Abstract
Changes in the cellular environment modulate protein energy landscapes to drive important biology, with consequences for signaling, allostery, and other vital processes. The effects of ubiquitination are particularly important because of their potential influence on degradation by the 26S proteasome. Moreover, proteasomal engagement requires unstructured initiation regions that many known proteasome substrates lack. To assess the energetic effects of ubiquitination and how these manifest at the proteasome, we developed a generalizable strategy to produce isopeptide-linked ubiquitin within structured regions of a protein. The effects on the energy landscape vary from negligible to dramatic, depending on the protein and site of ubiquitination. Ubiquitination at sensitive sites destabilizes the native structure and increases the rate of proteasomal degradation. Importantly, in well-folded proteins, ubiquitination can even induce the requisite unstructured regions needed for proteasomal engagement. Our results indicate a biophysical role of site-specific ubiquitination as a potential regulatory mechanism for energy-dependent substrate degradation.
Introduction:
A protein’s function and folding is defined by its energy landscape, which encompasses all the accessible conformations, their relative populations, and rates of interconversion. This energy landscape is determined by a protein’s amino-acid sequence and environment, and small changes modulate this landscape. The phenotypic effects of these changes can range from undetectable to pathological1-4. Posttranslational modifications (PTMs) are one important environmental change that affect the energy landscape. Many PTMs have been shown to affect protein structure and function5, and the attachment of ubiquitin to lysine side chains is particularly interesting, as one of its most important roles is to target proteins for degradation by the 26S proteasome.
The ubiquitin-proteasome system (UPS) is responsible for the majority of protein turnover in eukaryotic cells. Ubiquitin is an 8.5 kDa protein appended to other proteins (substrates) through an isopeptide bond between its C-terminus and the amino group of substrate lysines. Ubiquitin itself contains seven lysines, such that additional ubiquitin molecules can be added to form chains of different lengths, linkages, and topologies. The 26S proteasome is the executor of the UPS, using ubiquitin receptors to selectively bind ubiquitinated substrates and degrade them in an ATP-dependent manner. The degradation activity resides in the proteasome’s 20S core particle, whose proteolytic sites are sequestered inside a central cavity. Substrates are delivered to the 20S core particle through the 19S regulatory particle (RP), which caps one or both sides of the barrel-shaped 20S core. RP recruits ubiquitinated substrates, mechanically unfolds them with its AAA+ (ATPase Associated with diverse cellular Activities) motor, and translocates the unstructured polypeptides into the core particle for proteolysis.
Although ubiquitination is best known for its association with proteasomal degradation, it is involved in a wide array of other cellular processes6. Therefore, the proteasome must carefully differentiate between ubiquitinated proteins that should be degraded and those ubiquitinated for other purposes. Failure of the proteasome to properly regulate substrate selection results in aberrant degradation, wasted energy, and collapse of proteostasis.
Conformational properties also affect whether a protein is degraded by the proteasome. In order to engage with the proteasomal AAA+ motor for unfolding and translocation, a substrate needs an unstructured initiation region long enough to enter the central pore and interact with conserved pore loops of the ATPase hexamer7,8. Nonetheless, a significant percentage of proteasome clients lack an obvious unstructured region9. For some substrates, another AAA+ translocase, Cdc48 (also known as p97 or VCP), has been implicated in preparing them through partial or complete unfolding for subsequent proteasomal engagement10-12.
An exciting possibility is that ubiquitination itself can modulate the landscape and expose an unstructured region for initiation of proteasomal degradation. Molecular dynamics studies suggest that ubiquitination may destabilize proteins, principally through a decrease in substrate conformational entropy13,14. If true, does this destabilization populate a proteasome-engageable unstructured region? Purification of ubiquitin-conjugated substrates with native isopeptide bonds has been a challenging hurdle15, and the experimental characterization of ubiquitin-mediated changes in protein energetics has therefore been limited to artificial, non-physiological ubiquitin-attachment strategies16 or heterogeneous samples17. Thus, the potential energetic effects of substrate ubiquitination on proteasomal degradation remain completely unknown.
Here, we developed a generalizable protocol to generate milligram quantities of homogeneously mono-ubiquitinated proteins. In this system, ubiquitin is attached via a native isopeptide linkage to a lysine within a folded protein domain. Using several single-lysine variants of the small protein barstar, we show that mono-ubiquitination induces site-specific local and global energetic changes that can lead to significant protein destabilization. Furthermore, these energetic modulations can affect proteasomal processing. Substrate variants destabilized by mono-ubiquitination display enhanced proteasomal degradation rates when appended with an unstructured region for initiation. Importantly, in the absence of these unstructured regions, ubiquitin-induced energetic changes can transiently expose flexible initiation regions, presumably by allowing access to high-energy, partially-unstructured states that are proteasome-engageable. Our data establish a connection between ubiquitin-induced changes in substrate energetics and proteasomal processing. We propose that modulation of substrate energy landscapes by site-specific ubiquitination can play a consequential role for substrate engagement and degradation by the proteasome.
Results:
Generalizable strategy for site-specific ubiquitination
Traditional spectroscopic studies of protein energetics and dynamics require large amounts of homogeneous sample, yet such quantities are not feasible using established strategies of ubiquitin attachment9,15. Furthermore, many approaches employed for artificial ubiquitin attachment require harsh chemical conjugation conditions and result in non-physiological linkages. Here, we used a biochemically reconstituted enzymatic ligation and deubiquitination strategy to overcome these technical obstacles and produce ubiquitin-substrate conjugates with native isopeptide bonds.
Substrate proteins were expressed as C-terminal fusions to maltose binding protein (MBP) with a connecting linker containing a PPPY recognition sequence for the yeast HECT E3-ubiquitin ligase Rsp518. Since substrates lacking MBP were less efficiently conjugated with ubiquitin, we believe MBP acts as a scaffold to promote productive E3-substrate interaction, as previously described19,20. Substrates also contain a single cysteine for fluorescein-maleimide labeling (Fig. 1a).
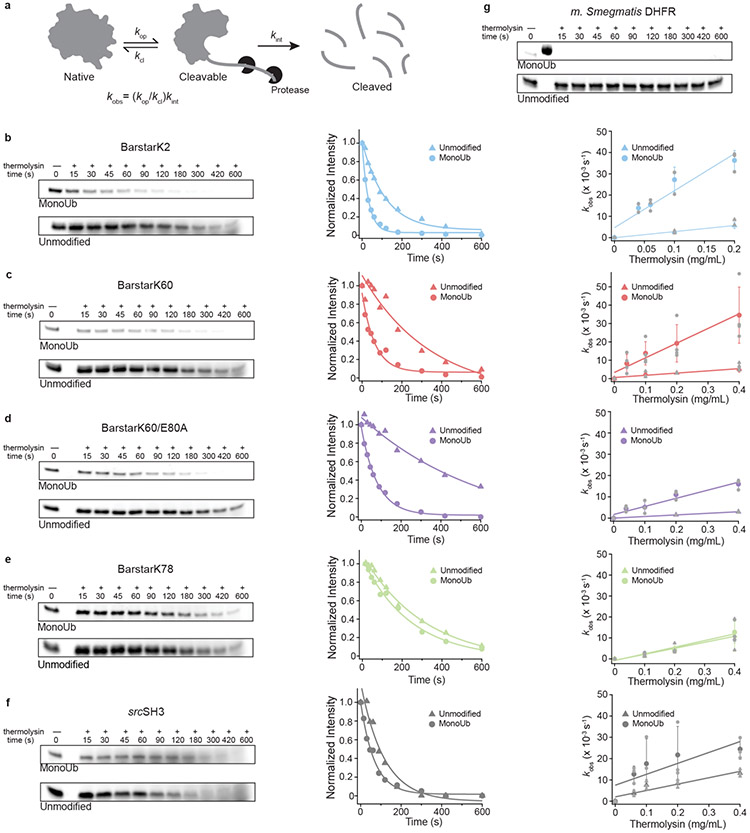
Fig. 1 ∣. Generation of substrates with isopeptide-linked ubiquitin in structured regions and equilibrium unfolding studies.
(a) Schematic of ubiquitination machinery and substrate design. A His6-MBP scaffold with PPPY Rsp5-binding motif for enzymatic ubiquitination is fused to the N-terminus of a single-lysine substrate. (b) Representative size exclusion chromatography trace for methylated monoUb-barstar and Coomassie-stained gel of selected size exclusion fractions. (c-f) Ribbon diagrams of barstar (green: PDB: 1BTA) showing the position of ubiquitinated lysines in red and urea-induced unfolding transition (n=1) of unmodified (triangles) and methylated monoUb-barstar (circles). (g) Ribbon diagram of M. smegmatis DHFR homolog from M. tuberculosis (orange: PDB: 1DG8) and srcSH3 (grey: PDB: 1SRL) showing ubiquitinated lysine positions in red.
After purification, efficient in vitro poly-ubiquitination was achieved using a reconstituted system with mouse Uba1 ubiquitin-activating enzyme (E1), yeast Ubc4 ubiquitin conjugase (E2), and yeast Rsp5 ubiquitin ligase (E3) (Fig. 1a). Treatment with the K63-specific deubiquitinase AMSH (Associated Molecule with the SH3 domain of STAM) collapses the heterogeneously poly-ubiquitinated substrates into lower molecular weight conjugates (Fig. 1a and Supplementary Fig. 1a-f). AMSH efficiently trims the Rsp5-generated, K63-linked ubiquitin chains, but is much slower in removing the proximal, substrate-attached ubiquitin moiety. Optimizing the AMSH amount and the duration of deubiquitination before quenching with EDTA allowed accumulation of the mono-ubiquitinated species. For experiments requiring large quantities, we generated mono-ubiquitinated substrates using methylated ubiquitin, which prevents chain formation and results in higher yield of modified protein. For both approaches, a two-step subtractive Ni2+-NTA purification followed by size-exclusion chromatography was sufficient to purify the mono-ubiquitinated substrate to homogeneity (Fig. 1b and Supplementary Fig. 2a-2e). Using this generalizable method, we attached ubiquitin to various single-lysine substrates (Fig. 1c-1h, Supplementary Fig. 1a-1f, and Supplementary Fig. 2a-2e) and scaled up to produce spectroscopic quantities.
