribe a functional long noncoding RNA termed lncRNA IFITM4P. Importantly, results showed that lncRNA IFITM4P was involved in innate antiviral immunity, which resembles some interferon-stimulated genes (ISGs). Furthermore, lncRNA IFITM4P was identified as a target of miR-24-3p and acts as a ceRNA to inhibit the replication of IAV through regulating the mRNA levels of IFITM1, IFITM2, and IFITM3. These data provide new insight into the role of a previously uncharacterized lncRNA encoded by a pseudogene in the host antiviral response and a better understanding of the IFITM antiviral network.
KEYWORDS: IFITM family, influenza A virus, innate immunity, lncRNA, pseudogene
INTRODUCTION
Influenza virus causes pandemics and seasonal epidemics worldwide, which poses a great threat to human beings (1–4). Influenza-associated respiratory diseases have been estimated to take 291,243 to 645,832 human lives annually (5). Besides deaths or symptoms directly caused by influenza infection, people with chronic disorders, such as cardiovascular disease, are particularly vulnerable to influenza virus infection (6). Human influenza A and B viruses are prevalent among human beings. However, most influenza A virus (IAV) genes have a higher evolutionary rate than that of influenza B virus (7–10). IAVs are subtyped according to their surface antigen hemagglutinin (HA) and neuraminidase (NA). Reassortment between distinct IAVs leads to the generation of new subtypes of IAV, which may cause the emergence of highly pathogenic strains that have pandemic potential (2, 11).
During IAV infection, the recognition of pathogen-associated molecular patterns (PAMPs) by pathogen recognition receptors (PRRs) like retinoic acid-inducible gene I (RIG-I) is the first step to activate host antiviral response. Transcription factors, including NF-κB and IRF3/7, are then activated, respectively, which together with other components, constitute an enhanceosome to trigger interferon (IFN) transcription (12, 13). IFNs bind to their corresponding receptors that activate Janus protein tyrosine kinase-signal transducer and the activator of transcription (JAK-STAT) signaling pathway to induce the expression of antiviral proteins encoded by interferon-stimulated genes (ISGs) (14). Moreover, initial activation of STAT1 could be mediated by the spleen tyrosine kinase (Syk) to induce ISGs downstream of RIG-I/MAVS pathway at an early stage of viral infection (15).
IFN-induced transmembrane (IFITM) proteins, which belong to the Dispanin/CD225 family, are well-characterized antiviral ISG products (16–18). The CD225 superfamily proteins harboring the CD225 domain can regulate vesicular membrane fusion (19). Human IFITM genes are located on chromosome 11 and are composed of four main functional members: IFITM1, IFITM2, IFITM3, and IFITM5. IFITM4P is a pseudogene located on chromosome 6. IFITM proteins share the same topology (16). IFITM1, IFITM2, and IFITM3 are basally and almost ubiquitously expressed in human tissues, while IFITM5 is primarily expressed in osteoblasts (20). Among those family members, IFITM1, IFITM2, and IFITM3 are termed immunity-related IFITMs (16, 21). Studies have revealed that IFITM proteins could restrict the replication of multiple pathogenic viruses, especially enveloped viruses (20, 22–27). IFITM3 has been reported to be an essential barrier against IAV infection both in vivo and in vitro (20, 24). IFITM proteins seem to restrict IAV entry through multiple ways, including impairing the formation of fusion pores, inhibiting viral membrane hemifusion, and promoting membrane rigidity, thereby preventing virion-host membrane fusion and trapping the endocytosed virions within the vesicles (20, 25, 28, 29). Emerging evidence indicates that some viruses can be engaged by IFITM3 and shuttled to lysosomes (30). IFITM3 is able to amplify phosphatidylinositol 3-kinase (PI3K) signaling in B cells as a PIP3 scaffold (31). Additional evidence suggests that IFITM proteins are also involved in diverse biological processes, such as bone mineralization and germ cell specification (32, 33).
Long noncoding RNAs (lncRNAs) are defined as transcripts longer than 200 nucleotides (nt) with no protein coding potential. Increasing data indicate that they are involved in modulating multiple kinds of cellular processes, including antiviral innate immunity through diverse mechanisms (34). For example, lincRNA-Cox2 is able to modulate the Pam3CSK4-induced inflammatory response, and it has been shown that lncRNA can mediate both activation and repression of immune response genes (35). We have also reported that a lncRNA named negative regulator of antiviral response (NRAV) negatively regulates host antiviral responses through suppressing transcription of several critical ISGs by affecting the histone modification of those ISGs (36). Another virus-induced lncRNA, termed lncRNA ACOD1, promotes viral replication by modulating cellular metabolism (37). However, only a small number of functional lncRNAs involved in antiviral responses have been well studied so far, and the functions of most identified lncRNAs remain largely uncharacterized (38). In-depth mechanisms underlying regulation of gene expression by lncRNAs are largely unexplored.
Here, we unveiled that IFITM4P, a pseudogene from the widely reported IFITM family, transcribed an lncRNA that can function as an important regulator of the host antiviral immunity. Our study identified lncRNA IFITM4P as a competing endogenous RNA (ceRNA) that was involved in innate immunity against viral infection by cross talking with other IFITM family members. These data provide a new line of evidence for the function of a pseudogene in the host-virus interaction.
RESULTS
The transcript of IFITM4P is identified as an lncRNA that can be induced by multiple viruses.
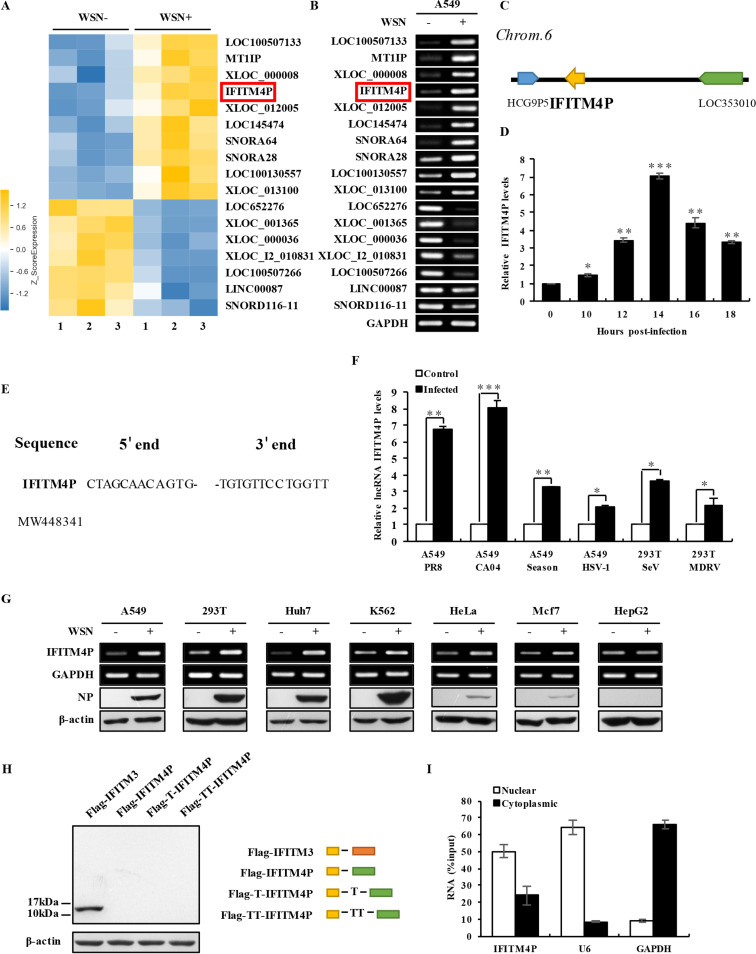
To investigate the roles of host lncRNAs in IAV infection and pathogenesis, cDNA microarray analysis was performed on human A549 cells infected or not infected with influenza virus A/WSN/33 (H1N1) for 12 h as described previously (http://www.ncbi.nlm.nih.gov/geo; GEO accession no. GSE58741) (39). Among 1,051 differentially expressed lncRNAs, 10 upregulated and 7 downregulated (fold change of >2; P < 0.05) lncRNAs were selected and analyzed (Fig. 1A). The differential expression of these lncRNAs was then confirmed by reverse transcription-PCR (RT-PCR) (Fig. 1B). Due to significant changes of its expression in IAV-infected cells and membership in the IFITM family, IFITM4P was chose for further investigation. IFITM4P, a pseudogene belonging to IFITM family, unlike other family members, is located on chromosome 6 (Fig. 1C). To further investigate the expression of IFITM4P during IAV infection, quantitative RT-PCR (qRT-PCR) was performed to detect the IFITM4P RNA levels at indicated infection time points. Results showed that lncRNA IFITM4P was induced by the viral infection in a time-dependent manner, and its levels reached the highest point at 14 h postinfection (Fig. 1D). Furthermore, we explored whether expression of lncRNA IFITM4P was elevated by a broad spectrum of viruses. Interestingly, we found that IFITM4P expression was significantly increased in 293T or A549 cells infected with various IAV strains, such as PR8, CA04, seasonal influenza A virus—Shanghai, and also other RNA viruses, like SeV and Muscovy duck reovirus (MDRV), as well as DNA viruses like herpes simplex virus 1 (HSV-1) (Fig. 1F). However, infection with pseudorabies virus (PRV), a DNA virus, did not induce the expression of IFITM4P in A549 cells (data not shown).
