SUMMARY
Frankliniella occidentalis (western flower thrips [WFT]) and Thrips tabaci (onion thrips [OT]) are insect species that greatly impact horticultural crops through direct damage and their efficient vectoring of tomato spotted wilt virus and iris yellow spot virus. In this study, we collected thrips of these species from 12 field populations in various regions in Italy. We also included one field population of Neohydatothrips variabilis (soybean thrips [ST]) from the United States. Total RNA data from high-throughput sequencing (HTS) were used to assemble the virome, and then we assigned putative viral contigs to each thrips sample by real-time reverse transcription-quantitative PCR (qRT-PCR). Excluding plant and fungal viruses, we were able to identify 61 viral segments, corresponding to 41 viruses: 14 were assigned to WFT, 17 to OT, and 1 to ST; 9 viruses could not be assigned to any species based on our stringent criteria. All these viruses are putative representative of new species (with only the exception of a sobemo-like virus that is 100% identical to a virus recently characterized in ST) and some belong to new higher-ranking taxa. These additions to the viral phylogeny suggest previously undescribed evolutionary niches. Most of Baltimore’s classes of RNA viruses were present (positive- and minus-strand and double-stranded RNA viruses), but only one DNA virus was identified in our collection. Repeated sampling in a subset of locations in 2019 and 2020 and further virus characterization in a subset of four thrips populations maintained in the laboratory allowed us to provide evidence of a locally persistent thrips core virome that characterizes each population.
IMPORTANCE Harnessing the insect microbiome can result in new approaches to contain their populations or the damage they cause vectoring viruses of medical, veterinary, or agricultural importance. Persistent insect viruses are a neglected component of their microbiota. In this study, for the first time, we characterize the virome associated with the two model systems for tospovirus-transmitting thrips species, of utmost importance for the direct and indirect damage they cause to a number of different crops. The thrips virome characterized includes several novel viruses, which in some cases reveal previously undescribed clades. More importantly, some of the viruses we describe are part of a core virome that is specific and consistently present in distinct geographical locations monitored over the years, hinting at a possible mutualistic symbiotic relationship with their host.
KEYWORDS: entomovirus, thrips, virome
INTRODUCTION
Thrips are taxonomically included in the order Thysanoptera. The order includes ca. 7,000 described species organized in two suborders: Tubulifera and Terebrantia (1). Thrips include species with different feeding behaviors: half of the described species feed on fungi, whereas the rest is mostly characterized by phytophagous behavior and feed on leaves, flowers and fruits; a few species are predators of other small arthropods (2). Most of the plant pest species are in the family Thripidae, which includes ca. 1,700 species. Phytophagous thrips can cause direct and indirect damage to the plants they feed on: indirect damage is greatly enhanced by their ability to transmit tospoviruses (genus Orthotospovirus, family Bunyaviridae). Currently, 14 species of thrips have been shown to transmit tospoviruses (3, 4). Tospoviridae are likely insect-infecting viruses that adapted to a plant host (5). The origin of the association between thrips and tospoviruses is not clear (6).
The two most important thrips species in the Mediterranean basin are Frankliniella occidentalis (Pergande) (western flower thrips [WFT]) and Thrips tabaci (Lindeman) (onion thrips [OT]) (7). WFT is native to the western United States and was first reported in the Mediterranean Basin in the late 1980s, but it is now present worldwide and is considered the most efficient vector of tomato spotted wilt virus (TSWV) and impatiens necrotic spot virus (INSV) in the Mediterranean Basin (7, 8). OT is instead native to the eastern Mediterranean, but it is now spread worldwide; in the Mediterranean basin, OT is now an efficient vector of iris yellow spot virus (IYSV). TSWV, INSV, and IYSV are endemically present in the Mediterranean Basin and Europe, and TSWV causes major loss in a number of horticultural crops, particularly because of resistance breaking strains in pepper and tomato (9). This necessitates new approaches in integrated pest management (IPM): in this regard, specific attention has been paid to the possibility of altering the microbiome associated with insects as a new tool for crop pest management (10).
For insect vectors of plant pathogens, an approach that does not put selective pressure on the insect population, but only on its vectoring competence, can be of great interest: a recent successful case study is that of the artificial introduction of a new Wolbachia strain to the brown planthopper Nilaparvata lugens (Stål), which resulted in the reduction of rice ragged stunt virus transmission (11). Other approaches can be envisioned based on the exploitation of the microbiome: specific virus-virus interactions could also be at the base of interference with vectoring ability, and synergistic or antagonistic relationships between resident/persistent insect viruses and the plant viruses they vector could indeed provide a new approach to contain tospovirus spread. In fact, viruses as elements of insect microbiomes are only minimally studied (12). After recent groundbreaking work showing the vast diversity of viruses present in insects and invertebrates (13–15), the importance of the insect microbiomes for virus evolution and biology is becoming apparent, but they are greatly understudied in their biological effects on the host. In this respect, up until very recently, viromes associated with thrips among the most important vectors of plant diseases have also been neglected: a list of viruses associated with Neohydatothrips variabilis (Beach) (soybean thrips [ST]), vector of soybean vein necrosis virus, has been only recently provided (16).
The purpose of this study was to characterize for the first time the virome associated with the two most efficient vectors of tospoviruses, WFT and OT, in the Mediterranean area, with an emphasis on their variability according to species, geographical location, and recurrence in different sampling efforts carried out between 2018 and 2020.
RESULTS
Taxonomic analysis of the meta-transcriptomes.
For each library, more than 100 million 150-bp paired-end reads were retrieved.
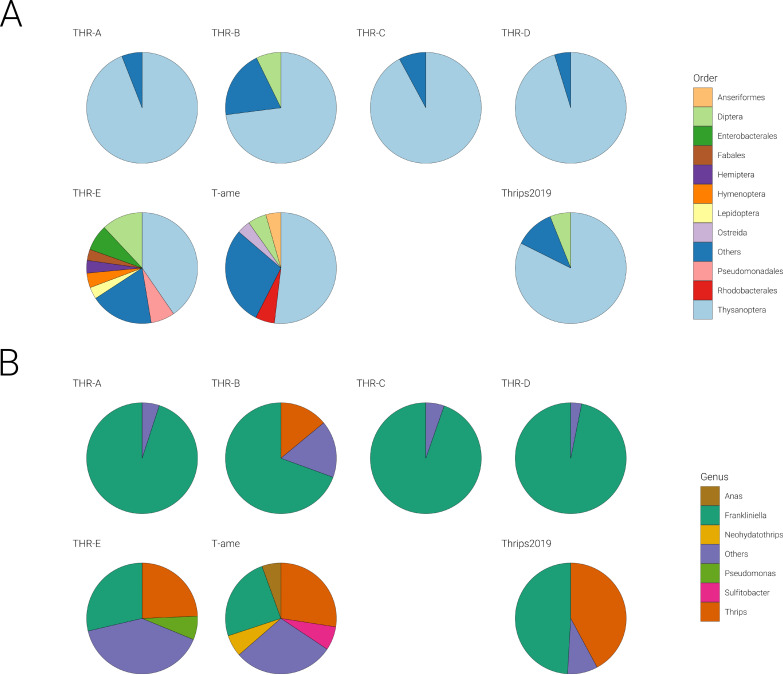
In order to check the taxonomic complexity of the samples, we performed a metatranscriptomic analysis using Kraken2. Results, processed with Pavian, show the sample composition of each library at different taxonomic levels: domain (data not shown), order (Fig. 1A), phylum (data not shown), and genus (Fig. 1B). At higher taxonomic ranks, all libraries (THR-A, THR-B, THR-C, THR-D, THR-E, T-ame, and Thrips2019) showed a clear prevalence (>90%) of Eukaryota. The lower taxonomic rank genus showed that in three libraries (THR-A, THR-C, and THR-D), the majority of sequences belong to Frankliniella, as expected. In the remaining libraries (THR-B, THR-E, T-ame, and Thrips2019), there is a mix of Frankliniella and Thrips genera, even if assignment of reads belonging to T. tabaci to the genus Thrips might be underestimated since only the Thrips palmi (Karny) genome is available in databases, and therefore, assignment of contigs/reads to F. occidentalis could be due to conservation of some sequences between the two genera and lack of a direct hit for T. tabaci. Moreover, the library THR-E contains many sequences that could not be assigned to thrips but are instead assigned to other genera, mostly of insects but also bacteria. In T-ame the genus Neohydatothrips, as expected, is also present. Referring to the order rank (Fig. 1A), two libraries (THR-E and T-ame) out of seven are more heterogeneous, including some other insect orders, among which are dipteran and hymenopteran (likely from some parasitoid insects).
FIG 1.
Pie charts represent the distribution of selected taxonomic groups across all pools, reporting the number of reads for each genus with more than 100,000 reads. (A) Number of reads for each order; (B) number of reads for each genus.
Viruses associated with thrips metatranscriptomic samples.
We discuss here all the viruses discovered over the years as true thrips viruses (confirmed in multiple instances in individual thrips checked for their taxonomic assignment); to be more conservative, the other viruses with less certain host assignment (or not reconfirmed in multiple assays on distinct individual thrips) are deposited in GenBank as “insect metagenomic” viruses, although some of them could indeed be thrips viruses.
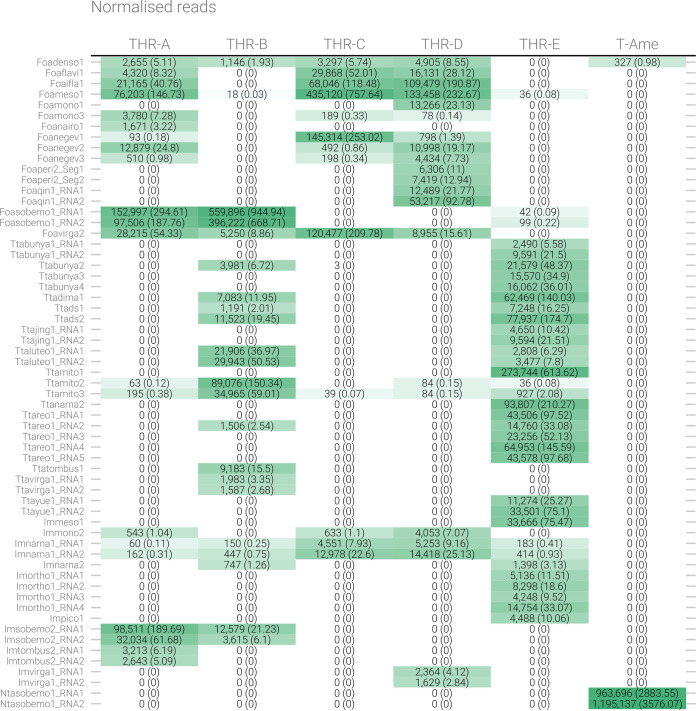
Overall, 95 viral contigs were identified (Tables 1 and 2), among which 13 were likely fungal viruses (based on their high identity percentages with confirmed mycoviruses present in databases), 17 were plant viruses (also based on their high identity percentages with confirmed viruses present in the databases), and 4 were likely endogenous viral elements in the host genome (positive PCR on DNA) (Table 1); 16 were uncertain insect host viral contigs (likely entomoviruses based on the first BLAST hit in databases), and 45 were confirmed thrips viral contigs corresponding to 32 viruses (some viral genomes are multisegmented) (Table 2). Within the 32 viruses with confirmed hosts, 14 infect WFT, 17 infect OT, and 1 infects the ST. Entomovirus contigs are reported in Fig. 2, where number of reads mapping to each contig and the normalized values are displayed (Fig. 2).
TABLE 1.
List of putative plant, fungal and endogenized viral contigs discovered applying our bioinformatic pipeline to RNA sequencing data of thrips samplesa
| Contig | Length (no. of bases) | Accession no. | NCBI BLASTx first hit | Identityb | Origin |
|---|---|---|---|---|---|
| T-Ame_DN12303 | 3,216 | QBX90567 | Cucumber mosaic virus | 100* | Plant virus |
| THR-B_DN28487 | 1,996 | AOX21989 | Iris yellow spot virus | 100* | Plant virus |
| THR-D_DN18945 | 7,762 | APG79622 | Tomato spotted wilt tospovirus | 99* | Plant virus |
| THR-E_DN20119 | 1,556 | AWK67805 | Iris yellow spot virus | 99* | Plant virus |
| THR-E_DN20826 | 1,091 | AWK67804 | Iris yellow spot virus | 99* | Plant virus |
| THR-E_DN23926 | 3,648 | ACJ04669 | Iris yellow spot virus | 99* | Plant virus |
| THR-B_DN30354 | 8,873 | YP_009241381 | Iris yellow spot virus | 99* | Plant virus |
| T-Ame_DN11632 | 3,025 | CCQ26876 | Cucumber mosaic virus | 99* | Plant virus |
| THR-A_DN12663 | 1,489 | YP_009551627 | Melon partitivirus | 64 | Plant virus |
| THR-A_DN9251 | 1,541 | YP_009551628 | Melon partitivirus | 41 | Plant virus |
| THR-E_Contig1 | 2,488 | QED45151 | Leek yellow stripe virus | 96* | Plant virus |
| THR-E_DN24091 | 10,357 | AGG18220 | Leek yellow stripe virus | 89 | Plant virus |
| THR-D_DN16018 | 2,950 | QPZ88447 | Tomato spotted wilt tospovirus | 95* | Plant virus |
| THR-C_DN21959 | 8,918 | AKM21265 | Tomato spotted wilt tospovirus | 100* | Plant virus |
| THR-A_DN26458 | 2,851 | ASU87377 | Bell pepper alphaendornavirus | 100* | Plant virus |
| THR-A_Contig1 | 9,812 | AJF48474 | Bell pepper alphaendornavirus | 100* | Plant virus |
| THR-C_DN28521 | 14,708 | AYR00620 | Bell pepper alphaendornavirus | 98* | Plant virus |
| T-Ame_DN13641 | 1,013 | NP_620728 | Ustilago maydis virus H1 | 70 | Fungal virus |
| T-Ame_DN4615 | 3,117 | NP_620728 | Ustilago maydis virus H1 | 48 | Fungal virus |
| THR-B_DN29920 | 1,119 | YP_009508064 | Heterobasidion partitivirus 8 | 35 | Fungal virus |
| THR-E_DN21437 | 3,110 | QDH89606 | Mitovirus sp. | 65 | Fungal virus |
| THR-E_DN23537 | 27,80 | QDB75006 | Acremonium sclerotigenum ourmia-like virus 1 | 60 | Fungal virus |
| THR-E_DN24038 | 2,138 | BBZ90081 | Red algae totivirus 1 | 30 | Fungal virus |
| THR-E_DN3125 | 1,465 | AWV67014 | Lysoka partiti-like virus | 47 | Fungal virus |
| THR-E_DN7093 | 1,673 | AWV67012 | Lysoka partiti-like virus | 65 | Fungal virus |
| THR-E_DN23526 | 2,468 | AKN79252 | Alternaria brassicicola mitovirus | 91* | Fungal virus |
| THR-E_DN14430 | 1,118 | YP_009182158 | Pleospora typhicola fusarivirus 1 | 86 | Fungal virus |
| THR-E_DN15320 | 2,534 | QGY72561 | Plasmopara viticola lesion associated ourmia-like virus 31 | 99* | Fungal virus |
| THR-E_DN23378 | 2,355 | QIR30262 | Plasmopara viticola lesion associated mitovirus 39 | 99* | Fungal virus |
| THR-B_DN23683 | 2,827 | QIR30272 | Plasmopara viticola lesion associated mitovirus 49 | 97* | Fungal virus |
| THR-B_DN27856 | 2,905 | QMP82309 | Hemipteran rhabdo-related virus OKIAV26 | 42 | Viral insertion |
| THR-D_DN18510 | 1,167 | QDZ71189 | Megastigmus ssRNA virus | 29 | Viral insertion |
| THR-E_DN22039 | 2,499 | QMP82403 | Hemipteran orthomyxo-related virus OKIAV188 | 55 | Viral insertion |
| THR-E_DN24098 | 10,345 | QQN90111 | Soybean thrips iflavirus 2 | 56 | Viral insertion |
Columns show contig identifier code, contig length (in bases), first viral hit accession number in NCBI:protein, first viral hit organism (NCBI BLASTx first hit), NCBI hit identity, and the origin of the viral contig.