Site-specific, ubiquitin-induced global energtic changes
The ability to purify milligram quantities of homogenously mono-ubiquitinated proteins enabled us to determine global stability changes using traditional chemically-induced equilibrium unfolding monitored by intrinsic fluorescence. The fluorescence signal arises exclusively from tryptophan residues in our substrates, as ubiquitin is tryptophan-free. For these studies, we used a well-established model protein, barstar from Bacillus amyloliquefaciens, in which all except one lysine were replaced by arginine to generate different single-lysine variants for site-specific ubiquitination.
Four single-lysine barstar variants were characterized: barstarK2, barstarK60, barstarK78, and barstarK60/E80A (where the position of the remaining lysine is denoted after barstar). We determined their global stabilities in both unmodified and purified mono-ubiquitinated forms by urea-induced chemical denaturation and fit the data using a two-state assumption and linear extrapolation (see Methods). The non-ubiquitinated versions of all single-lysine variants display only minor destabilization compared to wild-type barstar (ΔGunfolding = 5.0 +/− 0.5 kcal/mol and Cm = 4.7 +/− 0.2 M urea)21 (Fig. 1c-1f). In contrast, we observed dramatically different stabilities upon modification with mono-ubiquitin, indicating site-specific effects (Fig. 1c-1f, Table 1).
Table 1. Summary of thermodynamic values determined for all barstar variants and srcSH3 in this study.
Equilibrium denaturant-induced unfolding transitions were performed in a single trial (n=1), and values reported represent fit parameters. Native-state proteolysis experiments were performed in triplicate (n=3; barstarK2, barstarK78, and barstarK60/E80A) or quadruplicate (n=4; barstarK60), and values represent the mean. All reported errors represent S.E.M derived from curve fitting and propagated through all calculations.
| BarstarK2 | BarstarK60 | BarstarK78 | BarstarK60/E80A | SH3 | |
|---|---|---|---|---|---|
| Cm unmodified (M urea) | 4.97 +/− 0.28 | 4.41 +/− 0.55 | 4.65 +/− 1.19 | 6.02 +/− 0.38 | n.d. |
| Cm monoUb (M urea) | 2.52 +/− 0.16 | 2.51 +/− 0.34 | 4.24 +/− 1.05 | 3.72 +/− 0.26 | n.d. |
| ΔCm (unmodified-monoUb) (M urea) | 2.45 +/− 0.32 | 1.90 +/− 0.65 | 0.42 +/− 1.59 | 2.30 +/− 0.46 | n.d. |
| m-valueglobal unmodified (kcal/mol M) | −1.06 +/− 0.04 | −0.96 +/− 0.08 | −0.89 +/− 0.17 | −1.16 +/− 0.05 | n.d. |
| m-valueglobal monoUb (kcal/mol M) | −0.59+/− 0.02 | −0.68 +/− 0.07 | −0.48 +/− 0.09 | −0.70 +/− 0.04 | n.d. |
| Δm-valueglobal unmodified-monoUb (kcal/mol M) | −0.47 +/− 0.05 | −0.28 +/− 0.11 | −0.41 +/− 0.19 | −0.47 +/− 0.06 | n.d. |
| ΔGunfolding Unmodified# (kcal/mol) | −5.27 +/− 0.20 | −4.25 +/− 0.38 | −4.16 +/− 0.74 | −6.99 +/− 0.31 | n.d. |
| ΔΔGunfolding unmodified-monoUb* (kcal/mol) | −2.02 +/− 0.10 | −1.56 +/− 0.22 | −0.28 +/− 0.45 | −2.14 +/− 0.11 | n.d. |
| ΔGproteolysis unmodified (kcal/mol) | −2.72 +/− 0.02 | −3.24 +/− 0.11 | −2.72 +/− 0.06 | −3.52 +/− 0.01 | −2.70 +/− 0.12 |
| ΔGproteolysis monoUb (kcal/mol) | −1.66 +/− 0.10 | −2.12 +/− 0.06 | −2.66 +/− 0.09 | −2.56 +/− 0.08 | −2.38 +/− 0.23 |
| ΔΔGproteolysis unmodified-monoUb (kcal/mol) | −1.07 +/− 0.11 | −1.12 +/− 0.13 | −0.06 +/− 0.15 | −0.96 +/− 0.12 | −0.32 +/− 0.10 |
indicates that ΔGunfolding values were calculated using a two-state model with linear extrapolation.
indicates that ΔΔGunfolding values were calculated by multiplying the Cm from the denaturation curves by the average m-value for the unmodified and mono-ubiquitinated proteins.
Interestingly, all mono-ubiquitinated constructs show a small but notable decrease in m-value (the denaturant dependence of stability) compared to their unmodified counterparts (Table 1). m-values are known to correlate with the size of a protein or the non-polar surface area exposed during unfolding22, which may slightly change with the various ubiquitin attachments. Alternatively, these decreased m-values may indicate direct surface interactions with ubiquitin or a loss of two-state unfolding behavior, with the population of an unfolding intermediate23,24. Because this questions the validity of the two-state assumption used to calculate ΔGunfolding, we report the midpoints of the denaturation curves (Cm) for the unmodified and mono-ubiquitinated variants. BarstarK2 and barstarK60 were destabilized upon mono-ubiquitination (ΔCm of 2.5 M and 1.9 M urea, respectively, Fig. 1c and 1e). A stabilized mutant of barstarK60, barstarK60/E80A, exhibited nearly identical net destabilization upon mono-ubiquitination (ΔCm of 2.3 M urea, Fig. 1f). Conversely, mono-ubiquitination of barstarK78 caused only marginal destabilization (ΔCm of 0.42 M urea, Fig. 1d). To provide a sense for the thermodynamic changes associated with ubiquitination, we used the average m-value of the fits for unmodified and mono-ubiquitinated barstar variants to approximate ΔGunfolding (Table 1). Taken together, these results establish that the energetic effects of ubiquitin on a particular substrate can be highly site-specific, rather than broadly destabilizing.
Ubiquitination affects energetics of partial unfolding
While the above results demonstrate that mono-ubiquitin attached via a native isopeptide bond can site-specifically alter a substrate’s global stability, the globally unfolded state is unlikely to be the most relevant fluctuation for proteasomal degradation. Under cellular conditions, proteins sample partially-unfolded conformations more frequently than the globally-unfolded state. Furthermore, the proteasome does not require global unfolding for successful substrate engagement.
To assess the population of partially-unfolded states, we utilized a quantitative analysis of susceptibility to a soluble protease, thermolysin25,26. Because cleavage by soluble proteases requires regions of ~10-12 unstructured amino acids25, proteolysis of well-folded proteins under native conditions occurs via transient excursions to partially-unfolded, high-energy states (Fig. 2a). Typically, the lowest energy conformation that is competent for proteolytic cleavage (the “cleavable state”) predominates. Because thermolysin has low affinity for its substrates (Kd ~ 0.1-10 mM), proteolysis of the native state typically proceeds via an EX2-like kinetic regime, in which the proteolysis step itself, rather than the conformational change to the cleavable state, is rate-limiting25. As such, observed proteolysis rates are directly related to the free-energy difference between the native state and this cleavable state (ΔGproteolysis, Supplementary Fig. 3a). The ΔΔGproteolysis for the same protein in two different states (i.e. unmodified and mono-ubiquitinated) can be reliably determined25,26.
Fig. 2 ∣. Native-state proteolysis demonstrates the effects of mono-ubiquitination on the energetics of partial unfolding.
(a) Under native conditions, well-folded proteins are proteolyzed via transient excursions to partially-unfolded states. The observed rate of proteolysis, kobs, is proportional to the free energy of the conformational change from the native to partially-unfolded state (ΔGproteolysis). (b-f) Representative gels for native-state proteolysis and quantified band intensities for indicated substrate proteins at 0.2 mg/mL thermolysin. ΔΔGproteolysis upon mono-ubiquitination with non-methylated ubiquitin is calculated from the ratio of slopes of the mean kobs (n=3 for barstarK2, barstarK78, and barstarK60/E80A; or n=4 for barstarK60) against thermolysin concentration ranging from 0.04 to 0.4 mg/mL. Individual trial data are represented in light grey. Error bars represent the standard deviation of replicates. (g) Representative gels for native-state proteolysis of M. smegmatis DHFR at 0.2 mg/mL thermolysin (n=2). See Supplementary Fig. 5 and Source Data Fig. 2 for representative uncropped gels.
We measured the ΔGproteolysis for unmodified and mono-ubiquitinated (non-methylated) versions of all single-lysine barstar variants described above, as well as single-lysine srcSH3 and M. smegmatis DHFR (wildtype is single-lysine). AMSH concentration and reaction length were adjusted to yield a mixture of both unmodified and mono-ubiquitinated protein, which allowed their direct comparison within the same experiment. Importantly, although ubiquitin methylation has been observed to have various effects on the behavior and recognition of ubiquitin, we observed no difference compared to non-methylated ubiquitin in our biophysical measurements (Supplementary Fig. 4).
Unmodified barstarK2, barstarK60, and barstarK78 exhibit nearly identical proteolysis kinetics (Supplementary Fig. 3b,c) and are proteolyzed through sub-global unfolding (ΔGproteolysis < ΔGunfolding, Table 1). We observed a similar trend in the ΔΔGproteolysis values as for the global stabilities, with barstarK2 and barstarK60 showing significant changes in the population of the cleavable state (ΔΔGproteolysis = −1.1 kcal/mol, Fig. 2b,c, Supplementary Fig. 5a,c, and Table 1). BarstarK60/E80A exhibited a similar ΔΔGproteolysis (−0.96 kcal/mol, Fig. 2d and Supplementary Fig. 5d). Conversely, negligible ΔΔGproteolysis was detected for barstarK78, indicating no change in the energetics of partial unfolding upon mono-ubiquitination (Fig. 2e and Supplementary Fig. 5b).
