ABSTRACT
An RNA virus-based episomal vector (REVec) based on Borna disease virus 1 (BoDV-1) is a promising viral vector that achieves stable and long-term gene expression in transduced cells. However, the onerous procedure of reverse genetics used to generate an REVec is one of the challenges that must be overcome to make REVec technologies practical for use. In this study, to resolve the problems posed by reverse genetics, we focused on BoDV-2, a conspecific virus of BoDV-1 in the Mammalian 1 orthobornavirus. We synthesized the BoDV-2 nucleoprotein (N) and phosphoprotein (P) according to the reference sequences and evaluated their effects on the RNA polymerase activity of the BoDV-1 large protein (L) and viral replication. In the minireplicon assay, we found that BoDV-2 N significantly enhanced BoDV-1 polymerase activity and that BoDV-2 P supported further enhancement of this activity by N. A single amino acid substitution assay identified serine at position 30 of BoDV-2 N and alanine at position 24 of BoDV-2 P as critical amino acid residues for the enhancement of BoDV-1 polymerase activity. In reverse genetics, conversely, BoDV-2 N alone was sufficient to increase the rescue efficiency of the REVec. We showed that the REVec can be rescued directly from transfected 293T cells by using BoDV-2 N as a helper plasmid without cocultivation with Vero cells and following several weeks of passage. In addition, a chimeric REVec harboring the BoDV-2 N produced much higher levels of transgene mRNA and genomic RNA than the wild-type REVec in transduced cells. Our results contribute to not only improvements to the REVec system but also to understanding of the molecular regulation of orthobornavirus polymerase activity.
IMPORTANCE Borna disease virus 1 (BoDV-1), a prototype virus of the species Mammalian 1 orthobornavirus, is a nonsegmented negative-strand RNA virus that persists in the host nucleus. The nucleoprotein (N) of BoDV-1 encapsidates genomic and antigenomic viral RNA, playing important roles in viral transcription and replication. In this study, we demonstrated that the N of BoDV-2, another genotype in the species Mammalian 1 orthobornavirus, can participate in the viral ribonucleoprotein complex of BoDV-1 and enhance the activity of BoDV-1 polymerase (L) in both the BoDV-1 minireplicon assay and reverse genetics system. Chimeric recombinant BoDV-1 expressing BoDV-2 N but not BoDV-1 N showed higher transcription and replication levels, whereas the propagation and infectious particle production of the chimeric virus were comparable to those of wild-type BoDV-1, suggesting that the level of viral replication in the nucleus is not directly involved in the progeny virion production of BoDVs. Our results demonstrate a molecular mechanism of bornaviral polymerase activity, which will contribute to further development of vector systems using orthobornaviruses.
KEYWORDS: Borna disease virus, nucleoprotein, orthobornavirus, virus vector
INTRODUCTION
Borna disease virus 1 (BoDV-1) is a nonsegmented negative-strand RNA virus that belongs to the species Mammalian 1 Orthobornavirus (1). One of the unique properties of BoDV-1 is its ability to establish persistent infection in the host cell nucleus without integrating into the host genome (2, 3). Exploiting this unique property of BoDV-1, we previously developed an RNA virus-based episomal vector (REVec) system that enables stable and long-term gene expression in transduced cells (4, 5). In addition to long-term transgene expression both in vitro and in vivo (6–8), an REVec can efficiently transduce genes into stem cells, including human induced pluripotent stem cells (iPSCs), while maintaining their pluripotency (9, 10). While the REVec is a promising system for use in gene therapy and regenerative medicine, the onerous procedure of reverse genetics, which is required to produce an REVec, is one of the major challenges that must be overcome.
The genome of BoDV-1 encodes at least six viral proteins in the following 3′ to 5′ order: nucleoprotein (N), accessory protein (X), phosphoprotein (P), matrix protein (M), glycoprotein (G) and large protein (L), an RNA-dependent RNA polymerase (11–13). Among these proteins, N, P, and L are the minimum components of viral ribonucleoproteins (vRNPs) needed to transcribe and replicate viral RNA (14). To rescue the REVec by reverse genetics, a vector plasmid transcribing full-length antigenomic RNA of BoDV-1, which harbors an additional expression cassette between the P and M genes (4), and three helper plasmids expressing N, P, and L are cotransfected into 293T cells. Because the REVec cannot be rescued directly from transfected 293T cells, transfected cells are cocultured with Vero cells to propagate infection (15). However, several weeks of passages are required to rescue the high titer of the REVec. Thus, to improve reverse genetics systems, we and other groups have addressed several challenges thus far. For instance, Martin et al. utilized an RNA polymerase II promoter-driven vector and helper plasmids instead of T7 RNA polymerase promoter-driven plasmids (16). Ackermann et al. introduced two amino acid mutations, L1116R and N1398D, which are known to enhance the replication kinetics of BoDV-1, into the L gene of the full-length antigenome (17). Kojima et al. demonstrated that cultivation of cells at 40°C increased the rescue efficiency of the REVec (18). Although these challenges have contributed to improving reverse genetics systems to some extent, their efficacies remain limited, and extended time is still required to rescue the REVec.
BoDV-2 is another genotype belonging to the species Mammalian 1 orthobornavirus, and only one strain, No/98, has been isolated thus far (19). Among the strains of BoDV-1, genome sequences are highly conserved and almost 95% identical to each other, whereas strain No/98 is only approximately 80% identical to strain He/80, a major strain of BoDV-1 (20). While little is known about the virological properties of BoDV-2, it has been reported that BoDV-2 showed fast propagation in cultured Vero cells without causing cytopathogenic effects (19). Thus, we presume that BoDV-2 may provide clues for improving the reverse genetics system of BoDV-1.
In this study, to improve the reverse genetics of REVec, we investigated the effect of BoDV-2 proteins on BoDV-1 polymerase activity. We constructed expression plasmids for BoDV-2 N and P and utilized them in minireplicon assays using BoDV-1 L. Interestingly, we found that BoDV-2 N and P significantly enhanced the polymerase activity of the BoDV-1 minireplicon system. A series of single amino acid substitutions of the N and P proteins in BoDVs revealed amino acid residues that are critical for higher polymerase activity in BoDV-2 proteins. Furthermore, in reverse genetics, the rescue efficiency of the REVec was increased by using BoDV-2 N as a helper plasmid, which was sufficient to rescue the REVec directly from 293T cells without cocultivation with Vero cells. In addition, a chimeric REVec, which harbors BoDV-2 N instead of BoDV-1 N, produced high levels of viral mRNA and genomic RNA compared to the wild-type REVec, although the growth ability and titer of the chimeric REVec were not markedly increased, suggesting that increased transcription and replication are not necessarily linked to the production of infectious virus particles. Our results demonstrated a mechanism by which orthobornaviral N enhances viral polymerase activity, which will contribute to further development of the REVec system.
RESULTS
BoDV-2 N and P enhance the polymerase activity of BoDV-1 L.