Asterisks indicate all the blast hits with identity higher than 90.
TABLE 2.
List of insect-associated viral contigs discovered applying our bioinformatic pipeline to RNA sequencing data of thrips samplesa
| Contig | Length (no. of bases) | Accession no. | NCBI BLASTx first hitb | Identityc | GenBank accession no. | Suggested virus name | Paper ID | Origin |
|---|---|---|---|---|---|---|---|---|
| THR-D_DN15333 | 5,570 | ALV85426 | Diaphorina citri densovirus | 32 | MN764146 | Frankliniella occidentalis associated densovirus 1 | Foadenso1 | Thrips virus |
| THR-C_DN26815 | 19,679 | BAM99927 | Gentian Kobu-sho-associated virus | 37 | MN714664 | Frankliniella occidentalis associated flavi-like virus 1 | Foaflavi1 | Thrips virus |
| THR-A_DN28578 | 9,971 | QQN90111 | Soybean thrips iflavirus 2** | 65 | MN714670 | Frankliniella occidentalis associated iflavirus 1 | Foaifla1 | Thrips virus |
| THR-D_Contig1 | 19,113 | QPD01782 | Aphis citricidus meson-like virus | 33 | MN714663 | Frankliniella occidentalis associated mesonivirus 1 | Foameso1 | Thrips virus |
| THR-D_DN19783 | 14,297 | QLL27736 | Leveillula taurica associated rhabdo-like virus 1 | 30 | MN714688 | Frankliniella occidentalis associated mononegavirales virus 1 | Foamono1 | Thrips virus |
| THR-A_DN24521 | 12,116 | QPZ88392 | Soybean thrips rhabdo-like virus 3** | 40 | MN714690 | Frankliniella occidentalis associated mononegavirales virus 3 | Foamono3 | Thrips virus |
| THR-A_DN23655 | 4,933 | YP_009246486 | Yogue virus | 37 | MN764155 | Frankliniella occidentalis associated nairo-like virus 1 | Foanairo1 | Thrips virus |
| THR-C_DN25036 | 14,196 | YP_009342435 | Wuhan house centipede virus 1 | 25 | MN714666 | Frankliniella occidentalis associated negev-like virus 1 | Foanegev1 | Thrips virus |
| THR-A_DN26080 | 14,242 | AMO03221 | Buckhurst virus | 24 | MN714665 | Frankliniella occidentalis associated negev-like virus 2 | Foanegev2 | Thrips virus |
| THR-D_DN16267 | 8,294 | YP_009342435 | Wuhan house centipede virus 1 | 25 | MN714671 | Frankliniella occidentalis associated negev-like virus 3 | Foanegev3 | Thrips virus |
| THR-D_DN17549 | 9,618 | QQP18740 | Soybean thrips bunya-like virus 1** | 31 | MN764150 | Frankliniella occidentalis associated peribunyavirus-like virus 2 segm1 | Foaperi2_Seg1 | Thrips virus |
| THR-D_DN17549 | 4,566 | BBD75426 | Ixodes scapularis bunyavirus | 28 | MN764149 | Frankliniella occidentalis associated peribunyavirus-like virus 2 segm2 | Foaperi2_Seg2 | Thrips virus |
| THR-D_DN15448 | 5,780 | YP_009342465 | Wuhan insect virus 15 | 28 | MN764147 | Frankliniella occidentalis associated qin-like virus1 RNA1 | Foaqin1_RNA1 | Thrips virus |
| THR-D_DN15448 | 2,630 | YP_009342457 | Wuhan insect virus 15 | 27 | MN764148 | Frankliniella occidentalis associated qin-like virus1 RNA2 | Foaqin1_RNA2 | Thrips virus |
| THR-A_DN15734 | 2,856 | QHA33888 | Atrato Sobemo-like virus 2 | 51 | MN725049 | Frankliniella occidentalis associated sobemo-like virus 1 RNA1 | Foasobemo1_RNA1 | Thrips virus |
| THR-A_DN15734 | 1,604 | AWY11068 | Motts Mill virus | 38 | MN725050 | Frankliniella occidentalis associated sobemo-like virus 1 RNA2 | Foasobemo1_RNA2 | Thrips virus |
| THR-D_DN18360 | 12,138 | YP_009337819 | Hubei virga-like virus 12 | 38 | MN714667 | Frankliniella occidentalis associated virga-like virus 2 | Foavirga2 | Thrips virus |
| T-Ame_DN13171 | 3,410 | QPZ88398 | Soybean thrips sobemo-like virus 4** | 99* | MN714676 | Neohydatothrips associated sobemo-like virus 1 RNA1 | Ntasobemo1_RNA1 | Thrips virus |
| T-Ame_DN13171 | 1,555 | YP_009337912 | Hubei diptera virus 13 | 42 | MN714681 | Neohydatothrips associated sobemo-like virus 1 RNA2 | Ntasobemo1_RNA2 | Thrips virus |
| THR-E_DN17816 | 7,251 | QQM16265 | Degsystermes virus | 37 | MN764152 | Thrips tabaci associated bunyavirales 1 RNA1 | Ttabunya1_RNA1 | Thrips virus |
| THR-E_DN17816 | 1,222 | AOY18790 | Bunyavirus sp. | 26 | MN764153 | Thrips tabaci associated bunyavirales 1 RNA2 | Ttabunya1_RNA2 | Thrips virus |
| THR-E_DN23749 | 10,255 | APG79357 | Xingshan nematode virus 3 | 40 | MN764156 | Thrips tabaci associated bunya-like virus 2 | Ttabunya2 | Thrips virus |
| THR-E_DN23726 | 4,940 | APG79254 | Shayang ascaridia galli virus 1 | 38 | MN764154 | Thrips tabaci associated bunya-like virus 3 | Ttabunya3 | Thrips virus |
| THR-E_DN24108 | 4,897 | QIM73965 | Riboviria sp. | 36 | MN764151 | Thrips tabaci associated bunya-like virus 4 | Ttabunya4 | Thrips virus |
| THR-E_DN24112 | 12,836 | APG78763 | Hubei dimarhabdovirus virus 4 | 73 | MN714687 | Thrips tabaci associated dimarhabdovirus 1 | Ttadima1 | Thrips virus |
| THR-E_DN24095 | 7,929 | QQP18723 | Soybean thrips-associated dsRNA virus-1** | 39 | MN764138 | Thrips tabaci associated dsRNA virus 1 | Ttads1 | Thrips virus |
| THR-E_DN23955 | 8,740 | QQP18723 | Soybean thrips-associated dsRNA virus-1** | 56 | MN764139 | Thrips tabaci associated dsRNA virus 2 | Ttads2 | Thrips virus |
| THR-E_DN19401 | 3,243 | QPZ88376 | Soybean thrips virus 1** | 40 | MN764158 | Thrips tabaci associated jingmen-like virus 1 RNA1 | Ttajing1_RNA1 | Thrips virus |
| THR-E_DN19401 | 3,089 | QPZ88383 | Soybean thrips virus 2** | 36 | MN764159 | Thrips tabaci associated jingmen-like virus 1 RNA2 | Ttajing1_RNA2 | Thrips virus |
| THR-E_DN7390 | 3,135 | QQM16241 | Wifsystermes virus | 38 | MN714677 | Thrips tabaci associated luteo-like virus 1 RNA1 | Ttaluteo1_RNA1 | Thrips virus |
| THR-E_DN7390 | 1,541 | AKH40292 | Motts Mill virus | 40 | MN714682 | Thrips tabaci associated luteo-like virus 1 RNA2 | Ttaluteo1_RNA2 | Thrips virus |
| THR-E_DN24087 | 1,803 | DAD49837 | Lutzomyia longipalpis mitovirus 1 | 41 | MN714680 | Thrips tabaci associated mitovirus 1 | Ttamito1 | Thrips virus |
| THR-B_DN29294 | 2,816 | QDH89080 | Mitovirus sp. | 57 | MN714678 | Thrips tabaci associated mitovirus 2 | Ttamito2 | Thrips virus |
| THR-B_DN29604 | 3,063 | AWY10987 | Sclerotinia sclerotiorum mitovirus 29 | 63 | MN714686 | Thrips tabaci associated mitovirus 3 | Ttamito3 | Thrips virus |
| THR-E_DN24029 | 1,156 | QED42987 | Entomophthora narnavirus C | 47 | MN714683 | Thrips tabaci associated narna-like virus 2 | Ttanarna2 | Thrips virus |
| THR-E_DN18567 | 4,450 | Q02119 | Rice dwarf virus (isolate Akita) | 29 | MN764140 | Thrips tabaci associated reovirus 1 RNA1 | Ttareo1_RNA1 | Thrips virus |
| THR-E_DN18567 | 4,049 | NP_620541 | Rice ragged stunt virus | 23 | MN764141 | Thrips tabaci associated reovirus 1 RNA2 | Ttareo1_RNA2 | Thrips virus |
| THR-E_DN18567 | 3,679 | QIJ56929 | Scaphoideus titanus reo-like virus 1 | 31 | MN764142 | Thrips tabaci associated reovirus 1 RNA3 | Ttareo1_RNA3 | Thrips virus |
| THR-E_DN18567 | 3,207 | QHA33826 | Atrato reo-like virus | 25 | MN764143 | Thrips tabaci associated reovirus 1 RNA4 | Ttareo1_RNA4 | Thrips virus |
| THR-E_DN18567 | 2,522 | QIJ56928 | Scaphoideus titanus reo-like virus 1 | 31 | MN764144 | Thrips tabaci associated reovirus 1 RNA5 | Ttareo1_RNA5 | Thrips virus |
| THR-B_DN22880 | 4,173 | QQP18784 | Soybean thrips tombus-like virus 3 | 53 | MN714675 | Thrips tabaci associated tombus-like virus 1 | Ttatombus1 | Thrips virus |
| THR-B_DN23975 | 6,965 | YP_009337819 | Hubei virga-like virus 12 | 39 | MN714672 | Thrips tabaci associated virga-like virus 1 RNA1 | Ttavirga1_RNA1 | Thrips virus |
| THR-B_DN23975 | 3,565 | YP_009336553 | Hubei virga-like virus 9 | 26 | MW459159 | Thrips tabaci associated virga-like virus 1 RNA2 | Ttavirga1_RNA2 | Thrips virus |
| THR-E_DN21965 | 5,712 | YP_009337854 | Shahe yuevirus-like virus 1 | 25 | MN764157 | Thrips tabaci associated yue-like virus 1 RNA1 | Ttayue1_RNA1 | Thrips virus |
| THR-E_DN21965 | 1,853 | No significant similarity found | 0 | MW297844 | Thrips tabaci associated yue-like virus 1 RNA2 | Ttayue1_RNA2 | Thrips virus | |
| THR-E_DN23836 | 18,939 | YP_009026378 | Casuarina virus | 28 | MN714662 | Insect metagenomics mesonivirus 1 | Immeso1 | Insect virus |
| THR-D_DN18103 | 11,139 | YP_009505536 | Piry virus | 41 | MN714689 | Insect metagenomics mononegavirales virus 2 | Immono2 | Insect virus |
| THR-D_DN17285 | 3,058 | QNQ74064 | Plasmopara viticola lesion associated orfanplasmovirus 2 | 29 | MN764145 | Insect metagenomics narna-like virus 1 RNA1 | Imnarna1_RNA1 | Insect virus |
| THR-D_DN17285 | 2,461 | YP_001497520 | Paramecium bursaria Chlorella virus NY2A | 43 | MW297845 | Insect metagenomics narna-like virus 1 RNA2 | Imnarna1_RNA2 | Insect virus |
| THR-E_DN23507 | 2,646 | YP_009333179 | Beihai barnacle virus 10 | 42 | MN714679 | Insect metagenomics narna-like virus 2 | Imnarna2 | Insect virus |
| THR-E_DN1552 | 2,411 | QMP82343 | Hemipteran orthomyxo-related virus OKIAV188 | 32 | MN764160 | Insect metagenomics orthomyxo-like virus 1 RNA1 | Imortho1_RNA1 | Insect virus |
| THR-E_DN1552 | 2,263 | QMP82299 | Hemipteran orthomyxo-related virus OKIAV188 | 42 | MN764161 | Insect metagenomics orthomyxo-like virus 1 RNA2 | Imortho1_RNA2 | Insect virus |
| THR-E_DN1552 | 1,733 | QMP82317 | Hemipteran orthomyxo-related virus OKIAV188 | 34 | MN787040 | Insect metagenomics orthomyxo-like virus 1 RNA3 | Imortho1_RNA3 | Insect virus |
| THR-E_DN1552 | 1,075 | AAN34384 | Human immunodeficiency virus 1 | 31 | MW297846 | Insect metagenomics orthomyxo-like virus 1 RNA4 | Imortho1_RNA4 | Insect virus |
| THR-E_DN23568 | 9,979 | QKN89031 | Riboviria sp. | 35 | MN714669 | Insect metagenomics picorna-like virus 1 | Impico1 | Insect virus |
| THR-A_DN18812 | 3,345 | YP_009330120 | Hubei sobemo-like virus 25 | 76 | MN725051 | Insect metagenomics sobemo-like virus 2 RNA1 | Imsobemo2_RNA1 | Insect virus |
| THR-A_DN18812 | 1,549 | AKP18619 | Humaita-Tubiacanga virus | 46 | MN725052 | Insect metagenomics sobemo-like virus 2 RNA2 | Imsobemo2_RNA2 | Insect virus |
| THR-A_DN28235 | 3,634 | AXA52557 | Linepithema humile C virus 1 | 53 | MN787041 | Insect metagenomics tombusbipa-like virus 1 RNA1 | Imtombus2_RNA1 | Insect virus |
| THR-A_DN28235 | 2,271 | AXA52559 | Linepithema humile C virus 1 | 33 | MN787042 | Insect metagenomics tombusbipa-like virus 1 RNA2 | Imtombus2_RNA2 | Insect virus |
| THR-D_DN14008 | 6,340 | QDZ71189 | Megastigmus ssRNA virus | 35 | MN714674 | Insect metagenomics virga-like virus 1 RNA1 | Imvirga1_RNA1 | Insect virus |
| THR-D_DN14008 | 2,038 | YP_009337242 | Hubei virga-like virus 11 | 48 | MW459158 | Insect metagenomics virga-like virus 1 RNA2 | Imvirga1_RNA2 | Insect virus |
Columns show contig identifier code, contig length (in bases), first viral hit accession in NCBI:protein, first viral hit organism (NCBI BLASTx first hit), NCBI hit identity, new discovered virus identifier (ID) (GenBank accession no.), suggested virus name, the virus abbreviation (ID) in the paper, and the origin of the viral contig.
Double asterisks indicate all the hits from reference 16.
Asterisks indicate all the blast hits with identity higher than 90.
FIG 2.
Heat map table showing the number of reads mapping on the identified viruses in each library. Numbers in parentheses correspond to normalized reads per kilobase per million mapped reads (RPKM). The intensity of the green color reflects the number of reads.
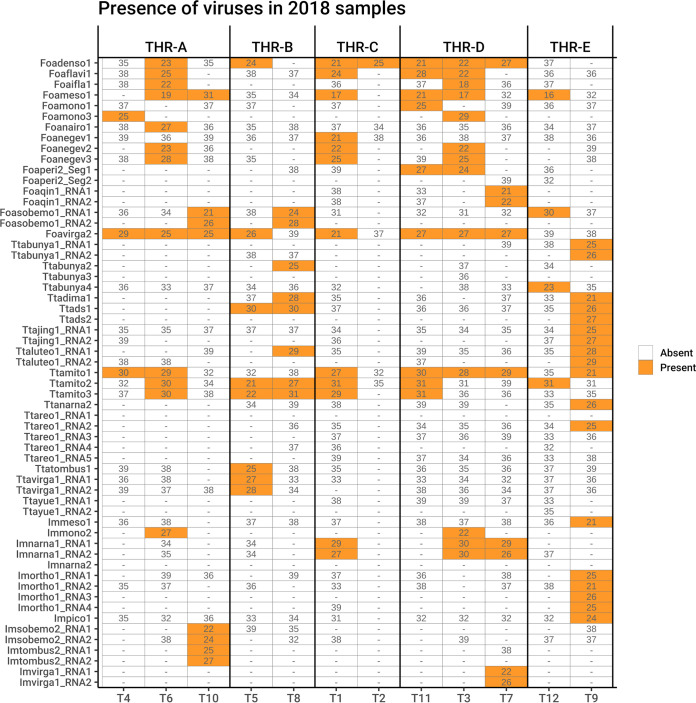
Each virus contig was tracked to each sample by real-time reverse transcription-quantitative PCR (qRT-PCR), and threshold cycle (CT) values were used as an approximate indication of relative accumulation of viral RNA; CT values above 31 were not considered reliable, and so samples with amplifications after that cycle were considered negative (Fig. 3).