These variable effects on ΔGproteolysis were recapitulated with other proteins. A single-lysine srcSH3 domain variant showed little ΔΔGproteolysis (−0.32 kcal/mol, Fig. 2f and Supplementary Fig. 5e), which is particularly interesting because the srcSH3 domain is smaller than ubiquitin (64 aa vs 76 aa). In contrast, the naturally single-lysine M. smegmatis DHFR (159 aa) shows the most drastic changes upon mono-ubiquitination (Fig. 2g and Supplementary Fig. 5f) and is completely proteolyzed within the dead time of the experiment (15 seconds), despite very little cleavage on this timescale for unmodified DHFR. Interestingly, monoUb-DHFR is still capable of binding methotrexate, albeit with greatly reduced affinity, suggesting that the native state is populated (Supplementary Fig. 2e and Supplementary Fig. 3d). Nevertheless, even in the presence of 500 μM methotrexate, the mono-ubiquitinated variant is completely proteolyzed within the dead time of the experiment (Supplementary Fig. 6).
Fast proteasomal degradation of Ub-destabilized proteins
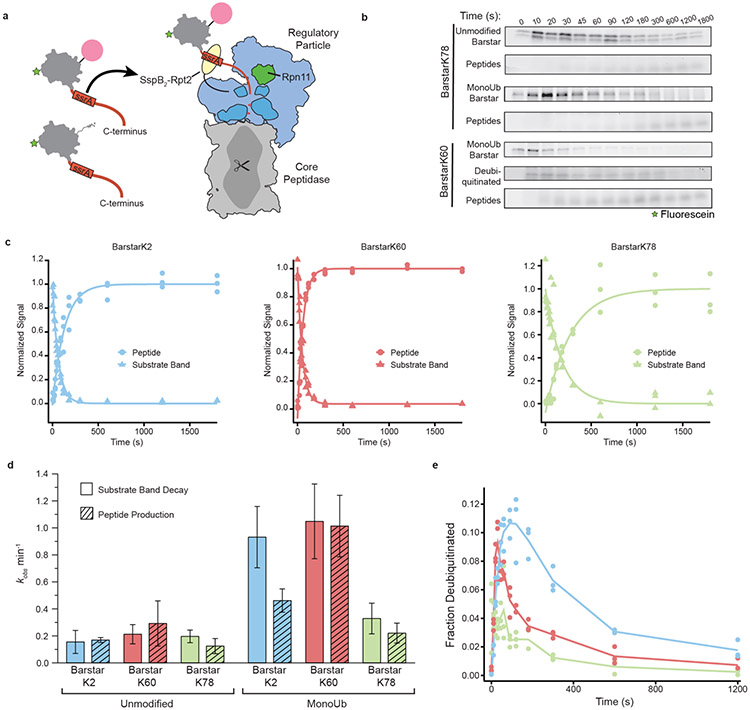
The ability of the proteasome’s AAA+ motor to unfold proteins is paramount to successful clearance of substrates and has been proposed as the rate-limiting step for degradation27. Therefore, we asked whether ubiquitin-mediated substrate destabilization conferred an increase to the proteasomal degradation rate. In order to compare directly mono-ubiquitinated substrates to their non-ubiquitinated counterparts, we used a system for ubiquitin-independent substrate delivery to the proteasome. In this system, a permutant of the bacterial SspB2 adaptor protein fused to the N-terminus of the Rpt2 ATPase in the proteasomal AAA+ motor recruits substrates that contain an ssrA sequence on a sufficiently long unstructured tail region for engagement28 (Fig. 3a and Supplementary Fig. 7a). All substrates delivered in this manner are engaged equally, and thus, observed changes in degradation rate can be attributed to differences in substrate energetics. These experiments were performed at substrate concentrations saturating for SspB2 binding, but well below the Kd of mono-ubiquitin for proteasomal ubiquitin receptors29 to rule out contributions of ubiquitin to substrate recruitment and engagement. Proteasome-mediated degradation under single-turnover conditions (Supplementary Fig. 7b) was monitored by SDS-PAGE, and rates were determined based on both the disappearance of full-length substrate and the appearance of peptide products (Fig. 3b and Supplementary Fig. 7c).
Fig. 3 ∣. Mono-ubiquitin mediated substrate destabilization directly modulates degradation rate.
(a) Schematic of ubiquitin-independent substrate delivery system, where substrates contain a flexible C-terminal tail with an ssrA-recognition motif that binds an SspB2-dimer (yellow) fused to the base AAA+ ATPase. Core particle is represented in gray, regulatory particle in blue, Rpn11 in green, the AAA+ ATPase motor in dark blue, pore loops in red, substrate in gray, with a green star representing fluorescein, red representing the unstructured tail, and ubiquitin in pink. (b) Representative fluorescein-scanned SDS-PAGE gels showing disappearance of unmodified barstarK78 or mono-ubiquitinated (monoUb) barstarK78 and K60 with concomitant peptide production during proteasomal degradation upon ubiquitin-independent delivery. The transient appearance of a deubiquitinated species for monoUb-barstarK60 is shown and quantified in e. (c) Normalized fractional signal plotted as individual points (n=3) of mono-ubiquitinated substrate band decay and peptide production. Lines represent fit of mean values (n=3) to Equation 3. (d) Calculated rates for proteasomal degradation derived by curve fitting to the mean (n=3) and associated fitting errors (S.E.M.) from b and c. (e) Fraction of total signal of deubiquitinated species plotted against time as mean (line) and individual data points (dots; n=3). See Supplementary Fig. 7 and Source Data Fig. 3 for uncropped gel images.
All ubiquitinated and non-ubiquitinated barstar variants were processed by the proteasome. As expected, all showed anti-correlated substrate depletion and peptide formation with fast kinetics that were dependent on the presence of RP and ATP (Fig. 3b,c, Supplementary Fig. 7c,d, and Supplementary Fig. 8). Degradation rates thereby correlated with the stability changes described above. All non-ubiquitinated barstar variants displayed similar degradation kinetics, with an observed rate (kobs) of 0.1 - 0.3 min−1 (Fig. 3d). As previously documented, full-length, unmodified substrate bands appeared as doublets27. MonoUb-barstarK78 showed comparable kinetics, consistent with the negligible stability change upon ubiquitination for this variant (Fig. 3d). However, for the highly destabilized monoUb-barstar variants, degradation kinetics were substantially increased (kobs = 1.04 min−1 for monoUb-barstarK60 and 0.93 min−1 for monoUb-barstarK2), suggesting that ubiquitin-mediated substrate destabilization increases the rate of unfolding by the proteasome.
For monoUb-barstarK60, we obtained similar results when following the substrate decay versus peptide production (Fig. 3c,d). For the monoUb-barstarK2 variant, however, these two processes were decoupled, with the mono-ubiquitinated species disappearing two times faster than the appearance of peptide products (Fig. 3c,d). This apparent decoupling may originate from differences in the temporal order of deubiquitination and unfolding. All variants showed a transient appearance of deubiquitinated species (Fig. 3e), accounting for ~10% of the total substrate intensity for barstarK2 and barstarK60 at their peak. However, the deubiquitinated barstarK60 species was short-lived (peaked at 30 s, negligible at 3 mins), while the barstarK2 species persisted for ~5 mins. Differences in the placement of ubiquitin relative to the substrate-engagement site (the C-terminal appended tail) may alter the timing of deubiquitination relative to crossing the unfolding barrier. In the native barstar structure, the N- and C-termini are located in close proximity (Fig. 1c-e and Fig. 3a, PDB: 1BTA). Engagement via the fused C-terminal tail may therefore place the K2-ubiquitin in close proximity to the proteasome’s deubiquitinase (Rpn11), allowing deubiquitination immediately after engagement and before unfolding. If deubiquitination occurs prior to substrate unfolding, the destabilizing effect conferred by ubiquitin is lost, resulting in a lower rate of peptide production compared to the disappearance of the ubiquitinated substrate. Other ubiquitination sites (such as K60 or K78) might require substrate unfolding and translocation to occur first to position the ubiquitin-modified lysine for deubiquitination. These data therefore support the correlation between a substrate’s thermodynamic stability and its rate of proteasomal degradation, and extend this hypothesis to include ubiquitin attachment as a mode of site-specific destabilization of substrate proteins.
Ubiquitination can induce a proteasome-engageable region
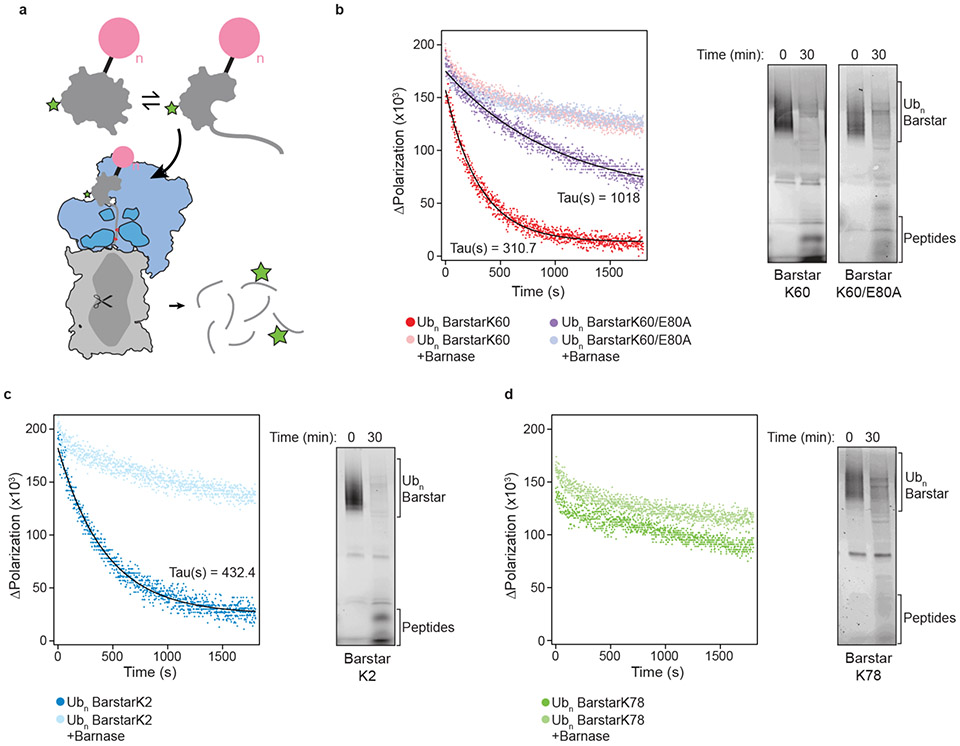
We next investigated the effect of ubiquitin-induced energetic changes on substrate engagement by the proteasomal AAA+ motor. Numerous studies have demonstrated the role of an unstructured initiation or engagement region7,8,30, yet a substantial fraction of cellular proteasomal substrates appear to lack such flexible segments9, begging the question of how their degradation is initiated. While other unfoldases, like Cdc48/p97 may generate disordered regions10,12,31, it is also possible that for some proteins ubiquitin-mediated conformational changes are sufficient to expose the obligate unstructured segments. To test this hypothesis, we poly-ubiquitinated our panel of single-lysine barstar variants (Ubn-barstar) and assayed the proteasome’s ability to recognize these substrates via its endogenous ubiquitin receptors and degrade them in an ATP-dependent manner (Fig. 4a). Native-state proteolysis experiments showed that these poly-ubiquitinated barstar variants have similar energetic profiles as the mono-ubiquitinated variants (Supplementary Fig. 9a).
