ABSTRACT
Coxsackievirus B3 (CVB3) is an enterovirus belonging to the family Picornaviridae. Its 5′ untranslated region (UTR) contains a cloverleaf structure followed by an internal ribosome entry site (IRES). The cloverleaf forms an RNA-protein complex known to regulate virus replication, translation, and stability of the genome, and the IRES regulates virus RNA translation. For positive-strand RNA-containing viruses, such as members of the flaviviruses or enteroviruses, the genomic RNA is used for translation, replication, and encapsidation. Only a few regulatory mechanisms which govern the accessibility of genomic RNA templates for translation or replication have been reported. Here, we report the role of human antigen R (HuR) in regulating the fate of CVB3 positive-strand RNA into the replication cycle or translation cycle. We have observed that synthesis of HuR is induced during CVB3 infection, and it suppresses viral replication by displacing PCBP-2 (a positive regulator of virus replication) at the cloverleaf RNA. Silencing of HuR increases viral RNA replication and consequently reduces viral RNA translation in a replication-dependent manner. Furthermore, we have shown that HuR level is upregulated upon CVB3 infection. Moreover, HuR limits virus replication and can coordinate the availability of genomic RNA templates for translation, replication, or encapsidation. Our study highlights the fact that the relative abundance of translation factors and replication factors in the cell decides the outcome of viral infection.
IMPORTANCE A positive-strand RNA virus must balance the availability of its genomic template for different viral processes at different stages of its life cycle. A few host proteins are shown to be important to help the virus in switching the usage of a template between these processes. These proteins inhibit translation either by displacing a stimulator of translation or by binding to an alternative site. Both mechanisms lead to ribosome clearance and availability of the genomic strand for replication. We have shown that HuR also helps in maintaining this balance by inhibiting replication and subsequently promoting translation and packaging.
KEYWORDS: coxsackievirus B3, ELAV1, HuR, human antigen R, IRES-mediated translation, PCBP-2, RNA replication, template switching
INTRODUCTION
Coxsackievirus B3 (CVB3), an enterovirus belonging to the Picornaviridae family, is majorly responsible for virus-induced myocarditis, pancreatitis, and aseptic meningitis in humans. The viral genome consists of a single-stranded positive-sense RNA with a 5′ untranslated region (UTR) and a long open reading frame (ORF) followed by a 3′ UTR. The terminal cloverleaf structure within the 5′ UTR is a cis-acting element that interacts with the viral polymerase and the host protein, poly(rC) binding protein (PCBP-2), to form a ternary complex. While this interaction is known to be essential for both viral RNA translation and replication in poliovirus (1–3), this complex has only been shown to be important for replication in CVB3 (4, 5). As translation proceeds, viral proteins required for RNA synthesis accumulate. This results in the formation of replication complexes and a switch of genomic RNA template usage from translation to RNA replication. Few proteins such as PCBP-2 and hnRNP C1/C2 have been reported to regulate the availability of RNA templates for these processes. In poliovirus, PCBP-2 binds to the 5′ cloverleaf structure and stimulates translation as well as RNA synthesis; when it binds to the internal ribosome entry site (IRES) region, it stimulates only translation (3). However, as viral proteins are made, the binding of 3CD protein to the cloverleaf together with PCBP-2 inhibits translation and stimulates replication (6). Similarly, in the case of CVB3, PCBP-2 binds to both cloverleaf and IRES. The binding to IRES stimulates CVB3 RNA translation, and the interaction with cloverleaf stimulates negative-strand RNA synthesis (5, 7). A stage-specific role of PCBP-2 in regulating RNA template usage has also been reported. At mid-replication stage, the viral 3C protease cleaves PCBP-2. The cleaved PCBP-2 no longer binds to IRES, and translation is inhibited; however, it can bind to the cloverleaf with a higher affinity than the full-length PCBP-2 and thereby stimulates replication (3). In a similar way, another host protein, hnRNP C1/C2, binds to IRES and displaces polypyrimidine tract-binding protein (PTB). This leads to inhibition of translation and stimulation of replication, as more genomic RNA template is accessible to the replication machinery (8).
Previously, using RNA affinity chromatography and liquid chromatography-mass spectrometry (LC-MS), we have identified several host proteins that interact with this cloverleaf region of CVB3 RNA (9). In the screen, we found that an ELAV-like protein 1, also known as human antigen R (HuR), interacts with the cloverleaf structure of the viral RNA. HuR is a ubiquitously nuclear-expressed RNA-binding protein, which is known to stabilize several cellular RNAs by interacting with their 3′ UTRs and thereby influence their translation. Though reports indicate the role of HuR in some RNA viruses, its role in CVB3 life cycle has not been elucidated so far. HuR influences the formation of a ribonucleoprotein complex at the 3′ UTR of hepatitis C virus (HCV) RNA and aids its replication (10). In Sindbis virus infection, HuR interacts with the 3′ UTR and protects the viral genome from cellular mRNA decay machinery (11). HuR inhibits Zika virus translation and replication as well as virus particle production (12). Several RNA-binding proteins required during the RNA virus life cycle are nuclear proteins. Therefore, their cytoplasmic relocation is crucial for robust infection. For infections by hepatitis C virus (HCV) and several alphaviruses such as Sindbis, chikungunya, and Ross River viruses, HuR relocalizes from the nucleus to the cytoplasm (11, 13). However, the cytoplasmic relocalization of HuR is not a general reaction of the cell to all virus infections, as it is not observed in measles and dengue virus infections (13). HuR relocates to the cytoplasm during CVB3 infection also (14). However, the underlying mechanism for HuR relocalization in any viral infection has not been elucidated so far.
We found that HuR interacts with cloverleaf RNA and modulates CVB3 replication and translation. Also, the enhanced level of HuR protein during CVB3 infection is due to the downregulation of a HuR targeting microRNA, miR-125b-5p. Furthermore, we found that binding of HuR at the cloverleaf RNA of the viral genome displaces PCBP-2 and that it is also involved in regulation of the viral RNA life cycle. The binding of HuR to the cloverleaf enhances translation and inhibits negative-strand synthesis. Thus, we propose that the overlapping translation and replication signals within the cloverleaf region regulated by HuR function as a viral strategy to regulate replication, thereby making the genomic RNA template accessible for translation or virus packaging.
RESULTS
HuR specifically interacts with CVB3 cloverleaf RNA.
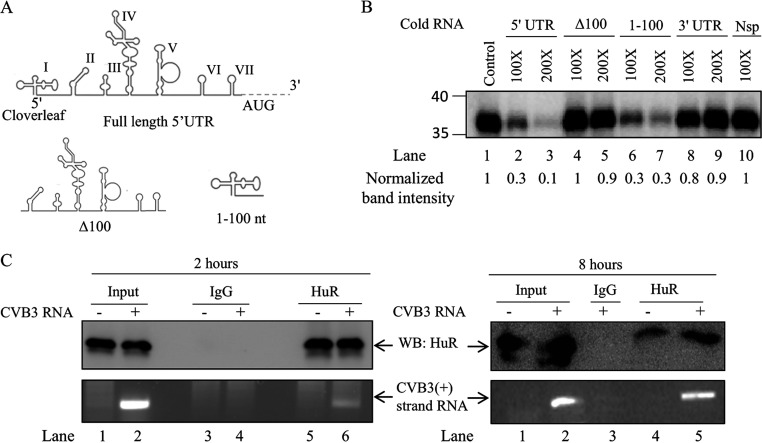
Previously, we have shown the interaction of several host proteins with CVB3 cloverleaf RNA by using an RNA affinity assay, and HuR was one of these proteins (9). To confirm the specificity of HuR interaction, and to determine if HuR interacts with any other noncoding region in the CVB3, an RNA competition UV cross-linking experiment was performed using a radiolabeled CVB3 5′ UTR RNA and recombinant HuR protein. Unlabeled CVB3 5′ UTR, Δ100 CVB3 5′ UTR (IRES region), 1 to 100 nucleotide (1-100 nt; cloverleaf region), and CVB3 3′ UTR RNA were used as competitors in this experiment (Fig. 1A and B). As observed in Fig. 1B, HuR interacted specifically with the cloverleaf structure within the CVB3 5′ UTR but not with Δ100 CVB3 5′ UTR and 3′ UTR. The specificity of interaction of HuR with CVB3 RNA was also confirmed using bovine serum albumin (BSA) as a nonspecific protein (data not shown). The interaction of HuR with CVB3 RNA at different time points within the cells was confirmed by an immunoprecipitation experiment. HeLa cells were transfected with the CVB3 replicon RNA, followed by immunoprecipitation using anti-HuR antibody. Anti-human IgG was used as a nonspecific antibody control. RNA was isolated from the immunoprecipitated RNP complexes, and the associated CVB3 sense-strand RNA was detected after 2 and 8 h posttransfection using reverse transcription-PCR (RT-PCR) (Fig. 1C). These results show that the HuR protein interacts with the sense strand of CVB3 RNA.