N, P, and L are the minimum components needed for the transcription and replication of BoDV (14). Thus, we first constructed expression plasmids using synthesized cDNA fragments for these genes in BoDV-2 strain No/98, referring to the genome sequence registered in NCBI GenBank (accession no. AJ311524). Western blotting analysis confirmed the expression of these viral proteins, except for BoDV-2 L (Fig. 1A). Although we attempted to detect the expression of BoDV-2 L using several clones, we could not confirm the expression of BoDV-2 L.
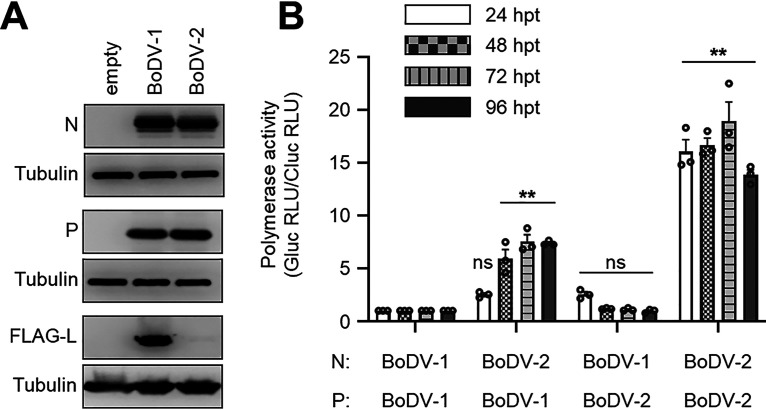
FIG 1.
BoDV-2 N and P enhance the polymerase activity of BoDV-1 L. (A) Detection of BoDV-2 N and P by Western blotting. Whole-cell lysates were prepared from 293T cells transfected with the indicated plasmid. The antibodies used in the analysis are indicated to the left of the panels. (B) Minireplicon assay using BoDV-2 N and P. The indicated N, P, and BoDV-1 L were used as helper plasmids. Polymerase activity was measured 24, 48, 72, and 96 hpt, and that of BoDV-1 N and P was presented as the relative value of 1.0 at each time point. The data are presented as the means ± standard error of the mean (SEM) of three independent experiments. One-way analysis of variance (ANOVA) and Dunnett’s multiple-comparison test were performed for statistical analysis at each time point. ns, not significant; **, P < 0.01. RLU, relative light unit.
Then, to examine the effect of BoDV-2 N and P on the polymerase activity of BoDV-1 L, we performed a minireplicon assay. Whereas replacement of BoDV-1 P with BoDV-2 P did not affect polymerase activity in any time points that we examined, replacement of BoDV-1 N with BoDV-2 N significantly enhanced polymerase activity from 48 h posttransfection (hpt) (Fig. 1B). Interestingly, replacement of both BoDV-1 N and P with BoDV-2 further enhanced polymerase activity at all time points (Fig. 1B). These results indicated that although BoDV-2 N alone can enhance the polymerase activity of BoDV-1 L, it is markedly enhanced when used in combination with BoDV-2 P.
Serine at position 30 of BoDV-2 N is critical for enhancing the polymerase activity of BoDV-1 L.
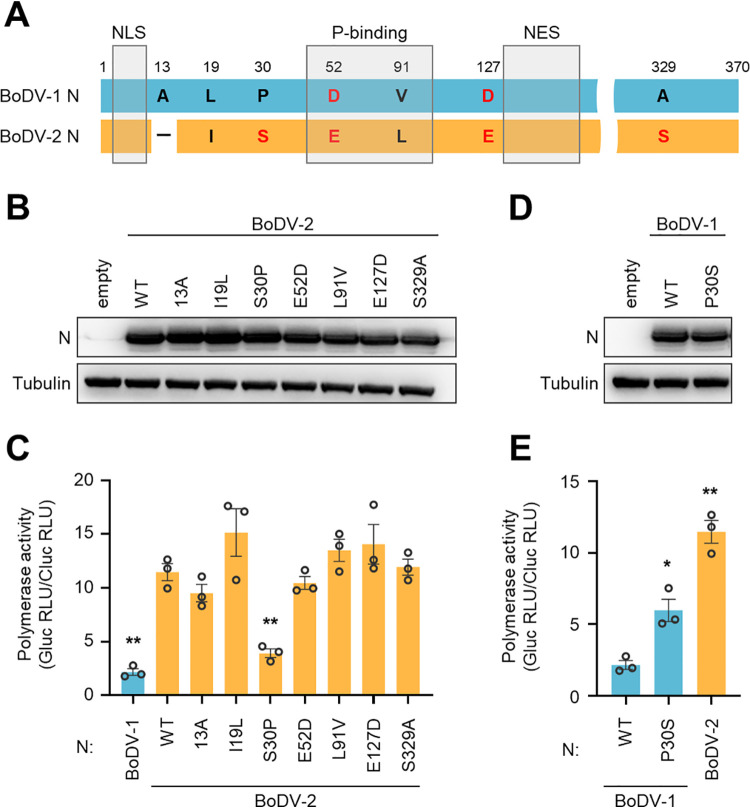
We next searched amino acid residues of BoDV-2 N that are critical for higher polymerase activity. Compared with BoDV-1 N, BoDV-2 N contains 6 amino acid differences and an amino acid deletion at position 13 (Fig. 2A). Whereas both the nuclear export signal (NES) and nuclear localization signal (NLS) are completely conserved in both BoDV-1 and BoDV-2 (21, 22), two differences are located in the N-terminal P-binding domain (23) (Fig. 2A). We constructed a series of plasmids expressing a single amino acid substitution based on BoDV-2 N and confirmed the expression of each by Western blotting analysis (Fig. 2B). A BoDV-1 minireplicon assay using BoDV-2 N mutants revealed that the polymerase activity was significantly reduced when serine at position 30 of BoDV-2 N was substituted with proline (Fig. 2C). In contrast, substitution of proline at position 30 of BoDV-1 N with serine significantly enhanced polymerase activity (Fig. 2D and E). These results suggested that serine at position 30 of BoDV-2 N is important for the enhancement of the polymerase activity of BoDV-1 L.
FIG 2.
Serine at position 30 of BoDV-2 N is critical for enhancing the polymerase activity of BoDV-1 L. (A) Schematic representation of BoDV Ns. The nature and position of amino acid mutations and deletion in BoDV-2 N are indicated. The hydrophobic and hydrophilic amino acid residues are represented in black and red, respectively. NLS, nuclear localization signal; NES, nuclear export signal; and P-bind, P-binding domain. (B and D) Detection of single amino acid-substituted BoDV-2 N (B) and BoDV-1 N (D) by Western blotting. Whole-cell lysates of 293T cells transfected with indicated plasmid were used. (C and E) Minireplicon assay using single amino acid-substituted BoDV-2 N (C) and BoDV-1 N (E). The indicated N, BoDV-2 P, and BoDV-1 L were used as helper plasmids. Polymerase activity was measured 48 hpt. The data are presented as the means ± SEM of three independent experiments. One-way ANOVA and Dunnett’s multiple-comparison test were performed for statistical analysis. *, P < 0.05; **, P < 0.01. RLU, relative light unit.
Alanine at position 24 of BoDV-2 P is critical for enhancing the polymerase activity of BoDV-1 L.