FIG 1.
Transcript of IFITM4P is identified as an lncRNA that can be induced by multiple viruses. (A) The differentially expressed lncRNAs in A549 cells infected or not infected with A/WSN/33 influenza virus were analyzed by using a cDNA microarray. Cells infected with WSN were collected at 12 h postinfection. Shown are representative lncRNAs whose expressions significantly changed after viral infection. IFITM4P is indicated in a red rectangle. (B) RT-PCR was performed to determine the expression of 17 lncRNAs in A549 cells infected or not infected with WSN (MOI of 0.5) for 14 h. IFITM4P is indicated in a red rectangle. (C) Shown is a paradigm of the genomic location of IFITM4P. (D) qRT-PCR was employed to detect RNA levels of IFITM4P in A549 cells infected with WSN (MOI of 0.5) for the indicated times. Data represent mean values ± SEM (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (E) 5′ and 3′ RACE analyses were performed. Shown are the 5′ end and 3′ end sequences of IFITM4P. (F) qRT-PCR was employed to detect the RNA levels of IFITM4P in A549 cells infected with A/Puerto Rico/8/1934 (PR8), A/California/04/2009 (CA04), A/Shanghai-Jiading/SWL1970/2015 (Season) or herpes simplex virus 1 (HSV-1), and in 293T cells infected with Sendai virus (SeV) or Muscovy duck reovirus (MDRV). Data represent mean values ± SEM (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (G) Human cell lines, including A549, 293T, Huh7, K562, HeLa, MCF7, and HepG2, were infected or not infected with WSN (MOI of 0.5) for 14 h. Expression of IFITM4P and IAV nucleoprotein (NP) in these cell lines was assessed by RT-PCR and Western blotting, respectively. (H) The full length of IFITM4P was cloned into pNL vector with an N-terminal Flag tag in three reading frames. Flag-IFITM3 served as the positive control. Yellow, orange, and green rectangles indicate Flag tag, IFITM3, and IFITM4P, respectively. Plasmids were transfected into 293T cells for 48 h, and cell lysates were subjected to Western blotting with anti-Flag antibody. (I) The RNA levels of IFITM4P in cytoplasmic and nuclear fractions from A549 cells were examined by qRT-PCR. U6, GAPDH, and total RNA levels served as the nuclear, cytoplasmic, and input RNA controls, respectively. Data are shown as percentage of input. Data represent mean values ± SEM. Shown are representatives from three independent experiments.
The levels of lncRNA IFITM4P induced by IAV strain WSN in other human cell lines were also examined. Increased expression of IFITM4P could be detected in several human cell lines infected with WSN, including A549, 293T, Huh7, K562, and HeLa. There was a rather moderate change of IFITM4P level in MCF7 cells after IAV infection. In HepG2 cells, however, the RNA level of IFITM4P was not obviously changed (Fig. 1G). Taken together, these data demonstrated that IFITM4P could be induced by infection of several types of viruses in some, but not all, human cell lines.
To determine the full length of lncRNA IFITM4P, 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE experiments were performed. The full length of lncRNA IFITM4P is 961 nt (Fig. 1E). Coding potential analyses tools, including NCBI ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder) and Coding Potential Calculator (CPC; http://cpc2.gao-lab.org/), were employed to ascertain whether lncRNA IFITM4P has protein coding potential. We noticed that all potential open reading frames (ORFs) of IFITM4P were less than 300 nt, and the CPC score for IFITM4P was minus, indicating that it is a noncoding RNA (data not shown). Next, we employed experimental translation tools in vitro and Western blotting to test the coding potential of IFITM4P. IFITM3 was used as a positive control. The band of Flag-tagged IFITM3 was observed at the position of right size, but no band was detected for Flag-tagged IFITM4P (Fig. 1H). Subcellular fractionation analysis showed that IFITM4P was localized in both cytoplasm and nucleus, but particularly in the nucleus (Fig. 1I). Collectively, these results suggest that lncRNA IFITM4P, the transcript of pseudogene IFITM4P, is a long noncoding RNA that can be induced by multiple viruses.
lncRNA IFITM4P can inhibit IAV and MDRV replication in vitro.
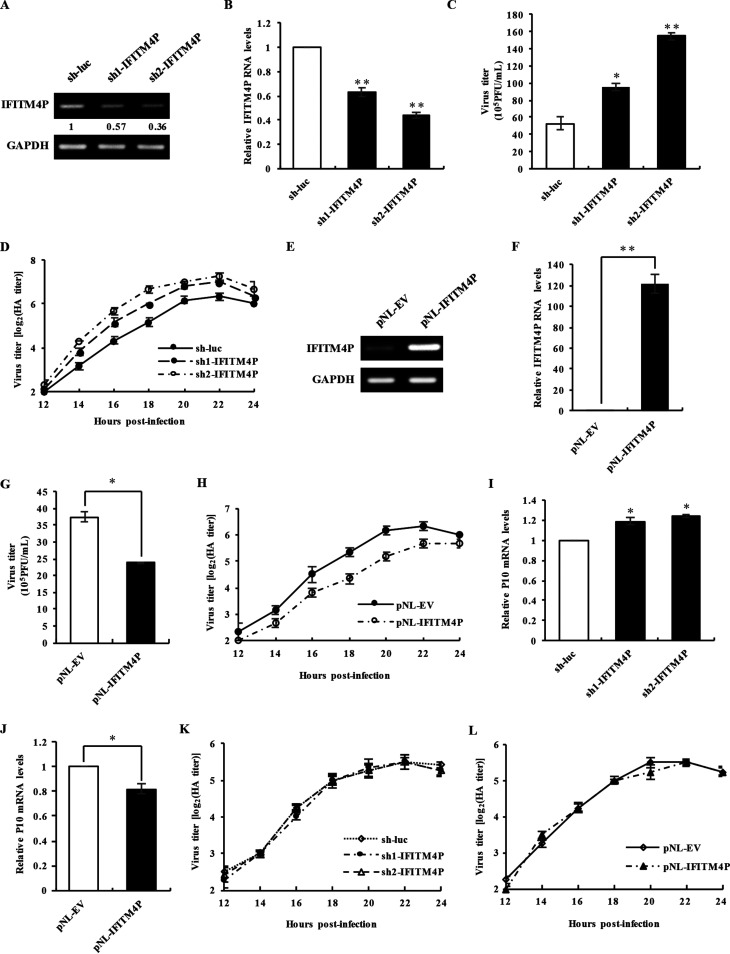
To investigate whether lncRNA IFITM4P is a functional transcript involved in viral infection, we examined effects of altered expression of lncRNA IFITM4P on host antiviral responses. For this, we designed two short hairpin RNAs (shRNAs) to disrupt lncRNA IFITM4P expression specifically in A549 cells (Fig. 2A and B). A plaque-forming assay (PFA) was applied to determine the differences in virus titers between lncRNA IFITM4P knockdown cells and control cells infected with IAV. It was noteworthy that silencing lncRNA IFITM4P remarkably promoted IAV replication in host cells (Fig. 2C). Similarly, the hemagglutination (HA) assay showed increased IAV titers in A549 cells with disrupted lncRNA IFITM4P expression (Fig. 2D).
FIG 2.
lncRNA IFITM4P can inhibit IAV and MDRV replication in vitro. (A and B) The knockdown efficiency of two shRNAs specifically targeting lncRNA IFITM4P in A549 cells was determined by RT-PCR (A) and qRT-PCR (B). Numbers indicate relative levels of lncRNA IFITM4P, which were quantitated by densitometry and normalized to GAPDH expression. The maximum value was regarded as 1. (C) Supernatants from lncRNA IFITM4P knockdown and control A549 cells infected with IAV were collected at 16 h postinfection (MOI of 0.5). IAV titers in the supernatants were examined by plaque-forming assay. (D) IAV titers in supernatants of infected lncRNA IFITM4P knockdown and control cells were measured at indicated time points by hemagglutination assay (MOI of 0.5). (E and F) The full length of lncRNA IFITM4P was cloned into the pNL vector, and its overexpression efficiency in A549 cells was assessed by RT-PCR (E) and qRT-PCR (F). (G) Cells were infected as described for panel C. IAV titers in supernatants from lncRNA IFITM4P-overexpressing and control cells were examined by plaque-forming assay. (H) The hemagglutination assay was performed to detect IAV replication in supernatants from lncRNA IFITM4P-overexpressing and control cells at indicated infection time points. (I and J) The mRNA levels of MDRV P10 in lncRNA IFITM4P knockdown and luciferase control cells (I) or lncRNA IFITM4P-overexpressing and control cells (J) were measured by qRT-PCR. Data represent mean values ± SEM (n = 3; *, P < 0.05; **, P < 0.01). (K and L) Sendai virus titers in supernatants derived from lncRNA IFITM4P knockdown (K) or overexpressing (L) and control 293T cells infected with Sev (MOI of 0.7) were measured at indicated time points by hemagglutination assay. Data represent mean values ± SEM. Shown are representatives from three independent experiments.