FIG 1.
HuR interacts with CVB3 cloverleaf RNA. (A) Schematic of CVB3 5′ UTR and its regions used in UV cross-linking assay (adopted from reference 9). Δ100 indicates the entire CVB3 5′ UTR without the cloverleaf region. (B) UV cross-linking assay with 32P-labeled full-length 5′ UTR of CVB3 genomic RNA alone or in the presence of indicated unlabeled RNA. For the competition, unlabeled full-length 5′ UTR (lanes 2 and 3), Δ100 (lanes 4 and 5), cloverleaf (1-100) RNA (lanes 6 and 7), CVB3 3′ UTR (lanes 8 and 9), or nonspecific RNA (lane 10) was used as indicated. Unlabeled nonspecific RNA of equivalent length was derived from the multiple-cloning site (MCS) region of the pGEMt vector. (C) CVB3 replicon RNA was transfected in HeLa cells. Two and eight hours posttransfection, HuR protein was immunoprecipitated using anti-HuR antibody or IgG isotype antibody (as a control). RNA was purified from the immunoprecipitated complexes, and viral positive-strand RNA was detected by RT-PCR.
Role of HuR in CVB3 RNA replication and translation.
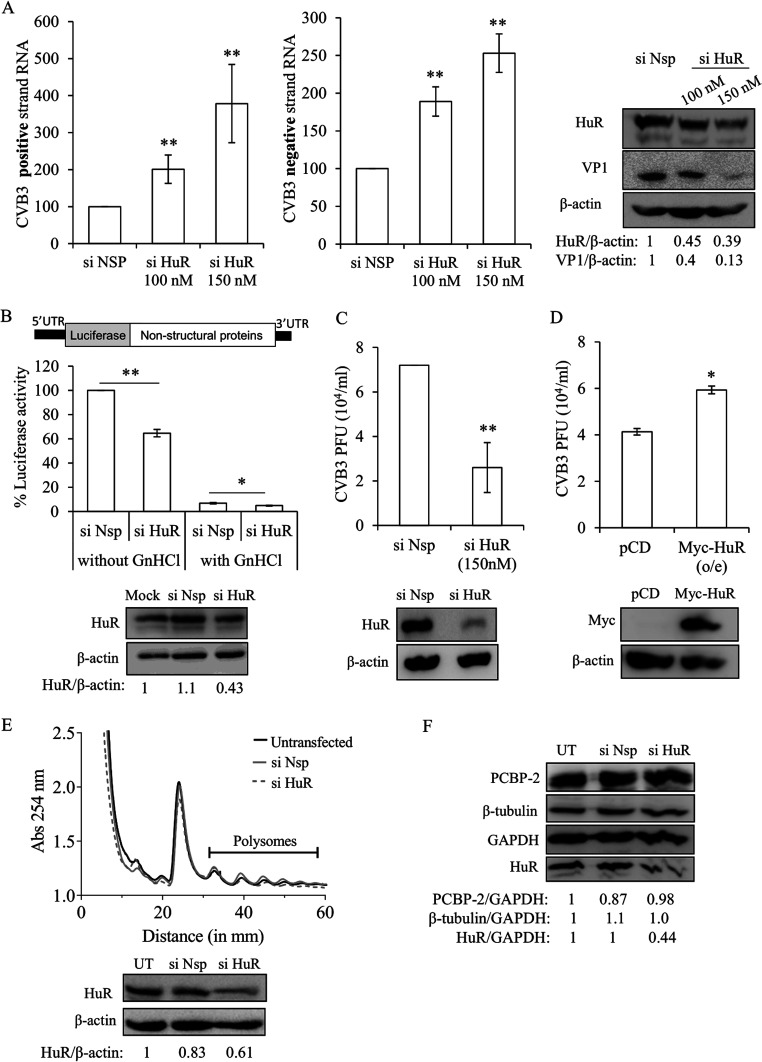
To determine the function of HuR in the CVB3 life cycle, partial silencing of HuR was done using small interfering RNA (siRNA) targeting the coding region of HuR. HeLa cells were transfected with CVB3 infectious RNA after partial silencing of HuR. Subsequently, the CVB3 positive and negative strands and viral protein levels were estimated using quantitative RT-PCR (qRT-PCR) and Western blotting, respectively. An increase in the CVB3 positive-strand as well as negative-strand RNA was observed upon silencing of HuR (Fig. 2A). A significant reduction in the viral protein levels was also observed, as shown in the Western blot in Fig. 2A. To further support our results, we carried out a luciferase assay following the transfection of CVB3 replicon RNA after partial silencing of HuR with or without guanidine hydrochloride (GnHCl) in the culture medium. A schematic of the CVB3 replicon RNA construct used in the experiment is depicted in Fig. 2B. GnHCl is known to inhibit CVB3 RNA replication by targeting the 2C protein. In the absence of GnHCl, the luciferase signal represents the sum of both viral translation and RNA replication by translation of newly synthesized RNA. In the presence of GnHCl, the luciferase signal indicates translation from the input replicon RNA. Thus, viral RNA translation or replication can be measured by comparing the difference in the luciferase signals in the presence and absence of GnHCl. In the absence of GnHCl, an approximately 36% decrease in the luciferase activity was observed upon partial knockdown of HuR compared to that in nonspecific-siRNA transfected cells. However, only a minor decrease in the luciferase signal was observed in GnHCl-treated cells (Fig. 2B). This indicates that silencing of HuR is involved in the enhancement of CVB3 replication, and, probably, the effect on translation is indirect. To rule out the possibility of the difference in luciferase values as a consequence of siRNA or plasmid transfection on cell viability, a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was performed. No significant effect on cell viability was observed upon silencing or overexpressing HuR (data not shown).
FIG 2.