Fig. 4 ∣. Ubiquitin-mediated destabilization of barstar is sufficient to expose a proteasome-engageable unstructured region.
(a) Schematic of degradation reaction, showing Ubn-substrate lacking an unstructured region at equilibrium with a partially-unfolded state, whereby the partially-unfolded state is competent for proteasome engagement, unfolding, and proteolysis. Core particle is represented in gray, regulatory particle in blue, the AAA+ ATPase motor in dark blue with pore loops in red, substrate in gray with a green star representing fluorescein, and ubiquitin in pink. Degradation can be monitored through the decrease in fluorescence polarization upon transition from a large poly-ubiquitinated substrate to peptides. (b-d) Left: fluorescence polarization kinetic measurements for single-turnover degradations of Ubn-barstar in absence or presence of saturating barnase, presented as individual data points (n=3), with lines representing fitting to Equation 3 and calculated time constants (Tau) shown. Right: fluorescein scan of SDS-PAGE gel with 30-minute endpoint samples for single-turnover Ubn-barstar degradations, showing conversion of substrate to peptides and/or deubiquitinated species. Uncropped gels are presented in Supplementary Fig. 9 and as Source Data Fig. 4.
Surprisingly, despite not harboring any obvious proteasome-engageable unstructured region, some poly-ubiquitinated single-lysine barstar variants were fully degraded by the 26S proteasome, whereas others were only slowly deubiquitinated (Supplementary Fig. 9b). Importantly, the degradation kinetics depend on the ubiquitination site and correlate with the thermodynamics reported above. To gain a quantitative understanding of the degradation kinetics, we utilized the fluorescein label on Ubn-barstar and monitored degradation through the decrease in fluorescence polarization (Fig. 4a). Under single-turnover conditions (confirmed by varying the proteasome concentration, Supplementary Fig. 9c), Ubn-barstarK60 and Ubn-barstarK2 showed exponential degradation kinetics, with time constants of approximately 310 s and 432 s, respectively (Fig. 4b,c). In contrast, Ubn-barstarK78 did not show measurable degradation, consistent with the hypothesis that site-specific, ubiquitin-mediated substrate destabilization determines whether an unstructured region for proteasome engagement is sufficiently populated (Fig. 4d). Furthermore, introducing the stabilizing mutation E80A to Ubn-barstarK60 substantially increased the degradation time constant to 1018 s (Fig. 4b).
To further support our hypothesis that the ubiquitin-mediated modulation of barstar’s energy landscape is the principal determinant for its degradability, we added saturating concentrations of barnase, the high-affinity ligand of barstar, to these reactions (Supplementary Fig. 9d). In all cases, barnase ablated substrate degradation. The remaining minimal decrease in fluorescence polarization could be attributed to minor degradation-independent deubiquitination (Fig. 4b-d). Addition of barnase has no effect on the degradation of a ubiquitinated titin substrate with a flexible initiation region (FAM-Titin-I27V13P,V15P-35mer-tail)27, confirming that the inhibition observed for the barstar variants was due to specific binding and stabilization of barstar’s folded state, rather than inhibitory interactions with the proteasome (Supplementary Fig. 9d).
In addition, we monitored degradation of the Ubn-barstar variants by the isolated core particle to verify that robust degradation requires the entire 26S proteasome and includes ubiquitin recognition, ATP-driven unfolding and translocation. The core particle can only hydrolyze unstructured polypeptides that diffuse into its central chamber to access the proteolysis sites. Indeed, the core particle only minimally cleaved the Ubn-barstarK2 and Ubn-barstarK60 species with low rates compared to the 26S holoenzyme (Supplementary Fig. 9c,e). Similar to the differences seen for the ATP-dependent degradation by the 26S proteasome, Ubn-barstarK78 displayed no core-particle mediated degradation, and Ubn-barstarK60/E80A was cleaved by the core much more slowly than Ubn-barstarK60.
Unlike our observations with the ubiquitin-independent delivery system, where we saw buildup of a deubiquitinated species for monoUb-barstarK2 (Fig. 3e), Ubn-barstarK2 did not populate a deubiquitinated species (Fig. 4c and Supplementary Fig. 9b). Because Ubn-barstarK2 lacks the appended unstructured C-terminal tail, it must engage via a partially-unfolded state, in which the ubiquitin attachment site may no longer be optimally positioned for Rpn11-mediated cleavage prior to unfolding. Moreover, given that this variant is ubiquitinated near the N-terminus, it must be engaged C-terminal to the ubiquitination site. This is confirmed by our observation that inhibition of Rpn11 deubiquitination by o-phenanthroline did not inhibit degradation of Ubn-barstarK2, but inhibited all other variants (Supplementary Fig. 10a). For Ubn-barstarK2, the polypeptide between the ubiquitin-attachment point, K2, and the fluorescein-labeled Cys82 (80 residues) is long enough to span the minimal distance between the entrance of the AAA+ pore and the proteolytic active sites (approximately 55 residues; Supplementary Fig. 10b). Rpn11-inhibited proteasomes can therefore move this substrate far enough into the 20S core for proteolysis near fluorescein, before translocation stalls on the K2-attached ubiquitin chain32.
Ub-induced unfolding is rate-limiting for degradation
The proteasomal degradation rates observed for poly-ubiquitinated barstar variants are notably lower than for barstar or other substrates with flexible tails27,33,34, suggesting that engagement of a spontaneously unfolding region represents the rate-limiting step for degradation. To probe this further, we turned to a proteasome variant, Rpn5-VTENKIF, whose mutations in the RP affect the conformational equilibrium of the proteasome and thereby hinder insertion of flexible segments into the AAA+ pore, making engagement rate-determining even for moderately stable substrates with unstructured tails34. Using Rpn5-VTENKIF proteasome, we see a three-fold (Ubn-barstarK2) and two-fold (Ubn-barstarK60) decrease in degradation rates (Supplementary Fig. 10c), suggesting that their slow degradation kinetics are indeed determined by slow engagement and not unfolding. This leads to the interesting conclusion that for well-folded substrates, exposure of a flexible segment through spontaneous unfolding determines the rate of degradation, providing an alternative means of regulation for proteasomal targeting.
Discussion:
Clearance of damaged, misfolded, and regulatory intracellular proteins is paramount for sustaining life and catalyzed largely by the UPS. While substrate energetics critically affect the degradation of various substrates7,27,35,36, the influence of the substrate-attached ubiquitin itself has been elusive. Here, we show that ubiquitin can mediate substrate destabilization with direct consequences for proteasomal degradation. To carry out these studies, we developed a generalizable system to produce ubiquitin-modified single-lysine proteins with native isopeptide bonds (Fig. 1a), achieving efficient ubiquitination for several different single-lysine substrates. We expect that this strategy will be useful to address a number of biological questions that are currently hampered by challenges in producing and purifying proteins with natively attached ubiquitin on structural domains15. Using these isopeptide-linked ubiquitinated substrates, we show that ubiquitin-mediated energetic effects can dictate how fast a protein is degraded and, surprisingly, whether a protein is susceptible to proteasomal degradation at all, thus providing an additional regulatory mechanism for clearance of a ubiquitinated substrate based on its conformational and energetic properties.
Consistent with this concept, we found that stabilizing the substrate via ligand binding (as in barstar:barnase) inhibits proteasomal processing. The engagement of these substrates appears to be rate-limiting and modulated directly by the accessibility of partially-unfolded, proteasome-engageable states. Thus, the overall context of the ubiquitinated protein with respect to cellular environment, binding partners, and perhaps other stabilizing or destabilizing PTMs can influence whether a ubiquitinated substrate is actually degraded.
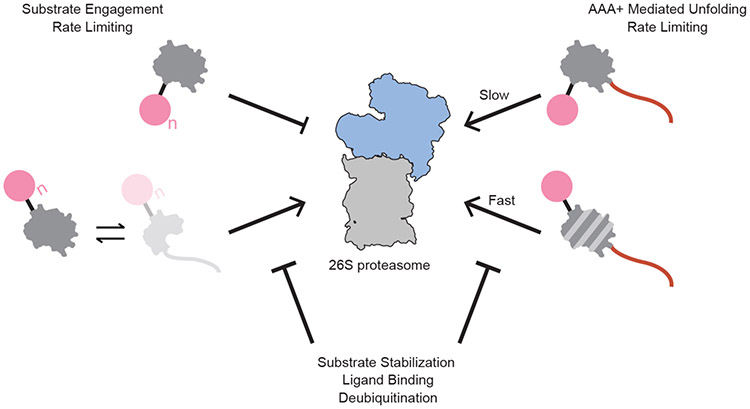
Based on our results, we can build a model for the effect of ubiquitin-mediated, site-specific changes in protein energy landscapes on proteasomal degradation (Fig. 5), in which: 1) a protein may or may not be engaged by the proteasome based on its altered energetics, and 2), the speed with which ubiquitinated substrates are degraded is related to the extent of ubiquitin-induced destabilization. Both aspects of proteasomal turnover are directly modulated by the increased sampling of partially-unfolded states and further influenced by other factors, such as stabilizing mutations or deubiquitination prior to substrate unfolding, either at the proteasome by Rpn11 or by a host of cellular deubiquitinases37.