Similarly, BoDV-2 P contains 7 amino acid differences compared with BoDV-1 P (Fig. 3A). While the NES is completely conserved between BoDV-1 and BoDV-2, the N-terminal NLS in BoDV-2 P contains an amino substitution at position 36 (24–26); however, the nuclear localization of BoDV-2 P, as well as its nuclear retention activity to N, seemed to be unaffected by the mutation (Fig. 3B). To determine which amino acid residues in BoDV-2 P are critical for the enhancement of polymerase activity by BoDV-2 N, we constructed a series of plasmids expressing a single amino acid substitution in BoDV-2 P and performed a minireplicon assay. The expression of each mutant P was confirmed by Western blotting analysis (Fig. 3C). When alanine at position 24 and arginine at position 48 of BoDV-2 P were substituted with glutamic acid and lysine, respectively, the polymerase activity was significantly reduced (Fig. 3D). Conversely, while the substitution of glutamic acid at position 24 of BoDV-1 P with alanine significantly enhanced polymerase activity, the substitution of lysine at position 48 of BoDV-1 P with arginine did not have the same effect (Fig. 3E and F). Although the reason why the R48K mutant of BoDV-2 P reduced the activity was obscured, it may be possible that the R to K change in BoDV-2 P affects its structure or interaction with other polymerase components, such as BoDV-2 N and BoDV-1 L. Considering that the K48R mutant of BoDV-1 P did not markedly enhance the polymerase activity, however, it was considered that an alanine residue at position 24 of BoDV-2 P plays a critical role in enhancing the polymerase activity of BoDV-1 L in combination with BoDV-2 N.
FIG 3.
Alanine at position 24 of BoDV-2 P is critical for enhancing the polymerase activity of BoDV-1 L. (A) Schematic representation of BoDV Ps. The nature and position of amino acid mutations in BoDV-2 P are indicated. The hydrophobic and hydrophilic amino acid residues are represented in black and red, respectively. NLS, nuclear localization signal; NES, nuclear export signal; N-bind, N-binding domain; X-bind, X-binding domain. (B) Intracellular localization of N and P. Vero cells were transfected with N alone (left) or in combination with P (right) and reacted with the antibodies indicated in the top of the panels. Bars, 20 μm. (C and E) Detection of single amino acid-substituted BoDV-2 P (C) and BoDV-1 P (E) by Western blotting. Whole-cell lysates of 293T cells transfected with the indicated plasmid were used. (D and F) Minireplicon assay using single amino acid-substituted BoDV-2 P (D) and BoDV-1 P (F). BoDV-2 N, the indicated P, and BoDV-1 L were used as helper plasmids. Polymerase activity was measured 48 hpt. The data are presented as the means ± SEM of three independent experiments. One-way ANOVA and Dunnett’s multiple-comparison test were performed for statistical analysis. ns, not significant; **, P < 0.01. RLU, relative light unit.
BoDV-2 N increases the rescue efficiency of the REVec.
The results suggested that the rescue efficiency of the REVec by reverse genetics may be increased by using BoDV-2 N and P as helper plasmids. To verify this hypothesis, we transfected a vector plasmid harboring the green fluorescent protein (GFP) gene between the P and M genes (Fig. 4A) with helper plasmids and assessed viral replication by measuring the fluorescence signal. As shown in Fig. 4B, the GFP signal was observed in 293T cells transfected with BoDV-2 N as a helper plasmid, but not with BoDV-1 N, 3 days posttransfection (dpt). Consistently, approximately 2-fold higher REVec genomic RNA was detected in the 293T cells transfected with BoDV-2 N than in the cells transfected with BoDV-1 N by quantitative reverse transcription-PCR (RT-qPCR) (Fig. 4C). Although the use of BoDV-2 P with BoDV-2 N markedly enhanced polymerase activity in the minireplicon assay (Fig. 1B), the combination of BoDV-2 N and P did not seem to enhance REVec replication in the reverse genetics.
FIG 4.
BoDV-2 N increases the rescue efficiency of the REVec. (A) Schematic representation of the REVec genome component. A foreign GFP gene is inserted between the P and M genes. (B) Expression of GFP in transfected 293T cells. Reverse genetics was performed using the indicated N, P, and BoDV-1 L as helper plasmids. Expression of GFP was observed 3 dpt. (C) Comparison of REVec genomic RNA in transfected 293T cells. Reverse genetics was performed using the indicated N, P and BoDV-1 L as helper plasmids. Total RNA was extracted 3 dpt, and the level of REVec genomic RNA was evaluated by RT-qPCR. 1, BoDV-1; 2, BoDV-2. (D and F) Titration assay of REVec rescued from transfected 293T cells. Reverse genetics was performed using the indicated N, P, and BoDV-1 L as helper plasmids. REVecs were prepared from transfected 293T cells 3 dpt and directly inoculated into Vero cells. The number of Vero cells expressing GFP was counted 3 dpi. (E) Spread of REVec infection in cocultured Vero cells. Reverse genetics was performed using the indicated N, BoDV-1 P, and BoDV-1 L, and Vero cells were cocultured with transfected 293T cells 3 dpt and passaged every 3 days. The spread of REVec infection was evaluated by the expression of GFP using a Tali image-based cytometer. The data are presented as the means ± SEM of three independent experiments. One-way ANOVA and Tukey’s multiple-comparison test (C and D), two-way ANOVA and Sidak’s multiple-comparison test (E), or one-way ANOVA and Dunnett’s multiple-comparison test (F) were performed for statistical analysis. ns, not significant; **, P < 0.01.
Because BoDV-2 N seems to enhance REVec replication in reverse genetics, we next examined whether BoDV-2 N can rescue the REVec directly in transfected 293T cells without cocultivation with Vero cells. We prepared the REVec from 293T cells 3 dpt and inoculated them into Vero cells. As shown in Fig. 4D, more than 30 GFP-positive Vero cells were observed 3 days postincubation (dpi) only when BoDV-2 N was used as a helper plasmid (Fig. 4D). Consistently, when Vero cells were cocultured with 293T cells transfected with BoDV-2 N, REVec infection spread faster than when Vero cells were cocultured with 293T cells transfected with BoDV-1 N (Fig. 4E). With the minireplicon assay, we identified two amino acid residues that are critical to enhanced polymerase activity, a serine residue at position 30 in BoDV-2 N and an alanine residue at position 24 in BoDV-2 P (Fig. 2C and 3D). Thus, we sought to determine whether these amino acid residues are also critical to increased rescue efficiency. While a mutation at position 30 of BoDV-2 N decreased rescue efficiency, a mutation at position 24 of BoDV-2 P did not (Fig. 4F), confirming that serine at amino acid position 30 of BoDV-2 N is critical to enhance the polymerase activity of BoDV-1 L and that BoDV-2 N alone is sufficient to increase the rescue efficiency of the REVec.
BoDV-2 N enhances viral transcription and replication of chimeric REVecs.