FIG 3.
Graphical representation of qRT-PCR results (expressed as CT number) showing the presence of each viral contig in each RNA sample. Above the heat map the library names are indicated, whereas below the table are the individual samples of the libraries.
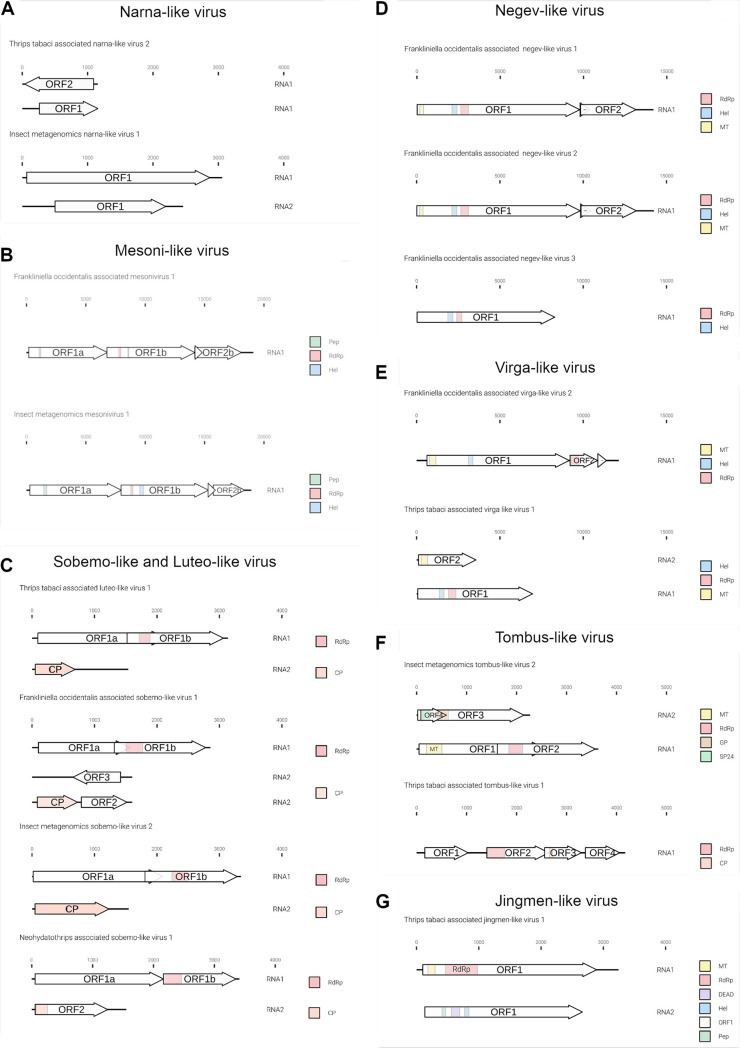
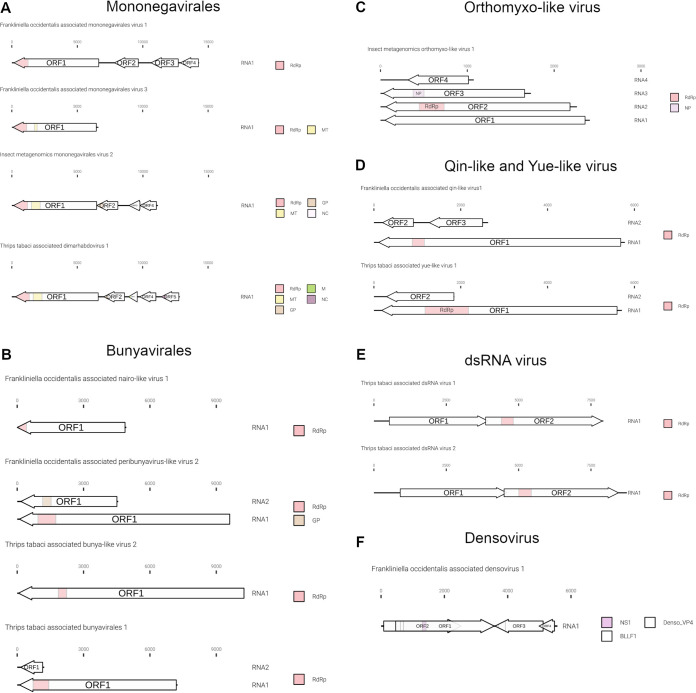
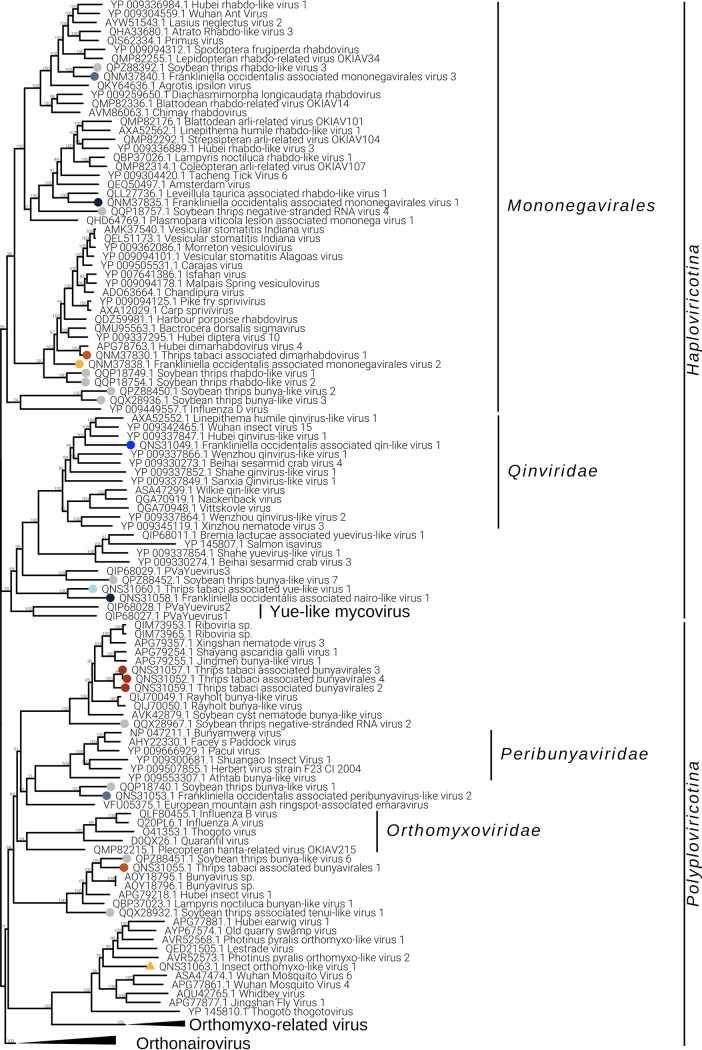
The genome organizations of the most abundant and previously undescribed viral contigs are displayed in Fig. 4 and 5.
FIG 4.
Graphical representation of the novel genome organizations of plus-strand RNA viruses associated with Frankliniella occidentalis and Thrips tabaci viromes. (A) New narna-like viruses; (B) new mesoni-like viruses; (C) Sobemo/luteo-like viruses; (D) new negev-like viruses; (E) new virga-like viruses; (F) new tombus-like viruses; (G) jingmen-like viruses. RdRP, RNA-dependent RNA polymerase; Pep, peptidase; Hel, helicase; CP, coat protein; MT, methyltransferase; GP, glycoprotein; SP24, putative virion membrane protein of plant and insect virus; DEAD, DEAD box helicases. Arrows represent open reading frames (ORFs).
FIG 5.
Graphical representation of the novel genome organizations of negative-strand, dsRNA virus and a densovirus-like sequence characterized from Frankliniella occidentalis and Thrips tabaci viromes. (A) Genomes in order Mononegavirales; (B) genomes in order Bunyavirales; (C) genome organization of an orthomyxo-like virus; (D) yue-like and qin-like virus genome organizations; (E) genome organization of two dsRNA viruses; (F) densovirus genome organization. MP, matrix protein domain; NS1, nonstructural protein 1; Denso_VP4, capsid protein VP4; BLLF1, envelope glycoprotein GP350. Arrows represent open reading frames.
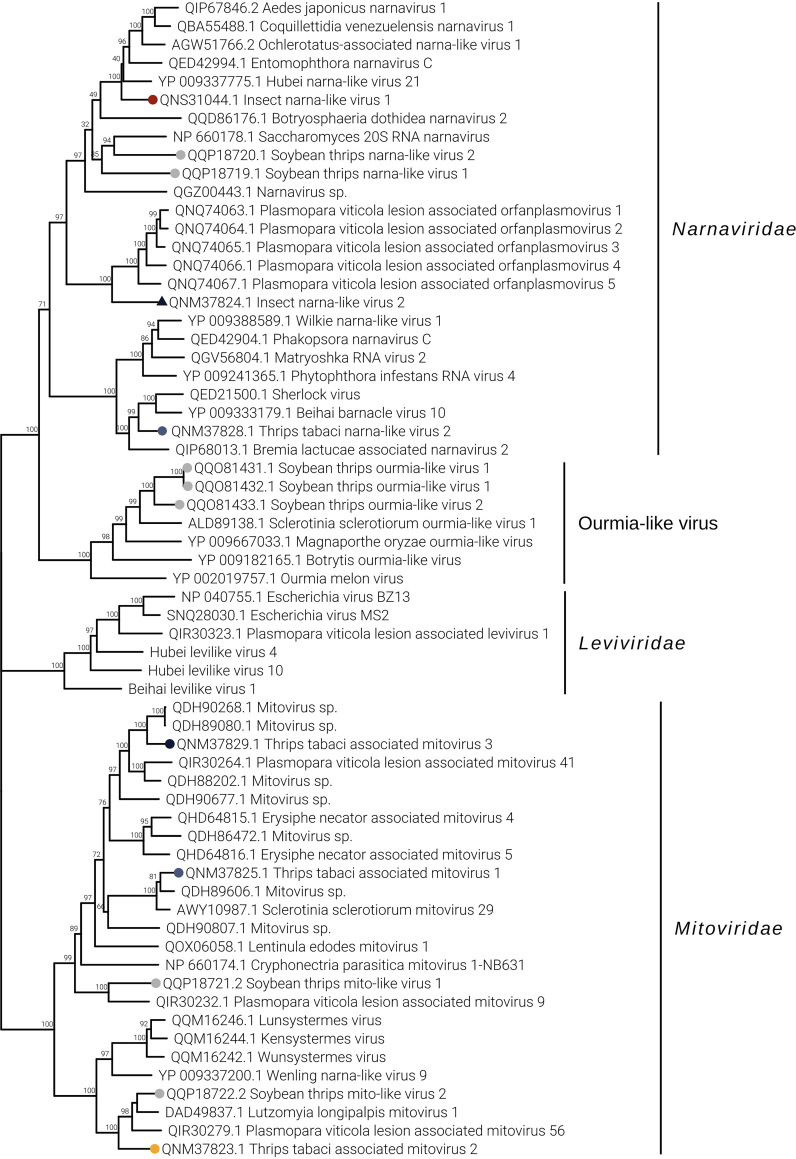
(i) Phylum Lenarnaviricota.
In our study, we identified three mitoviruses from T. tabaci, named Thrips tabaci associated mitoviruses 1, 2, and 3 (Ttamito1, Ttamito2, and Ttamito3) (Table 2). Based on read counts (Fig. 2), Ttamito1 is abundant in THR-E, and this observation is also confirmed by qRT-PCR results (Fig. 3), even if the qRT-PCR also detected the low-abundance presence of Ttamito1 in T4 to T6 (THR-A), T1 (THR-C), and T3, T7, and T11 (THR-D). Ttamito2 and Ttamito3 are more abundant in THR-B (Fig. 2) although present also in THR-A, THR-D, and THR-E. The qRT-PCRs confirmed the read count results, showing the same accumulation pattern (Fig. 3).
Three narnaviruses have also been identified: one from T. tabaci, named Thrips tabaci associated narnavirus 2 (Ttanarna2), and the remaining two from unconfirmed origin named insect metagenomics narnavirus 1 (Imnarna1) and insect metagenomics narnavirus 2 (Imnarna2), respectively (Table 2). The latter is present in low abundance in both THR-B and THR-E based on read counts, but we were not able to amplify it in qRT-PCR. Imanarna1, instead, is bipartite and mostly concentrated in THR-C and THR-D, based on read counts and qRT-PCR (Fig. 2 and 3). The second genomic RNA segment of Imanarna1 was detected not from BLAST similarity searches but from ORFans analysis linked to correlation in mapped-read abundance in different libraries and specific sample infection.
Genomic organization of mitoviruses show a monopartite genome, and all three have an RNA-dependent RNA polymerase (RdRP) domain (data not shown). While Ttamito2 and Ttamito3 have a genome size around 3 kb, Ttamito1 displays a genome shorter than 2 kb, with a complete open reading frame (ORF). On the other hand, none of the three narnaviruses identified in this study presents a conserved RdRP domain comparable to those already present in the databases, even if the BLAST search of the ORF1 of each virus returns as a first hit an RdRP of another virus. Sequence alignment shows that indeed the most conserved palm subdomains are partly conserved, including the GDD motif (data not shown). Moreover, Imnarna2 does not carry a complete ORF1, probably limited coverage did not allow to recover the complete genomic sequence. Interestingly, Ttanarna2 shows a very short genome and a putative ambisense ORF organization (Fig. 4A). Based on our phylogenetic analysis (Fig. 6), this virus lies in the same clade as Saccharomyces 20S RNA narnavirus (the reference virus for the Narnavirus genus). Imnarna2 clusters with the recently characterized orfanplasmoviruses (17), but in more basal position, while Imnarna1 does not. Mitoviruses are in a separate clade from the narnavirus, but while Ttamito1 falls in a clade that includes a putative insect virus (Wenling narna-like virus 9), Ttamito2 and Ttamito3 fall in clades with conserved fungal mitoviruses. Overall, the three mitoviruses we here describe are each in a distinct clade, possibly different viral genera.
FIG 6.
Lenarnaviricota phylogenetic tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. The model of substitution is VT+F+I+G4. The consensus tree is constructed from 1,000 bootstrap trees. The log likelihood of the consensus tree is −113395.268. At nodes are the percent bootstrap values. Different colors indicate different subgroups. Triangles indicate the insect metagenomics viruses, and circles indicate Thrips tabaci- or Frankliniella occidentalis-associated viruses. Viruses labeled with a gray circle are the ones identified in a recent soybean thrip virome characterization study (16).