Fig. 5 ∣. Model for the consequences of site-specific, ubiquitin-induced substrate energy landscape modulation on proteasomal degradation.
If ubiquitination occurs on a nonsensitive structured lysine, as in barstarK78, the substrate does not sufficiently populate a partially-unfolded, proteasome-engageable conformation. If ubiquitin-modification occurs on a sensitive lysine, as in barstarK2 and barstarK60, the otherwise well-folded substrate is sufficiently destabilized to populate partially-unfolded, proteasome-engageable conformations and is successfully degraded. The observed degradation kinetics thus appear dependent on the changes to the protein energy landscape upon ubiquitination. When substrates contain an unstructured proteasome-engageable region, ubiquitination at sensitive lysine positions allows for substantially faster degradation kinetics, while degradation kinetics of substrates with non-destabilizing ubiquitinations remain essentially unchanged. Successful proteasome engagement and degradation of ubiquitin-destabilized substrate proteins can be slowed or blocked by a number of energetically stabilizing events, including deubiquitination, ligand binding, or stabilizing mutation.
This model has implications for a number of different processes, including the engineering of substrate degradation via Proteolysis Targeting Chimeras (PROTACs)38. PROTACs are synthetic molecules containing two moieties, a ligand binding the target protein to be degraded and another ligand with affinity for an E3 ubiquitin ligase that facilitates ubiquitination of the target. The linker length between the two ligands has been found to affect whether the target protein is degraded39, likely because it determines which lysines on the target are ubiquitinated in a manner that facilitates delivery to the downstream processing enzymes (i.e. Cdc48/p97 and the proteasome)31,40, but also possibly depending on whether ubiquitination at these lysines destabilizes the target. Non-specific ligands that promiscuously bind to 50-100 protein kinases were found to facilitate the degradation of only a small subset of these kinases41,42, which could also be due to which lysines are ubiquitinated on the different targets and whether these ubiquitinations are sufficiently destabilizing to allow degradation.
While it is clear that ubiquitination has site-specific effects on the energy landscape, the mechanisms for ubiquitin-induced destabilization and the population of partially-unfolded conformers remains unknown. Potential mechanisms include destabilization from reduced conformational entropy in the substrate, a ubiquitin-induced entropic pulling force, direct substrate-ubiquitin interactions, or the ubiquitin-induced population of an intermediate state. There are no clear patterns regarding the region or type of secondary structure within the substrate that is energetically sensitive to the attachment of ubiquitin, nor are the effects correlated with the substrate size, as previously suggested16. It is reasonable to expect that the addition of a protein domain, such as ubiquitin, can alter the energetics and dynamics of a target protein in this manner. Biophysical studies of multidomain proteins have demonstrated that the stability of one domain can be modulated by the presence of another43. In differentially-linked polyubiquitin chains, the ubiquitin monomers themselves can have different thermodynamic and mechanical stabilities44,45. Studies on N-terminal ubiquitin fusions and disulfide-linked ubiquitin attachments have reported small changes in the midpoints for thermally-induced unfolding depending on the modification16.
Computational studies have postulated that ubiquitin-induced destabilization is a result of a decrease in a substrate’s overall conformational entropy14. Site-specific effects could be realized through the difference in the potential flexibility at the different sites. The local structure and packing at the three different ubiquitination sites in barstar, however, do not reveal any notable differences in the density of atomic contacts or number of contacting residues (PDB: 1BTA). Detailed calculations or experiments evaluating these potential changes in conformational entropy are needed to evaluate this hypothesis.
Our results do not yield specific information about a potential entropic pulling force. NMR studies of the protein FKBP12 with chemically conjugated ubiquitin demonstrated increased backbone flexibility16, which could be rationalized by an entropic pulling model whereby a highly stable protein fold, like ubiquitin, attached through a native isopeptide bond with many degrees of translational and rotational freedom, can provide a net pulling force on the substrate from the site of ligation46.
The energetic modulation may also arise from direct interactions between the ubiquitin and the substrate. Ubiquitin has multiple exposed hydrophobic patches, one near Ile44 and another at Ile36, which could potentially stabilize exposed hydrophobic residues on a partially-unfolded substrate. The Ile44 hydrophobic patch is known to interact with PCNA when in an N-terminal fusion47 and is responsible for the inter-ubiquitin interactions that give K48-linked ubiquitin chains their compact conformation48,49. Ubiquitin also contains an acidic patch that electrostatically interacts with some target proteins50. In sum, how exactly ubiquitin destabilizes the substrate protein remains unknown and will require further investigation.
Cellular proteostasis relies upon careful regulation of protein degradation via the UPS, and the consequences of aberrant degradation are severe. We find that ubiquitin directly modulates a protein’s conformational energy landscape, and these energetic changes play a pivotal role in regulating both 26S proteasome substrate selection and degradation kinetics. We conclude that ubiquitin signaling and proteasomal degradation overall are dependent on the biological and biophysical contexts of individual ubiquitinated proteins. A full understanding of the energetic effects contributed by a particular ubiquitination event is therefore crucial for building a complete model of how ubiquitin-mediated signals are transduced in vivo. We hope the tools and results presented herein can facilitate addressing these questions and be used to expand our model of the biophysical factors governing ubiquitin-mediated signaling.
Online Methods:
Protein Purification:
Preparation of substrate proteins
E. coli BL21 Rosetta 2 (DE3) cells were transformed with either pEC072 (single-lysine srcSH3), pEC074 (M. smegmatis DHFR), pEC076 (barstarK2), pEC062 (barstarK60), pEC081 (barstarK60/E80A), or pEC059 (barstarK78). Cells were then grown in 2 L LB Broth (Fisher) to 0.4 < OD600 < 0.8 and induced with 1 mM IPTG for 3 hours at 37°C. Bacteria were then pelleted and resuspended in 50 mM HEPES pH 7.0, 150 mM NaCl, 0.5 mM TCEP supplemented with 1X Halt™ protease inhibitor cocktail (Thermo) and benzonase (Novagen). Resuspended cells were lysed by sonication and the lysate was clarified by centrifugation at 20,000 rcf, 4°C, 30 minutes. The substrate was first purified by Ni2+-NTA affinity chromatography using its N-terminal His6 tag. Clarified lysate was allowed to batch bind to HisPur™ Ni2+-NTA resin (Thermo) washed with 50 mM HEPES pH 7.0, 150 mM NaCl, 25 mM imidazole, 0.5 mM TCEP and eluted with 50 mM HEPES pH 7.0, 150 mM NaCl, 500 mM imidazole, 0.5 mM TCEP. Concentration of protein in the eluate was then measured using UV-Vis absorption at 280 nm. Eluate was then labeled for 2 hours at room temperature with 5X molar excess fluorescein-maleimide dye (Thermo). The labeling reaction was quenched with 10X molar excess DTT and unreacted dye was removed using a S200 16/60 size exclusion column (GE) pre-equilibrated with 25mM HEPES pH 7.5, 150 mM KCl, and 15 mM MgOAc. Peak corresponding to the labeled, full length His-MBP substrate was collected, and quantified by UV-Vis absorption at 280 nm and 495 nm according to the manufacturer’s instructions before addition of 10% glycerol and flash freezing to store at −80°C for future use.
Preparation of substrate proteins with C-terminal ssrA tag/cyclin B engageable tail
E. coli BL21 Rosetta 2 (DE3) cells were transformed with either pEC098 (barstarK2), pEC093 (barstarK60), pEC097 (barstarK78). Cells were then grown in 2 L LB Broth (Fisher) to 0.4 < OD600 < 0.8 and induced with 1 mM IPTG for 3 hours at 37°C. Bacteria were then pelleted and resuspended in 50 mM HEPES pH 7.0, 150 mM NaCl, 0.5 mM TCEP supplemented with 1X Halt™ protease inhibitor cocktail (Thermo) and benzonase (Novagen). Resuspended cells were lysed by sonication and the lysate was clarified by centrifugation at 20,000 rcf, 4°C, 30 minutes. The substrate was first purified by Ni2+-NTA affinity chromatography using its N-terminal His6. Clarified lysate was allowed to batch bind to HisPur™ Ni2+-NTA resin (Thermo) washed with 50 mM HEPES pH 7.0, 150 mM NaCl, 25 mM imidazole, 0.5 mM TCEP and eluted with 50 mM HEPES pH 7.0, 150 mM NaCl, 500 mM imidazole, 0.5 mM TCEP + 1X Halt™ protease inhibitor cocktail. Eluate was diluted 1:2 with 50 mM HEPES pH 7.0, 150 mM NaCl, and 0.5 mM TCEP and 5 mM EDTA was added. Eluate was then batch bound to Strep-Tactin Superflow Plus resin (Qiagen), washed with 50 mM HEPES pH 7.0, 150 mM NaCl and eluted with 50 mM HEPES pH 7.0, 150 mM NaCl, 2.5mM desthiobiotin (Sigma). Eluate was labeled with 5X molar excess fluorescein-maleimide dye (Thermo). The labeling reaction was quenched with 10X molar excess DTT and unreacted dye was removed using a S200 16/60 size exclusion column (GE) pre-equilibrated with 25mM HEPES pH 7.5, 150 mM KCl, and 15 mM MgOAc. The peak corresponding to the labeled, full length, labeled His-MBP substrate was collected, and quantified by UV-Vis absorption at 280 nm and 495 nm according to the manufacturer’s instructions before addition of 10% glycerol and flash freezing to store at −80°C for future use.
Preparation of ubiquitin
E. coli BL21 Rosetta 2 (DE3) cells were transformed with pEC086. Cells were then grown in 2 L LB Broth (Fisher) to 0.4 < OD600 < 0.8 and induced with 1 mM IPTG for 3 hours at 37°C. Bacteria were then pelleted and resuspended in 20 mM sodium acetate pH 5.1 (pH adjusted with acetic acid). Resuspended cells were lysed by sonication and the lysate was clarified by centrifugation at 20,000 rcf, 4°C, 30 minutes. Clarified lysate was loaded onto a HiPrep™ SP XL 16/10 cation exchange column (GE) preequilibrated in 20 mM sodium acetate pH 5.1. Column was washed with 5 column volumes of 20 mM sodium acetate pH 5.1 and then eluted with a gradient of 20 mM sodium acetate to 500 mM sodium acetate pH 5.1. The peak corresponding to WT ubiquitin was collected and further purified by size exclusion on an S75 16/60 column (GE) preequilibrated with 50 mM Tris pH 7.5, 150 mM NaCl. Peak corresponding to WT ubiquitin was collected, and quantified by UV-Vis absorption at 280 nm before flash freezing to store at −80°C for future use.