Finally, to evaluate the effect of BoDV-2 N and P on viral growth ability, we generated chimeric REVecs, REVec-N2, REVec-XP2, and REVec-NXP2, which replaced the BoDV-1-derived N and/or P gene with those of BoDV-2 (Fig. 5A). Because the X gene that negatively regulates viral polymerase activity partially overlaps the P gene (27, 28), REVec-XP2 and REVec-NXP2 were designed to harbor the BoDV-2-derived X gene in the constructs. Although all chimeric REVecs were rescued successfully by reverse genetics, the recovery of REVec-XP2 and REVec-NXP2 was obviously delayed (Fig. 5B). Interestingly, the GFP fluorescence signal appeared to be brighter in the REVec-N2-infected cells than in the wild-type REVec-infected cells (Fig. 5B). To evaluate the growth ability of chimeric REVecs, Vero cells were inoculated with either a wild-type REVec or a chimeric REVec at a multiplicity of infection (MOI) of 1.0, and virus propagation was measured by GFP signal. While the growth ability of REVec-N2 was similar to that of the wild-type REVec, that of REVec-XP2 and REVec-NXP2 was significantly attenuated (Fig. 5C). To further analyze the transcription and replication of chimeric REVecs, the copy numbers of GFP mRNA and REVec genomic RNA were quantified by RT-qPCR. As shown in Fig. 5D and E, although the growth ability of REVec-N2 was similar to that of the wild-type REVec in Vero cells, REVec-N2 produced much higher levels of GFP mRNA and genomic RNA than the wild-type REVec in the cells. Intriguingly, we also found that the viral titers produced by the infected cells were comparable between the wild-type REVec and REVec-N2 (Fig. 5F). These results suggested that although BoDV-2 N can upregulate viral transcription and replication, it may not be linked to the production of infectious virus particles.
FIG 5.
BoDV-2 N enhances transcription and replication of chimeric REVecs. (A) Schematic representations of the genomic components of the chimeric REVecs. Genes derived from BoDV-1 and BoDV-2 are colored gray and orange, respectively. (B) Recovery of the chimeric REVecs. The recovery of wild-type and chimeric REVecs was confirmed by GFP expression in Vero cells after 2 weeks of cocultivation with transfected 293T cells. (C) Growth kinetics of the wild-type and chimeric REVecs. Vero cells were inoculated with each REVec at an MOI of 1.0 on day 1 and passaged every 3 days. The propagation of each REVec was evaluated by the expression of GFP using a Tali image-based cytometer. Comparison of GFP mRNA (D) and REVec genomic RNA (E) in infected Vero cells. Total RNA was extracted from infected Vero cells, and the level of REVec genomic RNA was evaluated by RT-qPCR. Relative values were corrected by infection ratio. (F) Titration assay of wild-type and chimeric REVecs. REVecs were prepared from infected Vero cells and directly inoculated into Vero cells. The relative value was corrected by infection ratio. The data are presented as the means ± SEM of three independent experiments. Two-way (C) or one-way (D to F) ANOVA and Dunnett’s multiple-comparison test were performed for statistical analysis. ns, not significant; *, P < 0.05; **, P < 0.01.
DISCUSSION
In this study, to improve the REVec system, we focused on BoDV-2, which was previously reported to show quick propagation in cultured Vero cells (19). First, we successfully increased the rescue efficiency of an REVec by using BoDV-2 N as a helper plasmid in reverse genetics. In addition to REVec infection quickly spreading in cocultured Vero cells, REVec was rescued directly in transfected 293T cells (Fig. 4D and E). Second, a chimeric REVec-N2, which harbored BoDV-2 N but not BoDV-1 N, transcribed more transgene mRNA than the wild-type REVec (Fig. 5D). Considering that the in vivo transduction efficiency of transmission-defective REVecs, such as ΔG-REVec (4) or ΔMG-REVec (5), which is ideal from a safety perspective, was quite low (8), REVec-N2 is a promising candidate to overcome this problem.
Although we demonstrated that BoDV-2 N upregulates the polymerase activity of BoDV-1 L, the detailed mechanism remains unclear. While BoDV-2 N contains 6 amino acid differences and an amino acid deletion not shared with BoDV-1 N (Fig. 2A), in addition to NLS and NES, which determine the intracellular localization of N (21, 22), four amino acid residues thought to interact with viral RNA (29) are completely conserved in BoDV-2 N. Moreover, a downstream start codon and splicing donor and acceptor sites that generate several isoforms of N are also completely conserved (30, 31). Whereas differences at amino acid positions 52 and 91 are located in the P-binding domain (23), they did not induce any deleterious effect on polymerase activity in the minireplicon assay (Fig. 2C). Similarly, either a deletion at amino acid position 13 or mutations at amino acid positions 19 and 127 slightly, but not significantly, affected the polymerase activity, suggesting that these differences are not critical for the activity (Fig. 2C). Conversely, our results demonstrated that a serine residue at position 30 of BoDV-2 N is critical for enhancement of the polymerase activity (Fig. 2C); however, to the best of our knowledge, no function for the amino acid residue at position 30 of BoDV N has been identified. Furthermore, some BoDV-1 field isolates also possess a serine residue at this position (32, 33). Further investigations, such as those directed to the interaction of BoDV-2 N with other viral proteins, especially L, might provide new insights into the mechanism of how BoDV N is involved in polymerase activity to regulate viral replication in the nucleus.
In this study, we demonstrated that although a chimeric REVec-N2 showed significantly higher transcription and replication levels than the wild-type REVec, it propagated with similar growth kinetics and produced subequal titers of infectious virus particles with the wild-type REVec (Fig. 5C to F). This observation suggested that upregulation of transcriptional activity does not necessarily increase the number of infectious virus particles in infected cells. Previous studies showed that BoDV-1 optimized the production of infectious virus particles by regulating the replication efficiency in the nucleus to maintain persistent infection (34–36). Our observation indicated, however, that polymerase activity is not the only factor controlling the level of infectious particle production. To assemble infectious particles, for instance, vRNPs must be transported from the nucleus to the cytoplasm, which requires coordinated functions among viral N, X, P, and M proteins (37). In chimeric REVecs, conspecific but nonsynonymous proteins may perturb this viral protein coordination while increasing polymerase activity. In addition, we recently reported that overexpression of the G envelope protein inhibits the maturation of G, resulting in a remarkable reduction in infectious virus particle formation (38). Thus, it is possible that a high level of transcriptional activity of REVec-N2 inhibits the appropriate maturation of G, inhibiting infectious virus production.