On the other hand, to define the role of lncRNA IFITM4P, we generated an A549 cell line stably overexpressing lncRNA IFITM4P (Fig. 2E and F). Both PFA and HA assay results showed that virus titer was remarkably decreased in the supernatant of lncRNA IFITM4P-overexpressing cells compared to that in control cells after WSN infection, suggesting that ectopic expression of lncRNA IFITM4P significantly impaired the replication of IAV in A549 cells (Fig. 2G and H).
We also utilized Muscovy duck reovirus (MDRV) and Sendai virus (SeV) to infect 293T cell lines aberrantly expressing lncRNA IFITM4P. Lentiviral shRNAs silencing lncRNA IFITM4P led to increased mRNA levels of MDRV P10 (Fig. 2I), whereas the P10 mRNA levels in 293T cells overexpressing lncRNA IFITM4P exhibited a moderate decline compared with the control (Fig. 2J). This finding indicates that lncRNA IFITM4P may inhibit the MDRV replication. However, altering IFITM4P expression had no significant effect on the replication of SeV in 293T cells (Fig. 2K and L). Together, the results reveal that lncRNA IFITM4P is a functional lncRNA that can restrict some viruses such as IAV and MDRV.
Expression of lncRNA IFITM4P is regulated by IFN signaling during viral infection.
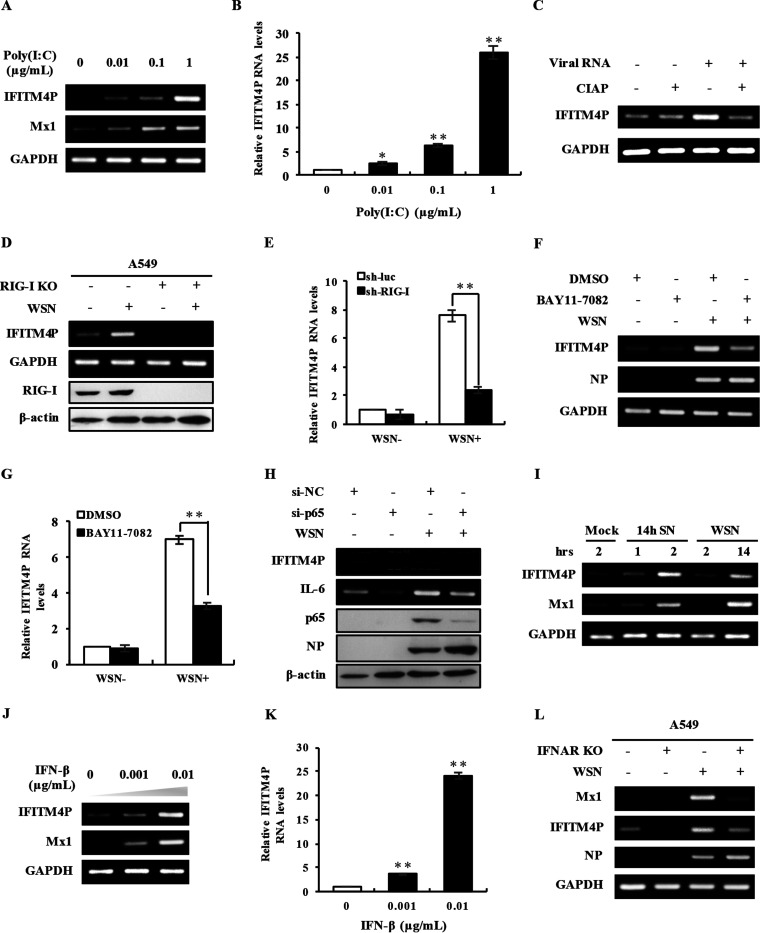
Since viral infection induced lncRNA IFITM4P expression immediately within a few hours postinfection, and increased lncRNA IFITM4P inhibited viral production, we speculated that lncRNA IFITM4P might be involved in antiviral innate immunity. Therefore, we asked whether innate immune signaling activated by invading viruses was responsible for the upregulation of lncRNA IFITM4P. To test this possibility, we transfected polyinosinic poly(C) [poly(I·C)], a synthetic analog of double-stranded RNA (dsRNA), into A549 cells to examine effect of dsRNA on the induction of lncRNA IFITM4P. Indeed, we observed that poly(I·C) significantly induced lncRNA IFITM4P expression in a dose-dependent manner (Fig. 3A and B). Viral RNA (vRNA) also triggered the expression of lncRNA IFITM4P, whereas vRNA deprived of 5′ phosphates after being treated with calf intestinal alkaline phosphatase (CIAP) showed an inefficiency in inducing lncRNA IFITM4P (Fig. 3C). In addition, silencing or knockout of RIG-I caused remarkable inhibition of lncRNA IFITM4P expression (Fig. 3D and E). Treatment with BAY 11-7082, an irreversible inhibitor of NF-κB, significantly decreased the IAV-induced lncRNA IFITM4P level in A549 cells compared with the control (Fig. 3F and G). Consistently, silencing the NF-κB p65 subunit with small interfering RNA (siRNA) led to a diminished amount of lncRNA IFITM4P in comparison with the control (Fig. 3H). Moreover, supernatants derived from WSN-infected A549 cells for 14 h were then used to stimulate fresh A549 cells. lncRNA IFITM4P expression level in A549 cells stimulated with the supernatants for 2 h was clearly elevated compared with that in the control cells, which was similar to that in A549 cells directly infected with WSN for 14 h (Fig. 3I).
FIG 3.
Expression of lncRNA IFITM4P is regulated by IFN signaling during viral infection. (A and B) A549 cells were transfected with poly(I·C) at the indicated concentrations for 4 h. The levels of lncRNA IFITM4P were examined by RT-PCR (A) and qRT-PCR (B). (C) Total RNAs were extracted from A549 cells infected or not infected with WSN for 14 h. The RNAs were then treated with or without calf intestine alkaline phosphatase (CIAP) and transfected into A549 cells for 4 h. Levels of lncRNA IFITM4P were examined by RT-PCR. (D and E) Knockout efficiency of RIG-I in A549 cells was assessed by Western blotting (D). RT-PCR (D) and qRT-PCR (E) were applied to examine lncRNA IFITM4P expression in RIG-I knockout or knockdown cells infected or not infected with WSN for 14 h (MOI of 0.5). (F and G) A549 cells were treated with BAY 11-7082 (8 μM) or the DMSO control 30 min before IAV infection. The RNA levels of lncRNA IFITM4P in A549 cells infected or not infected with WSN for 14 h were determined by RT-PCR (F) and qRT-PCR (G). (H) In A549 cells, knockdown efficiency of the NF-κB p65 subunit using siRNA was evaluated by Western blotting. The lncRNA IFITM4P levels in p65 knockdown or control cells infected or not infected with WSN for 14 h were detected by RT-PCR. (I) Native A549 cells were stimulated with supernatants from A549 cells infected or not infected with WSN for 14 h or were infected with WSN directly for 2 or 14 h. lncRNA IFITM4P expression was examined by RT-PCR. (J and K) A549 cells were treated with IFN-β at indicated concentrations for 2 h, and expression of lncRNA IFITM4P was examined by RT-PCR (J) and qRT-PCR (K). (L) lncRNA IFITM4P levels in IFNAR knockout or A549 control cells infected or not infected with WSN for 14 h were detected by RT-PCR. Data represent the mean values ± SEM (n = 3; *, P < 0.05; **, P < 0.01).
In addition, we noticed that lncRNA IFITM4P could be upregulated by IFN-β in a concentration-dependent manner (Fig. 3J and K). Consistently, knockout of IFNAR in A549 cells impaired the production of lncRNA IFITM4P to some extent (Fig. 3L). Taken together, these results suggest that like other IFITM family members, the expression of lncRNA IFITM4P is regulated through IFN signaling activated by viral infection.
Expression of lncRNA IFITM4P affects the mRNA levels of other IFITMs.
