ABSTRACT
Zebrafish provide an excellent model for in vivo cell biology studies because of their amenability to live imaging. Protein visualization in zebrafish has traditionally relied on overexpression of fluorescently tagged proteins from heterologous promoters, making it difficult to recapitulate endogenous expression patterns and protein function. One way to circumvent this problem is to tag the proteins by modifying their endogenous genomic loci. Such an approach is not widely available to zebrafish researchers because of inefficient homologous recombination and the error-prone nature of targeted integration in zebrafish. Here, we report a simple approach for tagging proteins in zebrafish on their N or C termini with fluorescent proteins by inserting PCR-generated donor amplicons into non-coding regions of the corresponding genes. Using this approach, we generated endogenously tagged alleles for several genes that are crucial for epithelial biology and organ development, including the tight junction components ZO-1 and Cldn15la, the trafficking effector Rab11a, the apical polarity protein aPKC and the ECM receptor Integrin β1b. Our approach facilitates the generation of knock-in lines in zebrafish, opening the way for accurate quantitative imaging studies.
KEY WORDS: Zebrafish, Knock-in, Epithelial, Morphogenesis, CRISPR, Quantitative imaging
Summary: Generation of endogenously tagged stable zebrafish knock-in lines is simplified by the integration of fluorescent protein cassettes with mRNA splicing elements into non-coding regions of genes.
INTRODUCTION
Protein visualization in zebrafish has traditionally relied on overexpression approaches based on microinjection of mRNA or random integration of small tagged transgenes (Kwan et al., 2007). These approaches are limited because overexpression seldom recapitulates endogenous levels and patterns. Bacterial artificial chromosome (BAC) transgenes (Bussmann and Schulte-Merker, 2011; Navis et al., 2013; Alvers et al., 2014; Rodriguez-Fraticelli et al., 2015; Fuentes et al., 2016) can recapitulate endogenous expression if they include necessary regulatory sequences. However, regulatory sequences can reside hundreds of kilobases away from the gene of interest, preventing their inclusion on a BAC transgene. Moreover, expression levels of BAC transgenes vary depending on the genomic insertion site (Fuentes et al., 2016) and copy number (Chandler et al., 2007). The most accurate way to recapitulate physiological gene expression is to tag the gene directly at its endogenous genomic locus to produce fusion proteins (Gibson et al., 2013), an approach referred to as knock-in (KI). KI has recently become feasible in cell lines and many model organisms (Dickinson et al., 2015; Koles et al., 2016; Dewari et al., 2018; Gao et al., 2019; Cronan and Tobin, 2019). In zebrafish, homology-based methods for generating KI lines have been described (Hoshijima et al., 2016; Wierson et al., 2020; Ranawakage et al., 2021). However, the available methods rely on inefficient DNA repair pathways, which are required for the precise integration of large DNA sequences (Peng et al., 2014). As a result, only a few KI zebrafish lines have been reported. N-terminal tagging has been difficult in zebrafish, with only insertion of small peptides (Hoshijima et al., 2016; Ranawakage et al., 2021) and one reported fluorescent protein tag (Wilson et al., 2021). To circumvent these challenges, we devised a simple KI approach in which precise integration is not needed for expression of endogenously tagged proteins. We targeted non-coding regions of genes, such as introns and 5′ untranslated regions (UTRs), for integration of targeting cassettes that code for fluorescent proteins. Imprecise integration events do not affect expression of tagged proteins because non-coding sequences targeted by CRISPR-Cas9 are removed from the transcripts during RNA splicing. We used this approach to generate stable zebrafish KI lines for several proteins tagged on their N or C terminus with fluorescent proteins. KI tagging in zebrafish allows the use of quantitative imaging studies, such as quantifying endogenous protein levels. As a proof of principle, we measured the concentration of endogenous eGFP-Rab11a molecules on apical vesicles in epithelial cells.
RESULTS AND DISCUSSION
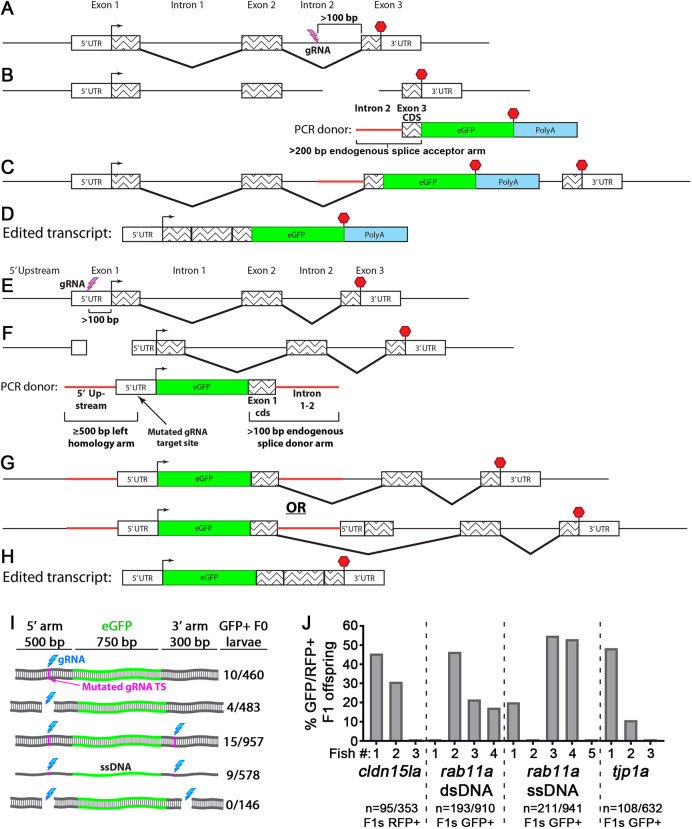
To establish KI methods in zebrafish, we sought to integrate fluorescent protein cassettes into non-coding regions of genes. For C-terminal tagging, we used CRISPR-Cas9 to induce a double strand DNA (dsDNA) break in the last intron (Fig. 1A) at least 100 base pairs (bp) upstream of the last exon to minimize RNA splicing interference (Fig. 1A). Together with Cas9 and guide RNA (gRNA), we co-injected linear dsDNA PCR donor amplicons spanning part of the gene's last intron and last exon coding sequence fused to a fluorescent protein coding sequence and polyadenylation (polyA) sequence (Fig. 1B). The intron serves as a splice acceptor element. Integration of the donor can proceed through non-homologous end joining (NHEJ) at the 5′ and 3′ ends (Fig. 1C). Expression of the modified transcript can tolerate errors, such as insertion-deletion mutations (INDELs) at integration boundaries, because these sequences are removed during RNA splicing (Fig. 1D). Because C-terminal tagging can impair function of many proteins, we also devised a strategy for N-terminal tagging by targeting non-coding regions upstream of the start codon (Fig. 1E). N-terminal PCR repair donors contain a 500 bp homology arm (the endogenous sequence upstream from the CRISPR-Cas9-induced dsDNA break), which is fused to the remaining upstream non-coding sequence, coding sequence of a fluorescent protein, coding sequence of the endogenous exon that harbors the start codon and a portion of the following intron as a splice donor element (Fig. 1F). To promote seamless integration at the 5′ end of the repair donor in the UTR, we mutated the gRNA target site in the repair donor (Fig. 1F). Integration of the 3′ end of the repair donor does not require precise insertion by homology-directed repair (HDR) because the intronic portion of the donor sequence is removed from transcripts during RNA splicing. (Fig. 1G,H).