Interestingly, in this study, BoDV-2 P plays inconsistent roles in minireplicon and reverse genetics systems; when used in combination with BoDV-2 N, BoDV-2 P enhanced the polymerase activity in the minireplicon assay (Fig. 1B), whereas it did not increase rescue efficiency of the REVec in reverse genetics (Fig. 4B to D). Moreover, chimeric REVecs that harbor BoDV-2 P, namely, REVec-XP2 and REVec-NXP2, showed severely attenuated growth ability and lower transcriptional activity compared with the wild-type REVec and chimeric REVec-N2 (Fig. 5C and D). Notably, these chimeric REVecs contain BoDV-2 X in the genome, because the X gene partly overlaps the P gene in different frames. Previous studies reported that BoDV-2 X reduced the polymerase activity of BoDV-1 L the same as BoDV-1 X in the minireplicon assay (27, 28). In addition, the N terminus of X, where several important functional domains, including the P-binding domain, are located, is highly conserved in BoDV-1 and BoDV-2 X (19, 39). Thus, BoDV-2 X alone cannot explain the observed properties of REVec-XP2 and REVec-NXP2. BoDV P serves as a molecular hub of vRNPs and interacts with N, X, P, M, and L via its binding domains (39–43). Because BoDV persists in the host nucleus, nucleocytoplasmic shuttling of vRNPs is important for regulating viral transcription, replication, and propagation. Previously, we demonstrated the possibility that X facilitates the nuclear export of vRNPs because P is translocated to the cytoplasm via interaction with X (24, 37). Thus, the inconsistent roles of BoDV-2 P observed in this study may be attributed to different interactions between X and P; BoDV-2 P did not interact with X in the minireplicon assay but interacted with BoDV-1 X in reverse genetics and with BoDV-2 X in chimeric REVecs. Conversely, it is unclear why REVec-NXP2 showed lower growth ability and infectious virus particle production than REVec-N2, but BoDV-2 X may reduce the polymerase activity of BoDV-1 L, in combination with BoDV-2 P. To verify this supposition, we recently recovered X-deficient recombinant viruses and are now performing experiments using exogenously expressed BoDV-2 X.
Finally, recent accumulating evidence showed that patients who die of encephalitis have BoDV-1-associated RNAs and proteins, indicating that BoDV-1 may be a lethal pathogenic agent in humans (44–48). Considering that both BoDV-1 and BoDV-2 are strains of Mammalian 1 orthobornavirus and that BoDV-2 strain No/98 was isolated from a pony showing severe and incurable neurological symptoms (19), it is possible that BoDV-2 causes fatal encephalitis in humans. Rubbenstroth et al. raised the alarm to the risk of human BoDV-2 infection and emphasized the need for more research (49). In this study, we constructed expression plasmids for BoDV-2 N, P, and L by referring to the genome sequence registered in NCBI GenBank, but the expression of BoDV-2 L was not confirmed (Fig. 1A). In a possible explanation, Pleschka et al. mentioned that some unanticipated errors may occur in cDNA cloning or sequencing (20). To further investigate the virological properties of BoDV-2, the development of a reverse genetics system for BoDV-2 is necessary. Thus, the nucleotide sequence of BoDV-2 strain No/98 should be reanalyzed and precisely determined.
In conclusion, we demonstrated that BoDV-2 N upregulates the polymerase activity of BoDV-1 L. By leveraging this property, we increased the rescue efficiency of the REVec and generated a chimeric REVec-N2 that induced a high level of transgene expression. We believe that this study will contribute not only to the further development of an REVec system but also to understanding the molecular mechanism of transcription and replication of bornaviruses.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney cells, 293T, were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Nacalai Tesque, Japan; no. 08456-36) supplemented with 10% fetal bovine serum (FBS). African green monkey kidney cells, Vero, were cultured in DMEM supplemented with 5% FBS.
Plasmid construction.
Plasmids expressing BoDV-2 N, P, and L were constructed by using artificially synthesized cDNA fragments generated by referring to the genome sequence of BoDV-2 strain No/98 registered in NCBI GenBank (accession no. AJ311524) at Thermo Fisher Scientific. Each gene was amplified by PCR and inserted into EcoRI and XhoI restriction enzyme recognition sites of the eukaryotic expression plasmid pCAGGS (50). Plasmids expressing BoDV-1 N, P, and L were constructed by using the previously described plasmids, pCA-N, pCXN2-P, and pCA-L, respectively (51). Each gene was amplified by PCR and inserted into EcoRI and XhoI restriction enzyme recognition sites of the eukaryotic expression plasmid pCAGGS (50). FLAG-tagged BoDV-1 L and BoDV-2 L were constructed by PCR mutagenesis. Plasmids expressing a series of single amino acid-substituted BoDV-1 N and P and BoDV-2 N and P were constructed by PCR mutagenesis. The previously described RNA polymerase II (Pol II)-driven BoDV-1 minigenome plasmid (52) was modified by PCR mutagenesis as follows. Two unavailing sequences, 12 nucleotides in the Gaussia luciferase gene and 17 nucleotides between the Gaussia luciferase gene and BoDV-1 trailer sequence and an SpeI restriction enzyme recognition site between the BoDV-1 leader sequence and Gaussia luciferase gene, were removed. A vector plasmid transcribing the BoDV-1 full-length antigenomic RNA harboring a foreign GFP gene between the P and M genes, pFct-BoDV P/M-GFP (4), was modified by PCR mutagenesis as follows. A hepatitis delta virus ribozyme sequence was optimized by removing an extra adenine from its 3′ terminus (53). In addition, to transcribe BoDV-1 antigenomic RNA harboring the correct terminal sequences (35), CTT at the 5′ terminus of the leader sequence was removed and thymine at the 3′ terminus of the trailer sequence was replaced with AACA. The resulting vector plasmid was designated pREVec. The chimeric vector plasmids pREVe-N2, pREVe-XP2, and pREVe-NXP2 were constructed by PCR mutagenesis.
Western blotting analysis.
Protein samples were prepared by lysing cells with 1× sample buffer (50 mM Tris-HCl, pH 6.8; 2% sodium dodecyl sulfate [SDS]; 5% wt/vol sucrose; and 5% 2-mercaptoethanol) followed by boiling at 95°C for 10 min. The proteins were separated by SDS-PAGE using ePAGEL (ATTO Corp., Japan; no. 2331720) and transferred to Trans-Blot Turbo Mini nitrocellulose membranes (Bio-Rad, USA; no. 1704156). The membranes were blocked with blocking buffer (Tris-buffered saline [TBS] containing 0.1% Tween 20 with 5% wt/vol skim milk). Then, the membranes were incubated with primary antibody (anti-BoDV N HB01, anti-BoDV P HB03, anti-FLAG M2 [Sigma-Aldrich, USA; no. F1804], or anti-tubulin [Sigma-Aldrich, USA; no. T5168]), followed by incubation with secondary antibody (horseradish peroxidase [HRP]-conjugated anti-mouse IgG [Jackson ImmunoResearch, USA; no. 715-035-150] or HRP-conjugated anti-rabbit IgG [Jackson ImmunoResearch, USA; no. 715-035-152]). The protein bands were detected with ECL prime Western blotting detection reagents (GE Healthcare, USA; no. RPN2232), and chemiluminescent signals were visualized by a LAS-4000 mini Lumino image analyzer (Fujifilm, Tokyo, Japan).
Minireplicon assay.
A minireplicon assay was performed as described previously (51) with some modifications. In brief, 293T cells seeded onto 48-well plates were transfected with 50 ng of minigenome plasmid, 50 ng of pCAGGS-N, 5 ng of pCAGGS-P, 50 ng of pCAGGS-L, and 5 ng of control plasmid expressing Cypridina luciferase using TransIT293 (Mirus, USA; no. MIR2700) according to the manufacturer’s specifications. At indicated time points, the Gaussia luciferase and Cypridina luciferase activity levels were measured with a Biolux Gaussia luciferase assay kit (New England BioLabs [NEB], USA; no. E3300) and Biolux Cypridina luciferase assay kit (NEB, USA; no. E3309), respectively, according to the manufacturers’ specifications. Polymerase activity was quantified by normalizing Gaussia luciferase activity to Cypridina luciferase activity.