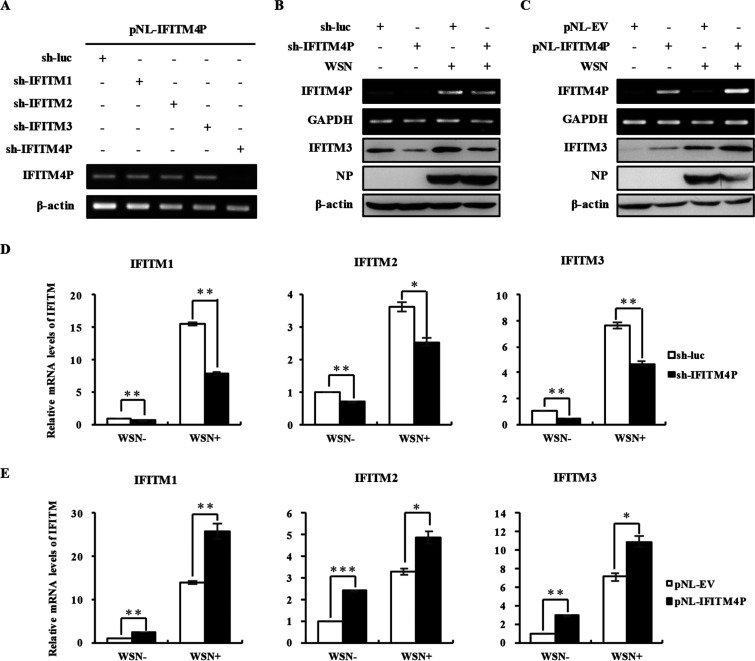
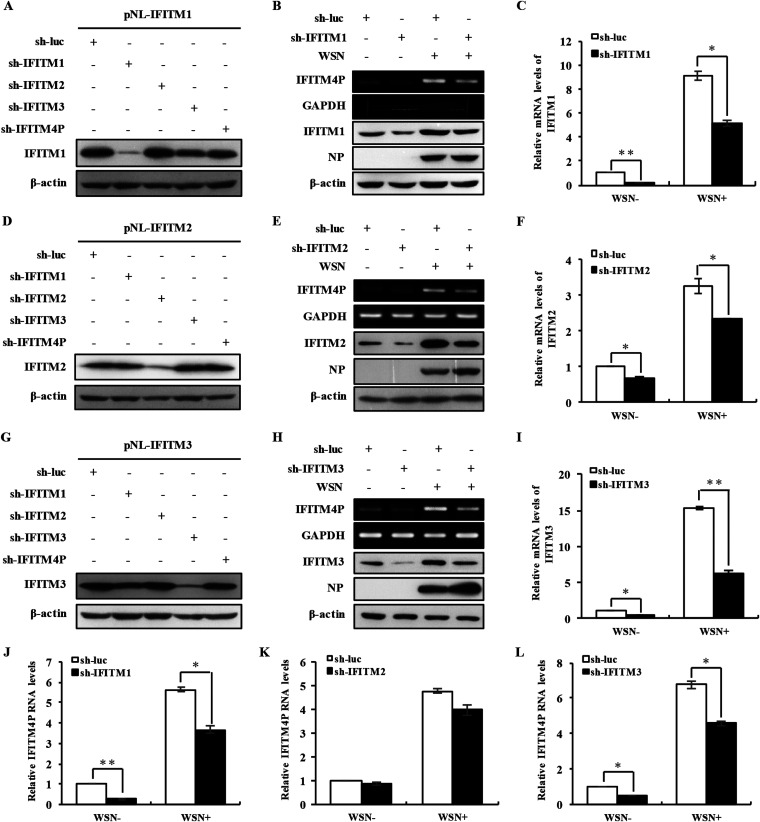
Some pseudogenes, by acting on homologous sequences, can affect expression of protein-coding genes. High homology does exist between IFITM4P and IFITM3; thereby, we wanted to explore if altering lncRNA IFITM4P expression could have any impact on IFITM3 level. Specific shRNAs were designed to target lncRNA IFITM4P, and the specificity of these shRNAs was further confirmed by coexpression of the IFITM family members and their shRNAs (Fig. 4A; also see Fig. 6). It was worth noting that disrupting lncRNA IFITM4P expression resulted in lower IFITM3 protein accumulation in cells either infected or not infected with IAV (Fig. 4B). In contrast, ectopic expression of IFITM4P in A549 cells notably upregulated IFITM3 protein levels (Fig. 4C). To further investigate whether this kind of effect happened only at the stage of translation, or occurred due to the changes of IFITM3 mRNA level, and whether other members of the IFITM family could also be regulated by lncRNA IFITM4P, three critical IFITM mRNA levels in cell lines with IFITM4P depleted or overexpressed were examined by qRT-PCR. Disrupting IFITM4P expression in A549 cells resulted in decreases in the mRNA levels of IFITM1, IFITM2, and IFITM3 (Fig. 4D), whereas upregulated mRNA levels of these IFITMs were observed in lncRNA IFITM4P-overexpressing cells infected or not infected with IAV (Fig. 4E). The results presented above revealed that lncRNA IFITM4P positively regulates the expression of other IFITM members.
FIG 4.
Expression of lncRNA IFITM4P affects the mRNA levels of other IFITMs. (A) pNL-IFITM4P plasmid (0.5 μg) and plasmid (4.5 μg) carrying shRNA targeting either IFITM1, IFITM2, IFITM3, or IFITM4P were cotransfected into 293T cells. lncRNA IFITM4P levels were determined by RT-PCR 48 h posttransfection. (B and C) Knockdown (B) and overexpression efficiency (C) of lncRNA IFITM4P in A549 cells were assessed by RT-PCR. Western blotting was employed to assess the IFITM3 protein expression in lncRNA IFITM4P knockdown (B) and overexpressing (C) cells infected or not infected with WSN (MOI of 0.5). (D) lncRNA IFITM4P knockdown or control cells were infected or not infected with WSN for 14 h. The mRNA levels of IFITM1, IFITM2, and IFITM3 were then determined by qRT-PCR. (E) Samples of A549 cells overexpressing lncRNA IFITM4P were obtained as described for panel D. The mRNA levels of IFITM1, IFITM2, and IFITM3 were determined by qRT-PCR. Data represent mean values ± SEM (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
FIG 6.
Silencing of IFITMs suppresses expression of lncRNA IFITM4P. (A) pNL-IFITM1 (0.5 μg) plasmid and plasmid (4.5 μg) carrying shRNA targeting IFITM1, IFITM2, IFITM3, or IFITM4P, were cotransfected into 293T cells. IFITM1 levels were examined by Western blotting 48 h posttransfection. (B and C) In A549 cells infected or not infected with WSN (MOI of 0.5), knockdown efficiency of IFITM1 was determined by Western blotting (B) and qRT-PCR (C). RT-PCR (B) was applied to detect lncRNA IFITM4P expression in IFITM1-silenced and control cells with or without viral infection for 14 h. (D) 293T cells were cotransfected with 0.5 μg pNL-IFITM2 plasmid and 4.5 μg plasmid carrying shRNA targeting IFITM1, IFITM2, IFITM3, or IFITM4P. Western blotting was used to assess the protein levels of IFITM2. (E and F) IFITM4P RNA levels in control and IFITM2 knockdown A549 cells infected or not infected with WSN (MOI of 0.5) for 14 h, were determined by RT-PCR. (G) 293T cells were cotransfected with 0.5 μg pNL-IFITM3 plasmid and 4.5 μg plasmid carrying shRNA targeting IFITM1, IFITM2, IFITM3, or IFITM4P. Western blotting was used to assess the protein levels of IFITM3. (H and I) RT-PCR was used to assess lncRNA IFITM4P levels in IFITM3 knockdown and control A549 cells with or without WSN infection (MOI of 0.5) for 14 h. (J to L) lncRNA IFITM4P levels in control and IFITM1 (J), IFITM2 (K), or IFITM3 (L) knockdown A549 cells infected or not infected with WSN, were determined by qRT-PCR. Data represent mean values ± SEM (n = 3; *P < 0.05; **P < 0.01).
lncRNA IFITM4P is a target of miR-24.