Role of HuR in CVB3 replication and translation. (A) HeLa cells were transfected with either siHuR or siNSP and, 24 h later, transfected with CVB3 infectious RNA. Cells were harvested 10 h posttransfection, and CVB3 positive-strand and negative-strand RNAs were measured by qRT-PCR. Graphs indicate the percent change in CVB3 RNA levels after siHuR transfection compared to that with siNSP. Error bars indicate the standard deviation from three independent experiments. HuR, CVB3 VP1, and β-actin protein levels are indicated in the immunoblots. (B) The effect of partial knockdown of HuR on CVB3 replication and translation was studied in the presence and absence of viral replication inhibitor, GnHCl (2 mM), using CVB3 subgenomic replicon RNA. A schematic of CVB3 replicon RNA is shown. HeLa cells were transfected with siHuR and, 24 h later, transfected with CVB3 replicon RNA. Cells were harvested 10 h post-replicon transfection, and luciferase activity was measured. HuR and β-actin protein levels are indicated in the immunoblots. (C and D) Role of HuR in CVB3 virion production. Partial silencing of HuR was done using siHuR, and overexpression was carried out using a Myc-tagged HuR construct. Twenty-four hours later, cells were infected with CVB3 virus at an MOI of 5. At 8 h p.i., the supernatant was collected and used to infect fresh cells. The number of plaques was quantified to estimate the virus titer. HuR, Myc, and β-actin protein levels are indicated in the immunoblots. (E) Uninfected HeLa cells were used for polysome profile analysis to check the effect of siHuR on global translation. The profiles for untransfected cells (black line), cells transfected with siNSP (gray line), and cells transfected with siHuR (gray dotted line) are shown. The polysomes peaks are indicated. (F) Specificity of siHuR used in silencing experiments. The off-target effect (if any) of siHuR was determined by checking the levels of other proteins such as PCBP-2 and β-tubulin, which are not the targets of siHuR. HeLa cells were transfected with siHuR, and 32 h posttransfection, total protein was isolated and levels of HuR, PCBP-2, β-tubulin, and GAPDH protein were detected. Western blots indicate the level of the above-described proteins. UT, untransfected cells. Error bars indicate standard deviation from three independent experiments. *, P < 0.05; **, P < 0.005 throughout. Two-tailed Student’s t test was used for statistical analysis.
To study the role of HuR on infectious virus particle production, we estimated the virus titer using a plaque assay after partial silencing or overexpression of HuR. The results indicate that the viral titer was significantly reduced upon partial silencing of HuR, and an increase in viral titer was observed upon overexpression of HuR. These results suggest that HuR is essential for virus production (Fig. 2C and D).
Since HuR is involved in the stability of several cellular mRNAs, we studied the effect of partial silencing of HuR on global translation using polysome profiling. Results show that partial silencing of HuR apparently does not alter the global translation (Fig. 2E). We checked the off-target activity of siRNA (if any) by analyzing the levels of other proteins such as PCBP-2, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-tubulin (Fig. 2F). Also, NCBI BLAST analysis of the siHuR sequence did not show other targets (data not shown).
HuR is upregulated in CVB3 infection.
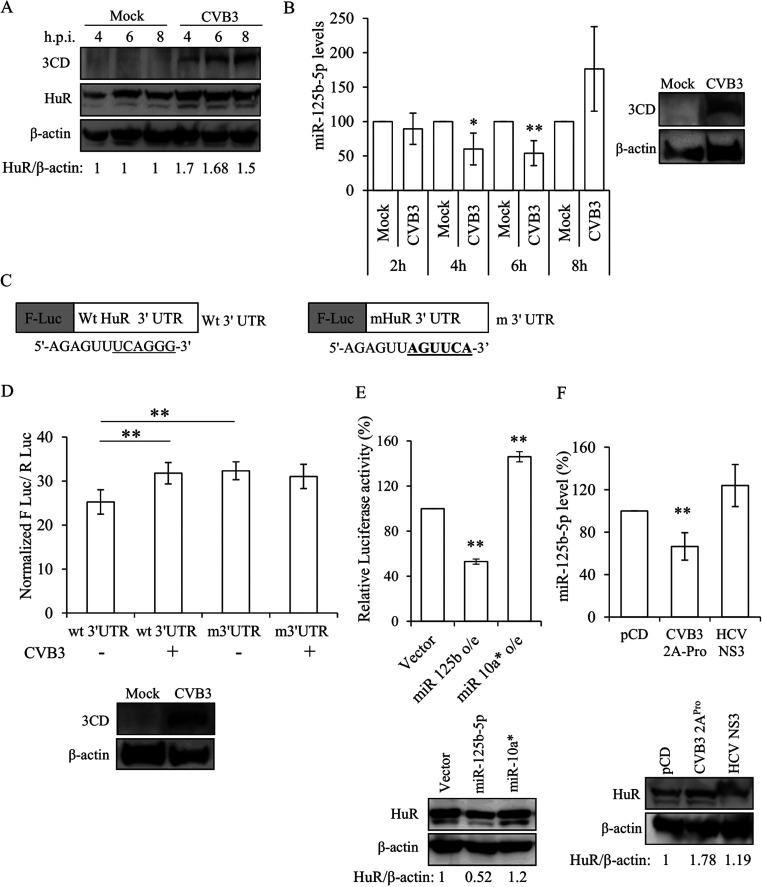
A time-dependent analysis of HuR showed that its protein level was induced upon virus infection. As observed in Fig. 3A, there was an increase in the HuR protein level at 4 h postinfection (h p.i.) of CVB3. An earlier report from our group has shown that in Huh 7.5 cells, miR-125b-5p interacts with the 3′ UTR of HuR and inhibits its translation (15). To investigate this, we checked the levels of miR-125b-5p in HeLa cells after CVB3 infection. The level of miR-125b-5p was found to be reduced by approximately 40% at 6 h p.i. of CVB3 at a multiplicity of infection (MOI) of 1 (Fig. 3B). To further validate these results, we used HuR 3′ UTR wild-type (WT) and mutant (mutation in miR-125b-5p binding site) luciferase constructs (Fig. 3C) and checked their activity in the presence of CVB3 (MOI of 1) at 6 h p.i. in HeLa cells. It was found that the luciferase activity of wild-type HuR 3′ UTR plasmid was significantly higher in the presence of CVB3. In addition, the HuR 3′ UTR mutant did not show an appreciable change in luciferase activity in the absence or presence of CVB3 infection (Fig. 3D, third and fourth bars). These results indicate that the increase in HuR protein upon CVB3 infection is probably due to reduced miR-125b-5p levels. To confirm its role, we overexpressed miR-125b-5p and miR-10a* followed by transfection with CVB3 replicon RNA. We observed a decrease in the HuR protein level and a concomitant decrease in luciferase activity upon overexpression of miR-125b-5p (Fig. 3E). miR-10a* was previously reported to stimulate virus translation; therefore, it was used as a positive control in this experiment. As reported previously (16), we also observed an increase in the luciferase activity of replicon upon overexpression of miR10a* (Fig. 3E). Since virus-encoded 2A protease is known to alter several host cellular processes, we checked the effect of CVB3-2A protease on the miR-125b-5p and HuR levels. We observed that the overexpression of 2A protease was sufficient to reduce miR-125b-5p and to induce HuR protein expression (Fig. 3F). HCV NS3 protease was used as a negative control in this experiment and did not show any significant effect on miR-125b-5p or HuR levels (Fig. 3F). These results confirm that HuR protein level is regulated by miR-125b-5p upon CVB3 infection.
FIG 3.
HuR is upregulated in CVB3 infection. (A) Western blot analysis of HuR protein levels at different time points post-CVB3 infection in HeLa cells. (B) miR-125b-5p levels at different time points post-CVB3 infection as determined by qRT-PCR. Western blot indicating the viral protein 3CD postinfection. (C) Schematic representation of F Luc-HuR 3′ UTR wild-type and mutant constructs, where F-Luc indicates the firefly luciferase gene. (D) HeLa cells were transfected with HuR 3′ UTR constructs (wild type or mutant) followed by CVB3 infection at an MOI of 1, and luciferase activity was measured at 6 h p.i. (E) miR-125b-5p and miR-10a* were overexpressed using respective overexpression plasmids followed by CVB3 replicon transfection, and luciferase activity was measured. Western blots indicate levels of HuR upon overexpression of miR-125b-5p and miR-10a*. (F) HeLa cells were transfected with CVB3 2A protease or HCV NS3 protease expression plasmids. After 24 h, levels of miR-125b-5p RNA and HuR protein were measured by qRT-PCR and immunoblotting, respectively. Error bars indicate standard deviation from the three independent experiments. *, P < 0.05; **, P < 0.005. Two-tailed Student’s t test was used for statistical analysis.