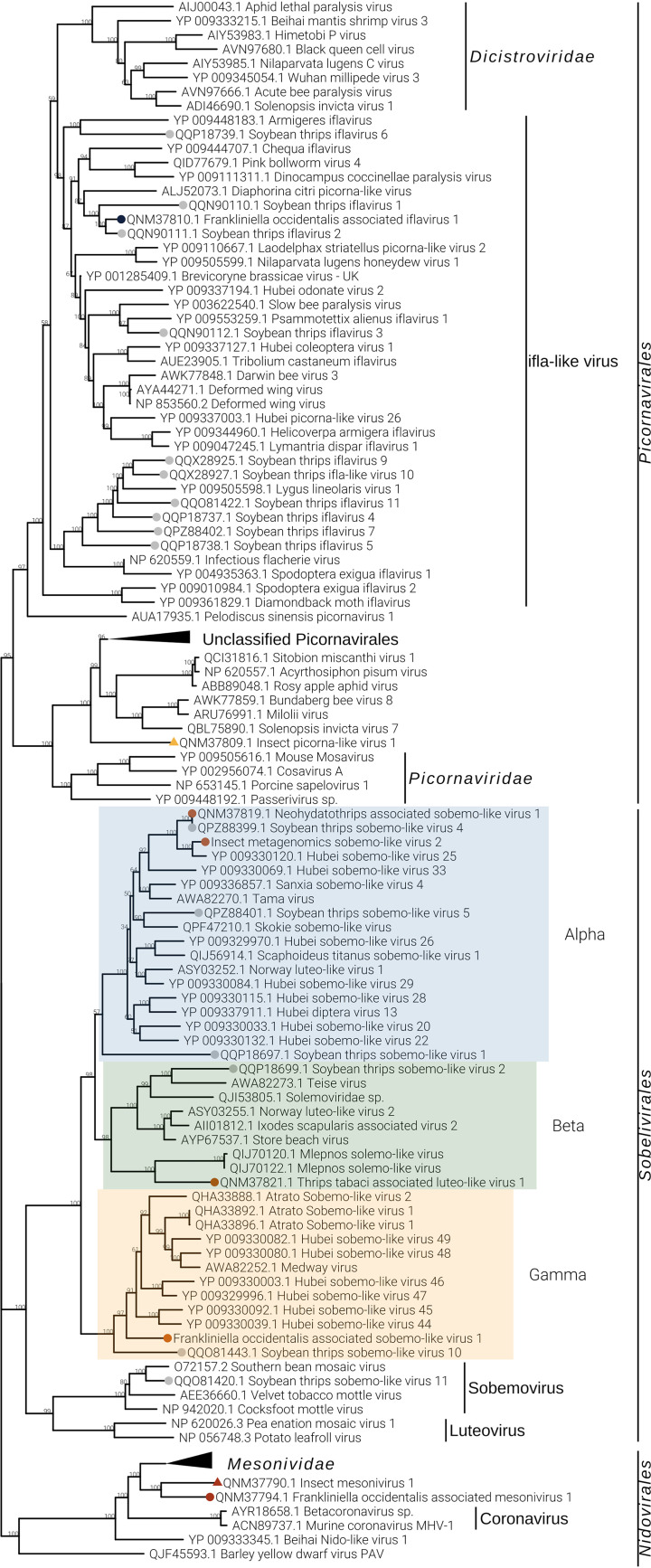
(ii) Phylum Pisuviricota.
Two virus contigs had as first hit a member of the Mesoniviridae family (Table 2) using BLASTx to search the databases. Mesoniviruses are viruses with a genome size around 20 kb, which is intermediate among the size ranges of nidoviruses. Typical mesoniviruses are monoparite, with a linear genome containing 7 ORFs (ORF1a, ORF1b, ORF2a, ORF2b, ORF3a, ORF3b, and ORF4), encoding an RdRP protein and other nonstructural (NS) proteins involved in RNA synthesis and at least two structural proteins. The primary identified hosts for mesoniviruses were insects; in fact, this family represents the first nidoviruses to be discovered in insects (18).
The two mesoni-like viruses identified in our study were named insect metagenomics mesonivirus 1 (Immeso1) and Frankliniella occidentalis associated mesonivirus 1 (Foameso1), both slightly shorter than 20 kb. Immeso1, 19 kb (GenBank accession no. MN714662), was found in only one pool (THR-E) from the north of Italy, while Foameso1, 19 kb (GenBank accession no. MN714663), was found in three different pools (THR-A, THR-C, and THR-D). Based on read counts (Fig. 2) and qRT-PCR (Fig. 3), we can infer that these mesoniviruses are highly concentrated in the samples. A phylogenetic tree (Fig. 7) derived from alignment of ORF1b (containing the RdRP) shows Immeso1 and Foameso1 clustering together in a well-supported branch (bootstrap value, 97) separated from the Mesoniviridae family clade, indicating these two viruses can be a new genus inside the Mesoniviridae family or possibly a new family in the order Nidovirales.
FIG 7.
Pisuviricota phylogenetic tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. The model of substitution is VT+F+I+G4. The consensus tree is constructed from 1,000 bootstrap trees. The log likelihood of the consensus tree is −504786.937. At nodes are the percent bootstrap values. Different colors indicate different subgroups. Triangles indicate the insect metagenomics viruses, and circles indicate Thrips tabaci- or Frankliniella occidentalis-associated viruses. Viruses labeled with a gray circle are the ones identified in a recent soybean thrip virome characterization study (16).
The genomic organizations of the two mesoniviruses are identical: both present four ORFs. ORF 2a and ORF2b do not carry any conserved domain present in the databases but are conserved, respectively, with the putative mesonivirus spike protein (S) and nucleocapsid protein (N). ORF1a has a peptidase domain and ORF1b contains the helicase (Hel) and RdRP domains (Fig. 4B). Typical mesoniviruses so far characterized from insects include 2 further ORFs (ORF3a and −3b) and often, but not always, a 4th ORF (18).
Two new sobemo-like viruses and one luteo-like virus have been discovered and named Foasobemo1, Imsobemo2, and Ttaluteo1, respectively. One sobemo-like virus was also identified in the sample from the United States and named Neohydatothrips associated sobemo-like virus 1 (Ntasobemo1). Read counts (Fig. 2) and qRT-PCR (Fig. 3) show the presence of Ttaluteo1 virus in library THR-E and, in particular, in sample T9; its presence in library THR-B seems to be strongly supported by the read counts. Foasobemo1 and Imsobemo2 are present in two pools out of five: THR-A and THR-B. Specifically, Foasobemo1 is present in samples T10 (THR-A) and T8 (THR-B) based on qRT-PCR results (Fig. 3), while Imsobemo2 is only amplified in T10 (THR-B) (Fig. 3). Both segments of Ntasobemo1 were present only in T-Ame sample at very high concentration (Fig. 3).
Phylogenetic analysis (Fig. 7) clearly shows that the three new Italian viruses from thrips are in distinct clades each defining a new viral family, while Ntasobemo1 clusters in a sister clade of Imsobemo2. These new viruses do not belong to either the Sobemovirus or Luteovirus genus. The phylogenetic tree reports three taxonomically classified sobemoviruses (cocksfoot mottle virus [NCBI:protein accession no. NP_942020.1], velvet tobacco mottle virus [AEE36660.1], and southern bean mosaic virus [O72157.2]) and three luteoviruses (barley yellow dwarf virus PAV [QJF45593.1], pea enation mosaic virus 1 [NP_620026.3], and potato leafroll virus [NP_056748.3]), and they form a distinct clade from the putative thrips-associated viruses we report. Foasobemo1, Imsobemo2, and Ttaluteo1 cluster in the same parent clade but form three distinct putative new families very well supported by the bootstrap values, labeled Alpha, Beta, and Gamma on the tree.
Viruses belonging to the Sobemovirus and Luteovirus genera are monopartite plant viruses with genomes of circa 4 kb and 5 kb, respectively. The four viruses we identified (Foasobemo1, Imsobemo2, Ttaluteo1, and Ntasobemo1) are instead bipartite insect viruses. Viruses present on the tree, like Scaphoideus titanus sobemo-like virus 1 (19) for clade Alpha, Wuhan heteropteran virus 2 (14) for clade Beta, and Hubei sobemo-like virus 47 (14) for clade Gamma, are also insect viruses with bipartite genomes, so official recognition and new names for these new taxa are needed.
Ttaluteo1 and Imsobemo2 show similar genomic organizations (Fig. 4C): RNA1 (∼3.2 kb) contains two ORFs translated via a −1 ribosomal frameshift, and one of them contains the RdRP motif (ORF1b). RNA2 (∼1.5 kb) contains the putative capsid protein. Foasobemo1 displays the same genomic organization for RNA1, while RNA2 shows the presence of the putative capsid protein plus two additional ORFs: ORF2 (∼0.7 kb) in the sense direction and ORF3 (∼0.7 kb) in the antisense direction. Neither of the two ORFs show conserved domains.
One ifla-like virus has been identified from F. occidentalis and named Foaifla1 (Frankliniella occidentalis associated ifla-like virus 1). The ifla-like virus shows a monopartite genome of about 10 kb, coding for a polyprotein with multiple domains. In the phylogenetic tree (Fig. 7), the taxonomically accepted iflavirus do not cluster with Foaifla1, but Foaifla1 shares a branch with another putative thrips virus from N. variabilis (16), supporting the contention that these are both indeed thrips viruses. The clade containing our identified virus also contains a mix of iflavirus and picornaviruses, which probably need to be accommodated in a new genus. Based on read counts and qRT-PCR, Foaifla1 is tracked to 3 pools (THR-A [T6], THR-C, and THR-D [T3]) (Fig. 2 and 3), but the qRT-PCR results only partially confirm the read count evidence: Foaifla1 was not amplified in THR-C library despite the fact that the reads count showed a high concentration, raising the possibility that a specific variant of this virus in the region where primers anneal makes this virus not detectable.
One picorna-like virus has been identified in THR-E pool (northwest of Italy), sample T9, named insect metagenomics picorna-like virus 1 (Impico1) (Fig. 3). Impico1 has a genome of ∼10 kb, slightly longer than a taxonomically characterized picornavirus but aligned with insect picorna-like viruses (14). The monopartite genome is similar to that of picorna-like viruses (Fig. 5) but shows an additional ORF with an arginine/serine-rich protein PNISR domain (PF15996). ORF1 contains two domains, i.e., a helicase domain and an RdRP domain, while ORF2 contains the coat protein domain. Phylogenetic analysis shows taxonomically accepted Picornaviridae (cosavirus A, mouse mosavirus, porcine sapelovirus 1, and Passerivirus sp.) not in the same clade as our newly identified virus (Fig. 7). So, based on genomic organization and phylogenetic tree topology, Impico1 virus requires the establishment of a new family to accommodate it.
(iii) Phylum Kitrinoviricota.
Frankliniella occidentalis associated flavi-like virus 1 (Foaflavi1) has been identified in all three pools containing F. occidentalis, always well represented by number of reads mapping the genome, especially in sample THR-C, where the read count is over 29,000 (Fig. 2). Real time RT-PCRs confirmed the presence of Foaflavi1 in all the above-mentioned pools (Fig. 3). Compared to typical Flavivirus, usually with a genome of about 10 to 11 kb, Foaflavi1 has a double-size genome, almost 20 kb, a feature shared with some recently characterized insect-specific flaviviruses identified in Apis mellifera L. and Diaphorina citri Kuwayama (20, 21). The genomic organization shows a monopartite, linear single-stranded RNA positive-sense [ssRNA(+)] genome, with a single ORF that produces a putative polyprotein (Fig. 5). MOTIF Search was not able to identify the RdRP signature, but the GDD motif is present and can be aligned to flavi-like virus palm domains (data not shown).
The phylogenetic analysis shows Foaflavi1 in a sister clade of members of the genus Flavivirus, with other flavi-like viruses from insects (Fig. 8).
FIG 8.
Kitrinoviricota phylogenetic tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. The model of substitution is Blosum62+I+G4. The consensus tree is constructed from 1,000 bootstrap trees. The log likelihood of the consensus tree is −529281.876. At nodes are the percent bootstrap values. Different colors indicate different subgroups. Triangles indicate the insect metagenomics viruses, and circles indicate Thrips tabaci- or Frankliniella occidentalis-associated viruses. Viruses labeled with a gray circle are the ones identified in a recent soybean thrip virome characterization study (16).
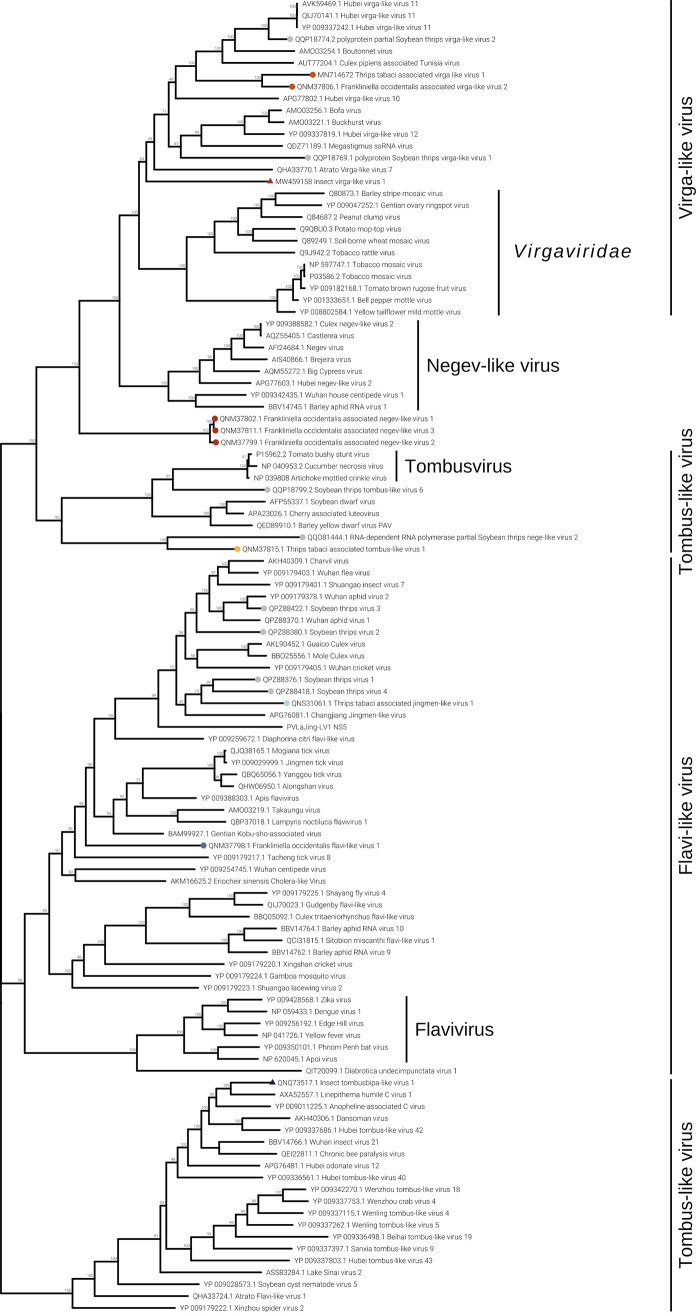
Three negev-like viruses have been identified in F. occidentalis: Frankliniella occidentalis associated negev-like virus 1 (Foanegev1), Frankliniella occidentalis associated negev-like virus 2 (Foanegev2), and Frankliniella occidentalis associated negev-like virus 3 (Foanegev3). Each virus has its peak concentration in a different pool (Fig. 2) and was tracked by qRT-PCR to distinct samples: Foanegev1 in THR-C (T1), Foanegev2 in THR-A (T6), and Foanegev3 in THR-D (T3); Foanegev1 is 10 and 30 times more concentrated than Foanegev2 and Foanegev3, respectively (Fig. 2). These three new viruses have been called, temporarily, negev-like viruses, but based on tree topology (Fig. 8), neither of these viruses is a real negevirus. Figure 4D shows that Foanegev1 and Foanegev2 have the same genomic organization and the same domain distribution, while Foanegev3 has a shorter genome, with one single ORF, and the methyltransferase (MT) domain is absent: likely this virus genome is not complete, since it is very similar and phylogenetically related to the other two that are present in the database. Both ORF2 and ORF3 do not have any reliable homologue in NCBInr (virus limited: taxid 10239) in a BLASTp (protein-protein BLAST) search with default parameters.
Three virga-like viruses were identified in our thrips metatranscriptomic analysis, Imvirga1, Foavirga2, and Ttavirga1 (Fig. 2 and Table 2): one putatively from F. occidentalis and one putatively from T. tabaci; a third one (Imvirga1, a bipartite virus), which could not be assigned to a specific host according to our more stringent criteria, has been identified only in sample T7 (THR-D) (Fig. 3). Conversely, Foavirga2, a monopartite virus, has been identified and amplified in all samples of the THR-A and THR-D libraries, but only in one sample for the libraries THR-B (T5) and THR-C (T1) (Fig. 3).
The virga-like viruses identified from T. tabaci displayed in pool THR-B a relative low level of read coverage for both its segments, and qRT-PCR confirmed their presence only in sample T5 (Fig. 2 and 3).
Two out of three virga-like viruses (Imvirga1 and Ttavirga1) show similar genome sizes, about 6.5 kb, for RNA1 and possess a second genomic segment (RNA2) of circa 3.5 and 2.5 kb in length, while Foavirga2 has a longer monopartite genome (∼12 kb). All the identified virgaviruses display 3 domains: RNA-dependent RNA polymerase (RdRP), MT, and Hel, even if their genomic organizations present significant differences. Ttavirga1 has the RdRP and the Hel domains on RNA1 and the MT domain on RNA2. Foavirga2 shows a genomic organization very similar to that of Ttavirga1, but on a single genomic segment, and it includes an ORF that carries the tobamovirus coat protein motif (ORF3) (Fig. 4E).