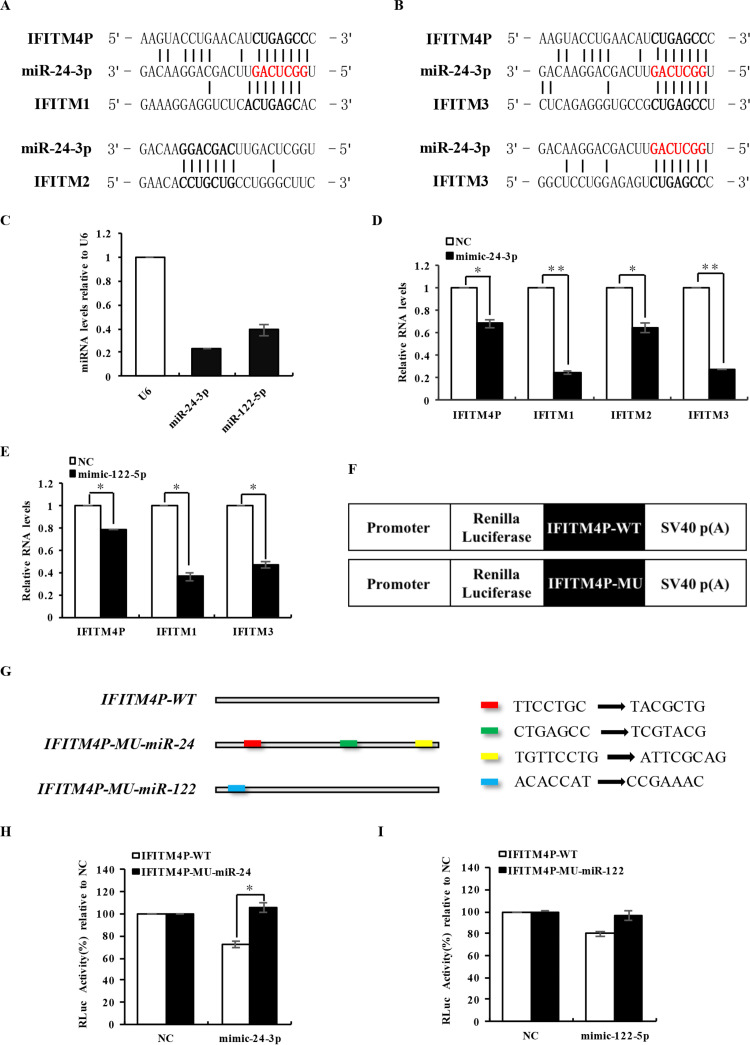
Our findings suggested a possibility that lncRNA IFITM4P might cross talk with its family members through an intermediate. To ascertain the underlying mechanism by which lncRNA IFITM4P might interact with its family members, we conducted a predictive bioinformatic analysis to identify whether there were any microRNAs (miRNAs) that might play a role in the process of IFITM4P-IFITM interaction. Previous studies and prediction results acquired from miRBase, TargetScanHuman, and PicTar were considered synthetically, and miR-24-3p, miR-122-5p and miR-130a-3p were sifted out as the most probable candidates (40–42). In particular, miR-24-3p was predicted to target IFITM1, IFITM2, IFITM3, and IFITM4P, and miR-122-5p might target IFITM1, IFITM3, and IFITM4P. Although miR-130a-3p was also predicted to likely target IFITM1, IFITM2, and IFITM4P, our experiments demonstrated that miR-130a-3p was unable to downregulate the RNA levels of IFITM4P significantly (data not shown). We found that IFITM1, IFITM2, IFITM3, and IFITM4P were able to match sequences of miR-24-3p and miR-122-5p, although they were located at different regions (Fig. 5A and B) (data not shown). Thus, we first examined the expression of miR-24-3p and miR-122-5p in A549 cells and found that they were abundant in A549 cells (Fig. 5C). We noticed that IAV infection slightly downregulated the endogenous miR-24-3p level, but it had little impact on miR-122-5p expression (data not shown). To determine the influence of miR-24-3p and miR-122-5p on expression of these IFITMs, A549 cells were transfected with the negative control or corresponding mimic, and the RNA levels of these IFITMs were examined. We observed that A549 cells treated with mimic-24-3p or mimic-122-5p had a lower level of lncRNA IFITM4P than that in control cells (Fig. 5D and E). Transfection of mimic-24-3p also led to a decline in the mRNA levels of IFITM1, IFITM2, and IFITM3 in A549 cells compared with the control (Fig. 5D). Similar results were obtained in A549 cells treated with mimic-122-5p, which showed that both IFITM1 and IFITM3 mRNA levels were downregulated by mimic-122-5p treatment (Fig. 5E). Conversely, RNA levels of IFITM1, IFITM2, IFITM3, and IFITM4P increased in A549 cells treated with specific chemically modified inhibitor to inhibit endogenous miR-24-3p (data not shown).
FIG 5.
lncRNA IFITM4P is a target of miR-24. (A and B) Shown are sequences of miR-24-3p binding sites in lncRNA IFITM4P, IFITM1, IFITM2, and IFITM3. (C) Levels of miR-24-3p and miR-122-5p were examined by qRT-PCR and compared with U6 in A549 cells. (D and E) A549 cells were transfected with negative-control and mimic-24-3p (D) or mimic-122-5p (E), the synthetic simulant of miR-24-3p or miR-122-5p. RNA levels of IFITM1, IFITM2, IFITM3, and lncRNA IFITM4P were examined in the A549 cells by qRT-PCR. (F) Wild-type lncRNA IFITM4P (IFITM4P-WT) or its mutants (IFITM4P-MU) were cloned downstream of the Renilla luciferase gene in the vector pRL-TK. A schematic diagram shows constructs generated for luciferase assays. (G) Shown is a diagram for the mutation sites of miR-24-3p and miR-122-5p response elements in lncRNA IFITM4P. (H and I) miR-24-3p response element-mutated lncRNA IFITM4P (IFITM4P-MU-miR-24) and miR-122-5p response element-mutated lncRNA IFITM4P (IFITM4P-MU-miR-122) in pRL-TK vector were, respectively, transfected into 293T cells, together with mimic-24-3p (H), mimic-122-5p (I), or the negative-control mimic (NC). The luciferase assay was performed as described in Materials and Methods. Data represent mean values ± SEM (n = 3; *, P < 0.05; **, P < 0.01).
To further verify the validity of predicted microRNA binding sites on lncRNA IFITM4P, wild-type IFITM4P (IFITM4P-WT) or its mutants devoid of the specific microRNA binding sites were cloned into the downstream of the Renilla luciferase gene in the pRL-TK vector (Fig. 5F). We constructed IFITM4P mutants carrying either the mutated three miR-24-3p binding sites shared by IFITM1, IFITM2, IFITM3, and lncRNA IFITM4P or the mutated miR-122-5p binding site in IFITM4P shared by IFITM1, IFITM3, and lncRNA IFITM4P (Fig. 5G). Plasmids were then transfected into 293T cells, respectively, together with the corresponding mimic or a negative control. As shown in Fig. 5H, mimic-24-3p suppressed the expression of the luciferase gene containing IFITM4P-WT but had no significant effect on the expression of the luciferase gene containing IFITM4P with mutation of miR-24-3p binding sites. Similar experiments were conducted in 293T cells transfected with IFITM4P carrying the miR-122-5p binding site mutation. However, only a slight difference in relative luciferase levels between IFITM4P-WT and its mutants was observed (Fig. 5I). These data suggest that mutations of the microRNA response elements (MREs) affect the capacity of lncRNA IFITM4P to bind with miR-24-3p. Additionally, the results imply that there might exist other unknown miR-122-5p binding sites in lncRNA IFITM4P.
Silencing IFITMs suppresses expression of lncRNA IFITM4P.
On the other hand, we asked whether altering the levels of the other IFITMs had any influence on lncRNA IFITM4P expression. To this end, we generated A549 cell lines stably expressing specific shRNAs targeting either IFITM1, IFITM2, or IFITM3. The knockdown specificity of these shRNAs was further verified by coexpression of the IFITMs and shRNAs (Fig. 6A and D and G). RT-PCR, qRT-PCR, and Western blotting were performed to evaluate the knockdown efficiency of these IFITMs in A549 cells (Fig. 6B, C, E, F, H, and I). Indeed, the amount of lncRNA IFITM4P was markedly reduced in IFITM1- and IFITM3-silenced cell lines (Fig. 6J and L). However, deficiency of IFITM2 only slightly reduced the level of lncRNA IFITM4P, which might be due to the weak ability of IFITM2 in binding miR-24-3p compared with IFITM1 and IFITM3 and the modest knockdown efficiency of the shRNA targeting IFITM2 (Fig. 6K). These data indicate that silencing of IFITM1 and IFITM3 can suppress the expression of lncRNA IFITM4P.
Overexpression of IFITM1 or IFITM3 significantly upregulates the levels of lncRNA IFITM4P.
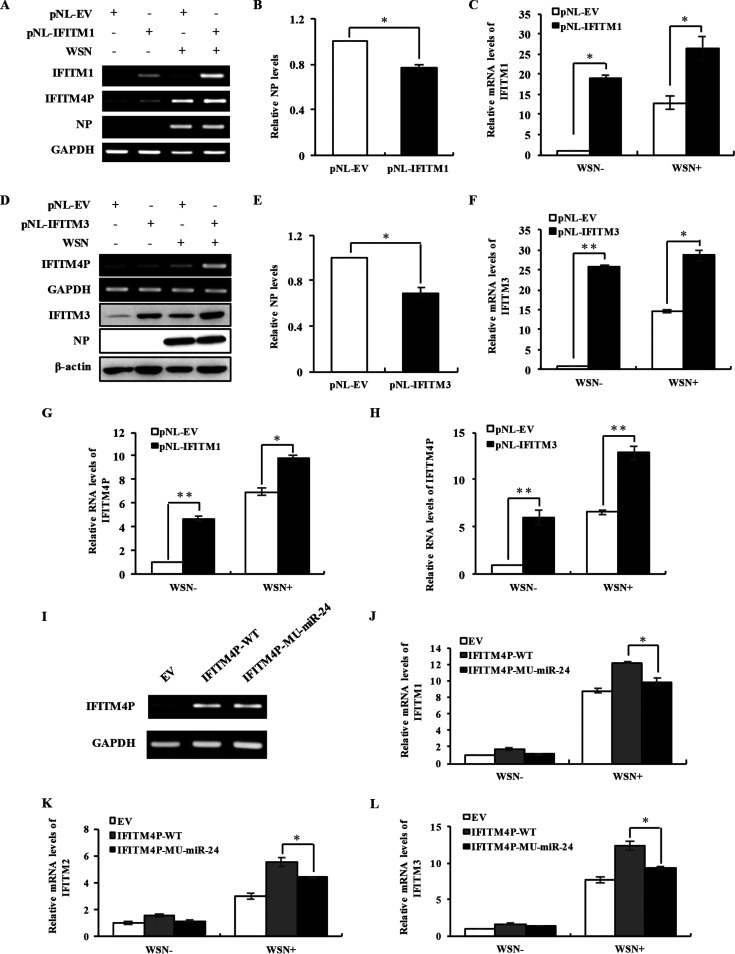
To further substantiate the effect of these IFITMs on lncRNA IFITM4P expression, we examined lncRNA IFITM4P levels in cell lines with IFITM1, IFITM2, and IFITM3 overexpressed. Ectopic expression efficiencies of the IFITMs were examined by RT-PCR, qRT-PCR, and Western blotting in A549 cells (Fig. 7A to F) (data not shown). As expected, overexpression of IFITM1 and IFITM3 in vitro, especially IFITM3, significantly enhanced lncRNA IFITM4P expression (Fig. 7G and H), while cells with increased IFITM2 expression displayed limited promotion of the lncRNA IFITM4P level (data not shown).