Immunofluorescence assay.
Cells cultured on coverslips (Matsunami Glass Ind., Japan; no. C015001) were fixed with 4% paraformaldehyde (Nacalai Tesque, Japan; no. 09154-85) for 10 min and permeabilized with phosphate-buffered saline (PBS) containing 0.5% Triton X-100 and 5% bovine serum albumin for 10 min. Then, the cells were incubated with primary antibody (anti-BoDV N HM132 or anti-BoDV P HB03), followed by incubation with secondary antibody (Alexa Fluor 488-conjugated anti-mouse IgG [Thermo Fisher Scientific, USA; no. A11029] or Alexa Fluor 555-conjugated anti-rabbit IgG [Thermo Fisher Scientific, USA; no. A21429]) and 300 nM 4′,6′-diamidino-2-phenylindole (DAPI) (Merck, Germany; no. 28718-90-3). The cells were mounted in ProLong Diamond antifade mountant (Thermo Fisher Scientific, USA; no. P36961). Fluorescence microscopy was performed using a Ti-E inverted microscope with a C2 confocal laser scanning system (Nikon, Japan).
Reverse genetics.
Reverse genetics was performed as described previously (4). In brief, 293T cells seeded onto 6-well plates were transfected with 2.0 μg of vector plasmid, 0.25 μg of pCAGGS-N, 0.025 μg of pCAGGS-P, and 0.25 μg of pCAGGS-L using TransIT-293 according to the manufacturer’s specifications. Three days posttransfection, the cells were cocultured with Vero cells to propagate infection or harvested to prepare REVecs.
REVec preparation.
REVecs were prepared as described previously (5). In brief, infected cells were trypsinized, centrifuged, and resuspended in DMEM. Then, the cells were sonicated and centrifuged at 1,200 × g for 25 min at 4°C. The supernatant contained REVecs.
Viral infection.
For the titration assay, Vero cells were inoculated with REVecs, followed by absorption at 37°C for 1 h. Then, the REVecs were replaced with culture medium, and the cells were incubated. Three days postinoculation, the number of GFP-positive cells was counted, and the virus titers were calculated. For the growth kinetics assay, Vero cells were inoculated with REVec at a multiplicity of infection (MOI) of 1.0, followed by absorption at 37°C for 1 h. Then, the REVecs were replaced with culture medium, and the cells were incubated. Cells were passaged every 3 days, and propagation of REVec was evaluated by expression of GFP using a Tali image-based cytometer (Thermo Fisher Scientific, USA).
Quantitative reverse transcription-PCR.
Total RNA was extracted using NucleoSpin RNA Plus (Macherey-Nagle, Germany; no. 740984) according to the manufacturer’s specifications. cDNA was synthesized from 1.0 μg of total RNA using a Verso cDNA synthesis kit (Thermo Fisher Scientific, USA; no. AB1453) and either an REVec genome-specific primer (5′-GGCCGTCATGGTGGCGAATGTTGCGTTAACAACAAACCAATCAT-3′) (8) or an anchored oligonucleotide deoxyribosylthymine (dT). Quantitative PCR (qPCR) was performed by a CFX Connect real-time system (Bio-Rad, USA) using 1.0 μl of synthesized cDNA as the template and Luna Universal qPCR master mix (NEB, USA; no. M3003) according to the manufacturer’s specifications. The following primers were used for qPCR: REVec genomic RNA (5′-GGCCGTCATGGTGGCGAATG and 5′-CGTCTCTTGGGTGGCATTGCG), GFP mRNA (5′-AGATCCGCCACAACATCGAG and 5′-TCTCGTTGGGGTCTTTGCTC), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (5′-ATCTTCTTTTGCGTCGCCAG and 5′-ACGACCAAATCCGTTGACTCC). The relative values for REVec genomic RNA and GFP mRNA were calculated by the relative quantification method using GAPDH mRNA as a reference.
Statistical analysis.
All statistical analyses were performed with GraphPad Prism 8 software. The tests used for each experiment are described in the figure legends.
ACKNOWLEDGMENTS
This study was supported in part by JSPS KAKENHI grant JP19J23468 (T.K.), JP18K19443 (M.H.), JP21H01199 (M.H.), JP19K22530 (K.T.), and JP20H05682 (K.T.); MEXT KAKENHI grant numbers JP16H06429, JP16K21723, and JP16H06430 (all to K.T.); JSPS Core-to-Core Program (K.T.); and the Joint Usage/Research Center Program on inFront and the Hakubi Project, Kyoto University.
Contributor Information
Keizo Tomonaga, Email: tomonaga@infront.kyoto-u.ac.jp.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Kuhn JH, Adkins S, Alioto D, Alkhovsky SV, Amarasinghe GK, Anthony SJ, Avsic-Zupanc T, Ayllon MA, Bahl J, Balkema-Buschmann A, Ballinger MJ, Bartonicka T, Basler C, Bavari S, Beer M, Bente DA, Bergeron E, Bird BH, Blair C, Blasdell KR, Bradfute SB, Breyta R, Briese T, Brown PA, Buchholz UJ, Buchmeier MJ, Bukreyev A, Burt F, Buzkan N, Calisher CH, Cao M, Casas I, Chamberlain J, Chandran K, Charrel RN, Chen B, Chiumenti M, Choi IR, Clegg JCS, Crozier I, da Graca JV, Dal Bo E, Davila AMR, de la Torre JC, de Lamballerie X, de Swart RL, Di Bello PL, Di Paola N, Di Serio F, Dietzgen RG, et al. 2020. 2020 Taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch Virol 165:3023–3072. 10.1007/s00705-020-04731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsumoto Y, Hayashi Y, Omori H, Honda T, Daito T, Horie M, Ikuta K, Fujino K, Nakamura S, Schneider U, Chase G, Yoshimori T, Schwemmle M, Tomonaga K. 2012. Bornavirus closely associates and segregates with host chromosomes to ensure persistent intranuclear infection. Cell Host Microbe 11:492–503. 10.1016/j.chom.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Horie M, Kobayashi Y, Suzuki Y, Tomonaga K. 2013. Comprehensive analysis of endogenous bornavirus-like elements in eukaryote genomes. Philos Trans R Soc Lond B Biol Sci 368:20120499. 10.1098/rstb.2012.0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daito T, Fujino K, Honda T, Matsumoto Y, Watanabe Y, Tomonaga K. 2011. A novel borna disease virus vector system that stably expresses foreign proteins from an intercistronic noncoding region. J Virol 85:12170–12178. 10.1128/JVI.05554-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujino K, Yamamoto Y, Daito T, Makino A, Honda T, Tomonaga K. 2017. Generation of a non-transmissive Borna disease virus vector lacking both matrix and glycoprotein genes. Microbiol Immunol 61:380–386. 10.1111/1348-0421.12505. [DOI] [PubMed] [Google Scholar]
- 6.Honda T, Yamamoto Y, Daito T, Matsumoto Y, Makino A, Tomonaga K. 2016. Long-term expression of miRNA for RNA interference using a novel vector system based on a negative-strand RNA virus. Sci Rep 6:26154. 10.1038/srep26154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai M, Ueda S, Daito T, Asada-Utsugi M, Komatsu Y, Kinoshita A, Maki T, Kuzuya A, Takahashi R, Makino A, Tomonaga K. 2018. Degradation of amyloid β peptide by neprilysin expressed from Borna disease virus vector. Microbiol Immunol 62:467–472. 10.1111/1348-0421.12602. [DOI] [PubMed] [Google Scholar]