Fig. 1.
Knock-in tagging in zebrafish using splice donor and acceptor arms. (A-D) C-terminal endogenous tagging strategy. An intron 5′ to the last exon is targeted for a dsDNA break (A) and integration of a PCR repair donor containing a 5′ splice acceptor element (B). mRNA splicing mediates expression of the tagged protein (C,D). (E-H) N-terminal endogenous tagging strategy. A non-coding region 5′ to the start codon is targeted for a dsDNA break (E) and integration of a PCR repair donor containing a 5′ homology arm and 3′ splice donor element (F). mRNA splicing mediates expression of the tagged protein (G,H). (I) One-cell-stage embryos were injected with different rab11a knock-in (KI) cocktails and visually screened. Cyan bolt, gRNA target site; magenta line, mutated PAM site in repair donor; ssDNA, single-stranded DNA. Numbers to the right indicate the proportion of GFP+ surviving larvae. (J) One-cell-stage embryos were injected with KI cocktails for different genes and then visually screened. GFP/RFP+ F0 fish were outcrossed, and F1 progeny were visually screened. Data plotted indicate the levels of mosaicism present in the F0 generation.
For some KI alleles, such as p2A-Cre, visual screening is not feasible in the absence of other markers. Therefore, we devised a slightly modified approach for inserting larger sequences. We constructed a donor plasmid in which an endogenous splice acceptor element and the coding sequence of the last exon is cloned upstream of a tag of interest followed by a polyA sequence. The splice acceptor element in the donor plasmid is targeted and linearized by the same gRNA target site as the endogenous genomic intron. Integration of the donor and expression of the modified transcript follows similar principles as described for PCR donors, but this approach allows for the addition of larger cassettes that include transgenesis markers. Insertions can be identified based on the expression of the transgenesis marker, but orientation of the insertion and expression of the tagged protein need to be confirmed by sequencing across the modified genomic region and by immunohistochemistry, respectively.
Using these approaches, we tagged several genes involved in epithelial morphogenesis. We injected KI cocktails containing Cas9 protein, gRNAs and repair donors into one-cell-stage embryos and visually screened embryos for fluorescence, comparing them with reported expression patterns of the endogenous transcripts (Howe et al., 2021). We consistently observed expression of the fusion proteins in 1-5% of the injected embryos. Injected embryos with expression showed varying levels of mosaicism (Fig. S1A-D), which, combined with restricted subcellular localization and endogenous levels of some proteins like aPKC, required us to use live confocal microscopy to identify embryos with KI insertions (Fig. S1A-C).
To optimize conditions for KI in zebrafish, we focused on rab11a because this gene exhibits widespread expression and KI insertion can be scored by visual screening (Fig. S1D). We prepared dsDNA or single-stranded DNA (ssDNA) repair donors for rab11a while modifying gRNA targeting. Although mutating the gRNA target site on the donor was not required for KI, this resulted in a >twofold increase in efficiency (Fig. 1I). However, inducing dsDNA breaks on both the 5′ and 3′ ends of the integration site did not enhance KI efficiency, nor did using ssDNA as repair donor instead of dsDNA (Fig. 1I).
To determine whether precise genomic integration is important for our KI approach, we obtained three stable alleles for eGFP-rab11a using different gRNA target sites and/or repair donors (Fig. S2A-C). Although the 5′ integration ends of the three alleles proceeded seamlessly, the lines showed distinct INDELs at the 3′ integration boundaries of intron 1. In addition, the 3′ non-coding donor sequence of the ssDNA-generated allele was 390 bp shorter than the other alleles (Fig. S2C). However, these sequence alterations had no detectable effect on eGFP-Rab11a protein expression levels, probably because the INDELs affect only non-coding regions. These findings indicate that KI integration sites in non-coding regions of genes, at least within introns, have minimal impact on expression of endogenously tagged proteins.
To determine the germline transmission efficiency of our approach, we targeted cldn15la, rab11a and tjp1a and raised to adulthood only F0 embryos that showed mosaic expression. We then outcrossed 3-5 F0 animals for each target to wild-type (WT) fish and determined the percentage of stably expressing F1 embryos. cldn15la and tjp1a, targeted for C-terminal tagging of the gene products, showed similar levels of germline transmission rates, with 26.9% and 17.1% of F1 progeny showing expression, respectively (Fig. 1J). We observed similar levels of efficiency despite using tdTomato (∼1400 bp) as a tag for cldn15la and eGFP (∼700 bp) for tjp1a, suggesting that insertion sizes in this range likely do not impact KI efficiency. For N-terminal tagging, we compared rab11a KI using dsDNA or ssDNA as donor sequences. However, using ssDNA as a repair donor did not improve the efficiency of obtaining stably expressing F1 embryos (Fig. 1J), indicating that simple dsDNA PCR donor amplicons are effective for KI in zebrafish.
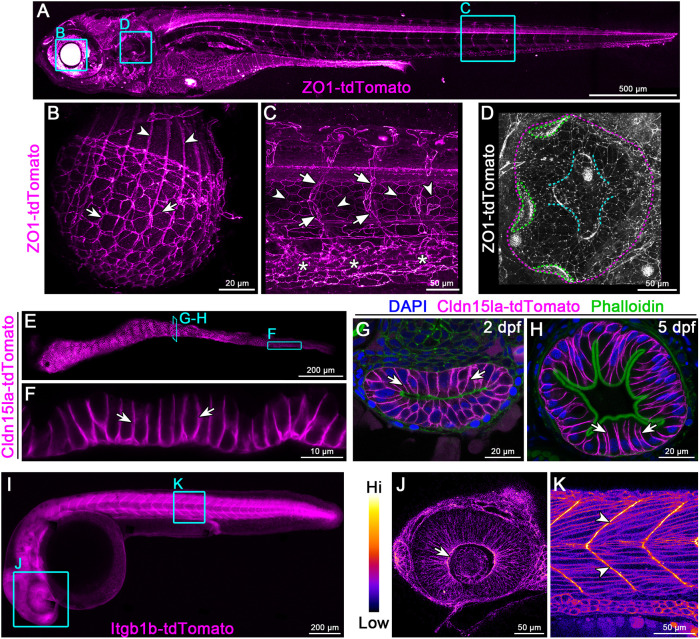
We next monitored expression and localization of endogenously tagged proteins in stably expressing larvae. ZO-1 (Tjp1a) is a peripheral component of tight junctions and is localized cortically in epithelial cells (Zihni et al., 2016). ZO-1-tdTomato showed widespread expression in epithelial organs, with enriched expression in the lens, floor plate, neural tube, vasculature and intestine (Fig. 2A). In the eye, ZO-1-tdTomato was present in the lens epithelium and lens fiber cells (Fig. 2B; Movie 1), whereas in the trunk it was enriched in the dorsal aorta, caudal vein, intersegmental vessels and notochord sheath cells (Fig. 2C; Movie 2). ZO-1-tdTomato also labeled epithelial cells throughout the otic capsule (Fig. 2D; Movie 3). We also generated a KI line expressing ZO-1-eGFP and when crossed to the ZO-1-tdTomato line, the two proteins showed identical expression patterns and perfect co-localization (Fig. S3A). In the epidermis, endogenously tagged ZO-1 was highly expressed in lateral line neuromasts, whereas in periderm cells it was enriched at tricellular junctions (Fig. S3B,C).