FIG 7.
Overexpression of IFITM1 or IFITM3 significantly upregulates the levels of lncRNA IFITM4P. (A to C) Overexpression efficiency of IFITM1 in A549 cells was determined by RT-PCR (A) and qRT-PCR (C). RT-PCR (A) was also employed to detect lncRNA IFITM4P expression in IFITM1-overexpressing and control cells with or without viral infection for 14 h. NP levels in panel A were quantitated by densitometry and normalized to GAPDH expression. The maximum value was regarded as 1 (B). (D to F) In A549 cells, the overexpression efficiency of IFITM3 was determined by Western blotting (D) and qRT-PCR (F). lncRNA IFITM4P expression in IFITM3-overexpressing and control cells infected or not infected with WSN for 14 h was determined by RT-PCR (D). NP levels in panel D were quantitated by densitometry and normalized to β-actin levels. The maximum value was regarded as 1 (E). (G and H) Levels of lncRNA IFITM4P in IFITM1 (G)- or IFITM3 (H)-overexpressing and control A549 cells infected or not infected with WSN were determined by qRT-PCR. (I to L) The mRNA levels of IFITM1 (J), IFITM2 (K), and IFITM3 (L) in IFITM4P-WT or IFITM4P-MU-miR-24-overexpressing and control A549 cells (I) with or without WSN infection (MOI of 0.5) were examined by qRT-PCR. Data represent mean values ± SEM (n = 3; *, P < 0.05; **, P < 0.01).
Since altering levels of other IFITMs could consistently modulate lncRNA IFITM4P, we further investigated whether changes in miR-24 response elements of lncRNA IFITM4P could influence members of the IFITM family. To this end, wild-type lncRNA IFITM4P (IFITM4P-WT) and its mutant devoid of the miR-24-3p binding sites (IFITM4P-MU-miR-24) were cloned into the pNL vector. Overexpression effects of IFITM4P-WT and IFITM4P-MU-miR-24 were confirmed in A549 cells by RT-PCR (Fig. 7I). Next, we compared the mRNA levels of the IFITMs in A549 cells with the overexpressed IFITM4P mutant or IFITM4P-WT. The results showed that the ectopic expression of IFITM4P-WT triggered increased expression of the IFITMs at mRNA levels, whereas mutation of the IFITM4P devoid of miR-24-3p response elements markedly impaired the ability of lncRNA IFITM4P to upregulate the levels of other IFITMs (Fig. 7J to L). Together, the experiments demonstrate that lncRNA IFITM4P can cross-regulate the expression of IFITM family members as a ceRNA to bind shared microRNAs, at least including miR-24-3p, during viral infection.
DISCUSSION
IFITM proteins have been reported to broadly inhibit the replication of multiple IAV strains and other pathogenic viruses (20, 23, 24, 43). Although numerous studies have been done to explore antiviral roles of the IFITM family, little information is available about IFITM4P, a pseudogene in this family. Here, we identified that IFITM4P, the RNA transcript of pseudogene IFITM4P, is an lncRNA. lncRNA IFITM4P, which was found to be expressed in various human cells, could be significantly upregulated by infection of several viruses, such as IAV, SeV, MDRV, and herpes simplex virus 1 (HSV-1). Notably, altered expression of IFITM4P had a profound effect on replication of some viruses, such as IAV and MDRV. For example, diminished expression of lncRNA IFITM4P significantly promoted the multiplication of WSN in A549 cells. Another finding is that lncRNA IFITM4P showed no effect on the replication of Sendai virus. This observation was consistent with previous studies claiming that IFITM1 and IFITM3 cannot inhibit the proliferation of SeV (44). However, more viruses should be tested in the future to determine whether lncRNA IFITM4P might be a broad-spectrum antiviral host factor.
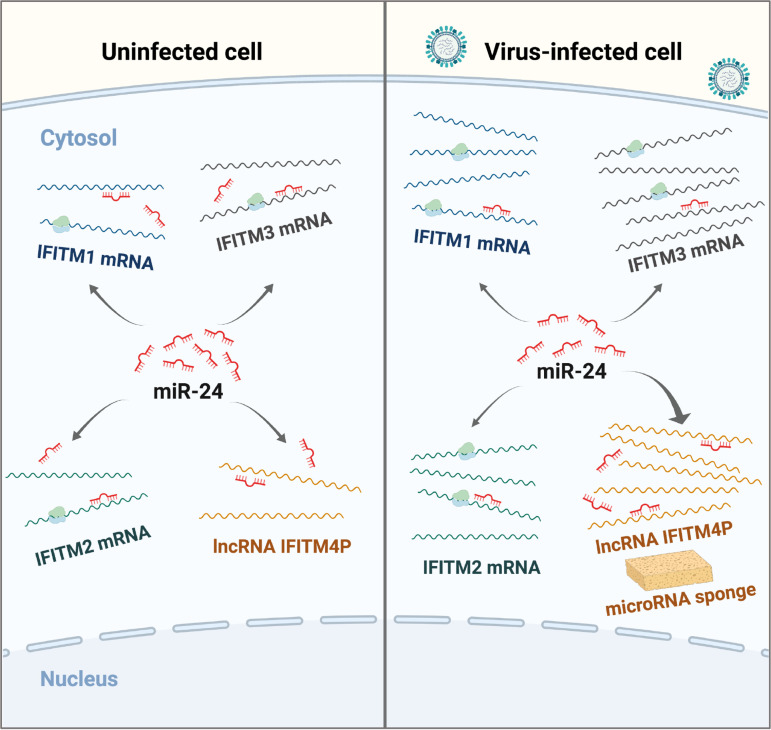
The results obtained from a series of experiments reveal that expression of lncRNA IFITM4P was regulated through an IFN signaling pathway during the viral infection, which is very similar to that of other IFITMs (20, 45). This finding led us to investigate further the function of IFITM4P in host innate immune response against viral infection. Interestingly, altering the expression of lncRNA IFITM4P could affect the mRNA levels of other IFITMs, suggesting that IFITM4P may play a role in antiviral immunity through cross talk with its family members. Furthermore, our data indicate that lncRNA IFITM4P acts as a ceRNA to cross-regulate the expression of other IFITMs (Fig. 8). MicroRNAs are approximately 22-nt endogenous RNAs that play important regulatory roles in various biological processes by targeting mRNAs for cleavage or translational repression (46–49). It has been proposed that microRNA targets may serve as competitive inhibitors of microRNA, keeping microRNAs from binding another target by sequestering them (50). Increasing evidence supports the ceRNA hypothesis, suggesting that RNA transcripts can communicate with and coregulate each other by competing for binding to shared microRNAs, and cross talk between the RNAs through miRNAs forms large-scale transcriptome regulatory networks (51, 52). Thus, altering the levels of one ceRNA would have impacts on expression of other RNAs that share the same MREs.
FIG 8.
lncRNA IFITM4P regulates innate antiviral responses by acting as a competing endogenous RNA. lncRNA IFITM4P is identified as a target of miR-24-3p that represses mRNA of IFITM1, IFITM2, and IFITM3. IFITM4P can cross-regulate the expression of IFITM family members as a competing endogenous RNA (ceRNA), leading to increased stability of these IFITM mRNAs, thereby regulating antiviral responses and virus replication in host cells. This graphic model was created with BioRender.com.