HuR relocates to cytoplasm during CVB3 infection.
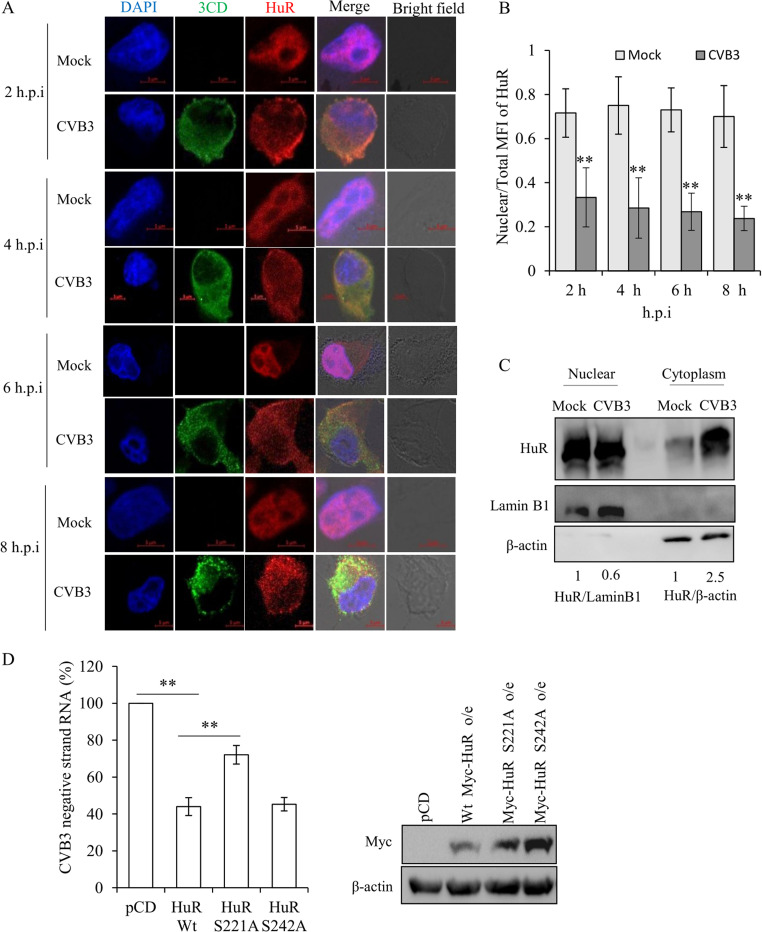
Since HuR is predominantly a nuclear protein and CVB3 replication and translation take place in the cytoplasm, the localization of HuR during CVB3 infection was studied by immunofluorescence. Our results indicated that the cytoplasmic abundance of HuR increases as early as 2 h p.i. compared to that in uninfected (mock) cells at 2 h (Fig. 4A and B). Similar results were obtained when HuR cytoplasmic abundance was analyzed by subcellular fractionation followed by Western blotting (Fig. 4C). Furthermore, the cytoplasmic relocalization of HuR is known to be regulated by its posttranslational modifications (PTMs), majorly phosphorylation. HuR contains three RNA recognition motifs (RRMs). The hinge region between RRM2 and RRM3 contains the HuR-nuclear shuttling (HNS) domain. This domain is important for the shuttling of HuR in and out of the nucleus. Several PTMs around the hinge region affect the nucleocytoplasmic shuttling of HuR (17). We generated HuR S221A and S242A mutants, which are known to be involved in cytoplasmic relocation of HuR under other pathological conditions (17), and analyzed the effect of overexpression of mutant HuR on CVB3 replication. We observed that overexpression of the wild-type HuR inhibited CVB3 RNA levels, as expected (Fig. 4D). Interestingly, when the HuR S221A mutant was overexpressed, the inhibitory effect was significantly reduced. However, the HuR S242A mutant inhibited CVB3 replication similarly to the wild-type HuR (Fig. 4D).
FIG 4.
Cytoplasmic abundance of HuR increases in the presence of CVB3. (A) Immunofluorescence microscopy of HeLa cells infected with CVB3 (MOI of 1) or mock infected at various time points postinfection. Green represents CVB3-3CD polymerase, red represents HuR, and blue represents cell nucleus. (B) Quantification of the confocal images. ImageJ software was used to calculate the mean intensities of HuR in the nucleus and total cell. The ratio of nuclear HuR intensity to total cell HuR intensity is represented in the graph as MFI (mean fluorescence intensity). For 2, 4, 6, and 8 h p.i., 28, 31, 35, and 42 cells were quantified, respectively, from three independent experiments. Error bars represent the standard deviation from three independent experiments. (C) Western blot analysis of cytoplasmic and nuclear extracts. HeLa cells were harvested at 8 h p.i. and processed for subcellular fractionation. β-actin and lamin B1 were used as cytoplasmic and nuclear markers, respectively. (D) HeLa cells were transfected with either wild-type or phospho-dead Myc-HuR mutants (S221A and S242A). After 24 h, CVB3 replicon RNA was transfected, and 10 h posttransfection, CVB3 negative strand levels were measured. Western blots indicate levels of Myc-tagged HuR upon overexpression.
Additionally, the effect of mutation on cytoplasmic relocation of HuR was studied using immunofluorescence. Interestingly, the S242A mutant showed complete relocation to the cytoplasm under CVB3 infection; however, the S221A mutant showed partial retention in the nucleus, suggesting possible involvement of other phosphorylation sites in relocalization (data not shown).
HuR-PCBP-2 interplay at the cloverleaf RNA regulates HuR replication.