Fig. 2.
Endogenous C-terminal tagging of ZO-1, Cldn15la and Integrin β1b. (A-D) Live 3D reconstructions of TgKI(tjp1a-tdTomato)pd1224 heterozygous larvae. B: arrows, lens epithelial cells; arrowheads, lens fiber cells. C: arrows, intersegmental vessels; arrowheads, notochord sheath cells; asterisks, caudal vein plexus. D: magenta dotted line, otic capsule; green dotted lines, cristae; cyan dotted lines, canals and septum. Animals are 7 dpf (A), 5 dpf (B) and 3 dpf (C,D). (E,F) Live imaging of the intestine of a 7 dpf TgKI(cldn15la-tdTomato)pd1249 heterozygous larva. Arrows, intestinal epithelial cell (IEC) basolateral membrane. (G,H) Transverse sections of the intestine at stages of lumen opening (2 dpf) (G) and onset of larval feeding (5 dpf) (H). Arrows, IEC basolateral membrane. (I-K) Live imaging of a 28 hpf TgKI(itgb1b-tdTomato)sk108 heterozygous embryo. J and K are pseudo-colored according to the look-up table (LUT) scale shown. J: arrow, optic cup basolateral membrane. K: arrowheads, myotendinous junctions. Cyan boxes are representative regions of interest panels indicated. Scale bars: 500 µm (A); 20 µm (B,G,H); 50 µm (C,D,J,K); 200 µm (E,I); 10 µm (F).
To specifically label intestinal epithelial cells (IECs), we targeted cldn15la, which encodes a member of the claudin family of tetraspanin membrane proteins that form tight junctions. Cldn15la is an atypical claudin that is not restricted to tight junctions and is instead localized along basolateral membranes (Alvers et al., 2014). Similar to the transgenic TgBAC(cldn15la-GFP)pd1034 allele (Fig. S4A,B) (Alvers et al., 2014), endogenously tagged Cldn15la-tdTomato was expressed in all IECs (Fig. 2E,F), where it localized to basolateral membranes throughout gut development (Fig. 2G,H). Of note, unlike the TgBAC(cldn15la-GFP)pd1034 allele, which is homozygous lethal at embryonic stages, homozygous TgKI(cldn15la-tdTomato)pd1249 larvae are indistinguishable from WT siblings.
To visualize cell-ECM adhesions with an endogenous protein in live zebrafish, we generated an itgb1b-tdTomato KI line (Fig. 2I). Consistent with previous studies (Martinez-Morales et al., 2009; Sidhaye and Norden, 2017), Itgb1b-tdTomato was clearly enriched at the basal membrane of optic cup cells at 28 h post-fertilization (hpf) (Fig. 2J). We also confirmed prominent enrichment of Itgb1b-tdTomato in myotendinous junctions at the somite boundaries (Jülich et al., 2005) (Fig. 2K). These observations suggest that itgb1b-tdTomato faithfully reports the localization of Itgb1b in live zebrafish.
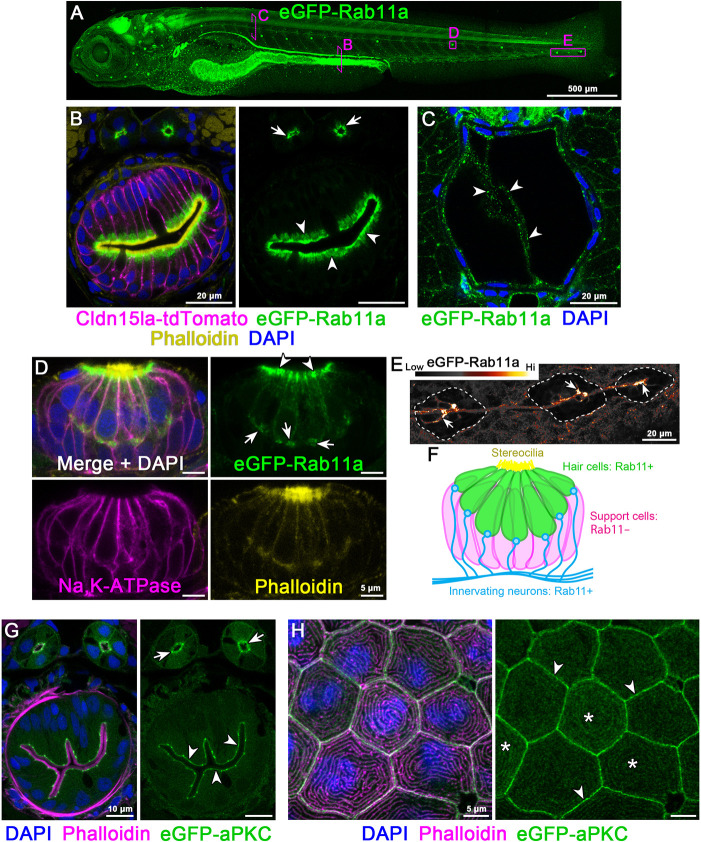
To establish a zebrafish model for membrane trafficking, we generated a KI line expressing N-terminally tagged eGFP-Rab11a. Live imaging revealed that eGFP-Rab11a was nearly ubiquitously expressed and enriched in epithelial organs (Fig. 3A; Movie 4). In the posterior intestine, eGFP-Rab11a was highly expressed and localized apically in lysosome-rich enterocytes (LREs) (Park et al., 2019) (Fig. 3B). Apical localization of eGFP-Rab11a in the intestine resembles that of endogenous Rab11a by immunostaining (Levic et al., 2020) (Fig. S4C,D). eGFP-Rab11a was detected in transverse sections of skeletal muscle, notochord sheath cells and notochord vacuolated cells (Fig. 3C), where live imaging revealed dynamic movement of eGFP-Rab11a (Movie 5). Within lateral line neuromasts, eGFP-Rab11a expression was restricted to hair cells, where it was enriched apically near stereocilia of the apical membrane (Fig. 3D) and was also present at basal puncta that may represent contact sites from innervating neurons (Fig. 3D). Accordingly, we detected enriched expression of eGFP-Rab11a in tracts and projections of neurons that underlie neuromasts (Fig. 3E,F).