Preparation of barnase
E. coli BL21 Rosetta 2 (DE3) cells were transformed with pEC099. Cells were then grown in 2 L LB Broth (Fisher) to 0.4 < OD600 < 0.8 and induced with 1 mM IPTG for 3 hours at 37°C. Bacteria were then pelleted and resuspended in 50 mM HEPES pH 7.0, 150 mM NaCl, 0.5 mM TCEP supplemented with 1X Halt protease inhibitor cocktail (Thermo) and benzonase (Novagen). Resuspended cells were lysed by sonication and the lysate was clarified by centrifugation at 20,000 rcf, 4°C, 30 minutes. The substrate was first purified by Ni2+-NTA affinity chromatography using its N-terminal His6. Clarified lysate was allowed to batch bind to HisPur™ Ni2+-NTA resin (Thermo) washed with 50 mM HEPES pH 7.0, 150 mM NaCl, 25 mM imidazole, 0.5 mM TCEP and eluted with 50 mM HEPES pH 7.0, 150 mM NaCl, 500 mM imidazole, 0.5 mM TCEP. HRV3C-protease was added and the cleavage reaction was allowed to proceed overnight at 4°C under dialysis to 50 mM HEPES pH 7.0, 150 mM NaCl, 0.5 mM TCEP. HRV3C-protease and His-MBP tags were removed using a subtractive Ni2+-NTA purification step. Flow through was further purified by size exclusion chromatography using a S75 16/60 column (GE). Peak corresponding to barnase was collected, and quantified by UV-Vis absorption at 280 nm before addition of 10% glycerol and flash freezing to store at −80°C for future use.
Preparation of ubiquitination enzymes
Ubiquitination machinery M. musculus mE1, S. cerevisiae Ubc4, and S. cerevisiae Rsp5 were purified as described previously using the same procedure27,33. E. coli BL21 Rosetta 2 (DE3) pLysS cells were transformed with pAM235 (mE1) or pAM236 (Ubc4) or pAM237 (Rsp5) and grown at 37°C in 6L of terrific broth (Novagen) until OD600 = 0.8 before expression was induced with 1 mM IPTG and allowed to continue overnight at 18°C. Cells were resuspended in 50 mM HEPES pH 7.6, 250 mM NaCl supplemented with protease inhibitors (pepstatin A, aprotonin, PMSF, and leupeptin), benzonase, and lysozyme (2 mg/mL) and stored at −80°C. Resuspended cells were thawed and lysed by sonication before lysate was clarified by centrifugation at 20,000 rcf for 30 mins at 4°C. Clarified lysate was batch bound to HisPur™ Ni2+-NTA resin (ThermoFisher) equilibrated with 50 mM HEPES pH 7.6, 250 mM NaCl for one hour at 4°C. Resin was washed in a gravity flow column with at least 50 mL of 50 mM HEPES pH 7.6, 250 mM NaCl, 20 mM imidazole before protein was eluted with 50 mM HEPES pH 7.6, 250 mM NaCl, 250 mM imidazole. Eluate was concentrated in an Amicon spin concentrator (Millipore) and loaded onto a Superdex200 16/60 size exclusion column (GE) equilibrated in 20 mM HEPES pH 7.6, 100 mM NaCl, 10% glycerol. Peak corresponding with target protein was collected, concentrated in Amicon spin concentrator (Millipore), quantified by absorbance at 280 nm, and flash frozen in liquid nitrogen for storage at −80°C.
Preparation of AMSH deubiquitinase
E. coli BL21 Rosetta 2 (DE3) pLysS cells were transformed with pAM241 and grown in 2 L of terrific broth (Novagen) at 37°C until OD600 = 0.6 after which expression was induced with 0.5 mM IPTG overnight at 18°C. Cells were resuspended in 50 mM HEPES pH 7.6, 250 mM NaCl supplemented with protease inhibitors (pepstatin A, aprotonin, PMSF, and leupeptin), benzonase, and lysozyme (2 mg/mL) and stored at −80°C. Resuspended cells were thawed and lysed by sonication before lysate was clarified by centrifugation at 20,000 rcf for 30 mins at 4°C. Clarified lysate was batch bound to HisPur™ Ni2+-NTA resin (ThermoFisher) equilibrated with 50 mM HEPES pH 7.6, 250 mM NaCl for one hour at 4°C. Resin was washed with 50 mM HEPES pH 7.6, 250 mM NaCl, 10 mM ATP (to remove contaminating DnaK), 20 mM imidazole. The His6 tag was cleaved from AMSH by HRV3C-protease overnight at 4°C and AMSH was clarified through an ortho Ni2+-NTA step using HisPur Ni2+-NTA resin (ThermoFisher). Protein was concentrated in Amicon spin concentrator (Millipore) before being loaded on a S75 16/60 size exclusion column (GE) equilibrated with 20 mM HEPES pH 7.6, 100 mM NaCl, 10% glycerol. Peak corresponding to AMSH was collected, concentrated in an Amicon spin concentrator (Millipore), quantified by absorbance at 280 nm, and flash frozen in liquid nitrogen for storage at −80°C.
Preparation of homogenous mono-ubiquitinated substrate proteins
Substrate proteins, ubiquitin, ubiquitination enzymes, and AMSH were prepared as described above. Ubiquitination reactions were set up in reaction buffer (50 mM HEPES pH 8.0, 150 mM NaCl, 5 mM MgCl2, and 5% glycerol) in 20 μL aliquots as follows: 5 μM Uba1 (E1), 5 μM Ubc4 (E2), 5 μM Rsp5 (E3), 20 μM substrate, 750 μM wild-type (non-methylated) ubiquitin or methylated ubiquitin, 5 mM ATP and incubated in a thermocycler for 3 hours at 25°C. 48 individual 20 μL reactions were performed for a typical prep. After three hours, reactions were pooled and HRV3C-protease was added and allowed to cleave overnight at 4°C. If wild-type (non-methylated) ubiquitin was used, reactions were then treated with 0.5 μM AMSH for 30 minutes at room temperature and quenched with 5 mM EDTA. His-tagged ubiquitination machinery and the His-MBP scaffold were then removed via a subtractive Ni2+-NTA affinity step using a 1 mL HisTrap HP column (GE) pre-equilibrated with 50 mM HEPES pH 7.0, 150 mM NaCl, 25 mM imidazole. This removed most, but not all, of the His-tagged ubiquitination machinery and ubiquitinated His-MBP substrate scaffold. Flow through was then concentrated and loaded onto an S75i 10/300 size exclusion column (GE) pre-equilibrated with 25 mM HEPES pH 7.5, 150 mM KCl, and 15 mM MgOAc. The peak corresponding to the mono-ubiquitinated substrate was collected, concentrated, and quantified by UV-Vis absorption at 280 nm and 495 nm according to the manufacturer’s instructions before addition of 10% glycerol and flash freezing to store at −80°C for future use.
Preparation of mono-ubiquitinated substrate proteins with C-terminal ssrA tag/engageable tail
Substrate proteins and ubiquitination enzymes were prepared as described above. Ubiquitination reactions were set up in reaction buffer (50 mM HEPES pH 8.0, 150 mM NaCl, 5 mM MgCl2, and 5% glycerol) in 20 μL aliquots as follows: 5 μM Uba1 (E1), 5 μM Ubc4 (E2), 5 μM Rsp5 (E3), 20 μM substrate, 500 μM methylated ubiquitin (Millipore), 5 mM ATP and incubated in a thermocycler for 3 hours at 25°C. 24 individual 20 μL reactions were performed for a typical prep. After three hours, reactions were pooled and HRV3C-protease was added and allowed to cleave for 30 minutes at room temperature. Ubiquitination enzymes and His-MBP were removed by batch binding to MagneHis™ (Promega) magnetic Ni2+-NTA resin for 1 hour at 4°C. Resin was pelleted in a magnetic tube rack, and the supernatant was collected for gel based single-turnover ubiquitin-independent degradation assays.
Preparation of proteasome lid subcomplex
Lid subcomplex was recombinantly expressed and purified as described previously27. E. coli BL21-star(DE3) (Invitrogen) cells were transformed with pAM80, pAM85, and pAM86 for lid. pAM80 encodes for Sem1 and rare tRNA codons, pAM85 encodes Rpn5, MBP-HRV3C-Rpn6, Rpn8, Rpn11, and Rpn9, and pAM86 encodes Rpn3, His6-HRV3C-Rpn12, and Rpn7. Cells were grown in 2 L of terrific broth (Novagen) at 37°C until 1.0 < OD600 < 1.5 after which expression was induced with 1 mM IPTG at 16°C for overnight. Bacteria were pelleted and resuspended in 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol and supplemented with protease inhibitors (aprotonin, pepstatinA, leupeptin, and PMSF or AEBSF), benzonase (Novagen), and 2 mg/mL lysozyme and stored at −80°C. Resuspended cells were lysed by sonication and the lysate was clarified by centrifugation at 20,000 rcf, 4°C, 30 minutes. Lid was first purified by Ni2+-NTA affinity chromatography via His6-HRV3C-Rpn12 using a 5mL HisTrap HP (GE) column, washed with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 20 mM imidazole and eluted with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 250 mM imidazole. Eluate was further purified via MBP-HRV3C-Rpn6 and amylose resin (NEB) and eluted with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 10 mM maltose. Amylose eluates were cleaved with HRV3C-protease overnight at 4°C before being loaded onto a Sup6i 10/300 size exclusion column (GE) pre-equilibrated with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 0.5 mM TCEP. Peak corresponding to fully assembled lid was collected, concentrated, and quantified by UV-Vis spectroscopy before being flash frozen and stored at −80°C for future use.