- 8.Komatsu Y, Tanaka C, Komorizono R, Tomonaga K. 2020. In vivo biodistribution analysis of transmission competent and defective RNA virus-based episomal vector. Sci Rep 10:5890. 10.1038/s41598-020-62630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda Y, Makino A, Matchett WE, Holditch SJ, Lu B, Dietz AB, Tomonaga K. 2016. A novel intranuclear RNA vector system for long-term stem cell modification. Gene Ther 23:256–262. 10.1038/gt.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komatsu Y, Takeuchi D, Tokunaga T, Sakurai H, Makino A, Honda T, Ikeda Y, Tomonaga K. 2019. RNA virus-based episomal vector with a fail-safe switch facilitating efficient genetic modification and differentiation of iPSCs. Mol Ther Methods Clin Dev 14:47–55. 10.1016/j.omtm.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briese T, de la Torre JC, Lewis A, Ludwig H, Lipkin WI. 1992. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc Natl Acad Sci USA 89:11486–11489. 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briese T, Schneemann A, Lewis AJ, Park YS, Kim S, Ludwig H, Lipkin WI. 1994. Genomic organization of Borna disease virus. Proc Natl Acad Sci USA 91:4362–4366. 10.1073/pnas.91.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomonaga K, Kobayashi T, Ikuta K. 2002. Molecular and cellular biology of Borna disease virus infection. Microbes Infect 4:491–500. 10.1016/s1286-4579(02)01564-2. [DOI] [PubMed] [Google Scholar]
- 14.Perez M, Sanchez A, Cubitt B, Rosario D, de la Torre JC. 2003. A reverse genetics system for Borna disease virus. J Gen Virol 84:3099–3104. 10.1099/vir.0.19467-0. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu Y, Tomonaga K. 2020. Reverse genetics approaches of Borna disease virus: applications in development of viral vectors and preventive vaccines. Curr Opin Virol 44:42–48. 10.1016/j.coviro.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Martin A, Staeheli P, Schneider U. 2006. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J Virol 80:5708–5715. 10.1128/JVI.02389-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackermann A, Staeheli P, Schneider U. 2007. Adaptation of Borna disease virus to new host species attributed to altered regulation of viral polymerase activity. J Virol 81:7933–7940. 10.1128/JVI.00334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima S, Honda T, Matsumoto Y, Tomonaga K. 2014. Heat stress is a potent stimulus for enhancing rescue efficiency of recombinant Borna disease virus. Microbiol Immunol 58:636–642. 10.1111/1348-0421.12193. [DOI] [PubMed] [Google Scholar]
- 19.Nowotny N, Kolodziejek J, Jehle CO, Suchy A, Staeheli P, Schwemmle M. 2000. Isolation and characterization of a new subtype of Borna disease virus. J Virol 74:5655–5658. 10.1128/jvi.74.12.5655-5658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pleschka S, Staeheli P, Kolodziejek J, Richt JA, Nowotny N, Schwemmle M. 2001. Conservation of coding potential and terminal sequences in four different isolates of Borna disease virus. J Gen Virol 82:2681–2690. 10.1099/0022-1317-82-11-2681. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi T, Kamitani W, Zhang G, Watanabe M, Tomonaga K, Ikuta K. 2001. Borna disease virus nucleoprotein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. J Virol 75:3404–3412. 10.1128/JVI.75.7.3404-3412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi T, Shoya Y, Koda T, Takashima I, Lai PK, Ikuta K, Kakinuma M, Kishi M. 1998. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology 243:188–197. 10.1006/viro.1998.9049. [DOI] [PubMed] [Google Scholar]
- 23.Berg M, Ehrenborg C, Blomberg J, Pipkorn R, Berg AL. 1998. Two domains of the Borna disease virus p40 protein are required for interaction with the p23 protein. J Gen Virol 79:2957–2963. 10.1099/0022-1317-79-12-2957. [DOI] [PubMed] [Google Scholar]
- 24.Yanai H, Kobayashi T, Hayashi Y, Watanabe Y, Ohtaki N, Zhang G, de la Torre JC, Ikuta K, Tomonaga K. 2006. A methionine-rich domain mediates CRM1-dependent nuclear export activity of Borna disease virus phosphoprotein. J Virol 80:1121–1129. 10.1128/JVI.80.3.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoya Y, Kobayashi T, Koda T, Ikuta K, Kakinuma M, Kishi M. 1998. Two proline-rich nuclear localization signals in the amino- and carboxyl-terminal regions of the Borna disease virus phosphoprotein. J Virol 72:9755–9762. 10.1128/JVI.72.12.9755-9762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwemmle M, Jehle C, Shoemaker T, Lipkin WI. 1999. Characterization of the major nuclear localization signal of the Borna disease virus phosphoprotein. J Gen Virol 80:97–100. 10.1099/0022-1317-80-1-97. [DOI] [PubMed] [Google Scholar]
- 27.Schneider U, Naegele M, Staeheli P, Schwemmle M. 2003. Active Borna disease virus polymerase complex requires a distinct nucleoprotein-to-phosphoprotein ratio but no viral X protein. J Virol 77:11781–11789. 10.1128/jvi.77.21.11781-11789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poenisch M, Unterstab G, Wolff T, Staeheli P, Schneider U. 2004. The X protein of Borna disease virus regulates viral polymerase activity through interaction with the P protein. J Gen Virol 85:1895–1898. 10.1099/vir.0.80002-0. [DOI] [PubMed] [Google Scholar]
- 29.Hock M, Kraus I, Schoehn G, Jamin M, Andrei-Selmer C, Garten W, Weissenhorn W. 2010. RNA induced polymerization of the Borna disease virus nucleoprotein. Virology 397:64–72. 10.1016/j.virol.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Pyper JM, Gartner AE. 1997. Molecular basis for the differential subcellular localization of the 38- and 39-kilodalton structural proteins of Borna disease virus. J Virol 71:5133–5139. 10.1128/JVI.71.7.5133-5139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima S, Sato R, Yanai M, Komatsu Y, Horie M, Igarashi M, Tomonaga K. 2019. Splicing-dependent subcellular targeting of Borna disease virus nucleoprotein isoforms. J Virol 93:e01621-18. 10.1128/JVI.01621-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolodziejek J, Dürrwald R, Herzog S, Ehrensperger F, Lussy H, Nowotny N. 2005. Genetic clustering of Borna disease virus natural animal isolates, laboratory and vaccine strains strongly reflects their regional geographical origin. J Gen Virol 86:385–398. 10.1099/vir.0.80587-0. [DOI] [PubMed] [Google Scholar]