Fig. 3.
Endogenous N-terminal tagging of Rab11a and aPKC. (A) Live 3D reconstruction of 5 dpf TgKI(eGFP-rab11a)pd1244 heterozygous larva. Magenta boxes show representative regions of interest for panels B-E. (B) Transverse section through the posterior mid-intestine (LREs, arrowheads) and pronephric ducts (arrows). (C) Transverse section through the notochord. Arrowheads, notochord vacuolated cells. (D) Whole-mount image of a neuromast. Arrowheads, apical cytoplasm; arrows, basal cytoplasm. (E) Live image of neurons innervating lateral line neuromasts (dotted line). Image is pseudo-colored according to the LUT scale shown. Arrows, neuronal tracts. (F) Schematic of Rab11a expression within neuromasts. (G,H) Transverse sections of TgKI(eGFP-prkci)pd1260 heterozygous larvae. (G) Transverse section of the mid-intestine. Arrowheads, IEC apical cortex; arrows, pronephric duct apical cortex. (H) Localization of eGFP-aPKC in periderm cells. Arrowheads, cell cortex; asterisks, apical microridges. Scale bars: 500 µm (A); 20 µm (B,C,E); 5 µm (D,H); 10 µm (G).
As a marker for the apical polarity complex we targeted aPCK (encoded by prkci; also known as has) (Horne-Badovinac et al., 2001; Peterson et al., 2001). In polarized IECs and pronephric duct cells (PN), eGFP-aPKC was enriched at the apical cortex (Fig. 3G). This localization closely resembled that of endogenous aPKC in WT larvae by immunostaining (Fig. S4E,F). Consistent with previous studies (Raman et al., 2016; Magre et al., 2019), eGFP-aPKC was present at apical microridges in periderm cells of the epidermis (Fig. 3H).
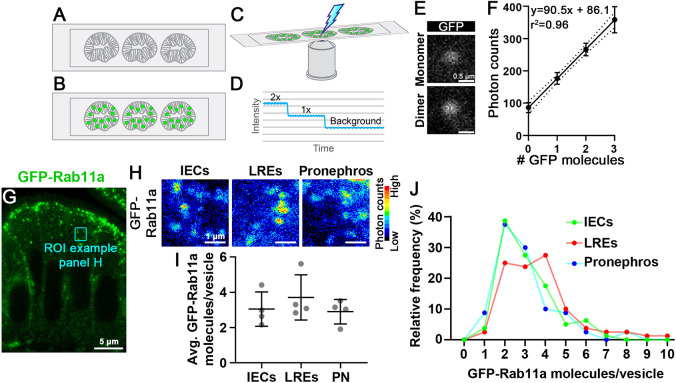
Having shown that proteins can be tagged endogenously, we tested whether our KI lines can be used to determine their relative cellular abundance. To explore this question, we adapted a microscopy-based approach to measure the concentration of eGFP molecules on diffraction-limited vesicles on zebrafish tissue sections to estimate the number of molecules per vesicle. Although this technique is commonly used in in vitro models (Clayton, 2018; Marques et al., 2019; Escamilla-Ayala et al., 2020), quantitative imaging of animal tissues can be obscured by factors such as autofluorescence and light scattering. To establish baseline standards, we measured the photon emission of purified eGFP on zebrafish tissue sections. We collected intestinal sections of eGFP-negative larvae (Fig. 4A), incubated a solution of purified eGFP at low concentration and then crosslinked eGFP particles to the tissue surface by fixation (Fig. 4B). We collected accumulated photon counts of eGFP particles and then photobleached them (Fig. 4C). During photobleaching we monitored signal intensity and inferred the original number of eGFP molecules in the particle based on the signal decay profile (Fig. 4D). Using this approach, we identified eGFP particles containing 1-3 molecules that exhibited a linear increase in photon emission (Fig. 4E,F). Next, we prepared eGFP-Rab11a KI larvae and performed single particle imaging of diffraction-limited apical vesicles on tissue sections (Fig. 4G,H) using identical processing and imaging conditions as those described above. Although Rab11a expression levels vary by more than twofold at the mRNA levels in LREs versus IECs (Park et al., 2019), endogenously tagged eGFP-Rab11a concentration on apical vesicles did not change in proportion (Fig. 4I). However, the relative distribution profile for LREs did reveal an increase in the fraction of vesicles containing >3 molecules (Fig. 4J), possibly reflecting a pool of vesicles that function in protein uptake in these specialized enterocytes (Park et al., 2019).
Fig. 4.
Tissue-specific Rab11a expression levels do not strongly affect its concentration on apical vesicles of the intestine or pronephros. (A-F) Single particle imaging of zebrafish tissue sections. Intestinal sections of GFP-negative larvae (A) are incubated with purified eGFP (B), and photon counts are collected and photobleached to background-level intensity (C). eGFP particle concentrations are inferred by the decay profile (D). (E,F) Single particle photon count imaging and linear regression analysis of purified eGFP photon emission from intestinal sections. (G) Transverse section of TgKI(eGFP-rab11a)pd1244 intestinal epithelial cells (IECs). Cyan box shows a representative region of interest of apical vesicles. (H) Pseudo-colored photon count images of apical vesicles of IECs, lysosome-rich enterocytes (LREs) and pronephric duct epithelial cells (PN). (I) Average eGFP-Rab11a concentration values from apical vesicles. Data points are mean values from tissue sections of individual larvae. Error bars show s.d. n=4 larvae for each organ (20 vesicles per animal). Data were not significantly different (one-way ANOVA). (J) Relative frequency plot of the data used for I. LREs versus IECs, P<0.05; LREs versus PN, P<0.01; IECs versus PN, not significant (one-way ANOVA). n=80 vesicles per organ. Scale bars: 0.5 µm (E); 5 µm (G); 1 µm (H).
