Abstract
The family Marnaviridae comprises small non-enveloped viruses with positive-sense RNA genomes of 8.6–9.6 kb. Isolates infect marine single-celled eukaryotes (protists) that come from diverse lineages. Some members are known from metagenomic studies of ocean virioplankton, with additional unclassified viruses described from metagenomic datasets derived from marine and freshwater environments. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Marnaviridae, which is available at ictv.global/report/marnaviridae.
Keywords: ICTV Report, Marnaviridae, taxonomy
Virion
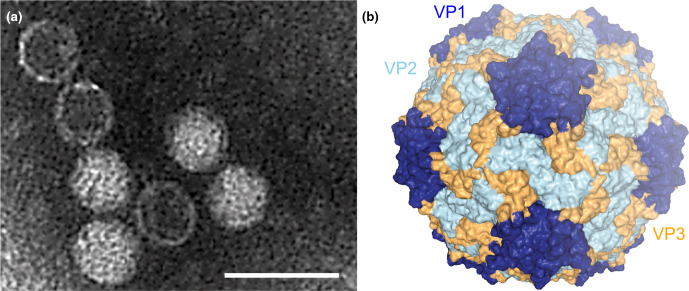
Structurally characterized members have polyhedral virions, 22–35 nm in diameter, without envelopes or discernible projections (Table 1, Fig. 1). There are four conserved structural proteins, VP1–4. Capsid protein and virion structures have been resolved by cryo-electron microscopy for Chaetoceros tenuissimus RNA virus type II (species Chaetenuissarnavirus II) [1].
Table 1.
Characteristics of members of the family Marnaviridae
|
Example: |
Heterosigma akashiwo RNA virus (AY337486), species Heterosigma akashiwo RNA virus, genus Marnavirus |
|---|---|
|
Virion |
Non-enveloped, 22–35 nm with four structural proteins |
|
Genome |
8.6–9.6 kb of positive-sense, non-segmented RNA |
|
Replication |
Cytoplasmic, involving RNA-directed RNA polymerase and helicase; cytolytic |
|
Translation |
Directly from genomic RNA containing one or more internal ribosomal entry sites |
|
Host range |
Single-celled eukaryotes (protists) from marine environments |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Picornavirales; >5 genera and >15 species |
Fig. 1.
Marnaviridae virion structure. (a) Negatively stained transmission electron micrograph of Heterosigma akashiwo RNA virus virions (scale bar=50 nm). (b) Cryo-electron microscopy reconstructed capsid structure at 3.1 Å resolution of Chaetoceros tenuissimus RNA virus type II (species Chaetenuissarnavirus II, genus Sogarnavirus) (PDB structure 6SHL viewed in NGL-WebGL). The three major capsid proteins are represented in dark blue (VP1), light blue (VP2) and yellow (VP3).
Genome
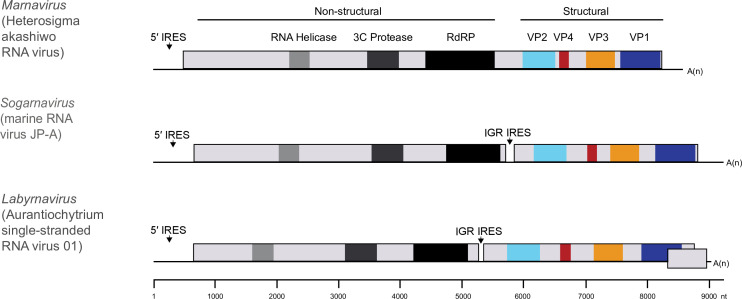
Members possess monopartite positive-sense RNA genomes of 8.6–9.6 kb [2] containing one (Locarnavirus, Marnavirus) or two ORFs (Locarnavirus, Kusarnavirus, Bacillarnavirus, Salisharnavirus, Sogarnavirus); members of the genus Labyrnavirus have a third small overlapping open reading frame (ORF) at the 3′-end of the genome (Fig. 2). Virus genomes encode conserved helicase, RNA-directed RNA polymerase and structural protein domains. Some genomes also encode regions that resemble the 3C cysteine proteinases of members of the family Picornaviridae. Predicted secondary structures within the 5′-non-coding regions and intergenic regions suggest the presence of internal ribosome entry sites. A 3′-poly(A) tail terminates the genome of those viruses that have been isolated.
Fig. 2.
Genome architectures of viruses in the family Marnaviridae. IRES, internal ribosome entry site. RdRP, RNA-directed RNA polymerase. There is a putative IRES in the intergenic region (IGR) of the di-cistronic genomes.