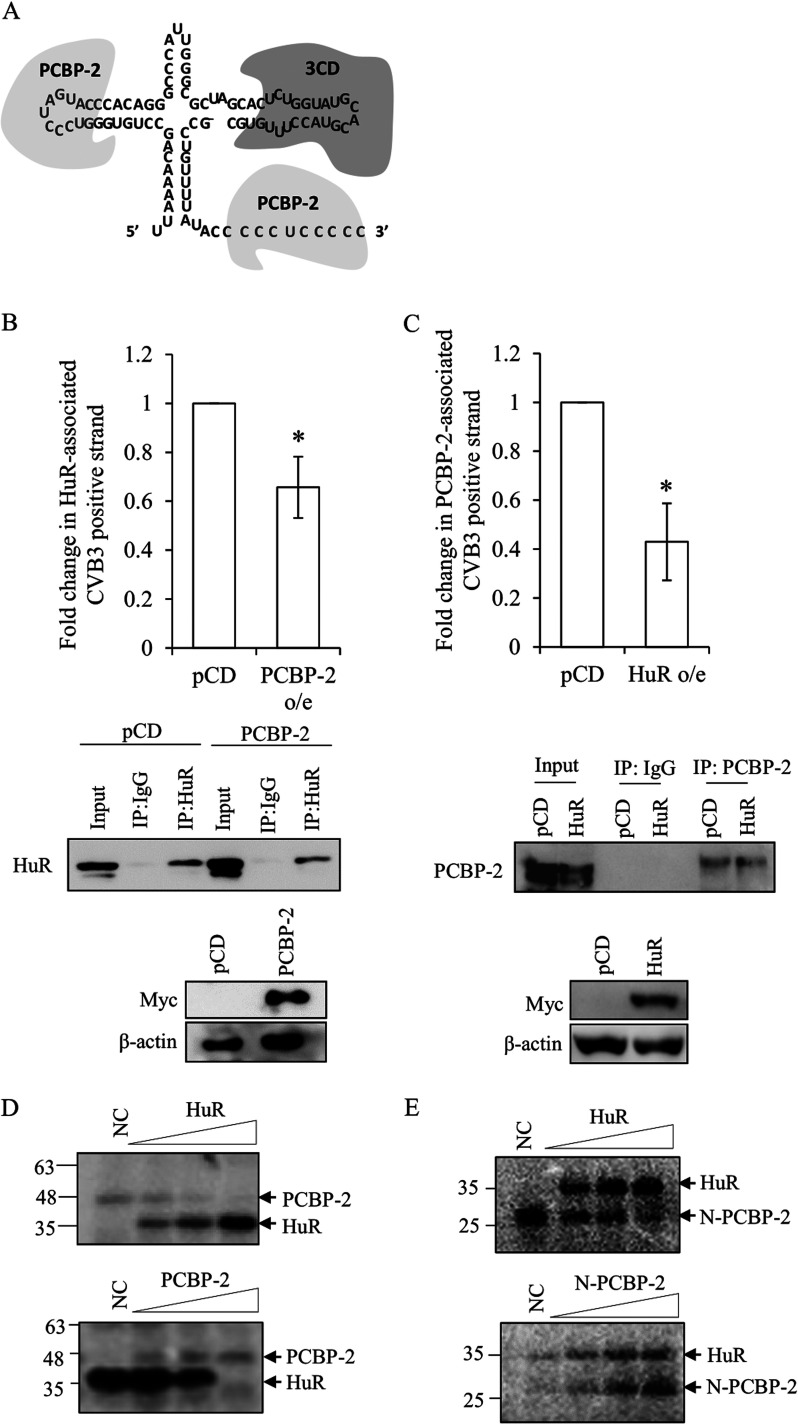
As the cloverleaf structure within the stem-loop I of the 5′ UTR RNA forms a ribonucleoprotein complex consisting of the cellular and viral proteins involved in viral RNA replication (Fig. 5A), we investigated the interaction of the host cellular protein PCBP-2 and HuR at the cloverleaf RNA.
FIG 5.
Interplay of viral and host proteins at cloverleaf region of CVB3. (A) Schematic of viral (3CD) and host protein (PCBP-2) binding at the cloverleaf structure. (B) RNA immunoprecipitation experiment was carried out with anti-HuR antibody upon overexpression of PCBP-2 or vector control (pcDNA3). HeLa cells were transfected with a plasmid expressing Myc-PCBP-2 or pcDNA3 vector followed by CVB3 replicon RNA transfection. At 10 h post-CVB3 replicon RNA transfection, the cells were processed for RNA immunoprecipitation with the anti-HuR antibody. Fold change in CVB3 positive-strand RNA associated with HuR upon PCBP-2 overexpression compared to that with the pcDNA3 vector is indicated. Anti-Myc antibody was used to detect overexpression of PCBP-2 protein. (C) RNA immunoprecipitation experiment was carried out with anti-PCBP-2 antibody upon overexpression of Myc-tagged HuR or vector control (pcDNA3). HeLa cells were transfected with a plasmid expressing Myc-HuR or pcDNA3 vector followed by CVB3 replicon RNA transfection. At 10 h post-CVB3 replicon RNA transfection, cells were processed for RNA immunoprecipitation with anti-PCBP-2 antibody. Fold change in CVB3 positive-strand RNA associated with PCBP-2 upon Myc-tagged HuR overexpression compared to that with pcDNA3 vector is indicated. Anti-Myc antibody was used to detect overexpression of HuR protein. Error bar indicates standard deviation from three independent experiments. *, P < 0.05. Two-tailed Student’s t test was used for statistical analysis. (D) In vitro UV cross-linking competition between HuR and full-length PCBP-2 was carried out using radiolabeled cloverleaf (1-100) RNA with increasing concentrations of either recombinant HuR or full-length PCBP-2 protein. (E) In vitro UV cross-linking competition between HuR and N-terminal PCBP-2 was carried out using radiolabeled cloverleaf (1-100) RNA with increasing concentration of HuR or N-terminal PCBP-2 protein.
To find out whether the interplay between HuR and PCBP-2 with the viral RNA occurs within cells, both proteins were overexpressed individually in HeLa cells followed by transfection with the CVB3 replicon RNA. The cell lysate was prepared 10 h posttransfection and immunoprecipitated with anti-HuR (for cells with PCBP-2 overexpression [o/e]) or anti-PCBP-2 (for cells with HuR o/e) antibody to estimate the association of HuR or PCBP-2 with CVB3 RNA (Fig. 5B and C). The results indicated that the association of PCBP-2 with the CVB3 RNA decreases significantly upon overexpression of HuR, and similarly, the association of HuR with the cloverleaf RNA reduces significantly upon overexpression of the PCBP-2 protein. This suggests that HuR and PCBP-2 compete to bind to the CVB3 genomic strand RNA.
To further understand the competition between these two proteins at the cloverleaf region, in vitro competition UV cross-linking was performed using the cloverleaf RNA as a probe. The results indicated that both HuR and full-length PCBP-2 can displace each other at the cloverleaf RNA (Fig. 5D). We further studied the in vitro competition between the N-terminal PCBP-2 and HuR for binding to the cloverleaf RNA. The results indicated that HuR can out compete the N-terminal PCBP-2 at the cloverleaf region (Fig. 5E). However, upon increasing the N-terminal PCBP-2, an increased association of HuR with the cloverleaf RNA was observed.
DISCUSSION
In the enterovirus life cycle, the genomic positive-strand RNA acts as a template to synthesize proteins as well as complementary negative-strand RNA. Both processes are mutually exclusive, and the mRNP complex assembled on the UTRs of the viral genomic RNA determines whether it would undergo translation to produce viral proteins or be used as a template to synthesize negative-strand RNAs. For a successful viral life cycle, it is critical to have a balanced rate of replication and translation. Perturbation in any one of the processes could directly or indirectly affect the other. Certain host factors, such as PCBP-2, bind to the viral RNA and are involved in both translation and RNA replication. The binding of full-length PCBP-2 at the IRES stimulates translation (3), whereas binding to the cloverleaf region enhances the synthesis of negative-strand RNA (5). Similarly, it has been shown previously that hnRNP C1/C2 also promotes negative-strand synthesis for CVB3 by inhibiting translation and reducing the ribosome flux on the positive-strand RNA (8).
The virus also has strategies to optimize the replication and translation rate in a stage-specific manner. For example, during the mid-phase of the replication cycle, the virus-encoded 3C protease cleaves PCBP-2; the cleaved PCBP-2 can no longer bind to IRES; thereby, viral translation is inhibited, making the genomic template available for replication (3, 18).
In this study, we have shown that HuR is another host factor that guides the template toward the replication cycle or translation cycle. The reduction in the virus titer could be due to a decrease in viral protein levels upon partial silencing of HuR. However, overexpression of HuR was found to increase virus titer. This could be due to the role of HuR in other viral processes such as encapsidation, which needs to be explored. CVB3 also induces the level of the HuR protein and relocates it to the cytoplasm. Our results suggest that interaction of HuR with the cloverleaf region of the CVB3 genomic RNA leads to the inhibition of replication and consequently makes the positive-strand RNA available for translation. HuR displaces PCBP-2 at the cloverleaf RNA, disrupting the ternary complex and hence inhibiting replication (Fig. 6A). However, cleaved PCBP-2 was found to enhance the association of HuR at the cloverleaf in in vitro experiments. It is possible that the binding sites of HuR and cleaved PCBP-2 at the cloverleaf are distinct; moreover, because of its smaller size, cleaved PCBP-2 may not hinder the binding site of HuR. Additionally, binding of cleaved PCBP-2 may bring the conformational changes that facilitate HuR binding. The importance of this in vitro observation needs to be further studied.