Here, we describe a simple and effective KI approach for zebrafish. Although KI zebrafish lines have been generated to study promoter activity (Kimura et al., 2014; Hoshijima et al., 2016; Li et al., 2019), there are few published examples of C-terminally tagged KI fusion lines (Cronan and Tobin, 2019) and only one N-terminally tagged line (Wilson et al., 2021). This scarcity may reflect the highly error-prone nature of HDR in zebrafish. Recently an endogenous tagging approach using short homology arms was reported for medaka (Seleit et al., 2021 preprint). Although it is unclear whether this approach can generate seamless integrations to tag endogenous proteins in zebrafish, the modifications reported for medaka KI (Seleit et al., 2021 preprint) may help to further optimize endogenous tagging in zebrafish. Our approach differs from existing methods because integration errors, such as INDELs, in non-coding regions can mediate expression of the tagged protein as integration boundaries are excluded by RNA splicing. A similar endogenous tagging approach was recently reported for cultured mammalian cells (Zhong et al., 2021). With our approach, we sought to minimize integration of undesired plasmid elements by using PCR donor amplicons that only encode relevant functional elements. Following this approach, we generated stable zebrafish KI lines for several integral membrane and membrane-associated proteins that are crucial for epithelial development and cell physiology, and we used one of these lines to quantify the concentration of eGFP-Rab11a molecules on apical vesicles in different epithelial organs. The endogenously tagged zebrafish lines presented here improve accuracy and allow experimental approaches not feasible with traditional transgenic lines. These include the abilities to recapitulate expression patterns of endogenous genes and to precisely quantify protein levels using single particle imaging or related techniques (Wang et al., 2018). They also can facilitate uncovering protein interaction networks and dynamics without overexpression artifacts (Ahmed et al., 2018) and acute manipulation of protein function using conditional loss-of-function approaches (Daniel et al., 2018; Yamaguchi et al., 2019). One factor that can impact expression levels of endogenously tagged proteins is the 3′ UTR used in the repair donor sequence. The 3′ tagged lines shown here have exogenous polyA sequences that may alter transcript stability. Endogenous 3′ UTR and polyA sequences can be substituted in 3′ repair donors to recapitulate mRNA stability levels more accurately. By contrast, genes tagged with 5′ insertions, such as eGFP-rab11a, will result in transcripts containing the endogenous 3′ UTR that should provide endogenous expression levels. Finally, because our KI approach relies on splice donor and acceptor elements, our method can be adapted to generate internal insertions at precise codon positions for targets such as membrane proteins containing signal peptide sequences that cannot be tagged at their C termini.
MATERIALS AND METHODS
Zebrafish maintenance
Zebrafish (Danio rerio) were used in accordance with Duke University Institutional Animal Care and Use Committee (IACUC) guidelines and NYU School of Medicine under the approval from protocol number 170105-02. Zebrafish stocks were maintained and bred as previously described (Westerfield, 2007). Genotypes were determined by PCR and DNA sequencing or phenotypic analysis. Male and female breeders from 3-18 months of age were used to generate fish for all experiments. We used 1-7 days post-fertilization (dpf) zebrafish larvae from the Ekkwill (EK) or AB/TL background in this study. Strains generated in this study are: TgKI(tjp1a-tdTomato)pd1224, TgKI(tjp1a-eGFP)pd1252, TgKI(cldn15la-tdTomato)pd1249, TgKI(itgb1b-tdTomato)sk108, TgKI(eGFP-rab11a)pd1244 and TgKI(eGFP-prkci)pd1260. Embryos and larvae were anesthetized with 0.4 mg/ml MS-222 (Sigma-Aldrich, A5040) dissolved in embryo media for handling when necessary.
Generation of C-terminal PCR donors
We first generated a series of donor vectors to expedite production of C-terminal KI constructs. In the pUC19 vector backbone, we constructed a multiple cloning site, a fluorescent protein coding sequence or other tag lacking the start codon (eGFP, mLanYFP, mScarlet, tdTomato, p2A-QF2, p2A-eGFP, p2A-mScarlet or p2A-Venus-PEST), a stop codon, a second multiple cloning site and the zebrafish ubb polyA sequence. This fragment was flanked by forward and reverse PCR primer sites to generate PCR donor amplicons for all targets using the same primers (pUC19_forward 5′-GCGATTAAGTTGGGTAACGC-3′ and pUC19_reverse 5′-TCCGGCTCGTATGTTGTGTG-3′). A gene fragment spanning from the middle of the last intron through the last coding sequence codon of the exon was cloned into the donor vector in frame with the fluorescent protein coding sequence using the following primers: tjp1a-forward 5′-CTTGCTAGCAGTTTCGATGACCACAGGGT-3′, tjp1a-reverse 5′-CCTCTCGAGGAAATGGTCAATAAGCACAGACA-3′, cldn15la-forward 5′-CTTCCGCGGGTTTCACGTCAGAAATTGTCGG-3′, cldn15la-reverse 5′-CTTCTCGAGGACGTAGGCTTTGGATGTTTC-3′. PCR donor amplicons were purified using the Nucleospin Gel and PCR Clean-up kit (Machery-Nagel, distributed by Takara Bio USA). PCR products were not gel purified. The final product was dried on column at 60°C for 10 min and then eluted with water and stored at −20°C. A detailed protocol for endogenous tagging is provided in the supplementary Materials and Methods. C-terminal donor vectors have been deposited to Addgene (Addgene plasmids 174021, 174022, 174023, 174024, 174025, 174026, 174027, 174028, 174029, 174030; https://www.addgene.org/Michel_Bagnat/).
Generation of N-terminal PCR donors for rab11a
A gene fragment spanning from 446 base pairs upstream from the 5′ UTR through 491 base pairs downstream of the end of exon 1 was cloned into pCS2+ with the following primers: rab11a-forward 5′-CTTCTCGAGGAACTTACGAGCTGGATTTGTGC-3′ and rab11a-reverse 5′-CTTTCTAGATGACAGCGTCGGTCACAGTT-3′. A small multiple cloning site was added before the start codon of exon 1 by site-directed mutagenesis (Q5 SDM Kit, New England Biolabs) using the primers rab11a-MCS-SDM-forward 5′-TACTAGTTCCATGGGGACACGAGACGAC-3′ and rab11a-MCS-SDM-reverse 5′-AGACCGGTAGGCTCGATCAAAACAAAAGCGC-3′. eGFP was cloned into the multiple cloning site using the primers GFP-forward 5′-CTTACCGGTGCCGCCACCATGGTGAGCAAGGGCGAGGA-3′ and GFP-reverse 5′-CTTACTAGTCTTGTACAGCTCGTCCATGCC-3′. The gRNA target sites used for genomic targeting were mutated in the donor plasmid using site-directed mutagenesis with the primers: gRNA-1-SDM-forward 5′-AACAGCGAACTGTCGCCTCCACTTTCCTT-3′, gRNA-1-SDM-reverse 5′-ATCTCCGCTGTAGCACTGCAGTCTGTCTGT-3′, gRNA-2-SDM-forward 5′-ACTCGAGCAGAGCAAACAAACTCCTGCTCTTC-3′, gRNA-2-SDM-reverse 5′-CGAGCTAGCATATTAGCTGGCCTTTACTGT-3′. PCR donors were generated as described above using the primers: rab11a-donor-forward 5′-GAACTTACGAGCTGGATTTGTGC-3′ and rab11a-donor-reverse 5′-CTTTCTAGATGACAGCGTCGGTCACAGTT-3′. For ssDNA production in Fig. 1, PCR of the same donor plasmid was performed using the same primers, but the forward primer was phosphorylated. After PCR ssDNA was generated using the Guide-it Long ssDNA Production System (Takara Bio USA) using the manufacturer's recommendations. ssDNA was purified using the Nucleospin Gel and PCR Clean-up kit with buffer NTC used as recommended by the manufacturer. ssDNA conversion was verified used gel electrophoresis and the product was stored at −80°C.