- 33.Dürrwald R, Kolodziejek J, Weissenböck H, Nowotny N. 2014. The bicolored white-toothed shrew Crocidura leucodon (Hermann 1780) is an indigenous host of mammalian Borna disease virus. PLoS One 9:e93659. 10.1371/journal.pone.0093659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosario D, Perez M, de la Torre JC. 2005. Functional characterization of the genomic promoter of Borna disease virus (BDV): implications of 3'-terminal sequence heterogeneity for BDV persistence. J Virol 79:6544–6550. 10.1128/JVI.79.10.6544-6550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider U, Schwemmle M, Staeheli P. 2005. Genome trimming: a unique strategy for replication control employed by Borna disease virus. Proc Natl Acad Sci USA 102:3441–3446. 10.1073/pnas.0405965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider U. 2005. Novel insights into the regulation of the viral polymerase complex of neurotropic Borna disease virus. Virus Res 111:148–160. 10.1016/j.virusres.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Honda T, Tomonaga K. 2013. Nucleocytoplasmic shuttling of viral proteins in Borna disease virus infection. Viruses 5:1978–1990. 10.3390/v5081978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai M, Fujita Y, Komorizono R, Kanda T, Komatsu Y, Noda T, Tomonaga K, Makino A. 2021. Optimal expression of the envelope glycoprotein of orthobornaviruses determines the production of mature virus particles. J Virol 95:e02221-20. 10.1128/JVI.02221-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwemmle M, Salvatore M, Shi L, Richt J, Lee CH, Lipkin WI. 1998. Interactions of the Borna disease virus P, N, and X proteins and their functional implications. J Biol Chem 273:9007–9012. 10.1074/jbc.273.15.9007. [DOI] [PubMed] [Google Scholar]
- 40.Malik TH, Kishi M, Lai PK. 2000. Characterization of the P protein-binding domain on the 10-kilodalton protein of Borna disease virus. J Virol 74:3413–3417. 10.1128/jvi.74.7.3413-3417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider U, Blechschmidt K, Schwemmle M, Staeheli P. 2004. Overlap of interaction domains indicates a central role of the P protein in assembly and regulation of the Borna disease virus polymerase complex. J Biol Chem 279:55290–55296. 10.1074/jbc.M408913200. [DOI] [PubMed] [Google Scholar]
- 42.Chase G, Mayer D, Hildebrand A, Frank R, Hayashi Y, Tomonaga K, Schwemmle M. 2007. Borna disease virus matrix protein is an integral component of the viral ribonucleoprotein complex that does not interfere with polymerase activity. J Virol 81:743–749. 10.1128/JVI.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker MP, Jordan I, Briese T, Fischer N, Lipkin WI. 2000. Expression and characterization of the Borna disease virus polymerase. J Virol 74:4425–4428. 10.1128/jvi.74.9.4425-4428.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korn K, Coras R, Bobinger T, Herzog SM, Lücking H, Stöhr R, Huttner HB, Hartmann A, Ensser A. 2018. Fatal encephalitis associated with Borna disease virus 1. N Engl J Med 379:1375–1377. 10.1056/NEJMc1800724. [DOI] [PubMed] [Google Scholar]
- 45.Schlottau K, Forth L, Angstwurm K, Höper D, Zecher D, Liesche F, Hoffmann B, Kegel V, Seehofer D, Platen S, Salzberger B, Liebert UG, Niller HH, Schmidt B, Matiasek K, Riemenschneider MJ, Brochhausen C, Banas B, Renders L, Moog P, Wunderlich S, Seifert CL, Barreiros A, Rahmel A, Weiss J, Tappe D, Herden C, Schmidt-Chanasit J, Schwemmle M, Rubbenstroth D, Schlegel J, Pietsch C, Hoffmann D, Jantsch J, Beer M. 2018. Fatal encephalitic Borna disease virus 1 in solid-organ transplant recipients. N Engl J Med 379:1377–1379. 10.1056/NEJMc1803115. [DOI] [PubMed] [Google Scholar]
- 46.Coras R, Korn K, Kuerten S, Huttner HB, Ensser A. 2019. Severe bornavirus-encephalitis presenting as Guillain-Barre-syndrome. Acta Neuropathol 137:1017–1019. 10.1007/s00401-019-02005-z. [DOI] [PubMed] [Google Scholar]
- 47.Liesche F, Ruf V, Zoubaa S, Kaletka G, Rosati M, Rubbenstroth D, Herden C, Goehring L, Wunderlich S, Wachter MF, Rieder G, Lichtmannegger I, Permanetter W, Heckmann JG, Angstwurm K, Neumann B, Markl B, Haschka S, Niller HH, Schmidt B, Jantsch J, Brochhausen C, Schlottau K, Ebinger A, Hemmer B, Riemenschneider MJ, Herms J, Beer M, Matiasek K, Schlegel J. 2019. The neuropathology of fatal encephalomyelitis in human Borna virus infection. Acta Neuropathol 138:653–665. 10.1007/s00401-019-02047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niller HH, Angstwurm K, Rubbenstroth D, Schlottau K, Ebinger A, Giese S, Wunderlich S, Banas B, Forth LF, Hoffmann D, Höper D, Schwemmle M, Tappe D, Schmidt-Chanasit J, Nobach D, Herden C, Brochhausen C, Velez-Char N, Mamilos A, Utpatel K, Evert M, Zoubaa S, Riemenschneider MJ, Ruf V, Herms J, Rieder G, Errath M, Matiasek K, Schlegel J, Liesche-Starnecker F, Neumann B, Fuchs K, Linker RA, Salzberger B, Freilinger T, Gartner L, Wenzel JJ, Reischl U, Jilg W, Gessner A, Jantsch J, Beer M, Schmidt B. 2020. Zoonotic spillover infections with Borna disease virus 1 leading to fatal human encephalitis, 1999–2019: an epidemiological investigation. Lancet Infect Dis 20:467–477. 10.1016/S1473-3099(19)30546-8. [DOI] [PubMed] [Google Scholar]
- 49.Rubbenstroth D, Schlottau K, Schwemmle M, Rissland J, Beer M. 2019. Human bornavirus research: back on track! PLoS Pathog 15:e1007873. 10.1371/journal.ppat.1007873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 51.Yanai H, Hayashi Y, Watanabe Y, Ohtaki N, Kobayashi T, Nozaki Y, Ikuta K, Tomonaga K. 2006. Development of a novel Borna disease virus reverse genetics system using RNA polymerase II promoter and SV40 nuclear import signal. Microbes Infect 8:1522–1529. 10.1016/j.micinf.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Reuter A, Horie M, Höper D, Ohnemus A, Narr A, Rinder M, Beer M, Staeheli P, Rubbenstroth D. 2016. Synergistic antiviral activity of ribavirin and interferon-α against parrot bornaviruses in avian cells. J Gen Virol 97:2096–2103. 10.1099/jgv.0.000555. [DOI] [PubMed] [Google Scholar]
- 53.Chadalavada DM, Cerrone-Szakal AL, Bevilacqua PC. 2007. Wild-type is the optimal sequence of the HDV ribozyme under cotranscriptional conditions. RNA 13:2189–2201. 10.1261/rna.778107. [DOI] [PMC free article] [PubMed] [Google Scholar]