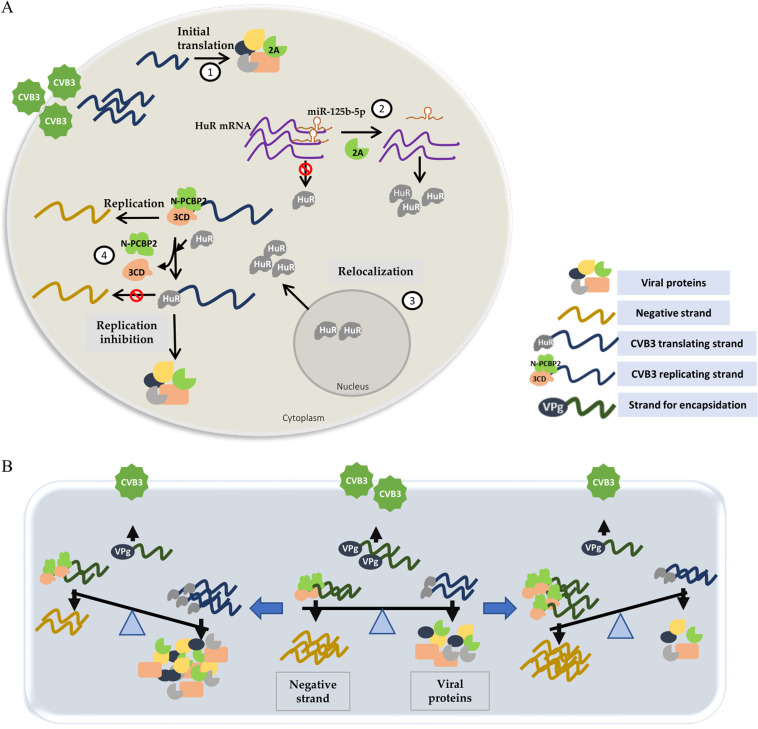
FIG 6.
(A) Model for HuR-mediated inhibition of CVB3 replication. (1) After entry into the cell cytoplasm, the virus life cycle begins by IRES-mediated translation of positive-strand RNA, producing structural and nonstructural proteins. (2) Virus-encoded 2A protease leads to a reduction in the levels of miR-125b-5p (it targets HuR and inhibits its translation), causing increased levels of HuR protein. (3) HuR protein, which is predominantly nuclear, relocates to the cytoplasm. (4) Increased cytoplasmic HuR displaces N-PCBP-2 from cloverleaf RNA and inhibits virus replication. This leads to an increased genomic RNA template availability for translation. (B) A balance between various processes is required for an efficient virus life cycle: positive-strand RNA viruses use their genomic templates for three different processes, i.e., translation, replication and encapsidation. The virus uses several mechanisms that aid in maintaining a balance of template usage at different stages of its life cycle (middle). Disproportionate competition between host factors (either by altered affinity or abundance) perturbs this balance and shifts it toward either translation or replication (left and right).
Our results strongly suggest that HuR does not directly influence translation and does not directly interact with IRES, providing evidence regarding the competition between replication factors and translation factors for genomic RNAs (Fig. 6B). Thus, our study reveals a fundamental aspect of positive-strand RNA genome-containing viruses where the same template is used for both replication and translation.
Furthermore, we also studied the regulation of HuR by the virus during infection. We have demonstrated that the level of HuR protein is induced by virus-encoded 2A protease through the reduction in the level of miR-125b-5p, which targets HuR mRNA. Additionally, HuR is relocated to the cytoplasm during the CVB3 life cycle; this is essential for the balance between viral replication and translation.
Phosphorylation at the S242 and S221 site of HuR is known to be involved in its cytoplasmic relocation under pathological conditions. However, we found that S242 mutation did not affect relocation, while the S221 mutant showed only partial relocation upon CVB3 infection (data not shown). It is possible that the HuR relocation could be influenced by additional sites. Also, the effect of S221A mutation on CVB3 replication might be due to its altered interaction with CVB3 RNA, as the 221 amino acid position is very close to the RNA-binding domain of HuR, which could be further investigated in future.
Previously, we reported the role of HuR in hepatitis C virus, where HuR is essential for virus replication. Here, we have demonstrated the opposite role of HuR during CVB3 infection. Thus, our study also reveals how the same cellular protein can be involved in different virus infections and exhibit different functions. HuR can be involved in the virus life cycle by either enhancing viral RNA stability, promoting virus replication or translation, inhibiting virus replication, or regulating the balance between availability of templates between translation and replication, as shown in the present study. This indicates the diversity in the function of the same host protein in different virus infections (10–12).
MATERIALS AND METHODS
Plasmids and RNA.
CVB3 replicon RNA was prepared from pRibCB3-T7-luc vector (a kind gift from Frank van Kuppeveld) and used for viral replication/translation studies. In CVB3 replicon RNA, the structural genes are replaced by the firefly luciferase gene. CVB3 1-100 nt and Δ100 5′ UTR, cloned in the pcDNA3 vector, were linearized with XbaI and used for in vitro transcription to prepare the cloverleaf RNA and Δ100 CVB3 5′ UTR RNA, respectively, as described earlier (9). For UV cross-linking experiments, [α-32P]UTP (obtained from the Board of Radiation and Isotope Technology, India) body-labeled RNA probes were prepared using T7 RNA polymerase (Promega). To prepare nonspecific RNAs used in the competition UV cross-linking assays, the pGEMt-easy vector was linearized with SalI enzyme and used for in vitro transcription by T7 RNA polymerase. The pET28a-HuR plasmid, as described earlier (10), was used to express and purify recombinant HuR protein. To purify recombinant PCBP-2, pET28a-PCBP-2 plasmid was obtained by subcloning the PCBP-2 gene from the pQE30-PCBP-2 construct. pQE30-PCBP-2 was a kind gift from Bert Semler. To purify the N-terminal PCBP-2, a stop codon was generated after amino acid 253 in the pET28a-PCBP-2 plasmid. The resulting plasmid pET28a-PCBP-2-ΔKH3 encodes amino acids 1 to 253 of PCBP-2 (N-PCBP-2) and produces a protein of ∼26 kDa. The pcDNA3-Myc-HuR, as described earlier (10), was used to overexpress Myc-tagged HuR, and pcDNA3-Myc-PCBP-2 construct (a kind gift from Peng Jin) was used to overexpress the Myc-tagged PCBP-2 protein in HeLa cells.
Virus preparation and infection.
The pRiB-T7-CB3 plasmid containing CVB3 cDNA (a kind gift from Nora Chapman) was used to prepare the CVB3 infectious RNA. The infectious RNA was transfected to HeLa cells, and the virus was purified from the cell culture supernatant. The virus titer was calculated by performing a plaque assay in Vero cells, and plaque-forming units per milliliter were estimated. For experiments, HeLa cells were transfected with siHuR or siNSP, and 24 h later, the cells were infected with CVB3 at a multiplicity of infection (MOI) of 5. The cell supernatant was harvested 8 h postinfection (h p.i.) and used to infect fresh cells, and the number of virus particles was estimated by plaque assay.
Cell lines, transfections, and luciferase assay.