Generation of N-terminal PCR donors for prkci
A gene fragment spanning from 488 base pairs upstream from the 5′ UTR through 61 base pairs downstream from exon 1 was cloned into pDONR221 using a BP reaction (Thermo Fisher Scientific) using the primers prkci-BP-forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCTATCTAGGTATATGGGCCCTC-3′ and prkci-BP-reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGCAATCCTGAGAATAAGTGAGA-3′. The gRNA target site in intron 1 was mutated by PCR during initial cloning (the reverse cloning primer was mutagenic) and the gRNA target site sequence was verified independently in a population of WT fish. Next, a multiple cloning site was inserted before the start codon of exon 1 using the primers prkci-MCS-SDM-forward 5′-TACTAGTTCCATGCCCACGCTGCGGGAC-3′ and prkci-MCS-SDM-reverse 5′-AGACCGGTAGGTATGGACTATCCGTACTCCTGCTAGC-3′. eGFP was cloned to the site using the primers GFP-forward 5′-CTTACCGGTGCCGCCACCATGGTGAGCAAGGGCGAGG-3′ and GFP-reverse 5′-CTTACTAGTCTTGTACAGCTCGTCCATGCC-3′. PCR donors were generated as described above using the primers prkci-forward-donor 5′-TATCTAGGTATATGGGCCCTC-3′ and prkci-reverse-donor 5′-GCAATAGTGCGAATAAGTGAGA-3′.
Generation of plasmid donors for itgb1b
A genomic fragment spanning the last 29 bp of itgb1b exon 8 to the end of itgb1b exon 9 was cloned into the pUC19 plasmid (exon numbering is based on transcript ID: ENSDART00000161711.2). A linker sequence coding for amino acids GGPVAT was inserted after the codon for the last amino acid of itgb1b and the fragment was fused to the tdTomato coding sequence followed by the SV40 polyA signal sequence. A gRNA target site was designed to target both the donor plasmid, thereby linearizing it in the intron, and the endogenous genomic intron.
Production of gRNA
gRNA target sites were identified using CRISPRscan (Moreno-Mateos et al., 2015) and gRNAs were synthesized by in vitro transcription using an oligo-based template method (Yin et al., 2015) using the MEGAshortscript T7 Transcription Kit (Thermo Fisher Scientific). gRNAs were precipitated by ammonium acetate/isopropanol, resuspended in water and stored at −80°C. For itgb1b, crRNA and the tracrRNA were purchased from IDT, and the cRNA was designed using the Custom Alt-R CRISPR-Cas9 guide RNA Design Tool (IDT). gRNA target sites used in this study were: cldn15la, 5′-GTTTCACGTCAGAAATTGTCGGG-3′ and 5′-GGATTTCTCTAGATTATGACCGG-3′; prkci, 5′-GCATTCTCACTTATTCTCAACGG-3′; rab11a, 5′-GGCAGCGGAGAGGACAGCGACGG-3′ and 5′-CCGGCTAGCTCACTTCGAGCACC-3′; tjp1a, 5′-TGCGAATAGGGGTTGATAATGGG-3′ and 5′-GAGTTTCGATGACCACAGGGTGG-3′; crRNA for itgb1b, 5′-GGAGGTCTTGATGTAGGATT-3′.
Microinjections and visual screening
Early one-cell-stage embryos were injected with 1-2 nl of a KI cocktail consisting of gRNA (final concentration 30-50 pg/nl), dsDNA or ssDNA PCR donors (final concentration 5-10 pg/nl) Cas9 protein tagged with a nuclear localization sequence (PNA Bio CP-01) (final concentration 300-500 pg/nl), and phenol red (final concentration 0.05%). We observed mortality rates of 10-20% for dsDNA-injected embryos and 40-50% for ssDNA-injected embryos. For itgb1b, the injection mix containing Cas9-NLS protein, crRNA, tracrRNA and the plasmid harboring the itgb1b targeting cassette was heat-activated at 37°C and injected into one-cell-stage WT embryos. Embryos were visually screened daily between 1-5 dpf for fluorescence using an Axio Zoom V16 microscope (Zeiss). Embryos suspected of showing fluorescence were mounted in 0.7% low melting point agarose and imaged by confocal microscopy on a Leica SP8 microscope using an HC FLUOTAR VISIR 25×/0.95 NA water immersion objective (Leica). Positive embryos were recovered from anesthesia and raised to adulthood.
Isolation of stable alleles
Injected embryos showing expression of fluorescently tagged proteins were raised and crossed to WT fish. The positive F1 embryos were raised and crossed to WT fish. The integration site was then sequenced (supplementary files 1-8) and the lines were designated allele numbers. With the exception of Fig. S1, all imaging data presented are from stable animals of the F1 or greater generation.
Imaging and image processing
All imaging was performed on a Leica SP8 confocal microscope. Live imaging was conducted with a FLUOTAR VISIR 25×/0.95 NA or HC PL APO CS2 20×/0.75 water immersion objectives (Leica), and cross-sections with an HC PL APO CS2 63×/1.40 oil immersion objective (Leica). All fluorescent proteins for KI lines were imaged directly without additional antibody labeling. Whole animals were imaged in tiling mode and the data were stitched in Leica LAS software. Imaging data were processed in ImageJ/FIJI (National Institutes of Health) to prepare 3D reconstructions using native plugins. To enhance visualization of some images, data were pseudo-colored using default lookup tables (LUTs) in ImageJ/FIJI. LUTs are described in the figure legends and scales shown in figure panels where appropriate. Post-processing for linear changes in brightness were performed in Photoshop using the levels tool.
Single particle imaging
We fixed 6 dpf GFP-negative larvae in 4% paraformaldehyde in PBS (pH 7.5) overnight at 4°C, rinsed in PBS and then embedded in 5% low melting point agarose. Then, 200 µm sections were collected using a Leica VT1000S vibratome (Levic et al., 2020). Sections were incubated with 340 ng/ml purified eGFP, which was generated as previously described (Park et al., 2019), overnight at 4°C. The solution was then gently aspirated and sections were fixed in 4% paraformaldehyde in PBS (pH 7.5) for 30 min at room temperature. Sections were rinsed in PBS and then mounted on glass slides in 90% glycerol buffered with 10 mM Tris (pH 8.0) with 1% N propyl-gallate added. Sections were imaged near the coverslip surface with a Leica SP8 confocal microscope using an HC PL APO CS2 63×/1.40 oil immersion objective. Excitation was performed with a 20 mW 488 nm laser operating at 0.2% power, and scans were performed at 400 Hz with a pixel size of 50 nm. Emission spectra were collected from 498-550 nm using a HyD detector operating in photon counting mode with 10× line accumulation and at 10% gain. Experimental samples (eGFP-Rab11a larvae) were processed identically. Raw 12-bit images were analyzed in ImageJ/FIJI. Photon counts of 5 pixel2 regions of interest of eGFP particles were collected and analyzed by linear regression using GraphPad Prism. Photon counts experimental samples were interpolated from the linear regression analysis of purified eGFP particles to infer the number of eGFP molecules per vesicle.