Our phylogenetic analysis indicates that the viruses we characterized from thrips are clearly only distantly related to the plant family Virgaviridae and probably need establishment of a new family taxon to be accommodated (Fig. 8).
Members of the family Tombusviridae are worldwide-spread viruses, causing several diseases in plants, but tombus-like viruses are also present in arthropods (14). Thrips tabaci associated tombus-like virus 1 (Ttatombus1) has been identified in THR-B pool (sample T10), while insect metagenomics tombus-like virus 2 (Imtombus2) has been identified in pool THR-A (Fig. 2 and 3). Official members of the family Tombusviridae are monopartite viruses with a genome between 4 kb and 5.4 kb. Ttatombus1 has these features but clusters in a sister clade of plant tombusviruses (Fig. 8) and presents a slightly different genomic organization. Tomato bushy stunt virus (reference genome for tombusvirus) has five ORFs. Ttatombus1 (Fig. 4F) has four ORFs, and the RdRP domain is located on ORF2. ORF3 contains the coat protein domain. ORF1 and ORF4 do not contain known motifs based on MOTIF Search analysis and the default BLASTp on the NCBInr database (June 2020) limited to viruses (taxid 10239) did not find significant similarities. Imtombus2 is instead bipartite, and the first match for RNA1 in NCBInr (Oct 2018) is Linepithema humile C virus 1 from Argentine ant (22). Phylogenetic analysis shows the two tombus-like viruses to cluster in different well-separated clades. In particular, Ttatombus1 clusters in a sister group of real tombusvirus, while Imtombus2 clusters with mostly unclassified viruses (Fig. 8). Both viruses need a new family to accommodate them in the current viral taxonomical framework.
Jingmen viruses are segmented positive-sense single-stranded RNA viruses associated with arthropods (23). In our study, we identified one jingmen-like virus in pool THR-E, and we called it Thrips tabaci associated jingmen-like virus 1 (Ttajing1), and the closest hits in the databases are soybean thrips-associated viruses (16). RNA1 and RNA2 have been associated based on read counts (Fig. 2), qRT-PCR results (Fig. 3), and 5′ and 3′ end alignments (data not shown). The viral genomic organization is shown in Fig. 4G; on RNA1 two domains are present, a methyltransferase domain and an RdRP domain, while on RNA2, we identified three domains: a helicase domain, a peptidase domain, and a DEAD domain. A phylogenetic tree (Fig. 8) shows the known jingmen viruses (infecting ticks) clustering in a separate branch from Ttajing1_RNA1 virus. Moreover, Ttajing1_RNA1 virus is basal to the cluster of insect-infecting jingmen-like viruses.
(iv) Phylum Negarnaviricota.
Members of the order Mononegavirales have been identified in all five pools: one virus is associated with T. tabaci samples (Thrips tabaci associated dimarhabdovirus 1 [Ttadima1]), and two viruses are associated with F. occidentalis (Frankliniella occidentalis associated mononegavirales virus 1 and Frankliniella occidentalis associated mononegavirales virus 3 [Foamono1 and Foamono3]) (Fig. 2). One could not be confirmed to be associated with any thrips species according to our strict criteria, so it has been called insect metagenomics mononegavirales virus 2 (Immono2). Based on read counts, three out of four mononegaviruses are not highly concentrated, apart from Ttadima1 in pool THR-E (Fig. 3).
Foamono1 and Immono2 viruses have quite similar genomic organizations (Fig. 5A): they code for four ORFs, and for each of them, the RdRP domain is present in ORF1. While for Foamono1 we were able to identify only the RdRP domain, for Immono2 and Ttadima1 we identified several domains. The Foamono3 genome segment displays only 1 ORF, coding for the putative RdRP and a putative methyltransferase. The Ttadima1 genome has 5 ORFs coding for an RdRP (ORF1), a methyltransferase (ORF1), a glycoprotein (ORF2), a matrix protein (ORF3), and a nucleocapsid (ORF5).
Phylogenetic analysis shows that Foamono3 groups with an N. variabilis mononegavirus, which has a larger genomic segment that includes at least three extra ORFs (16) (Fig. 9); it is therefore likely that Foamono3 is an incomplete genome. The other three identified members of Mononegavirales are in a sister clade of Foamono3 but on different, well-sustained branches, indicating that these viruses could represent different subfamilies/genera.
FIG 9.
Negarnaviricota phylogenetic tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. The model of substitution is VT+F+G4. The consensus tree is constructed from 1,000 bootstrap trees. The log likelihood of the consensus tree is −526045.530. At nodes are the percent bootstrap values. Different colors indicate different subgroups. Triangles indicate the insect metagenomics viruses, and circles indicate Thrips tabaci- or Frankliniella occidentalis-associated viruses. Viruses labeled with a gray circle are the ones identified in a recent soybean thrip virome characterization study (16).
Members of the order Bunyavirales are single-stranded, segmented, linear negative-sense viruses infecting plants, fungi, vertebrates, and invertebrates. The genome size and organization are family dependent. We identified six bunyavirus RdRPs, two in WFT and the remaining four in OT. For Frankliniella occidentalis associated nairovirus 1 (Foanairo1), present only in THR-A, we identified one segment of 4.9 kb; for Frankliniella occidentalis associated peribunyavirus 1 (Foaperi1), present in THR-D, we assembled two segments of 9.6 kb and 4.5 kb (Fig. 5B). We also identified four OT-associated bunyavirus, named Ttabunya1, Ttabunya2, Ttabunya3, and Ttabunya4, all of them identified in pool THR-E (Fig. 2 and 3). Among them, only for Ttabunya1 could we find a second genomic segment (7.2-kb RNA1 and 1.2-kb RNA2) (Fig. 5B). Ttabunya2 has the longest genomic segment, with 10.2 kb (Fig. 5B), while Ttabunya3 and Ttabunya4 have very similar genome sizes, with 4.9 kb and 4.8 kb, respectively, that possibly represent incomplete virus segments.
Phylogenetic analysis (Fig. 9) of the identified Bunyavirales shows Ttabunya2, Ttabunya3, and Ttabunya4 clustering closely together with a bootstrap value of 100. Foaperi1 lies in a sister group of Peribunyaviridae, together with another soybean thrips-associated virus (16), indicating that the virus is only distantly related to the family Peribunyaviridae and that these viruses likely belong to a specific clade of thrips-infecting viruses. Ttabunya1 clusters with several insect bunyaviruses with a well-sustained bootstrap value, and Ttabunya4 can be found in a sister branch. Finally, Foanairo1 clusters in the orthonairovirus branch of the tree, supporting its relatedness to the Orthonairovirus genus, but it is basal to a group of insect-infecting nairo-like viruses that requires a possible new taxon.
One orthomyxo-like virus was identified in our data set and named insect metagenomic orthomyxo-like virus 1 (Imortho1) (Table 2 and Fig. 2). The genome is quadripartite (Fig. 5C), with 2,411 bases for RNA1, 2,263 bases for RNA2, 1,733 bases for RNA3, and 1,075 bases for RNA4. The RdRP domain is located on RNA2 and the nucleoprotein domain on RNA3, while RNA1 and RNA4 did not show any known domain. The four segments have been assigned to the same virus based on 5′ and 3′ end alignment (data not shown), which shows a high conservation in nucleotide sequences. The presence of the virus is limited to one pool (THR-E); moreover, the four genomic segments are present only in sample T9 (Fig. 3).
Phylogenetic analysis shows Imortho1 to be in a separate sister clade from taxonomically accepted orthomyxoviruses, indicating the need to define a new family to accommodate our virus and several others named orthomyxo-like viruses that have insects as true hosts (Fig. 9).
The bisegmented Frankliniella occidentalis associated qinvirus 1 (Foaqin1; putatively belongs to order Muvirales, family Qinviridae, genus Yingvirus) has been identified in a single sample (T7) of a unique pool (THR-D) with quite a high concentration based on read counts (Fig. 2) and qRT-PCR (Fig. 3). The results of qRT-PCR clearly show the presence of the two segments not only in the same pool but also in the same sample (T7) at a high concentration. Foaqin1 genomic organization (Fig. 5D) shows that RNA1 has one ORF coding for a putative RdRP, while RNA2 segment codes for two distinct ORFs, but they do not have conserved domains. Thrips tabaci associated yuevirus 1 (Ttayue1) shows a concentration similar, in terms of read counts, to that of Foaqin1 (Fig. 2). The genomic organization presents a bipartite genome with RNA1 coding for an RdRP and an RNA2 without any conserved domain (Fig. 5D). The conserved putative catalytic domain of Ttayue1 is SDD, corresponding to the yuevirus catalytic domain, while that of the qinvirus is IDD, the one present in Foaqin1 (data not shown).
Both the yueviruses and qinviruses characterized from invertebrates have a second genomic segment coding for a protein of unknown function, conserved among each of the two groups of viruses (14); indeed, we could find the second associated segment through a similarity search for the thrips qinvirus, while for the yue-like thrips virus, we could identify the second segment only through a careful analysis of ORFan sequences and correlation of abundance of reads mapping to distinct libraries and codetection of both segments in the same samples (Fig. 3).
Although the RdRP domain was not found in the ORF1 of Ttayue1, the BLAST hits are all RdRP (BLASTp program, database NCBInr limited to viruses [taxid 10239] in August 2020), so this sequence was used to generate the phylogenetic tree with RNA1 of Foaqin1, containing the RdRP domain (Fig. 9). The analysis placed the two viruses in two sister clades, each branch associated with a confirmed viral family: in the case of Foaquin1, we can support the taxonomical assignment to Qinviridae family, while Ttayue1 is only distantly related to previously described insect yuevirus and even more distantly related to the recently discovered yue-like mycoviruses (17).
(v) Phylum Duplornaviricota.
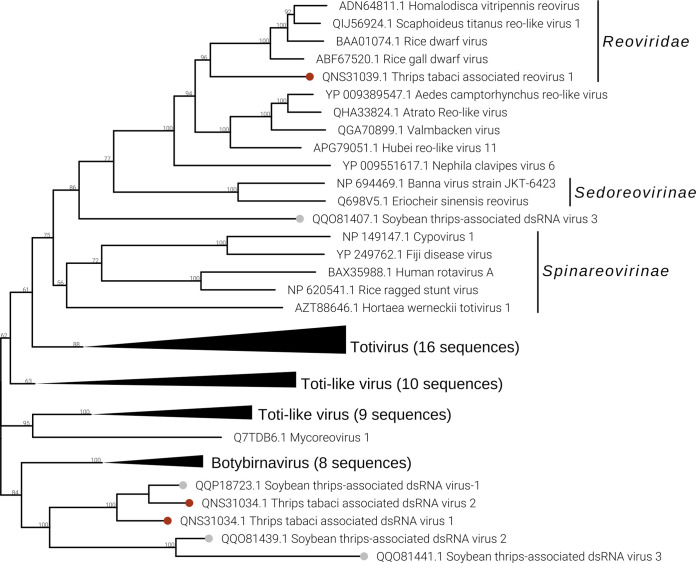
In both sample pools THR-B and THR-E, we identified two double-stranded RNA (dsRNA) viruses named Thrips tabaci associated dsRNA viruses 1 and 2 (Ttads1 and Ttads2). The virus Ttads2 was in each pool, 10 times more expressed than Ttads1, based on read counts (Fig. 2), while the qRT-PCR amplified both viral contigs only in sample T9 (THR-E) (Fig. 3). Both viruses present an ORF2 coding for a putative RdRP protein with the closest BLAST hit to soybean thrips infecting dsRNA viruses (16), while the ORF1 does not contain any known domains based on MOTIF Search, but a BLAST search finds as first hit a proline-alanine-rich protein of Scaphoideus titanus toti-like virus 1 (NCBI:protein accession no. QIJ56902; 26% identity and 58% query coverage). The genomic organization is very similar to that of members of the genus Totivirus, presenting a putative −1 frameshift between ORF1 and ORF2 (Fig. 5E). The phylogenetic analysis showed the two viruses to be closely related to each other (Fig. 10). The closest virus on the tree is Scaphoideus titanus toti-like virus 1 (19), a toti-like virus recently identified in the Flavescence dorée phytoplasma vector Scaphoideus titanus (Ball). All the viruses present in the same clade, with a bootstrap value of 94, are viruses having an insect as a host.
FIG 10.
Duplornaviricota phylogenetic tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. The model of substitution is VT+F+G4. The consensus tree is constructed from 1,000 bootstrap trees. The log likelihood of the consensus tree is −180450.432. At nodes are the percent bootstrap values. Different colors indicate different subgroups. Triangles indicate the insect metagenomics viruses, and circles indicate Thrips tabaci- or Frankliniella occidentalis-associated viruses. Viruses labeled with a gray circle are the ones identified in a recent soybean thrip virome characterization study (16).
Five segments of a single putative multisegmented reovirus have been identified in pool THR-E (data not shown). The virus, named Thrips tabaci associated reovirus 1 (Ttareo1), has the identified genome total size of 17,907 bp, with segment sizes ranging between 4,450 bp and 2,522 bp. Three conserved domains have been identified in the five segments, the RdRP on RNA1, the peptidase on RNA3, and the outer capsid protein P3 on RNA4 (data not shown). The last corresponds to the PF09231 PFam family, named Rice dwarf virus p3. Based on read counts, the five segments are only present in THR-E, apart from Ttareo1 RNA2, which seems to be also present in THR-B (Fig. 2); Ttareo1 RNA2 was amplified in sample T9 (Fig. 3).
Phylogenetic analysis (Fig. 10) of the Ttareo1 RdRP shows the virus clusters in a sister clade of the Spinareovirinae subfamily and in the same clade as the Sedoreovirinae subfamily, but on a deep branch, indicating that Ttareo1 can represent a member of a new Reoviridae subfamily.
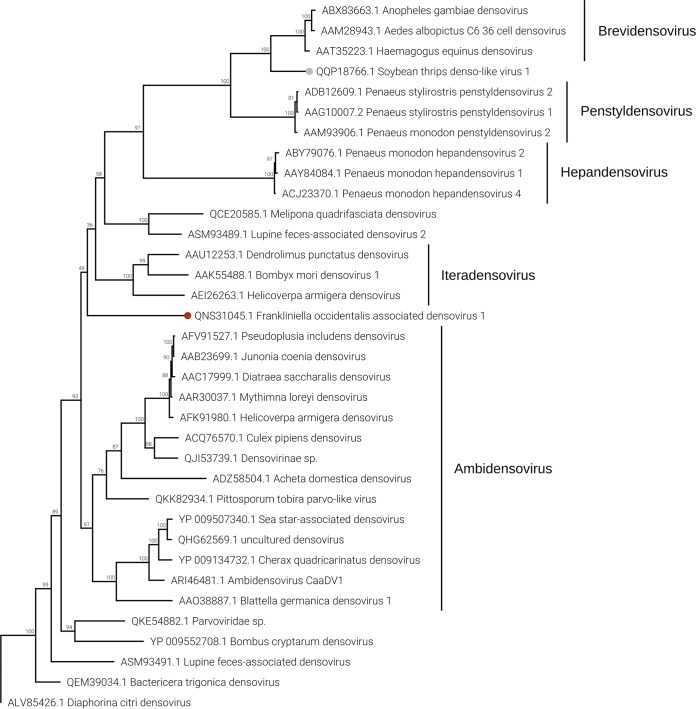
(vi) DNA virus.
In four pools out of five, we were able to detect the presence of a densovirus (Frankliniella occidentalis associated densovirus 1 [Foadenso1]) associated with F. occidentalis: pools THR-A, THR-C, and THR-D contain only the WFT, while THR-B contains a mix of WFT and OT (Fig. 2). Pool THR-B shows the lowest concentration of Foadenso1 reads count due to the sample contamination with OT. The qPCR results reveal a high concentration of Foadenso1 in T5 (CT of 24) but its absence in T8, the other sample in THR-B pool (Fig. 3).