HeLa cells, maintained in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) with 10% fetal bovine serum (Gibco, Invitrogen), were used for all experiments. For viral translation and replication studies, the CVB3 replicon RNA was transfected in Opti-MEM (Invitrogen, Life Technologies) using Lipofectamine 2000 (Invitrogen, Life Technologies). Four hours posttransfection, the medium was replaced by DMEM containing 10% FBS. The cells were harvested at 10 h posttransfection and processed for luciferase assay using a luciferase reporter assay system (Promega) or processed in TRI reagent (Sigma) for total RNA isolation according to the manufacturer’s protocol. For partial silencing experiments, the cells were transfected using Lipofectamine 2000 transfection reagent in Opti-MEM (Invitrogen). The above-indicated amount of siHuR (Dharmacon) or nonspecific siRNA (Dharmacon) or lipofectamine alone was transfected 24 h prior to CVB3 RNA transfection. The siRNA sequences used against HuR are as follows: 5′-GGUGAAGUUGAAUCUGCAAAACUTA-3′ and 5′-UAAGUUUUGCAGAUUCAACUUCACCAA-3′.
UV-induced cross-linking of proteins with RNA and immunoprecipitation assays.
α-32P body-labeled RNA probes were mixed with purified recombinant protein for 30 min at 30°C in 1× RNA binding buffer (20 mM HEPES [pH 7.6], 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.2 mM EDTA, and 2 mM dithiothreitol [DTT]) and then UV irradiated (254 nm) for 20 min at 4°C. The mixture was treated with RNase A at 37°C to digest the unprotected RNAs. The complexes were separated on 10% SDS-PAGE gels and visualized by phosphorimaging. For the competitive UV cross-linking assay, a molar excess of unlabeled RNAs, where indicated, was added along with the labeled RNA probe.
For RNA immunoprecipitation (RIP), HeLa cells were transfected with CVB3 replicon RNA or mock transfected and harvested 10 h posttransfection in 1× polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES [pH 7.0], 0.05% NP-40, 1 mM DTT, and 100 U/ml RNasin). The cells were centrifuged, and the protein in the supernatant was quantified. The proteins were precleared by mixing with protein G-Sepharose beads for 1 h at 4°C. The samples were centrifuged for 2 min at 1,500 × g, and the supernatant was discarded. Separately, the protein G-Sepharose beads were incubated with 1 μg of anti-HuR antibody (Santa Cruz Biotechnology) or anti-IgG antibody (Calbiochem) overnight at 4°C in 1× polysome lysis buffer. Equal amounts of precleared lysate from mock or CVB3 RNA-transfected cells were incubated with protein G beads bound to HuR antibody, protein G beads bound to IgG antibody, or only protein G beads for 4 h at 4°C. The bead-bound RNA-protein complex was centrifuged and resuspended in 1× polysome lysis buffer, and an aliquot (20%) was separated for Western blotting. To the remaining lysate, 0.1% SDS and 30 μg proteinase K were added and incubated at 55°C for 30 min. To this, 3 volumes of TRI reagent were added, and the RNA was isolated followed by qRT-PCR using CVB3-specific primers.
Protein purification.
The recombinant HuR, PCBP-2, and N-PCBP-2 proteins were purified from pET28a-HuR, pET28a-PCBP-2, and pET28a-PCBP2-ΔKH3 constructs, respectively. The expression of recombinant PCBP-2 and N-PCBP-2 was induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) at an optical density of 0.6 at 660 nm and grown for another 16 h at 25°C. The cells were pelleted, resuspended in lysis buffer (50 mM Tris, [pH 8.0], 300 mM NaCl, and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]), and disrupted on ice by sonication. All subsequent steps were carried out at 4°C. The lysates were cleared by centrifugation at 10,000 rpm for 20 min and incubated with a Ni-nitrilotriacetic acid (Ni-NTA)-agarose slurry (Qiagen) with rocking for 4 h at 4°C. The lysate was loaded onto a column, and the flowthrough was discarded. The column was washed with 50 ml of wash buffer (lysis buffer containing 40 mM imidazole). The bound protein was eluted with 600 μl of elution buffer containing 300 mM imidazole. The eluted protein fractions were dialyzed at 4°C for 3 to 5 h in dialysis buffer (50 mM Tris [pH 8.0], 100 mM KCl, 5 mM β-mercaptoethanol [β-ME], and 10% glycerol), aliquoted, and stored at −70°C. The method followed for the purification of recombinant HuR was as described earlier (10).
Western blot analysis.
Proteins from the cell lysate supernatant were quantified using Bradford reagent (Bio-Rad), and equal amounts of protein were separated on 12% denaturing PAGE gels. The separated proteins were transferred to a nitrocellulose membrane (Pall Biosciences) and analyzed using antibodies as described in Results. Mouse monoclonal anti-HuR antibody (Calnexin) and mouse monoclonal anti-hnRNPE2/PCBP-2 (Santa Cruz Biotechnology) were used to analyze the samples followed by horseradish peroxidase (HRP)-conjugated secondary antibody (goat raised anti-rabbit and goat raised anti-mouse IgGs; Sigma). Monoclonal anti-β-actin antibody (HRP conjugated, obtained from Sigma) was used to detect β-actin. Mouse monoclonal anti-β-tubulin (Sigma) and mouse monoclonal anti-GAPDH (Imgenex) were used to detect tubulin and GAPDH levels, respectively. The protein antibody complexes were visualized by chemiluminescence using Immobilon Western assay solution (EMD Millipore). Cytoplasmic and nuclear lysates were prepared from the cells using an NXTRACT kit (Sigma) according to the manufacturer’s protocol.
In vitro transcription.
RNAs corresponding to 1-00 nt and Δ100 CVB3 5′ UTR were prepared using linearized plasmid constructs under T7 promoters in runoff transcription reactions. Linear DNA templates were extracted from the gel and purified using a gel extraction kit (Qiagen). The linear DNA templates were used as the template for the synthesis of 32P-labeled RNA or unlabeled RNA using T7 RNA polymerase (Fermentas) and [32P]UTP (PerkinElmer Life Sciences). In vitro transcription was performed under standard conditions using 2 μg of linear template DNA (Fermentas protocol). The precipitated RNA was resuspended in 25 μl of nuclease-free water. The incorporated radioactivity was measured using a liquid scintillation counter.
RNA isolation, qPCR, and semiquantitative reverse transcription-PCR.
Total RNA was isolated from HeLa cells using TRI reagent (Sigma). CVB3 positive strand, negative strand, and HuR levels were quantified by qRT-PCR using a DyNAmo HS SYBR green qPCR kit (Finnzymes). cDNA was synthesized using a Moloney murine leukemia virus (M-MuLV) reverse transcriptase at 42°C for 1 h (Promega) using 1 μg of total RNA by adding primers targeting CVB3 positive- or negative-strand RNA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA in the same reaction mixture. To determine the HuR levels, HuR-specific primers were used. For each reaction, 2 μl of cDNA was added. The following primer sequences were used in the present study: CVB3-F, 5′-GAATGCGGCTAATCCTAACTGC-3′; CVB3-R, 5′-GCTCTATTAGTCACCGG ATGGC-3′; (HuR mRNA) HuR-F, 5′-CCCAGGATGAGTTACGAAGCC-3′; HuR-R, 5′-GC CTCAAGCCGTTCAGCGTGT-3′; GAPDH F, 5′-CAGCCTCAAGATCATCAGCAAT-3′; and GAPDH R, 5′-GGTCATGAGTCCTTCCACGA-3′. The miR-125b-5p primer sequence is as described earlier (15). To calculate the fold change, the comparative threshold cycle (CT) method was used (2−ΔΔCT), and the values were normalized to GAPDH values.
Indirect immunofluorescence.