Replication
Viral proteins are synthesized as part of one or two polyproteins. Regardless of whether the genomes are mono- or di-cistronic, the non-structural proteins are encoded in the 5′-region and the structural proteins are encoded in the 3′-region. Replication is cytolytic and various cytopathic effects have been observed [3–8]. Membrane vesicle structures associated with the endoplasmic reticulum [3] are likely to represent sites of RNA synthesis.
Taxonomy
Current taxonomy: www.ictv.global/taxonomy. The genus Marnavirus includes the species Heterosigma akashiwo RNA virus, whose only member has a mono-cistronic genome and infects a harmful algal bloom-forming raphidophyte. Labyrnavirus includes the species Aurantiochytrium single-stranded RNA virus 01, whose only member has a di-cistronic genome and a third small overlapping ORF, which is transcribed as a sub-genomic RNA during infection but may not be functional [9]. The host is Aurantiochytrium sp., a marine thraustochytrid. Locarnavirus includes the species Jericarnavirus B, Sanfarnavirus 1, Sanfarnavirus 2 and Sanfarnavirus 3, members of which were discovered through metagenomic analysis of marine virioplankton. Both mono- and di-cistronic genome organizations are found. Kusarnavirus includes the species Astarnavirus, whose only member infects the diatom Asterionellopsis glacialis and has a di-cistronic genome. Bacillarnavirus includes the species Rhizosolenia setigera RNA virus 01, Chaetoceros tenuissimus RNA virus 01 and Chaetoceros socialis forma radians RNA virus 1, members of which infect diatoms and have di-cistronic genomes. Salisharnavirus includes the species Britarnavirus 1, Britarnavirus 4, Palmarnavirus 128 and Palmarnavirus 473, members of which were discovered by metagenomics and have di-cistronic genomes. Sogarnavirus includes the species Chaetenuissarnavirus II, Chaetarnavirus 2, Britarnavirus 2, Britarnavirus 3, Palmarnavirus 156 and Jericarnavirus A. Members have di-cistronic genomes and have been isolated from diatoms or discovered by metagenomics.
Resources
Full ICTV Report on the family Marnaviridae: https://www.ictv.global/report/marnaviridae
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Elliot Lefkowitz, Peter Simmonds, F. Murilo Zerbini, Donald B. Smith, Richard Orton and Sead Sabanadzovic.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Munke A, Kimura K, Tomaru Y, Okamoto K. Capsid structure of a marine algal virus of the order Picornavirales . J Virol. 2020;94:e01855–19. doi: 10.1128/JVI.01855-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vlok M, Lang AS, Suttle CA. Application of a sequence-based taxonomic classification method to uncultured and unclassified marine single-stranded RNA viruses in the order Picornavirales . Virus Evol. 2019;5:vez056. doi: 10.1093/ve/vez056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, et al. Characterization of HaRNAV, a single‐stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae) . J Phycol. 2003;39:343–352. [Google Scholar]
- 4.Nagasaki K, Tomaru Y, Katanozaka N, Shirai Y, Nishida K, et al. Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera . Appl Environ Microbiol. 2004;70:704–711. doi: 10.1128/AEM.70.2.704-711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomaru Y, Takao Y, Suzuki H, Nagumo T, Nagasaki K. Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis . Appl Environ Microbiol. 2009;75:2375–2381. doi: 10.1128/AEM.02580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takao Y, Nagasaki K, Mise K, Okuno T, Honda D. Isolation and characterization of a novel single-stranded RNA Virus infectious to a marine fungoid protist, Schizochytrium sp. Appl Environ Microbiol. 2005;71:4516–4522. doi: 10.1128/AEM.71.8.4516-4522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomaru Y, Toyoda K, Kimura K, Hata N, Yoshida M, et al. First evidence for the existence of pennate diatom viruses. ISME J. 2012;6:1445–1448. doi: 10.1038/ismej.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomaru Y, Tarutani K, Yamaguchi M, Nagasaki K. Quantitative and qualitative impacts of viral infection on a Heterosigma akashiwo (Raphidophyceae) bloom in Hiroshima Bay, Japan. Aquat Microb Ecol. 2004;34:227–238. doi: 10.3354/ame034227. [DOI] [Google Scholar]
- 9.Takao Y, Mise K, Nagasaki K, Okuno T, Honda D. Complete nucleotide sequence and genome organization of a single-stranded RNA virus infecting the marine fungoid protist Schizochytrium sp. J Gen Virol. 2006;87:723–733. doi: 10.1099/vir.0.81204-0. [DOI] [PubMed] [Google Scholar]