Foadenso1 has a genome size around 5.5 kb (excluding the terminal inverted repeats, which we could not clone). The monopartite genome has four ORFs: ORF1 codes for a putative nonstructural protein (NS1), ORF2 codes for a putative viral late glycoprotein (BLLF1), also termed gp350/220, ORF3 codes for a putative densovirus capsid protein (Denso_VP4), and ORF4 has no conserved functional domain (Fig. 5F).
Phylogenetic analysis of NS1 proteins (Fig. 11) shows Foadenso1 virus to be an outgroup to the Densovirinae subfamily clade. Also, the two closest homologues to Foadenso1, based on NCBI BLAST results (BLASTp, 2 August 2020)—Diaphorina citri densovirus and lupine feces-associated densovirus 2—are far from Foadenso1 and lie on a different branch of the tree with the real Densovirinae subfamily viruses. The phylogenetic tree seems to indicate that Foadenso1 is related only to the Densovirinae subfamily and can be a member of a new Parvoviridae subfamily. The other subfamilies of the Parvoviridae family—Hamaparvovirinae and Parvovirinae—are in different clades of the tree.
FIG 11.
Densovirus phylogenetic tree computed by IQ-TREE stochastic algorithm to infer phylogenetic trees by maximum likelihood. The model of substitution is LG+F+I+G4. The consensus tree is constructed from 1,000 bootstrap trees. The log likelihood of the consensus tree is −35692.033. At nodes are the percent bootstrap values. A red circle shape is the label for the virus identified in this work. The virus labeled by a gray circle is the one identified in a recent soybean thrip virome characterization study (16).
Endogenized virus fragments.
Among the contigs we identified as viral, four contigs were amplified by PCR also from DNA, suggesting their possible integration in the genome of insect hosts (Table 1). BLAST analysis of the available full genomes of WFT, using as queries all the virus contigs we identified, confirmed the presence of the fragment corresponding to contig THR-D_DN18510, a fragment of a positive-strand virga-like virus, while fragment THR-B_DN27856 has partial hits in the Thrips palmi genome and is a fragment of a putative rhabdo-related sequence. Interesting, contrary to what has been reported so far in most analyses, where only small segments of the virus have been endogenized, here we report the assembly of a full-length iflavirus-like sequence, with high similarity to soybean thrips iflavirus 2 (contig THR-E_DN24098); its presence in only two populations of OT (data not shown) could hint at the possibility that the DNA we amplified corresponds to a partial cDNA resulting from host reverse transcriptase activity corresponding to a full-length replicating virus as observed in other entomovirus-host or mycovirus-host systems (24, 25).
Persistence of viruses in specific thrips populations.
(i) Field populations. To check the persistence of the identified viruses from WFT and OT populations in different years, some of the locations (fields) inspected in 2018 were sampled also in 2019 and 2020. Virus fragments were amplified using a qRT-PCR approach (qRT-PCR was used solely for detection and not for quantification). Figure 12A contains the correspondence between the 2018 samples and those found in the following years.
FIG 12.
Graphical representation of qRT-PCR results showing the presence of each viral contig in each RNA sample. Above the heat map are indicated the sample names. (A) Virus persistence over the studied years; on the x axis are the years of sample collection. (B) Virus persistence in mass reared populations in controlled environment; on the x axis are the names of the rearing subpopulations.
Few viruses were consistently detected over the 3 years (Foadenso1 in the field corresponding to sample T1, Foamono3 in the field corresponding to sample T4, Ttabunya1 in sample T9, and Foamono1 in the field corresponding to sample T11). As a representative example, in WFT from pepper in Piedmont (T1), 10 viruses were present in at least 2 years out of 3 and 7 were present in only 1 year. This mix of occasionally found and consistently found viruses in the same field over the years remains true also for the other four populations surveyed.
For the thrips population collected on onion in Jordan in 2019, results of qRT-PCR on all viruses of our 2018 library showed the presence of the Ttadima1, which in Italy was found in samples T9 and T12.
(ii) Laboratory-reared populations.
Four populations sampled in 2019 (T12, T9, T11, and T4) were maintained in the laboratory and reared on healthy bean pods changed at each generation, without infestations from other insects, to assess the effect of living in captivity on the insect virome and to exclude that viruses detected in thrips might come from ingested plant sap, where a mix of other insects might feed under natural conditions. Results are summarized in Fig. 12B. In sample T4, Foaperi2 was found in the field in 2019 and was consistently detected in all four cohorts (i.e., individuals from the same rearing jar), confirming its persistent presence in WFT at high prevalence. In contrast, Foavirga2 was not detected in the 2019 field population but was then identified in rearing populations, but segregating among cohorts. Foadenso1 was present both in 2019 field samples and in all the cohorts tested from population T11; Foamono1 was amplified only in two cohorts out of five. The two T9 cohorts confirmed the presence of Ttanarna2 consistently, while Ttads1 and Foasobemo1 were present in only one of the two cohorts, confirming a tendency to segregate among individual thrips. Finally, sample T12 has lost all the viruses identified in the population from the field in 2019, except for Foaflavi1.
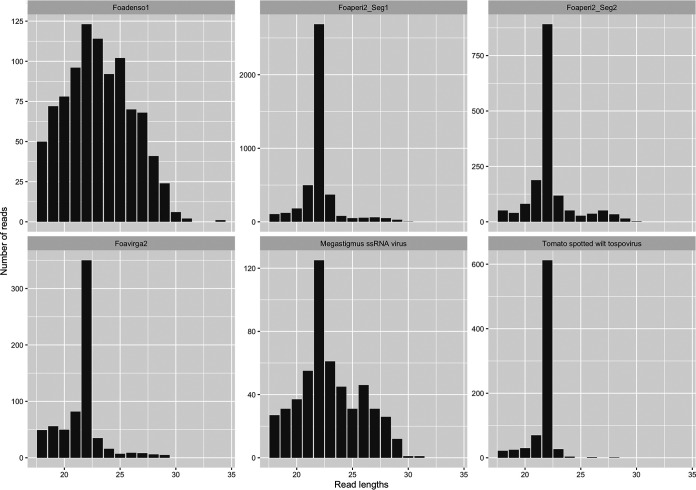
As a confirmation of the persistence of specific viromes in some populations, we report here the small RNA (sRNA) sequencing of a population reared in a controlled environment for two successive years, corresponding to sample T1 but collected in 2013 (Table 3 and Fig. 13): in that population, maintained over many generations, we could report the presence of both segments of the peribunya-like virus, the densovirus, and the Foavirga2 virus. Interestingly, the 22-nucleotide (nt)-long sRNA is the prominent size in length distributions of the sRNA mapping to replicating entomovirus with an RNA genome, while in the case of densovirus and the endogenized virus, lengths of sRNA are more uniformly distributed. A 26-nt secondary peak is also present. To our knowledge this is the first description of small RNA distribution of entomovirus from thrips (Fig. 13).
TABLE 3.
Small RNA mapping results
| Virus | Length of contig | No. of reads | NCBI hit | Host |
|---|---|---|---|---|
| Foaperi2_Seg1 | 9,618 | 4,290 | Frankliniella occidentalis associated peribunyavirus-like virus 2 segm1 | Thrips virus |
| Foaperi2_Seg2 | 4,897 | 1,587 | Frankliniella occidentalis associated peribunyavirus-like virus 2 segm2 | Thrips virus |
| Foadenso1 | 5,570 | 939 | Frankliniella occidentalis associated densovirus 1 | Thrips virus |
| Foavirga2 | 12,138 | 673 | Frankliniella occidentalis associated virga-like virus 2 | Thrips virus |
| THR-D_DN18510 | 1,167 | 529 | Megastigmus ssRNA virus | Viral insertion |
| THR-D_DN18945 | 7,762 | 515 | Tomato spotted wilt tospovirus S + M segment | Plant virus |
| THR-C_DN21959 | 8,918 | 278 | Tomato spotted wilt orthotospovirus L segment | Plant virus |
| Foanairo1 | 4,933 | 45 | Frankliniella occidentalis associated nairo-like virus 1 | Thrips virus |
| Foaflavi1 | 19,679 | 31 | Frankliniella occidentalis associated flavi-like virus 1 | Thrips virus |
| T-Ame_DN4615 | 3,117 | 28 | Ustilago maydis virus H1 | Fungal virus |
| THR-E_DN20119 | 1,556 | 17 | Iris yellow spot virus | Plant virus |
| T-Ame_DN13641 | 1,013 | 13 | Ustilago maydis virus H1 | Fungal virus |
| Foamono1 | 14,297 | 11 | Frankliniella occidentalis associated mononegavirales virus 1 | Thrips virus |
FIG 13.
Small RNA read length distributions on selected viruses.
DISCUSSION
Insects were among the first organisms that were recognized as holobionts, particularly since some of them have specific organs that accommodate bacterial obligatory symbionts (26–28). The mutualistic symbiotic relationship between insects and a subset of their microbiota can be harnessed, resulting in detrimental effects on insect fitness or in their vectoring capacity. A striking example of a translational application of such approach is the recently reported successful artificial infection of the hemipteran planthopper N. lugens with a Wolbachia strain (wStri) from Laodelphax striatellus (Fallén): the new bacterial association with the insect host inhibited both infection and transmission of rice rugged stunt virus, therefore offering a clear proof of concept for a new strategy to prevent plant virus infection through alteration of the insect vector microbiota (11). Such a possible approach in agriculture follows previous successful approaches to interfere with mosquito-borne arbovirus (particularly dengue flavivirus) through Wolbachia-based population replacement or population suppression strategies (29–31).
In this respect, the microbiota associated with insects is not limited to bacteria, and insect-specific viruses (ISV) are also persistently present in insect populations (32, 33). The possibility of exploiting this specific interaction to prevent arbovirus transmission (again, mostly of human-infecting flaviviruses) has been envisioned and has been shown to be based on superinfection exclusion (34); an example of successful interference with transmission is that of the ISV Nhumirim virus blocking West Nile virus transmission in mosquitoes (35). These premises encouraged us to pursue the first step (virome characterization) of a similar strategy for the containment of thrips and the viruses they vector because of the need for new approaches to limit their great direct and indirect economical damage (36).
The different genomic components of the thrips holobiont are beginning to be unveiled: recently, the genome of WFT and that of the melon thrips (T. palmi) were characterized (37, 38); the bacteria associated with thrips were also partially characterized in the case of WFT (39–41). Regarding the viral component of the thrips microbiome, knowledge is still very limited: a recent work described viruses associated with soybean thrips samples from various U.S. regions (16); furthermore, four genomovirus genomes were amplified from Echinothrips americanus (Morgan), a pest thrips sampled in Florida (42). Here, we investigated the virome associated with two important thrips vectors, WFT and OT, with samples collected mostly in Italy, where WFT is an invasive species of Nearctic origin introduced in the late 1980s (43), whereas OT is endemic to the Mediterranean basin (7).
Considering important vectors of plant pathogens (bacteria, phytoplasmas, and viruses), the viromes associated with some important species have been reported (19, 44–47).
Our work includes samples mostly from Italy (with only two samples from the United States and one from Jordan), and this allowed us to show variation in resident viromes even in a relatively small geographic area (samples inside Italy were collected less than 1,000 km apart). We found more viral species in T. tabaci (even if with a smaller number of samples), and this could be due to bottlenecks associated with the recent invasion of the WFT that could explain its lower diversity than of an endemic species (OT); nevertheless, we cannot see the dramatic reduction of viruses associated with other invasive species sampled in newly invaded areas (19, 48).
Our work describes new viruses that are diverse enough from existing viruses to warrant new higher-rank taxonomic recognition: a first example is the different genomic organization of the mesoni-like viruses characterized from thrips in comparison with those previously characterized from other insects (18).
Another interesting new virus deserving further taxonomic attention is most closely related to the recently characterized orfanplasmoviruses (17) tentatively named insect metagenomics narna-like virus 2: the virus we found was very abundant in two WFT populations from Piedmont and Sicily. We could find for the first time a second RNA associated with the orfanplasmo-like virus through a further search in the ORFan protein-coding segments from our libraries. However, we searched also Plasmopara viticola assemblies from our previous work (17) by BLAST using as a query the putative protein encoded by RNA2, but we were not able to retrieve homologues, suggesting that they are too distant to be retrieved by homology searches (a contention supported by the little similarity also present at the RdRP level). The presence of narna-like viruses in metazoans (other than in fungi and apicomplexan), including insects, was recently confirmed (49–51).
Even if we cannot exclude that the mito-like and narna-like viruses we included as likely insect-associated viruses could be indeed fungal viruses, their relatively high abundance in some samples and the very low abundance of true mycoviruses and fungal reads suggest that they could indeed be entomoviruses.
The tombus/sobemo-like viruses we identified are also new entomoviruses belonging to clades that need a taxonomic classification and that have not been characterized biologically or ecologically.
The same lack of biological characterization is also true for the numerous virga-like viruses from insects that constitute new virus clades; one of them (Foavirga2) is present in our laboratory-reared populations, allowing us to attempt a more complete biological characterization in the near future. The negev-like viruses we found are instead a completely new virus clade representing a distinct phylogenetic branch, possibly thrips specific, that requires taxonomic accommodation. A negev-like virus was also found in the virome of soybean thrips (16), but the genome is much shorter (9.3 kb, compared to over 14 kb) than those of the WFT-infecting negev-like viruses.
The jingmen-like virus for which we characterized two segments belongs to a well-established clade of insect-infecting multipartite viruses (opposed to the original tick-infecting jingmen viruses): the best characterized of these viruses is the multisegmented Guaico culex virus (52).
Insect-infecting negative-strand viruses are very common and have been well characterized from many insect taxonomic groups (15). Among those putatively infecting thrips, a knowledge gap that needs to be filled is related to the characterization of the putative peribunya-like virus from F. occidentalis, a virus that is persistent in a population we reared in a controlled environment for many years and that can be found in populations in the field. All the closely related viruses were characterized from metagenomic studies: in fact, only two segments were identified, and a putative nucleocapsid protein was never identified, not even for Ixodes scapularis bunyavirus, a virus that was characterized from a tick cell line (53). Lack of a nucleocapsid is unlikely, since in Bunyavirales the nucleocapsid plays a role in replication.
Other expanding groups of negative-strand RNA viruses are those in the order Muvirales (qin-like viruses) and in the order Goujianvirales (yue-like viruses). To our knowledge, none of the qin-like or yue-like insect viruses has been characterized biologically, and given the relatively high accumulation in some thrips populations, we could suggest thrips as a good model system to study them.
The only DNA virus we found in our thrips metatranscriptome is a densovirus-like sequence that was always associated with some WFT populations in Italy. This virus is basal to a still-not-well-characterized group of insect-infecting denso-like viruses, among which the best characterized is the Diaphorina citri densovirus (54). We could not find evidence of the presence of an ssDNA genomovirus-like sequence as shown instead for E. americanus (42).
In general, our work has shown that viruses infecting thrips are mostly species specific. In fact, even if the populations of WFT and OT we sampled are in some cases geographically very close (a few kilometers apart), there is only limited, unconfirmed evidence of common viruses between the two species (as an example, the Foamesoni1 in WFT populations T1, T4, and T11 and OT populations T9 and T12). Also, we can for the first time show a regional specificity of viromes in a thrips species. Some of these viruses are likely part of a “core virome,” since we can see their high titer after rearing in controlled environment over time, as in the case of Foperi2, Foadenso1, and Foavirga2. All three viruses were present in a population from pepper from Piedmont which we have used in a number of leaf disk assays in 2012-2013 (55, 56) and that are still present in the same area, after almost 10 years.
We developed an interactive map available at https://thrips-virusmap.herokuapp.com/ as a webpage or at https://hub.docker.com/repository/docker/chiapellom/virusmap as a docker image that allows one to see the geographical distribution of each virus in our set of samples over time. We also have preliminary evidence that some viruses are maintained over time in populations reared in controlled environments, whereas some other are lost: in analogy to what has been demonstrated for Aedes species (57), we also can hypothesize the more constant presence of cryptic viruses that can occasionally be integrated by pathogenic viruses.
The value of insect virome characterization through next-generation sequencing (NGS) for surveillance of viruses of vertebrates and plants potentially threatening their health has been underlined previously (16): in our case, we also found evidence of plant viruses of interest, but none of them is new for the locations monitored, in contrast to new orthotospovirus and new tenuivirus found in soybean thrips samples (16). The presence of putative mycoviruses in our samples can be due to fungi being part of the thrips-associated microbiome (no specific study has investigated fungi in thrips), or to the presence of entomopathogenic fungal species in some individuals, but since the discovery that also some partiti-like sequences can directly infect insects (58), we cannot disregard the possibility that some of them are indeed true thrips viruses.