Preparation of proteasome base subcomplex and SspB2-fused base subcomplex
Base subcomplex was recombinantly expressed and purified as described previously51. E. coli BL21-star(DE3) (Invitrogen) cells were transformed with pAM81, pAM83, and pAM82 for wild-type base or pAM81, pAM83, and pAM210 for SspB2-Rpt2 base. pAM82 encodes for Rpt1, Rpt2, Rpt3, Rpt4, Rpt5, and Rpt6, pAM210 encodes Rpt1, SspB2-Rpt2, Rpt3, Rpt4, Rpt5, and Rpt6, pAM81 encodes Rpn1, Rpn2, and Rpn13, and pAM83 encodes rare tRNA codons and base chaperones (Nas6, Nas2, Rpn14, and Hsm3). Cells were grown in 3 L of terrific broth (Novagen) at 37°C until 0.6 < OD600 < 0.8 after which expression was induced with 1 mM IPTG at 30°C for 5 hours followed by 16°C overnight expression. Bacteria were pelleted and resuspended in 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 1 mM ATP and supplemented with protease inhibitors (aprotonin, pepstatinA, leupeptin, and PMSF or AEBSF), benzonase (Novagen), and 2 mg/mL lysozyme and stored at −80°C. Resuspended cells were lysed by sonication and the lysate was clarified by centrifugation at 20,000 rcf, 4°C, 30 minutes. Base was first purified by Ni2+-NTA affinity chromatography via His6-Rpt6 using a 5mL HisTrap HP (GE) column, washed with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 1 mM ATP, 20 mM imidazole and eluted with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 1 mM ATP, 250 mM imidazole. Eluate was further purified via FLAG-Rpt1 and anti-FLAG M2 affinity resin (Sigma) and eluted with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 1 mM ATP, 0.15 mg/mL FLAG peptide (Genscript). FLAG eluates were loaded onto a Sup6i 10/300 size exclusion column (GE) pre-equilibrated with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 1 mM ATP, 0.5 mM TCEP. Peak corresponding to fully assembled base was collected, concentrated, and quantified by Bradford assay (BioRad) using BSA (Sigma) as a standard before being flash frozen and stored at −80°C for future use.
Preparation of proteasome core particle
20S core particle from S. cerevisiae was purified as described previously52 from yeast strain yAM54 bearing 3X-FLAG-Pre1. yAM54 cells were grown in 3 L of YPD at 30°C until saturation (3 days). Cells were pelleted and resuspended in 60 mM HEPES pH 7.6, 500 mM NaCl, 10 mM MgCl2, 5% glycerol, plunged into liquid nitrogen and subsequently stored at −80°C. Frozen resuspended cells were lysed using a 6875 Freezer Mill Dual Chamber Cryogenic grinder (SPEX Sample Prep). Lysate was diluted in 60 mM HEPES pH 7.6, 500 mM NaCl, 10 mM MgCl2, 5% glycerol and clarified by centrifugation at 20,000 rcf, 4C, 45 minutes. Base was first purified by anti-FLAG affinity chromatography using anti-FLAG M2 affinity resin (Sigma), exhaustively washed with 60 mM HEPES pH 7.6, 500 mM NaCl, 10 mM MgCl2, 5% glycerol, and eluted with 60 mM HEPES pH 7.6, 500 mM NaCl, 10 mM MgCl2, 5% glycerol, 0.15 mg/mL FLAG peptide (Genscript). Eluate was loaded onto a Sup6i 10/300 size exclusion column (GE) pre-equilibrated with 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 0.5 mM TCEP. Peak corresponding to fully assembled core was collected, concentrated, and quantified by UV-Vis spectroscopy before being flash frozen and stored at −80°C for future use.
Determination of global substrate stability by intrinsic tryptophan fluorescence
Two 5 μM protein stocks were prepared: A no denaturant protein stock and a high urea protein stock both in 25 mM HEPES pH 7.5, 150 mM KCl, and 15 mM MgOAc. The exact urea concentration in the high denaturant stock was determined by taking the refractive index. Samples with a range of urea concentrations were prepared by serial dilution of the two stocks and allowed to equilibrate at room temperature overnight. Measurements were then performed at 25°C using a PTI Quantamaster Fluorometer (Horiba). Tryptophan fluorescence was excited at 295 nm and a 10 second kinetic read of fluorescence emission at both 330 nm and 350 nm was performed at each denaturant concentration. Samples were recovered from the cuvette after each measurement and the exact urea concentration was determined by taking the refractive index. The signal was averaged over each 10 second period and reported as a ratio of average signal 330/average signal 350. Ratios were then normalized using equation 1 and each mono-ubiquitinated and unmodified variant were globally fit with linked baselines to a two state folding model (equation 2) using Igor Pro 7, which allowed determination of the Cm, ΔGunfolding, and m-value.
| (1) |
| (2) |
Parameter definitions:
m1=folded intercept, m2 = unfolded intercept, m3 = ΔGunfolding, m4 = m-value, m5= folded baseline slope, m6=unfolded baseline slope
Determination of substrate native-state energetics by native-state proteolysis
Ubiquitinated substrate sample prep was performed as described above except that AMSH deubiquitinase was allowed sufficient time to leave a mixed population of unmodified and mono-ubiquitinated species. Additionally, the final size exclusion step was omitted. Protein stocks were prepared in a 2 mL volumetric flask with final buffer of 25 mM HEPES pH 7.5, 150 mM KCl, and 15 mM MgOAc. Samples were allowed to equilibrate at room temperature in the dark overnight. Native-state proteolysis experimental protocol was adapted from previous work25. The equilibrated stock was divided into 200 μL aliquots and thermolysin protease (stock concentration 10 mg/mL) was added to a final concentration of 0.04 to 0.4 mg/mL. Time points (15 μL) were taken at (no protease control, 0:15, 0:30, 0:45, 1:00, 1:30, 2:00, 3:00, 5:00, 7:00, and 10:00) from the reactions and quenched in 2.5 μL of 0.5 M EDTA. 2.5 μL of 6X SDS-PAGE loading buffer was added to each sample and time points were run out on a 12% NuPAGE Bis-Tris™ gel (Invitrogen) in 1X MES running buffer (50 mM MES, 50 mM Tris Base, 0.1% SDS, 1 mM EDTA). Gels were imaged using a BioRad ChemiDoc™ and color inverted using the “Invert” command in ImageJ for ease of viewing and analysis. Band intensities of the unmodified and mono-ubiquitinated substrate bands were then quantified using ImageJ. SH3 and mono-ubiquitinated SH3 gels were quantified in ImageQuant (GE Healthcare) with a rolling ball background subtraction because proteolysis products could comigrate near full length protein. Band intensities were normalized to the no protease lane and fit to a first order exponential (equation 3) using IgorPro 7 to calculate the observed proteolysis kinetics (kobs). For a given substrate, kobs was determined at several thermolysin concentrations and plotted against protease concentrations. ΔΔGproteolysis was calculated from the slope of the linear fit to thermolysin vs. kobs. using equation 4 and equation 5. Individual ΔGproteolysis could also be calculated using equation 6 and the measured kcat/KM of thermolysin for a generic protein of 99,000 M−1s−1 25.
| (3) |
| (4) |
| (5) |
| (6) |
Cy5-methotrexate binding to DHFR by fluorescence polarization
Equilibrium binding of Cy5-methotrexate to M. smegmatis DHFR was assessed by monitoring the increase in fluorescence polarization of Cy5-methotrexate upon binding DHFR. 50 nM Cy5-methotrexate was incubated with increasing concentration of unmodified or mono-ubiquitinated DHFR (quantified by fluorescein fluorescence using a standard curve) in 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 5% glycerol, 0.5 mM TCEP for 20 minutes at room temperature to reach equilibrium. Fluorescence polarization was monitored for 5 minutes on a Synergy Neo2 multi-mode plate reader. Time points were averaged and normalized to Cy5-methotrexate in the absence of DHFR. For the unmodified DHFR, Kd was determined by fitting the change in fluorescence polarization as a function of DHFR concentration to simple single site binding model53 (Equation 7).
| (7) |
Proteasome Degradation Assays:
Preparation of polyubiquitinated barstar variants
Barstar ubiquitination was performed exactly as above except that AMSH removal of K63-linked polyubiquitin chains was omitted. Ortho-Ni2+ purified ubiquitinations were subsequently separated by size-exclusion chromatography on an S200i 10/300 (GE Healthcare) and 0.5 mL fractions were assessed for degradable species by incubating with proteasome under single turnover conditions at 30°C for 30 minutes and analyzing products by SDS-PAGE (Supplementary Fig. 9b).
Gel-based single-turnover ubiquitin-independent degradation assay
2X stocks of substrate (300 nM final) were prepared in assay buffer (60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 0.5 mM TCEP, 5 mM ATP, 5% glycerol, 1 mg/ml BSA). 2X proteasome stocks were performed by reconstituting recombinant lid (5 μM final), recombinant SspB2-Rpt2 base (5 μM final), recombinant Rpn10 (5 μM final), and core particle (2.5 μM final) in assay buffer with an ATP-regeneration system (creatine kinase, creatine phosphate, and 5 mM ATP) and allowed to assemble for 3 minutes at room temperature. Reactions were performed in technical triplicate at 30°C in a thermocycler and initiated by mixing equivolume (12.5 μL) of 2X substrate with 2X proteasome. Time points (1.2 μL) were taken at (0:10, 0:20, 0:30, 0:45, 1:00, 1:30, 2:00, 3:00, 5:00, 10:00, 15:00, 20:00, 30:00 min) from the reactions and quenched in 5 μL of 2X SDS-PAGE loading buffer (125 mM TrisHCl pH 6.8, 20% glycerol, 4% SDS). Gel samples were separated by electrophoresis on 4-20% TGX gels (Bio-Rad) before fluorescence imaging on a typhoon variable mode scanner (GE) with 50 μm per pixel density. Images were quantified in ImageQuant (GE) by normalizing band intensity of each species per total lane intensity to account for loading variation. Quantified species were plotted as percent total signal (Supplementary Fig. 7c) and fit to a single exponential equation (Equation 3) in IgorPro7. For degradations performed with ATPγS, proteasomes were assembled in ATP for 3 minutes at room temperature, then ATPγS was added (5 mM final) for 3 minutes at room temperature prior to substrate addition. For degradations using only the core particle, core particle was added to 900 nM final with substrate and incubated at 30°C for the indicated time points. Time points of 0, 10:00, and 30:00 minutes were quenched in SDS-PAGE loading buffer for trials involving core particle only or ATPγS inhibited proteasome and separated by SDS-PAGE on 4-20% and assess qualitatively.