By the same token, the high sequence identity between IFITM4P and other IFITM family members, especially IFITM3, implies that the same MREs may exist on their RNAs. Bioinformatic analysis further suggest that miR-24, miR-122, and miR-130a binding sites are among the list. Consistently, serial analysis and experimental observations indicated that miR-24-3p and miR-122-5p were effective in binding and regulating the RNAs of IFITMs. Notably, IFITM1, IFITM3, and IFITM4P harbor complementarity to both miR-24 and miR-122, while IFITM2 only has complementarity to miR-24. It was shown that the repression effect of miR-24 on IFITM2 was weaker than those on IFITM1 and IFITM3, suggesting a weaker interaction between IFITM2 and miR-24. In addition, it seems that the shRNA targeting IFITM2 had distinct knockdown efficiency in different cell lines, as much higher knockdown efficiency was observed in 293T cells than that in A549 cells. These may be the reasons why the weaker cross-regulation was observed between IFITM2 and IFITM4P compared with IFITM1/3 and IFITM4P.
In this study, miR-130a-3p is not further discussed because it appeared unable to target IFITM4P. However, we noticed that it could remarkably decrease the mRNA levels of both IFITM1 and IFITM2 in vitro. Interaction between miR-130a and IFITM1 has also been reported previously (53). We consider the effect of miR-130a on IFITM2 could also be noteworthy in future studies. On the other hand, in order to ensure that lncRNA IFITM4P inhibited IAV replication through the interaction with its family members, we merely mutated miR-24-3p or miR-122-5p binding sites in lncRNA IFITM4P to verify its effect on other IFITMs. The results showed that mutation of miR-24-3p response elements in lncRNA IFITM4P clearly impaired its capacity to regulate the mRNA levels of other IFITM family members, suggesting that the MREs are the key binding sites on lncRNA IFITM4P for cross talk with other IFITMs. Similar experiments were performed to determine the miR-122-5p binding site in lncRNA IFITM4P, but only a slight change in lncRNA IFITM4P capacity was observed, indicating that there might exist other unknown miR-122-5p binding sites in lncRNA IFITM4P. This remains to be further defined.
We also observed that, compared to the IFITM4P overexpression group, loss of lncRNA IFITM4P in A549 cells had more significant effects on IAV titers. A possible explanation is that lncRNA IFITM4P was originally expressed at a relatively high basal level, and impaired lncRNA IFITM4P expression renders microRNAs acting on lncRNA IFITM4P binding to the mRNAs of other IFITMs. However, excessive lncRNA IFITM4P sponged microRNAs to the greatest extent, and the finiteness of endogenous microRNAs limited the effectiveness of overexpressed lncRNA IFITM4P.
Based on these findings, we conclude that lncRNA IFITM4P, as a ceRNA, is involved in the innate immunity against viral infection through the lncRNA IFITM4P–miR-24-3p–IFITM1/2/3 regulatory network. However, although we inferred that lncRNA IFITM4P functions as a ceRNA to regulate other IFITM family members, this may not be the only mechanism by which lncRNA IFITM4P works to inhibit the viral replication. Investigation of other functions of lncRNA IFITM4P and the precise mechanisms by which lncRNA IFITM4P regulates biological processes remains an ongoing task.
Additionally, the number of IFITM genes and pseudogenes varies, depending on species, suggesting there may be some species-specific gene duplications (54). Thus, it is likely that more functional IFITM pseudogenes may exist in humans, nonhuman primates, and other species, and their functions may be similar to or different from that of the pseudogene IFITM4P. These deserve further investigations in the future.
MATERIALS AND METHODS
Cell lines and cell culture.
The A549, 293T, K562, HeLa, and MDCK cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). The Huh7, Mcf7, and HepG2 cell lines were purchased from the National Infrastructure of Cell Line Resource (Beijing, China; http://cellresource.cn). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or RPMI 1640 (Gibco, USA) supplemented with 10% or other required concentrations of fetal bovine serum (Gibco, USA), penicillin (100 U/ml), and streptomycin (100 U/ml) at 37°C under a humidified 5% CO2 atmosphere as previously described (36).
Viruses and viral infection.
Influenza A virus strains, including A/WSN/1933 (H1N1), A/Puerto Rico/8/1934 (H1N1), A/California/04/2009 (H1N1), A/Shanghai-Jiading/SWL1970/2015 (H1N1) (a seasonal H1N1 influenza A virus, provided by Dayan Wang from the Chinese National Influenza Center, Beijing, China), and Sendai virus (SeV) were propagated in specific-pathogen-free (SPF) embryonated chicken eggs as previously described (55). Herpes simplex virus 1 (HSV-1) and Muscovy duck reovirus (MDRV) were propagated in Vero cells as previously described (36). PR8, CA04, seasonal influenza A virus—Shanghai, pseudorabies virus (PRV), and HSV-1 were used to infect A549 cells. SeV and MDRV were employed to infect 293T cells. Cells were incubated with virus for 1 h and cultured in DMEM for the indicated times.
Cell stimulation.
Reagents, including poly(I·C), dimethyl sulfoxide (DMSO) (Sigma-Aldrich, Germany), BAY 11-7082 (Sigma-Aldrich, Germany), and recombinant human IFN-β (PeproTech, USA) were purchased, and cells were treated following the manufacturer’s instructions.
Virus titer assay.
Cell culture supernatant was harvested at indicated time points after virus infection. Virus titers in the supernatant after serial dilutions were determined by a standard hemagglutination (HA) assay or a plaque-forming assay (PFA) using MDCK cells as described previously (56).
5′ RACE and 3′ RACE.
The 5′ and 3′ RACE analyses were performed using the SMARTer RACE 5′/3′ kit (Clontech, USA) as per the manufacturer’s instructions. The purified RACE PCR products were cloned into pRACE vector (Clontech, USA) and sequenced as described previously (31).
Subcellular fractionation.
A549 cells were harvested and lysed with TD buffer (25 mM Tris-HCl, 100 mM NaCl, 5 mM KCl, and 0.7 mM Na2HPO4) containing 1% NP-40. The lysates were then centrifuged under 12,000 × g for 5 min at 4°C. The supernatant was collected as the cytoplasmic fraction. The nuclear precipitates were washed with TD buffer containing 0.5% NP-40 and centrifuged in order to clear away cytoplasm remnants as described previously (57).
MicroRNA detection.
MicroRNAs were extracted using the RNAmisi miRNA isolation kit (Aidlab, Beijing, China). cDNAs were generated using an miRNA cDNA synthesis kit (Aidlab, Beijing, China) and examined with an miRNA real-time PCR assay kit (Aidlab, Beijing, China) according to the user’s manuals. U6 small nuclear RNA (snRNA) served to normalize microRNA levels.
Primers and shRNAs.
The following primers were used: IFITM1-F (5′-CAA CAT CCA CAG CGA GAC-3′), IFITM1-R (5′-TCA CAG AGC CGA ATA CCA-3′), IFITM2-F (5′-CGG CCA GCC TCC CAA CTA C-3′), IFITM2-R (5′-GCT GGG CCT GGA CGA CCA A-3′), IFITM3-F (5′-GGC CAG CCC CCC AAC TAT-3′), IFITM3-R (5′-CCA TAG GCC TGG AAG ATC AGC-3′), IFITM4P-F (5′-TGC CCA AAC CTT CTT CAT TCC-3′), and IFITM4P-R (5′-TGG GGG TTC ATG AAG AGG ATG-3′). The shRNAs targeting IFITM4P are sh1-IFITM4P (ATG TCC ACC ATG ATC CAT ATC TG) and sh2-IFITM4P (GCT CAC CAT CAT CAT CCT AGT).
Antibodies and Western blotting.
The following antibodies were used in this study for Western blotting: anti-IFITM1 (Proteintech, 60074-1-Ig), anti-IFITM2 (Proteintech, 66137-1-Ig), anti-IFITM3 (Proteintech, 11714-1-AP), anti-Flag (Sigma, F1804 M2), and anti-β-actin (Santa Cruz, sc-1616). For Western blotting, cell lysates were separated by SDS-PAGE, transferred onto a 0.45-μm-pore nitrocellulose membrane, and probed with indicated antibodies as previously described (58).
Luciferase assay.
lncRNA IFITM4P (IFITM4P-WT) and its mutant devoid of specific miRNA-binding sites (IFITM4P-MU-miR-24) were cloned into the 3′ untranscribed region (UTR) of the Renilla luciferase gene in the vector pRL-TK (Promega, Madison, WI, USA). Then, the plasmids were transfected into 293T cells, together with specific miRNA mimics or a negative-control mimic (RiboBio, Guangzhou, China). A firefly luciferase gene in the vector pGL3-control (Promega, Madison, WI, USA), which provides an internal control that serves as the baseline response, was used as a reference for transfection efficiency. Luciferase assays were performed using the dual-luciferase reporter assay system kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Luciferase levels were detected by a Modulus single-tube multimode reader (Promega, Madison, WI, USA). The relative luciferase expression equals the expression of Renilla luciferase divided by the levels of firefly luciferase (59).
Statistical analysis.
Comparison between groups was made using Student's t test. Data represent mean values ± the standard error of the mean (SEM). Differences were considered statistically significant at P < 0.05.