Given the recent publications of the virome associated with another thrips species (ST) from populations in 8 states in the United States in 20 different locations (16), we can compare the results: given the different size of the area surveyed and the number of samples for a total of more than 15,000 individual thrips in the ST study and around 4,000 in our 2018 survey, the total numbers of virus contigs identified in the two studies are similar (181 contigs for ST and 95 in our study). Nevertheless, the only common viral sequence is indeed related to the single sample of ST in our study, where the same sobemo-like sequence was identified; we associated with this virus a second RNA, which escaped detection in the work from Thekke-Vetil and collaborators (16). Therefore, even in this case, we have no evidence of cross-species infection among three distinct species: in the comparison with ST, the distant geographical area surveyed could also play a role. Nevertheless, some commonalities among the thrips viromes so far characterized can be drawn: in fact, we also have more plus-strand RNA viruses than minus-strand and dsRNA viruses, and, finally, the same class of DNA virus is the least represented one (denso-like viruses). Furthermore, phylogenetic analysis shows that some clades are indeed enriched of thrips viruses, showing some commonalities among the different viruses infecting thrips possibly resulting in thrips-specific clades.
A number of studies have shown different competence in tospovirus transmission comparing different populations of the same vector species (59–62), pointing to great variability that likely has a strong genetic component as previously shown both in the thrips vector (63–65) and the tospovirus (55, 66). An additional hypothesis is that interactions with a resident virome could also play an important role in different vectoring capacity: in fact, the coinfection of two viruses in the same insect could be synergistic, antagonistic, or neutral. Both antagonistic and synergistic relationships can result in lower tospovirus transmission, either because of direct interference with tospovirus accumulation (antagonism) or because synergism can tip the balance of neutrality of single infection toward detrimental effects and population collapse when one or both viruses increase their titer considerably. A greater titer in itself might not explain specific local interactions as recently reported for TSWV-OT interaction (67). Other finely tuned molecular interactions playing different roles in distinct insect immune pathways could also come into play, as recently shown for Argentine ants (68), and this should be studied on a case-by-case virus-virus interaction.
MATERIALS AND METHODS
Preliminary field collection of thrips populations in 2018.
An initial sampling effort was organized in Italy and, to a lesser extent, in the United States during summer 2018. Thrips populations were sampled in seven Italian regions (Piedmont, Veneto, Liguria, Emilia Romagna, Campania, Puglia, and Sicily) spanning from north to south and from west to east. Thrips populations (200 to 250 individuals) were collected from a variety of crops: tomato (2 samples), leek (2 samples), onion (2 samples), pepper (2 samples), strawberry (2 samples), white African daisy/Dimorphoteca sp. (1 sample), and watermelon (1 sample). Sampling was performed by scouting flowers and leaves on a white tray (250 by 350 mm) and transferring the fallen adults into glass vials (diameter, 24 mm; length, 120 mm) with an insect aspirator. In some cases, the whole plants were collected in plastic bags and further processed in the laboratory as described above. In total, 12 individual field populations were collected (Table 4). A subsample of 30 adult thrips belonging to each population was examined under a stereomicroscope for their identification (OT or WFT). Two populations were sampled from the United States (pool T-ame): one from a field (ST) and one from a greenhouse (WFT).
TABLE 4.
Sample collection
| Sample | Thrips species | Plant host | Region | Pool |
|---|---|---|---|---|
| T4 | F. occidentalis | Watermelon | Emilia Romagna | THR-A |
| T6 | F. occidentalis | Tomato | Campania | THR-A |
| T10 | F. occidentalis | Tomato | Puglia | THR-A |
| T5 | F. occidentalis/Thrips tabaci | Onion | Veneto | THR-B |
| T8 | Thrips tabaci | Onion | Piedmont | THR-B |
| T1 | F. occidentalis | Pepper | Piedmont | THR-C |
| T2 | F. occidentalis | Strawberry | Veneto | THR-C |
| T3 | F. occidentalis | Pepper | Veneto | THR-D |
| T7 | F. occidentalis | Pepper | Sicily | THR-D |
| T11 | F. occidentalis | Dimorphoteca | Liguria | THR-D |
| T9 | Thrips tabaci | Leek | Liguria | THR-E |
| T12 | Thrips tabaci | Leek | Piedmont | THR-E |
| T13 | Neohydatothrips variabilis | Green bean | USA (lab) | T-ame |
| T14 | F. occidentalis/Thrips tabaci | Soybean | USA (field) | T-ame |
Field collection and laboratory rearing of selected thrips populations in 2019-2020.
In summer 2019 and 2020, thrips populations (500 to 1,000 individuals) were collected in five locations corresponding to five samples from 2018—T1 (WFT, pepper, Piedmont), T4 (WFT, watermelon, Emilia-Romagna), T9 (OT, leek, Liguria), T11 (WFT, Dimorphoteca, Liguria), and T12 (OT, leek, Piedmont)—and a new one was added from Jordan (OT, onion, Jordan Valley). Sampling was performed as described above. All field-collected adults were cold anesthetized and identified under a stereomicroscope; only adults that belonged to the species F. occidentalis or T. tabaci were used for further analyses and mass rearing. To maintain populations in purity in a controlled environment, mass rearing was established on a subset of these populations, starting from at least 300 adults for every field-collected population. Laboratory populations were reared on green pea pods, Pisum sativum (L.), inside 1-liter glass jars in growth chambers at 20 to 26°C with a 16-h photoperiod. Every 2 to 3 days some green pea pods were replaced, and granulated pollen was regularly added as a dietary supplement.
Total RNA extraction and sample pooling.
Total RNA from each thrips population was extracted using TRIzol as previously described (55). RNA extracts were quantified with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA). To reduce sequencing costs, the RNA extracted from individual populations was pooled to have five final samples with the same final concentration (7 ng/μl).
RNA sequencing and bioinformatics pipeline.
After rRNA depletion with Ribo-Zero Gold human/mouse/rat (Epicentre Biotechnologies, Madison, WI), cDNA library preparation (Illumina Inc., San Diego, CA; TrueSeq Stranded) and sequencing were carried out by Macrogen Inc. (Seoul, Republic of Korea).
Bioinformatics workflow was divided into five main steps: (i) read quality check and filtering, (ii) assembly of clean reads into contigs, (iii) identification of viral sequences, (iv) “uni-contig” production, and (v) mapping of reads on the viral genomes. Step i was performed using BBTools (v 38.70) (69), in order to remove Illumina adaptor sequences and artifacts and short and residual ribosomal sequences. In step ii, clean reads were used as input for Trinity software (v 2.3.2) (70) for de novo assembly. In step iii, DIAMOND (v 0.9.21.122) (71) was used to perform a match between assembled contigs and a custom viral database using BLASTx (taxonomy identifiers for viruses used to create the custom viral database are txid1925802, txid1921431, txid1917979, txid1915204, txid1922240, txid2732900, txid2732416, and txid2732396). After visual inspection of alignments obtained with BLASTx, selected candidate viral contigs were aligned by BLAST against the NCBI nonredundant protein database (version September 2020) to discriminate between already discovered viruses, host sequences present in viral genomes, and true new virus sequences. CAP3 (default parameters) (72) was used to further assemble selected contigs. Step iv aimed to reduce the redundancy of viruses present in different libraries. After concatenating the contigs from all libraries, individual contigs from each library were aligned by BLAST against it. Results were processed, and contigs with identity over 90% and length over 1,000 nucleotides were grouped and considered a single virus contig in our final list. The longest contig of each group (named “uni-contig”) was selected as the representative contig and deposited in NCBI. Uni-contig selection was performed by custom R scripts. Step v was performed using bowtie2 (73) and a custom R script.
ORFan contig detection.
Each assembled library was aligned with DIAMOND against the NCBI nonredundant whole database (version September 2020). Every contig with a significative BLAST hit was discarded, while the contigs without an NCBInr BLAST hit, longer than 1 kbp, and encoding a protein of at least 15 kDa were kept. This set of contigs defined the “ORFan” sequences. ORFan contigs were mapped with reads considering their orientation, and contigs that showed only positive or only negative reads were discarded, since a typical feature of replicating viruses is the presence of both minus- and plus-sense genomic template for replication (17).
Genome organization.
The NCBI ORF finder tool (https://www.ncbi.nlm.nih.gov/orffinder/) was used for open reading frame prediction, while MOTIF Search (https://www.genome.jp/tools/motif) was used for functional domain search. Default parameters were applied for both software.
Taxonomic analysis of the metasample.
In order to evaluate the taxonomic complexity of the metasamples present in our libraries, we used Kraken2 (74) in combination with Pavian (75) and a custom R script to create visualization plots.
Phylogenetic analysis of viral sequences.
RNA-dependent RNA polymerase (RdRP) proteins from all identified viruses and at least the closest 10 homologues from NCBI databases were used for phylogenetic analysis. Representative viruses, based on International Committee on Taxonomy of Viruses (ICTV) classification, were also added when needed to support the subgroups in the phylogenetic tree. The phylogenetic analysis was performed using ggtree R package (76). Msa R package (10.18129/B9.bioc.msa) was used to align the sequences using the MAFFT algorithm with default parameters (77). The RdRP alignments were then processed by IQ-TREE software (78) to obtain phylogenetic trees using the default parameters (combine ModelFinder, tree search, ultrafast bootstrap, and SH-aLRT test) and the maximum likelihood (ML) model. Treeio and ggtree R packages were used for tree data import, manipulation, and display (76, 79). The accession numbers of the proteins and the corresponding virus names and acronyms are displayed on the trees.
Virus names.
Viruses identified in this paper have been named using the following criteria: (i) the first part of the name is the main taxon present in the metasample source of the virus, or when such assignment is not reconfirmed by further more specific assays, the term “insect associated” was used; (ii) the second part of the name identifies the virus taxonomic group, if the virus taxonomic assignment was clear; and (iii) the last part of the name is a sequential number. For example, the contig THR-A_DN23655 has as first match Artashat orthonairovirus and the corresponding virus was named Frankliniella occidentalis associated (part i) nairo-like virus (part ii) 1 (part iii).
Association between thrips viruses and specific thrips samples by qRT-PCR.
Real-time reverse transcription-quantitative PCR (qRT-PCR) analyses with virus-specific primers (see Table S1 in the supplemental material) were performed to associate any specific virus assembled in silico with the original specific RNA sample (a cohort of five thrips/sample). DNA copies were produced using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific, Waltham, MA) following manufacturer instructions. qRT-PCR was performed using a CFX Connect real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) and iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories). The same primer pairs specific for each virus contig were used in qPCR to check for possible endogenization events using total nucleic acids as the template.
Small RNA high-throughput sequencing from a WFT population.
A population of WFT, initially sampled from pepper in Piedmont in 2014, from the same field corresponding to sample T1, was maintained in purity until 2017. Total RNA was extracted as described above, and small RNAs were purified and sequenced by the BGI Group company as described previously (80). Reads were mapped to the viral contigs resulting from our virome characterization as described above. Size distribution of the sRNA reads was displayed as described previously (80).
Data availability.
All raw reads have been deposited in the Sequence Read Archive (SRA) under BioProject number PRJNA637687 and BioSample numbers SAMN15150576 to SAMN15150580. All the insect-associated viral contigs have been deposited in GenBank under accession numbers MN714662 to MN714667, MN714669 to MN714672, MN714674 to MN714683, MN714686 to MN714690, MN725049 to MN725052, MN764138 to MN764161, MN787040 to MN787042, MW297844 to MW297846, MW459158, and MW459159 (Table 2). Script and fasta files of clean small RNA reads are available at the following link: https://bit.ly/3m9Yw5Q.
ACKNOWLEDGMENTS
We extend grateful thanks to Andrea Delliri for his technical assistance and to Giovanni Zanini and Teresa Cosmi from the Phytosanitary Inspection Service of Veneto region for helping to identify thrips populations in their region.
This work was fully supported by VIROPLANT, a project that received funding from the European Union’s Horizon H2020 Research and Innovation Program, grant agreement number 773567.
Footnotes
Supplemental material is available online only.