Fluorescence polarization single-turnover ubiquitin-independent degradation assay
For ubiquitin-independent degradations assessed by fluorescence polarization, reactions were initiated with equivolume (2.5 μL) addition of substrate to proteasome directly within a 384-well black bottom plate (Corning) and fluorescence polarization was monitored in a Synergy Neo2 multimode plate reader (BioTek). Decreased fluorescence polarization over time as substrate was processed into peptides could also be fit to a single exponential model (Equation 3) in IgorPro7.
Fluorescence polarization single-turnover ubiquitin-dependent degradation assay of Ubn barstar variants
Substrates were prepared to 2X concentration (6 nM final) in assay buffer. Proteasome was reconstituted to 2X concentration in assay buffer (2.5 μM lid, base, and Rpn10 with 0.9 μM core particle) and allowed to assemble for 3 minutes at room temperature prior to reaction initiation. Reactions were initiated with appropriate dilution of 2X substate (2.5 μL) into 2X proteasome (2.5 μL) in a 384-well black bottom plate (Corning) and the decrease of fluorescence polarization over time was monitored on a Synergy Neo2 multimode plate reader (BioTek). Trials were repeated for n=3. Where exponential decay was observed, curves could be fit to a single exponential model (Equation 3) in IgorPro7. For reactions performed with core particle only, core particle was made to 2X concentration (1.8 μM) and added equivolume with 2X substrate (5 μL final) and fluorescence polarization was monitored as above. Single turnover conditions were verified by single reactions with doubled proteasome concentration by reconstituting proteasome to 4X concentration and diluting with equivolume substrate (2.5 μl each) to 2X proteasome and monitoring fluorescence polarization kinetics as described above. For degradations with o-phenanthroline inhibited proteasomes, proteasomes were allowed to assemble at 3X concentration for 3 minutes at room temperature between dilution with o-phenanthroline (30 mM stock in assay buffer; 5 mM final) to 2X concentration for 2 minutes before degradation initiation as described above. For degradations using only the core particle, core particle was added to 900 nM final with substrate as described above.
Fluorescence polarization single-turnover ubiquitin-dependent degradation assay of substrates in the presence of barnase
Substrates were prepared to 2X concentration (6 nM final) in assay buffer with barnase added in excess (20 μM final) and allowed to come to equilibrium for greater than 5 minutes at room temperature53 prior to degradation initiation. Degradations were performed exactly as described above. Saturation of barnase binding was assessed by doubling barnase concentration (40 μM final) and comparing fluorescence polarization kinetic differences. FAM-Titin-I27V13P,V15P ubiquitinated as described above was degraded in the presence or absence of 20 μM barnase with proteasome at the same concentration as described.
Supplementary Material
Acknowledgements:
We thank all members of the Marqusee and Martin labs for helpful discussions. We also thank B. Maguire and K. Dong for assistance with protein purification and troubleshooting expertise. We acknowledge support from the US National Institutes of Health: R01 – GM050945 (SM), R01-GM094497 (AM). SM is a Chan Zuckerberg Biohub investigator. AM is an investigator of the Howard Hughes Medical Institute.
Footnotes
Declaration of Interests:
The authors declare no competing financial or non-financial interests.
Data availability:
The data that support the findings of this study are available within the manuscript and its supplementary information or from the corresponding author upon reasonable request. All constructs generated for this study are also available from the corresponding author upon reasonable request.
References:
- 1.Raschke TM, Kho J & Marqusee S Confirmation of the hierarchical folding of RNase H: A protein engineering study. Nat. Struct. Biol 6, 825–830 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Kenniston JA, Burton RE, Siddiqui SM, Baker TA & Sauer RT Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J. Struct. Biol 146, 130–140 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Liu T, Whitten ST & Hilser VJ Ensemble-based signatures of energy propagation in proteins: A new view of an old phenomenon. Proteins Struct. Funct. Genet 62, 728–738 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Martin A, Baker TA & Sauer RT Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat. Struct. Mol. Biol 15, 139–145 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Xin F & Radivojac P Post-translational modifications induce significant yet not extreme changes to protein structure. Bioinformatics 28, 2905–2913 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Swatek KN & Komander D Ubiquitin modifications. Cell Res. 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prakash S, Tian L, Ratliff KS, Lehotzky RE & Matouschek A An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol 11, 830–837 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Yu H & Matouschek A Recognition of Client Proteins by the Proteasome. Annu. Rev. Biophys 46, 149–173 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Hagai T, Azia A, Tóth-Petróczy Á & Levy Y Intrinsic disorder in ubiquitination substrates. J. Mol. Biol 412, 319–324 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Godderz D et al. Cdc48-independent proteasomal degradation coincides with a reduced need for ubiquitylation. Sci. Rep 5, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuchiya H et al. In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol. Cell 66, 488–502.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Olszewski MM, Williams C, Dong KC & Martin A The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. Commun. Biol 2, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagai T & Levy Y Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding. Proc. Natl. Acad. Sci. U. S. A 107, 2001–2006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavrilov Y, Hagai T & Levy Y Nonspecific yet decisive: Ubiquitination can affect the native-state dynamics of the modified protein. Protein Sci. 24, 1580–1592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faggiano S & Pastore A The Challenge of Producing Ubiquitinated Proteins for Structural Studies. Cells 3, 639–656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morimoto D, Walinda E, Fukada H, Sugase K & Shirakawa M Ubiquitylation Directly Induces Fold Destabilization of Proteins. Sci. Rep 6, 39453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cundiff MD et al. Ubiquitin receptors are required for substrate-mediated activation of the proteasome’s unfolding ability. Sci. Rep 9, 14506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saeki Y, Isono E & Toh-E A Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 399, 215–227 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Kim HC, Steffen AM, Oldham ML, Chen J & Huibregtse JM Structure and function of a HECT domain ubiquitin-binding site. EMBO Rep. 12, 334–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamadurai HB et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife 2013, 1–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khurana Ritu; Hate Anita T; Nath Utpal; Udgaonkar JB pH dependence of the stability of barstar to chemical and thermal denaturation. Protein Sci. 4, 1133–1144 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers JK, Pace CN & Scholtz JM Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 4, 2138–2148 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolting B et al. The folding pathway of a protein at high resolution from microseconds to seconds. Proc. Natl. Acad. Sci 94, 826–830 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaidi FN, Nath U & Udgaonkar JB Multiple intermediates and transition states during protein unfolding. Nat. Struct. Biol 4, 1016–1024 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Park C & Marqusee S Probing the high energy states in proteins by proteolysis. J. Mol. Biol 343, 1467–1476 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Park C Mini Review P 117–128 Probing Transient Partial Unfolding in Proteins by Native-State Proteolysis. 117–128 (2014). [Google Scholar]
- 27.Bard JAM, Bashore C, Dong KC & Martin A The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 177, 286–298.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bashore C et al. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat. Struct. Mol. Biol 22, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chojnacki M et al. Polyubiquitin-Photoactivatable Crosslinking Reagents for Mapping Ubiquitin Interactome Identify Rpn1 as a Proteasome Ubiquitin-Associating Subunit. Cell Chem. Biol 24, 443–457.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C, Schwartz MP, Prakash S, Iwakura M & Matouschek A ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell 7, 627–637 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Twomey EC et al. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science (80-. ). 365, eaax1033 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De la Peña AH, Goodall EA, Gates SN, Lander GC & Martin A Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation. Science (80-. ). 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worden EJ, Dong KC & Martin A An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Mol. Cell 67, 799–811.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Greene ER et al. Specific lid-base contacts in the 26S proteasome control the conformational switching required for substrate engagement and degradation. Elife 1–27 (2019). doi: 10.1101/687921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichard EL et al. Substrate Ubiquitination Controls the Unfolding Ability of the Proteasome. J. Biol. Chem 291, jbc.M116.720151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Q et al. In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell 172, 696–705.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komander D & Rape M The Ubiquitin Code. Annu. Rev. Biochem 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto KM et al. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci 98, 8554–8559 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowak RP et al. Plasticity in binding confers selectivity in ligand-induced protein degradation article. Nat. Chem. Biol. 14, 706–714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith BE et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun 10, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang HT et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chem. Biol 25, 88–99.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bondeson DP et al. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol 25, 78–87.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batey S, Nickson AA & Clarke J Studying the folding of multidomain proteins. HFSP J. 2, 365–377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrion-Vazquez M et al. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol 10, 738–743 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Morimoto D et al. The unexpected role of polyubiquitin chains in the formation of fibrillar aggregates. Nat. Commun 6, 6116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sousa R & Lafer EM The Physics of Entropic Pulling: A Novel Model for the Hsp70 Motor Mechanism. Int. J. Mol. Sci 20, 2334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freudenthal BD, Gakhar L, Ramaswamy S & Washington MT Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat. Struct. Mol. Biol 17, 479–484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varadan R, Walker O, Pickart C & Fushman D Structural properties of polyubiquitin chains in solution. J. Mol. Biol 324, 637–647 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Eddins MJ, Varadan R, Fushman D, Pickart CM & Wolberger C Crystal Structure and Solution NMR Studies of Lys48-linked Tetraubiquitin at Neutral pH. J. Mol. Biol 367, 204–211 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Debelouchina GT, Gerecht K & Muir TW Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat. Chem. Biol 13, 105–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References:
- 51.Beckwith R, Estrin E, Worden EJ & Martin A Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat. Struct. Mol. Biol 20, 1164–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matyskiela ME, Lander GC & Martin A Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol 20, 781–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pollard TD MBOC technical perspective: A guide to simple and informative binding assays. Mol. Biol. Cell 21, 4061–4067 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the manuscript and its supplementary information or from the corresponding author upon reasonable request. All constructs generated for this study are also available from the corresponding author upon reasonable request.