For immunofluorescence staining, ∼0.1 × 106 HeLa ATCC cells were seeded on a coverslip in a 24-well plate for 16 h followed by CVB3 infection. After the designated time points, cells were processed for confocal microscopy. The cells were washed twice with phosphate-buffered saline (PBS) and fixed using 4% paraformaldehyde at room temperature for 30 min. Then, they were permeabilized with 0.1% Triton X-100 for 3 min at room temperature and incubated with 3% bovine serum albumin (BSA) at 37°C for 45 min. The cells were incubated with primary antibodies as indicted above for 1 h and 45 min at 4°C and then incubated with Alexa 633-conjugated anti-mouse or Alexa 488-conjugated anti-rabbit secondary antibody (Invitrogen) for 30 min. Images were taken using a Zeiss microscope, and image analysis was performed using the ZEN software tool.
MTT assay.
To study the effect of silencing and overexpression of HuR on cell viability and metabolic activity, a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay was carried out as described previously (19).
Polysome profiling.
HeLa cells were seeded in 100-mm dishes and transfected with siNSP or siHuR. After 32 h posttransfection, the cells were incubated with cycloheximide (100 μg/ml) for 10 min at 37°C. Cells were then washed once with ice-cold PBS containing cycloheximide and then once with hypotonic buffer (5 mM Tris-HCl [pH 7.5], 1.5 mM KCl, 5 mM MgCl2, and 100 μg/ml cycloheximide). Next, cells were scraped in ice-cold lysis buffer (5 mM Tris-HCl [pH 7.5], 1.5 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide, 1 mM DTT, 200 U/ml RNasin, 200 μg/ml tRNA, 0.5% Triton X-100, 0.5% sodium deoxycholate, and 1× protease inhibitor cocktail) and kept on ice for 15 min. After 15 min, the KCl concentration in the lysate was adjusted to a final concentration of 150 mM. Cells were then centrifuged at 3,000 × g for 8 min at 4°C, and the supernatant was collected and processed immediately or flash frozen and stored at −80°C for later use. Then, 1.5 μg of protein was loaded onto a 17% to 51% sucrose gradient and centrifuged at 36,000 rpm for 2 h at 4°C in an SW41 rotor (Beckman).
ACKNOWLEDGMENTS
We thank Frank van Kuppeveld and Nora Chapman for various constructs. We thank the Centre for Infectious Disease Research, Indian Institute of Science (IISc), for allowing use of the biosafety level 3 (BSL3) facility and the Department of Microbiology and Cell Biology, IISc, for use of the confocal facility. We also thank Deepak Saini, Department of Molecular Reproduction, Development and Genetics, IISc, for providing the HeLa cell line, Umesh Varshney, Department of Microbiology and Cell Biology, for allowing the usage of laboratory facilities for polysome analysis, and Harsha Raheja, our lab member, for useful suggestions.
Funding was provided by a D.S. Kothari fellowship from University Grants Commission, Government of India (to B.G. and P.B.). P.D. is supported by a research fellowship from the Council of Scientific and Industrial Research (CSIR). P.R. is supported by the Ministry of Human Resource Development (MHRD). Support to the department from the DST Fund for Improvement of Science and Technology Infrastructure (FIST) level II infrastructure and University Grants Commission Centre of Advanced Studies is acknowledged. Financial help from DBT-IISc Partnership Program is also acknowledged. S.D. acknowledges the J C Bose fellowship from Department of Science and Technology, Government of India. The work was also supported by a research grant from Department of Biotechnology, Government of India.
Contributor Information
Saumitra Das, Email: sdas@iisc.ac.in.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5'-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 2.Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. 2007. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J Virol 81:8919–8932. 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chase AJ, Daijogo S, Semler BL. 2014. Inhibition of poliovirus-induced cleavage of cellular protein PCBP2 reduces the levels of viral RNA replication. J Virol 88:3192–3201. 10.1128/JVI.02503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leveque N, Garcia M, Bouin A, Nguyen JHC, Tran GP, Andreoletti L, Semler BL. 2017. Functional consequences of RNA 5'-terminal deletions on coxsackievirus B3 RNA replication and ribonucleoprotein complex formation. J Virol 91:e00423-17. 10.1128/JVI.00423-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma N, Ogram SA, Morasco BJ, Spear A, Chapman NM, Flanegan JB. 2009. Functional role of the 5' terminal cloverleaf in coxsackievirus RNA replication. Virology 393:238–249. 10.1016/j.virol.2009.07.039. [DOI] [PubMed] [Google Scholar]
- 6.Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12:2293–2304. 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zell R, Ihle Y, Seitz S, Gundel U, Wutzler P, Gorlach M. 2008. Poly(rC)-binding protein 2 interacts with the oligo(rC) tract of coxsackievirus B3. Biochem Biophys Res Commun 366:917–921. 10.1016/j.bbrc.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 8.Dave P, George B, Balakrishnan S, Sharma DK, Raheja H, Dixit NM, Das S. 2019. Strand-specific affinity of host factor hnRNP C1/C2 guides positive to negative-strand ratio in coxsackievirus B3 infection. RNA Biol 16:1286–1299. 10.1080/15476286.2019.1629208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dave P, George B, Sharma DK, Das S. 2017. Polypyrimidine tract-binding protein (PTB) and PTB-associated splicing factor in CVB3 infection: an ITAF for an ITAF. Nucleic Acids Res 45:9068–9084. 10.1093/nar/gkx519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shwetha S, Kumar A, Mullick R, Vasudevan D, Mukherjee N, Das S. 2015. HuR displaces polypyrimidine tract binding protein to facilitate la binding to the 3' untranslated region and enhances hepatitis C virus replication. J Virol 89:11356–11371. 10.1128/JVI.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. 2010. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe 8:196–207. 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonenfant G, Williams N, Netzband R, Schwarz MC, Evans MJ, Pager CT. 2019. Zika virus subverts stress granules to promote and restrict viral gene expression. J Virol 93:e00520-19. 10.1128/JVI.00520-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickson AM, Anderson JR, Barnhart MD, Sokoloski KJ, Oko L, Opyrchal M, Galanis E, Wilusz CJ, Morrison TE, Wilusz J. 2012. Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J Biol Chem 287:36229–36238. 10.1074/jbc.M112.371203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S, Wang Y, Lin L, Si X, Wang T, Zhong X, Tong L, Luan Y, Chen Y, Li X, Zhang F, Zhao W, Zhong Z. 2014. Protease 2A induces stress granule formation during coxsackievirus B3 and enterovirus 71 infections. Virol J 11:192. 10.1186/s12985-014-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shwetha S, Sharma G, Raheja H, Goel A, Aggarwal R, Das S. 2018. Interaction of miR-125b-5p with human antigen R mRNA: mechanism of controlling HCV replication. Virus Res 258:1–8. 10.1016/j.virusres.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Tong L, Lin L, Wu S, Guo Z, Wang T, Qin Y, Wang R, Zhong X, Wu X, Wang Y, Luan T, Wang Q, Li Y, Chen X, Zhang F, Zhao W, Zhong Z. 2013. MiR-10a* upregulates coxsackievirus B3 biosynthesis by targeting the 3D-coding sequence. Nucleic Acids Res 41:3760–3771. 10.1093/nar/gkt058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grammatikakis I, Abdelmohsen K, Gorospe M. 2017. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA 8:e1372. 10.1002/wrna.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sean P, Nguyen JH, Semler BL. 2009. Altered interactions between stem-loop IV within the 5' noncoding region of coxsackievirus RNA and poly(rC) binding protein 2: effects on IRES-mediated translation and viral infectivity. Virology 389:45–58. 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciofani G, Danti S, D'Alessandro D, Moscato S, Menciassi A. 2010. Assessing cytotoxicity of boron nitride nanotubes: interference with the MTT assay. Biochem Biophys Res Commun 394:405–411. 10.1016/j.bbrc.2010.03.035. [DOI] [PubMed] [Google Scholar]