Contributor Information
M. Turina, Email: massimo.turina@ipsp.cnr.it.
Julie K. Pfeiffer, University of Texas Southwestern Medical Center
REFERENCES
- 1.Buckman RS, Mound LA, Whiting MF. 2013. Phylogeny of thrips (Insecta: Thysanoptera) based on five molecular loci. Syst Entomol 38:123–133. 10.1111/j.1365-3113.2012.00650.x. [DOI] [Google Scholar]
- 2.Lewis T. 1973. Thrips, their biology, ecology and economic importance. Academic Press, New York, NY. [Google Scholar]
- 3.Riley DG, Joseph SV, Srinivasan R, Diffie S. 2011. Thrips vectors of tospoviruses. J Integr Pest Manag 2:I1–I10. 10.1603/IPM10020. [DOI] [Google Scholar]
- 4.Rotenberg D, Whitfield AE. 2018. Molecular interactions between tospoviruses and thrips vectors. Curr Opin Virol 33:191–197. 10.1016/j.coviro.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Goldbach R, Peters D. 1996. Molecular and biological aspects of tospoviruses, p 129–157. In Elliot RM (ed), The Bunyaviridae. Plenum Press, New York, NY. [Google Scholar]
- 6.Mound LA. 2002. So many thrips-so few tospoviruses, p 15–18. In Thrips and tospoviruses: proceedings of the 7th International Symposium on Thysanoptera. Australian National Insect Collection, Canberra, Australia. [Google Scholar]
- 7.Turina M, Tavella L, Ciuffo M. 2012. Tospoviruses in the Mediterranean area. Adv Virus Res 84:403–437. 10.1016/B978-0-12-394314-9.00012-9. [DOI] [PubMed] [Google Scholar]
- 8.Kirk WDJ, Terry LI. 2003. The spread of western flower thrips Frankliniella occidentalis (Pergande). Agric Forest Ent 5:301–310. 10.1046/j.1461-9563.2003.00192.x. [DOI] [Google Scholar]
- 9.Turina M, Kormelink R, Resende RO. 2016. Resistance to tospoviruses in vegetable crops: epidemiological and molecular aspects. Annu Rev Phytopathol 54:347–371. 10.1146/annurev-phyto-080615-095843. [DOI] [PubMed] [Google Scholar]
- 10.Qadri M, Short S, Gast K, Hernandez J, Wong AC-N. 2020. Microbiome innovation in agriculture: development of microbial based tools for insect pest management. Front Sustain Food Syst 4:547751. 10.3389/fsufs.2020.547751. [DOI] [Google Scholar]
- 11.Gong J-T, Li Y, Li T-P, Liang Y, Hu L, Zhang D, Zhou C-Y, Yang C, Zhang X, Zha S-S, Duan X-Z, Baton LA, Hong X-Y, Hoffmann AA, Xi Z. 2020. Stable introduction of plant-virus-inhibiting Wolbachia into planthoppers for rice protection. Curr Biol 30:4837–4845.e5. 10.1016/j.cub.2020.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Gurung K, Wertheim B, Salles JF. 2019. The microbiome of pest insects: it is not just bacteria. Entomol Exp Appl 167:156–170. 10.1111/eea.12768. [DOI] [Google Scholar]
- 13.Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378. 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X, Qin X-C, Li J, Cao J-P, Eden J-S, Buchmann J, Wang W, Xu J, Holmes EC, Zhang Y-Z. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 15.Käfer S, Paraskevopoulou S, Zirkel F, Wieseke N, Donath A, Petersen M, Jones TC, Liu S, Zhou X, Middendorf M, Junglen S, Misof B, Drosten C. 2019. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog 15:e1008224. 10.1371/journal.ppat.1008224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thekke-Veetil T, Lagos-Kutz D, McCoppin NK, Hartman GL, Ju H-K, Lim H-S, Domier LL. 2020. Soybean thrips (Thysanoptera: Thripidae) harbor highly diverse populations of arthropod, fungal and plant viruses. Viruses 12:1376. 10.3390/v12121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiapello M, Rodriguez-Romero J, Ayllon MA, Turina M. 2020. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol 6:veaa058. 10.1093/ve/veaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zirkel F, Roth H, Kurth A, Drosten C, Ziebuhr J, Junglen S. 2013. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J Virol 87:6346–6358. 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ottati S, Chiapello M, Galetto L, Bosco D, Marzachi C, Abba S. 2020. New viral sequences identified in the Flavescence doree phytoplasma vector Scaphoideus titanus. Viruses 12:287. 10.3390/v12030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remnant EJ, Shi M, Buchmann G, Blacquiere T, Holmes EC, Beekman M, Ashe A. 2017. A diverse range of novel RNA viruses in geographically distinct honey bee populations. J Virol 91:e00158-17. 10.1128/JVI.00158-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumura EE, Nerva L, Nigg JC, Falk BW, Nouri S. 2016. Complete genome sequence of the largest known flavi-like virus, Diaphorina citri flavi-like virus, a novel virus of the Asian citrus psyllid, Diaphorina citri. Genome Announc 4:e00946-16. 10.1128/genomeA.00946-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viljakainen L, Holmberg I, Abril S, Jurvansuu J. 2018. Viruses of invasive Argentine ants from the European Main supercolony: characterization, interactions and evolution. J Gen Virol 99:1129–1140. 10.1099/jgv.0.001104. [DOI] [PubMed] [Google Scholar]
- 23.Qin X-C, Shi M, Tian J-H, Lin X-D, Gao D-Y, He J-R, Wang J-B, Li C-X, Kang Y-J, Yu B, Zhou D-J, Xu J, Plyusnin A, Holmes EC, Zhang Y-Z. 2014. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc Natl Acad Sci USA 111:6744–6749. 10.1073/pnas.1324194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh M-C. 2013. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol 14:396–403. 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 25.Turina M, Ghignone S, Astolfi N, Silvestri A, Bonfante P, Lanfranco L. 2018. The virome of the arbuscular mycorrhizal fungus Gigaspora margarita reveals the first report of DNA fragments corresponding to replicating non-retroviral RNA viruses in fungi. Environ Microbiol 20:2012–2025. 10.1111/1462-2920.14060. [DOI] [PubMed] [Google Scholar]
- 26.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 27.Zakharov IA, Goryacheva II. 2020. Hereditary symbionts: genomic integration. Russ J Genet 56:639–654. 10.1134/S1022795420060125. [DOI] [Google Scholar]
- 28.Gil R, Latorre A. 2019. Unity makes strength: a review on mutualistic symbiosis in representative insect clades. Life (Basel) 9:21. 10.3390/life9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu GJ, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O’Neill SL. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139:1268–1278. 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 30.Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. 2008. Wolbachia and virus protection in insects. Science 322:702. 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, Greenfield M, Durkan M, Leong YS, Dong Y, Cook H, Axford J, Callahan AG, Kenny N, Omodei C, McGraw EA, Ryan PA, Ritchie SA, Turelli M, O’Neill SL. 2011. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476:454–457. 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 32.Bolling BG, Weaver SC, Tesh RB, Vasilakis N. 2015. Insect-specific virus discovery: significance for the arbovirus community. Viruses 7:4911–4928. 10.3390/v7092851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nouri S, Matsumura EE, Kuo Y-W, Falk BW. 2018. Insect-specific viruses: from discovery to potential translational applications. Curr Opin Virol 33:33–41. 10.1016/j.coviro.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Laureti M, Paradkar PN, Fazakerley JK, Rodriguez-Andres J. 2020. Superinfection exclusion in mosquitoes and its potential as an arbovirus control strategy. Viruses 12:1259. 10.3390/v12111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goenaga S, Kenney JL, Duggal NK, Delorey M, Ebel GD, Zhang B, Levis SC, Enria DA, Brault AC. 2015. Potential for co-infection of a mosquito-specific flavivirus, Nhumirim virus, to block West Nile virus transmission in mosquitoes. Viruses 7:5801–5812. 10.3390/v7112911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pappu HR, Jones RAC, Jain RK. 2009. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res 141:219–236. 10.1016/j.virusres.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Rotenberg D, Baumann AA, Ben-Mahmoud S, Christiaens O, Dermauw W, Ioannidis P, Jacobs CGC, Vargas Jentzsch IM, Oliver JE, Poelchau MF, Rajarapu SP, Schneweis DJ, Snoeck S, Taning CNT, Wei D, Widana Gamage SMK, Hughes DST, Murali SC, Bailey ST, Bejerman NE, Holmes CJ, Jennings EC, Rosendale AJ, Rosselot A, Hervey K, Schneweis BA, Cheng S, Childers C, Simão FA, Dietzgen RG, Chao H, Dinh H, Doddapaneni HV, Dugan S, Han Y, Lee SL, Muzny DM, Qu J, Worley KC, Benoit JB, Friedrich M, Jones JW, Panfilio KA, Park Y, Robertson HM, Smagghe G, Ullman DE, van der Zee M, Van Leeuwen T, Veenstra JA, et al. 2020. Genome-enabled insights into the biology of thrips as crop pests. BMC Biol 18:142. 10.1186/s12915-020-00862-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo SK, Cao LJ, Song W, Shi P, Gao YF, Gong YJ, Chen JC, Hoffmann AA, Wei SJ. 2020. Chromosome-level assembly of the melon thrips genome yields insights into evolution of a sap-sucking lifestyle and pesticide resistance. Mol Ecol Resour 20:1110–1125. 10.1111/1755-0998.13189. [DOI] [PubMed] [Google Scholar]
- 39.de Vries EJ, Jacobs G, Breeuwer JAJ. 2001. Growth and transmission of gut bacteria in the western flower thrips, Frankliniella occidentalis. J Invertebr Pathol 77:129–137. 10.1006/jipa.2001.5010. [DOI] [PubMed] [Google Scholar]
- 40.De Vries EJ, Van De Wetering F, Van Der Hoek MM, Jacobs G, Breeuwer JAJ. 2012. Symbiotic bacteria (Erwinia sp.) in the gut of Frankliniella occidentalis (Thysanoptera: Thripidae) do not affect its ability to transmit tospovirus. Eur J Entomol 109:261–266. 10.14411/eje.2012.034. [DOI] [Google Scholar]
- 41.Facey PD, Meric G, Hitchings MD, Pachebat JA, Hegarty MJ, Chen XR, Morgan LVA, Hoeppner JE, Whitten MMA, Kirk WDJ, Dyson PJ, Sheppard SK, Del Sol R. 2015. Draft genomes, phylogenetic reconstruction, and comparative genomics of two novel cohabiting bacterial symbionts isolated from Frankliniella occidentalis. Genome Biol Evol 7:2188–2202. 10.1093/gbe/evv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraberger S, Polston JE, Capobianco HM, Alcala-Briseno RI, Fontenele RS, Varsani A. 2017. Genomovirus genomes recovered from Echinothrips americanus sampled in Florida, USA. Genome Announc 5:e00445-17. 10.1128/genomeA.00445-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reitz SR, Gao YL, Kirk WDJ, Hoddle MS, Leiss KA, Funderburk JE. 2020. Invasion biology, ecology and management of western flower thrips. Annu Rev Entomol 65:17–37. 10.1146/annurev-ento-011019-024947. [DOI] [PubMed] [Google Scholar]
- 44.Nouri S, Salem N, Nigg JC, Falk BW. 2015. Diverse array of new viral sequences identified in worldwide populations of the Asian citrus psyllid (Diaphorina citri) using viral metagenomics. J Virol 90:2434–2445. 10.1128/JVI.02793-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosario K, Capobianco H, Ng TFF, Breitbart M, Polston JE. 2014. RNA viral metagenome of whiteflies leads to the discovery and characterization of a whitefly-transmitted carlavirus in North America. PLoS One 9:e86748. 10.1371/journal.pone.0086748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wamonje FO, Michuki GN, Braidwood LA, Njuguna JN, Mutuku JM, Djikeng A, Harvey JJW, Carr JP. 2017. Viral metagenomics of aphids present in bean and maize plots on mixed-use farms in Kenya reveals the presence of three dicistroviruses including a novel Big Sioux River virus-like dicistrovirus. Virol J 14:188. 10.1186/s12985-017-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kondo H, Fujita M, Hisano H, Hyodo K, Andika IB, Suzuki N. 2020. Virome analysis of aphid populations that infest the barley field: the discovery of two novel groups of Nege/Kita-like viruses and other novel RNA viruses. Front Microbiol 11:509. 10.3389/fmicb.2020.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medd NC, Fellous S, Waldron FM, Xuereb A, Nakai M, Cross JV, Obbard DJ. 2018. The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus Evol 4:vey009. 10.1093/ve/vey009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goertz GP, Miesen P, Overheul GJ, van Rij RP, van Oers MM, Pijlman GP. 2019. Mosquito small RNA responses to West Nile and insect-specific virus infections in Aedes and Culex mosquito cells. Viruses 11:271. 10.3390/v11030271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richaud A, Frezal L, Tahan S, Jiang H, Blatter JA, Zhao G, Kaur T, Wang D, Felix M-A. 2019. Vertical transmission in Caenorhabditis nematodes of RNA molecules encoding a viral RNA-dependent RNA polymerase. Proc Natl Acad Sci USA 116:24738–24747. 10.1073/pnas.1903903116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandler JA, Liu RM, Bennett SN. 2015. RNA shotgun metagenomic sequencing of northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Front Microbiol 6:185. 10.3389/fmicb.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ladner JT, Wiley MR, Beitzel B, Auguste AJ, Dupuis AP, II, Lindquist ME, Sibley SD, Kota KP, Fetterer D, Eastwood G, Kimmel D, Prieto K, Guzman H, Aliota MT, Reyes D, Brueggemann EE, John LS, Hyeroba D, Lauck M, Friedrich TC, O'Connor DH, Gestole MC, Cazares LH, Popov VL, Castro-Llanos F, Kochel TJ, Kenny T, White B, Ward MD, Loaiza JR, Goldberg TL, Weaver SC, Kramer LD, Tesh RB, Palacios G. 2016. A multicomponent animal virus isolated from mosquitoes. Cell Host Microbe 20:357–367. 10.1016/j.chom.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakao R, Matsuno K, Qiu Y, Maruyama J, Eguchi N, Nao N, Kajihara M, Yoshii K, Sawa H, Takada A, Sugimoto C. 2017. Putative RNA viral sequences detected in an Ixodes scapularis-derived cell line. Ticks Tick Borne Dis 8:103–111. 10.1016/j.ttbdis.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Nigg JC, Falk BW. 2020. Diaphorina citri densovirus is a persistently infecting virus with a hybrid genome organization and unique transcription strategy. J Gen Virol 101:226–239. 10.1099/jgv.0.001371. [DOI] [PubMed] [Google Scholar]
- 55.Margaria P, Bosco L, Vallino M, Ciuffo M, Mautino GC, Tavella L, Turina M. 2014. The NSs protein of tomato spotted wilt virus is required for persistent infection and transmission by Frankliniella occidentalis. J Virol 88:5788–5802. 10.1128/JVI.00079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mautino GC, Sacco D, Ciuffo M, Turina M, Tavella L. 2012. Preliminary evidence of recovery from Tomato spotted wilt virus infection in Frankliniella occidentalis individuals. Ann Appl Biol 161:266–276. 10.1111/j.1744-7348.2012.00571.x. [DOI] [Google Scholar]
- 57.Shi CY, Zhao L, Atoni E, Zeng WF, Hu XM, Matthijnssens J, Yuan ZM, Xia H. 2020. Stability of the virome in lab- and field-collected Aedes albopictus mosquitoes across different developmental stages and possible core viruses in the publicly available virome data of Aedes mosquitoes. mSystems 5:e00640-20. 10.1128/mSystems.00640-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cross ST, Maertens BL, Dunham TJ, Rodgers CP, Brehm AL, Miller MR, Williams AM, Foy BD, Stenglein MD. 2020. Partitiviruses infecting Drosophila melanogaster and Aedes aegypti exhibit efficient biparental vertical transmission. J Virol 94:e01070-20. 10.1128/JVI.01070-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacobson AL, Booth W, Vargo EL, Kennedy GG. 2013. Thrips tabaci population genetic structure and polyploidy in relation to competency as a vector of tomato spotted wilt virus. PLoS One 8:e54484. 10.1371/journal.pone.0054484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacobson AL, Kennedy GG. 2013. Specific insect-virus interactions are responsible for variation in competency of different Thrips tabaci isolines to transmit different tomato spotted wilt virus isolates. PLoS One 8:e54567. 10.1371/journal.pone.0054567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van de Wetering F, van der Hoek M, Goldbach R, Mollema C, Peters D. 1999. Variation in tospovirus transmission between populations of Frankliniella occidentalis (Thysanoptera: Thripidae). Bull Entomol Res 89:579–588. 10.1017/S0007485399000735. [DOI] [Google Scholar]
- 62.Wetering F, Hoek M, Goldbach R, Peters D. 1999. Differences in tomato spotted wilt virus vector competency between males and females of Frankliniella occidentalis. Entomol Exp Appl 93:105–112. 10.1046/j.1570-7458.1999.00567.x. [DOI] [Google Scholar]
- 63.Rosa J, Kennedy GG. 2007. Thrips tabaci and tomato spotted wilt virus: inheritance of vector competence. Entomol Exp Appl 124:161–166. 10.1111/j.1570-7458.2007.00564.x. [DOI] [Google Scholar]
- 64.Halaweh N, Poehling HM. 2009. Inheritance of vector competence by the thrips Ceratothripoides claratris (Shumsher) (Thysanoptera: Thripidae). J Appl Entomol 133:386–393. 10.1111/j.1439-0418.2008.01357.x. [DOI] [Google Scholar]
- 65.Ogada PA, Debener T, Poehling H-M. 2016. Inheritance genetics of the trait vector competence in Frankliniella occidentalis (Western flower thrips) in the transmission of Tomato spotted wilt virus. Ecol Evol 6:7911–7920. 10.1002/ece3.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sin SH, McNulty BC, Kennedy GG, Moyer JW. 2005. Viral genetic determinants for thrips transmission of Tomato spotted wilt virus. Proc Natl Acad Sci USA 102:5168–5173. 10.1073/pnas.0407354102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linak JA, Jacobson AL, Sit TL, Kennedy GG. 2020. Relationships of virus titers and transmission rates among sympatric and allopatric virus isolates and thrips vectors support local adaptation. Sci Rep 10:7649. 10.1038/s41598-020-64507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lester PJ, Buick KH, Baty JW, Felden A, Haywood J. 2019. Different bacterial and viral pathogens trigger distinct immune responses in a globally invasive ant. Sci Rep 9:5780. 10.1038/s41598-019-41843-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bushnell B, Rood J, Singer E. 2017. BBMerge—accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 72.Huang XQ, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res 9:868–877. 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Breitwieser FP, Salzberg SL. 2020. Pavian: interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 36:1303–1304. 10.1093/bioinformatics/btz715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu G, Smith D, Zhu H, Guan Y, Lam T. 2017. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 77.Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trifinopoulos J, Lam-Tung N, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Lam T, Xu S, Dai Z, Zhou L, Feng T, Guo P, Dunn C, Jones B, Bradley T, Zhu H, Guan Y, Jiang Y, Yu G. 2020. Treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Mol Biol Evol 37:599–603. 10.1093/molbev/msz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Margaria P, Miozzi L, Rosa C, Axtell MJ, Pappu HR, Turina M. 2015. Small RNA profiles of wild-type and silencing suppressor-deficient tomato spotted wilt virus infected Nicotiana benthamiana. Virus Res 208:30–38. 10.1016/j.virusres.2015.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1<br>. Download JVI.00597-21-s0001.pdf, PDF file, 0.2 MB (163.6KB, pdf)
Data Availability Statement
All raw reads have been deposited in the Sequence Read Archive (SRA) under BioProject number PRJNA637687 and BioSample numbers SAMN15150576 to SAMN15150580. All the insect-associated viral contigs have been deposited in GenBank under accession numbers MN714662 to MN714667, MN714669 to MN714672, MN714674 to MN714683, MN714686 to MN714690, MN725049 to MN725052, MN764138 to MN764161, MN787040 to MN787042, MW297844 to MW297846, MW459158, and MW459159 (Table 2). Script and fasta files of clean small RNA reads are available at the following link: https://bit.ly/3m9Yw5Q.