Accession number(s).
The sequence of full-length IFITM4P has been submitted to GenBank under accession no. MW448341.
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (32030110) and National Key Research and Development Program of China (2016YFD0500206).
We thank all members of the J. L. Chen laboratory for helpful discussions and assistance.
Contributor Information
Ji-Long Chen, Email: chenjl@im.ac.cn.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Hutchinson EC. 2018. Influenza virus. Trends Microbiol 26:809–810. 10.1016/j.tim.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Medina RA, García-Sastre A. 2011. Influenza A viruses: new research developments. Nat Rev Microbiol 9:590–603. 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. 2006. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22. 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson NP, Mueller J. 2002. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med 76:105–115. 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 5.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Azziz-Baumgartner E, Cheng P-Y, Dawood F, Foppa I, Olsen S, Haber M, Jeffers C, MacIntyre CR, Newall AT, Wood JG, Kundi M, Popow-Kraupp T, Ahmed M, Rahman M, Marinho F, Sotomayor Proschle CV, Vergara Mallegas N, Luzhao F, Sa L, Barbosa-Ramírez J, Sanchez DM, Gomez LA, Vargas XB, Acosta Herrera A, Llanés MJ, et al. 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis MM, Taubert K, Benin AL, Brown DW, Mensah GA, Baddour LM, Dunbar S, Krumholz HM, Centers for Disease Control and Prevention and the Advisory Committee on Immunization. 2006. Influenza vaccination as secondary prevention for cardiovascular disease: a science advisory from the American Heart Association/American College of Cardiology. J Am Coll Cardiol 48:1498–1502. 10.1016/j.jacc.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Nobusawa E, Sato K. 2006. Comparison of the mutation rates of human influenza A and B viruses. J Virol 80:3675–3678. 10.1128/JVI.80.7.3675-3678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiromoto Y, Saito T, Lindstrom SE, Li Y, Nerome R, Sugita S, Shinjoh M, Nerome K. 2000. Phylogenetic analysis of the three polymerase genes (PB1, PB2 and PA) of influenza B virus. J Gen Virol 81:929–937. 10.1099/0022-1317-81-4-929. [DOI] [PubMed] [Google Scholar]
- 9.Lindstrom SE, Hiromoto Y, Nishimura H, Saito T, Nerome R, Nerome K. 1999. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J Virol 73:4413–4426. 10.1128/JVI.73.5.4413-4426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaoka Y, Gorman OT, Ito T, Wells K, Donis RO, Castrucci MR, Donatelli I, Webster RG. 1998. Influence of host species on the evolution of the nonstructural (NS) gene of influenza A viruses. Virus Res 55:143–156. 10.1016/s0168-1702(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 11.Petrova V, Russell C. 2018. The evolution of seasonal influenza viruses. Nat Rev Microbiol 16:47–60. 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- 12.Panne D. 2008. The enhanceosome. Curr Opin Struct Biol 18:236–242. 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5:730–737. 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 14.Loo YM, Gale M, Jr.. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Liao Y, Chen B, Chen Y, Yu Z, Wei H, Zhang L, Huang S, Rothman PB, Gao GF, Chen JL. 2021. Critical role of Syk-dependent STAT1 activation in innate antiviral immunity. Cell Rep 34:108627. 10.1016/j.celrep.2020.108627. [DOI] [PubMed] [Google Scholar]
- 16.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57. 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman K, Coomer CA, Majdoul S, Ding SY, Padilla-Parra S, Compton AA. 2020. Homology-guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state. eLife 9:e58537. 10.7554/eLife.58537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi G, Schwartz O, Compton AA. 2017. More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology 14:53. 10.1186/s12977-017-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coomer CA, Rahman K, Compton AA. 2021. CD225 proteins: a family portrait of fusion regulators. Trends Genet 37:406–410. 10.1016/j.tig.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey CC, Kondur HR, Huang IC, Farzan M. 2013. Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J Biol Chem 288:32184–32193. 10.1074/jbc.M113.514356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, Liang C, Casartelli N, Schwartz O. 2014. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 16:736–747. 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perreira JM, Chin CR, Feeley EM, Brass AL. 2013. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol 425:4937–4955. 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P, GenISIS Investigators, MOSAIC Investigators. 2012. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484:519–523. 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, Elledge SJ, Bavari S, Denison MR, Choe H, Farzan M. 2011. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7:e1001258. 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, Brass AL. 2011. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7:e1002337. 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mudhasani R, Tran JP, Retterer C, Radoshitzky SR, Kota KP, Altamura LA, Smith JM, Packard BZ, Kuhn JH, Costantino J, Garrison AR, Schmaljohn CS, Huang IC, Farzan M, Bavari S. 2013. IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J Virol 87:8451–8464. 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. 2014. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 10:e1004048. 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu SL. 2013. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9:e1003124. 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spence JS, He R, Hoffmann HH, Das T, Thinon E, Rice CM, Peng T, Chandran K, Hang HC. 2019. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol 15:259–268. 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Robinson ME, Ma N, Artadji D, Ahmed MA, Xiao G, Sadras T, Deb G, Winchester J, Cosgun KN, Geng H, Chan LN, Kume K, Miettinen TP, Zhang Y, Nix MA, Klemm L, Chen CW, Chen J, Khairnar V, Wiita AP, Thomas-Tikhonenko A, Farzan M, Jung JU, Weinstock DM, Manalis SR, Diamond MS, Vaidehi N, Müschen M. 2020. IFITM3 functions as a PIP3 scaffold to amplify PI3K signalling in B cells. Nature 588:491–497. 10.1038/s41586-020-2884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffatt P, Gaumond MH, Salois P, Sellin K, Bessette MC, Godin E, de Oliveira PT, Atkins GJ, Nanci A, Thomas G. 2008. Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res 23:1497–1508. 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 33.Lange UC, Saitou M, Western PS, Barton SC, Surani MA. 2003. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol 3:1. 10.1186/1471-213x-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisler S, Coller J. 2013. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14:699–712. 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O'Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. 2013. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341:789–792. 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. 2014. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16:616–626. 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, Xu J, Wang Y, Cao X. 2017. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358:1051–1055. 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Wu Z, Fu X, Han W. 2014. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res 762:1–21. 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Li F, Chen Y, Zhang Z, Ouyang J, Wang Y, Yan R, Huang S, Gao GF, Guo G, Chen JL. 2015. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res 43:10321–10337. 10.1093/nar/gkv1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal V, Bell GW, Nam J, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005. 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144. 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, Guo JT. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol 84:8332–8341. 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hach J, McMichael T, Chesarino NM, Yount JS. 2013. Palmitoylation on conserved and non-conserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J Virol 87:9923–9927. 10.1128/JVI.00621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman RL, Manly SP, McMahon M, Kerr IM, Stark GR. 1984. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38:745–755. 10.1016/0092-8674(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 46.Towler BP, Jones CI, Newbury SF. 2015. Mechanisms of regulation of mature miRNAs. Biochem Soc Trans 43:1208–1214. 10.1042/BST20150157. [DOI] [PubMed] [Google Scholar]
- 47.Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840. 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27:91–105. 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 50.Seitz H. 2009. Redefining microRNA targets. Curr Biol 19:870–873. 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 51.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358. 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tay Y, Rinn J, Pandolfi PP. 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505:344–352. 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhanja Chowdhury J, Shrivastava S, Steele R, Di Bisceglie AM, Ray R, Ray RB. 2012. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J Virol 86:10221–10225. 10.1128/JVI.00882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Compton AA, Roy N, Porrot F, Billet A, Casartelli N, Yount JS, Liang C, Schwartz O. 2016. Natural mutations in IFITM3 modulate post-translational regulation and toggle antiviral specificity. EMBO Rep 17:1657–1671. 10.15252/embr.201642771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei H, Wang S, Chen Q, Chen Y, Chi X, Zhang L, Huang S, Gao GF, Chen JL. 2014. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PLoS Pathog 10:e1003845. 10.1371/journal.ppat.1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S, Li H, Chen Y, Wei H, Gao GF, Liu H, Huang S, Chen JL. 2012. Transport of influenza virus neuraminidase (NA) to host cell surface is regulated by ARHGAP21 and Cdc42 proteins. J Biol Chem 287:9804–9816. 10.1074/jbc.M111.312959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon Y, Lee GG. 2012. Subcellular localization prediction through boosting association rules. IEEE/ACM Trans Comput Biol Bioinform 9:609–618. 10.1109/TCBB.2011.131. [DOI] [PubMed] [Google Scholar]
- 58.Chen JL, Limnander A, Rothman PB. 2008. Pim-1 and Pim-2 kinases are required for efficient pre-B-cell transformation by v-Abl oncogene. Blood 111:1677–1685. 10.1182/blood-2007-04-083808. [DOI] [PubMed] [Google Scholar]
- 59.Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y, Ouyang J, Zhang L, Tan H, Chen R, Huang S, Chen JL. 2015. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 34:1768–1779. 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]