Supplementary Material
Acknowledgements
We thank the Duke Zebrafish Core, Joseph Proietti and Sam Pirani for excellent fish care and maintenance. Jieun Esther Park provided purified eGFP. We also thank Xiaolei Wang, Ian Macara and Kristen Kwan for helpful discussions. The use of the NYULH DART Microscopy Laboratory (P30CA016087) is gratefully acknowledged.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.S.L., N.Y., H.K., M.B.; Methodology: D.S.L., N.Y.; Validation: D.S.L., N.Y.; Formal analysis: D.S.L.; Investigation: D.S.L., N.Y., S.W.; Resources: H.K., M.B.; Writing - original draft: D.S.L.; Writing - review & editing: N.Y., H.K., M.B.; Visualization: D.S.L.; Supervision: H.K., M.B.; Project administration: H.K., M.B.; Funding acquisition: H.K., M.B.
Funding
This work was supported by the National Institutes of Health (DK121007 and DK113123 to M.B.; NS102322 to H.K.) D.S.L was supported by a Duke University Training Grant in Digestive Diseases and Nutrition (DK007568). N.Y. was supported by a New York State Stem Cell Science institutional training grant (C322560GG) and by an American Heart Association fellowship (20PRE35180164). M.B. is a Howard Hughes Medical Institute Faculty Scholar. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199994
References
- Ahmed, S. M., Nishida-Fukuda, H., Li, Y., Mcdonald, W. H., Gradinaru, C. C. and Macara, I. G. (2018). Exocyst dynamics during vesicle tethering and fusion. Nat. Commun., 9, 5140. 10.1038/s41467-018-07467-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers, A. L., Ryan, S., Scherz, P. J., Huisken, J. and Bagnat, M. (2014). Single continuous lumen formation in the zebrafish gut is mediated by smoothened-dependent tissue remodeling. Development, 141, 1110-1119. 10.1242/dev.100313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann, J. and Schulte-Merker, S. (2011). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development, 138, 4327-4332. 10.1242/dev.068080 [DOI] [PubMed] [Google Scholar]
- Chandler, K. J., Chandler, R. L., Broeckelmann, E. M., Hou, Y., Southard-Smith, E. M. and Mortlock, D. P. (2007). Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm. Genome, 18, 693-708. 10.1007/s00335-007-9056-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, A. H. (2018). Fluorescence-based approaches for monitoring membrane receptor oligomerization. J. Biosci., 43, 463-469. 10.1007/s12038-018-9762-5 [DOI] [PubMed] [Google Scholar]
- Cronan, M. R. and Tobin, D. M. (2019). Endogenous tagging at the cdh1 locus for live visualization of E-Cadherin dynamics. Zebrafish, 16, 324-325. 10.1089/zeb.2019.1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, K., Icha, J., Horenburg, C., Müller, D., Norden, C. and Mansfeld, J. (2018). Conditional control of fluorescent protein degradation by an auxin-dependent nanobody. Nat. Commun., 9, 3297. 10.1038/s41467-018-05855-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewari, P. S., Southgate, B., Mccarten, K., Monogarov, G., O'Duibhir, E., Quinn, N., Tyrer, A., Leitner, M.-C., Plumb, C., Kalantzaki, M.et al. (2018). An efficient and scalable pipeline for epitope tagging in mammalian stem cells using Cas9 ribonucleoprotein. eLife, 7, e35069. 10.7554/eLife.35069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D. J., Pani, A. M., Heppert, J. K., Higgins, C. D. and Goldstein, B. (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics, 200, 1035-1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Ayala, A. A., Sannerud, R., Mondin, M., Poersch, K., Vermeire, W., Paparelli, L., Berlage, C., Koenig, M., Chavez-Gutierrez, L., Ulbrich, M. H.et al. (2020). Super-resolution microscopy reveals majorly mono- and dimeric presenilin1/γ-secretase at the cell surface. eLife, 9, e56679. 10.7554/eLife.56679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes, F., Reynolds, E., Lewellis, S. W., Venkiteswaran, G. and Knaut, H. (2016). A plasmid set for efficient bacterial artificial chromosome (BAC) transgenesis in zebrafish. G3 Genes, Genomes, Genetics, 6, 829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Hisey, E., Bradshaw, T. W. A., Erata, E., Brown, W. E., Courtland, J. L., Uezu, A., Xiang, Y., Diao, Y. and Soderling, S. H. (2019). Plug-and-play protein modification using homology-independent universal genome engineering. Neuron, 103, 583-597.e8. 10.1016/j.neuron.2019.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, T. J., Seiler, M. and Veitia, R. A. (2013). The transience of transient overexpression. Nat. Methods, 10, 715-721. 10.1038/nmeth.2534 [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac, S., Lin, D., Waldron, S., Schwarz, M., Mbamalu, G., Pawson, T., Jan, Y.-N., Stainier, D. Y. R. and Abdelilah-Seyfried, S. (2001). Positional cloning of heart and soul reveals multiple roles for PKCλ in zebrafish organogenesis. Curr. Biol., 11, 1492-1502. 10.1016/S0960-9822(01)00458-4 [DOI] [PubMed] [Google Scholar]
- Hoshijima, K., Jurynec, M. J. and Grunwald, D. J. (2016). Precise editing of the zebrafish genome made simple and efficient. Dev. Cell, 36, 654-667. 10.1016/j.devcel.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, D. G., Ramachandran, S., Bradford, Y. M., Fashena, D., Toro, S., Eagle, A., Frazer, K., Kalita, P., Mani, P., Martin, R.et al. (2021). The zebrafish information network: major gene page and home page updates. Nucleic Acids Res., 49, D1058-d1064. 10.1093/nar/gkaa1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jülich, D., Geisler, R. and Holley, S. A. (2005). Integrinα5 and Delta/Notch signaling have complementary spatiotemporal requirements during zebrafish somitogenesis. Dev. Cell, 8, 575-586. 10.1016/j.devcel.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Kimura, Y., Hisano, Y., Kawahara, A. and Higashijima, S. (2014). Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep., 4, 6545. 10.1038/srep06545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koles, K., Yeh, A. R. and Rodal, A. A. (2016). Tissue-specific tagging of endogenous loci in Drosophila melanogaster. Biology Open, 5, 83-89. 10.1242/bio.016089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, K. M., Fujimoto, E., Grabher, C., Mangum, B. D., Hardy, M. E., Campbell, D. S., Parant, J. M., Yost, H. J., Kanki, J. P. and Chien, C.-B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn., 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Levic, D. S., Ryan, S., Marjoram, L., Honeycutt, J., Bagwell, J. and Bagnat, M. (2020). Distinct roles for luminal acidification in apical protein sorting and trafficking in zebrafish. J. Cell Biol. 219, e201908225. 10.1083/jcb.201908225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Zhang, Y., Han, B., Li, L., Li, M., Lu, X., Chen, C., Lu, M., Zhang, Y., Jia, X.et al. (2019). One-step efficient generation of dual-function conditional knockout and geno-tagging alleles in zebrafish. eLife, 8, e48081. 10.7554/eLife.48081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magre, I., Fandade, V., Damle, I., Banerjee, P., Yadav, S. K., Sonawane, M. and Joseph, J. (2019). Nup358 regulates microridge length by controlling SUMOylation-dependent activity of aPKC in zebrafish epidermis. J. Cell Sci., 132, jcs224501. 10.1242/jcs.224501 [DOI] [PubMed] [Google Scholar]
- Marques, P. E., Nyegaard, S., Collins, R. F., Troise, F., Freeman, S. A., Trimble, W. S. and Grinstein, S. (2019). Multimerization and retention of the Scavenger receptor SR-B1 in the plasma membrane. Dev. Cell, 50, 283-295.e5. 10.1016/j.devcel.2019.05.026 [DOI] [PubMed] [Google Scholar]
- Martinez-Morales, J. R., Rembold, M., Greger, K., Simpson, J. C., Brown, K. E., Quiring, R., Pepperkok, R., Martin-Bermudo, M. D., Himmelbauer, H. and Wittbrodt, J. (2009). Ojoplano-mediated basal constriction is essential for optic cup morphogenesis. Development, 136, 2165-2175. 10.1242/dev.033563 [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos, M. A., Vejnar, C. E., Beaudoin, J.-D., Fernandez, J. P., Mis, E. K., Khokha, M. K. and Giraldez, A. J. (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods, 12, 982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis, A., Marjoram, L. and Bagnat, M. (2013). Cftr controls lumen expansion and function of Kupffer's vesicle in zebrafish. Development, 140, 1703-1712. 10.1242/dev.091819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J., Levic, D. S., Sumigray, K. D., Bagwell, J., Eroglu, O., Block, C. L., Eroglu, C., Barry, R., Lickwar, C. R., Rawls, J. F.et al. (2019). Lysosome-rich enterocytes mediate protein absorption in the vertebrate gut. Dev. Cell, 51, 7-20.e6. 10.1016/j.devcel.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y., Clark, K. J., Campbell, J. M., Panetta, M. R., Guo, Y. and Ekker, S. C. (2014). Making designer mutants in model organisms. Development, 141, 4042-4054. 10.1242/dev.102186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, R. T., Mably, J. D., Chen, J.-N. and Fishman, M. C. (2001). Convergence of distinct pathways to heart patterning revealed by the small molecule concentramide and the mutation heart-and-soul. Curr. Biol., 11, 1481-1491. 10.1016/S0960-9822(01)00482-1 [DOI] [PubMed] [Google Scholar]
- Raman, R., Damle, I., Rote, R., Banerjee, S., Dingare, C. and Sonawane, M. (2016). aPKC regulates apical localization of Lgl to restrict elongation of microridges in developing zebrafish epidermis. Nat. Commun., 7, 11643. 10.1038/ncomms11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranawakage, D. C., Okada, K., Sugio, K., Kawaguchi, Y., Kuninobu-Bonkohara, Y., Takada, T. and Kamachi, Y. (2021). Efficient CRISPR-Cas9-mediated knock-in of composite tags in zebrafish using long ssDNA as a donor. Front. Cell Dev. Biol., 8, 598634. 10.3389/fcell.2020.598634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli, A. E., Bagwell, J., Bosch-Fortea, M., Boncompain, G., Reglero-Real, N., Garcia-Leon, M. J., Andres, G., Toribio, M. L., Alonso, M. A., Millan, J.et al. (2015). Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat. Cell Biol., 17, 241-250. 10.1038/ncb3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleit, A., Aulehla, A. and Paix, A. (2021). Endogenous protein tagging in medaka using a simplified CRISPR/Cas9 knock-in approach. bioRxiv, 2021.07.29.454295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhaye, J. and Norden, C. (2017). Concerted action of neuroepithelial basal shrinkage and active epithelial migration ensures efficient optic cup morphogenesis. eLife, 6, e22689. 10.7554/eLife.22689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Yin, Y., Lau, S., Sankaran, J., Rothenberg, E., Wohland, T., Meier-Schellersheim, M. and Knaut, H. (2018). Anosmin1 shuttles Fgf to facilitate its diffusion, increase its local concentration, and induce sensory organs. Dev. Cell, 46, 751-766.e12. 10.1016/j.devcel.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield, M. (2007). The Zebrafish Book: A guide for the laboratory use of zebrafish (Danio rerio), 5th edn. University of Oregon Press. [Google Scholar]
- Wierson, W. A., Welker, J. M., Almeida, M. P., Mann, C. M., Webster, D. A., Torrie, M. E., Weiss, T. J., Kambakam, S., Vollbrecht, M. K., Lan, M.et al. (2020). Efficient targeted integration directed by short homology in zebrafish and mammalian cells. eLife, 9, e53968. 10.7554/eLife.53968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M. H., Ekker, S. C. and Farber, S. A. (2021). Imaging cytoplasmic lipid droplets in vivo with fluorescent perilipin 2 and perilipin 3 knock-in zebrafish. eLife, 10, e66393. 10.7554/eLife.66393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, N., Colak-Champollion, T. and Knaut, H. (2019). zGrad is a nanobody-based degron system that inactivates proteins in zebrafish. eLife, 8, e43125. 10.7554/eLife.43125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, L., Jao, L.-E. and Chen, W. (2015). Generation of targeted mutations in zebrafish using the CRISPR/Cas system. New York: Springer. [DOI] [PubMed] [Google Scholar]
- Zhong, H., Ceballos, C. C., Massengill, C. I., Muniak, M. A., Ma, L., Qin, M., Petrie, S. K. and Mao, T. (2021). High-fidelity, efficient, and reversible labeling of endogenous proteins using CRISPR-based designer exon insertion. eLife, 10, e64911. 10.7554/eLife.64911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihni, C., Mills, C., Matter, K. and Balda, M. S. (2016). Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol., 17, 564-580. 10.1038/nrm.2016.80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.