Abstract
Background
Lung function is a heritable complex phenotype with obesity being one of its important risk factors. However, knowledge of their shared genetic basis is limited. Most genome-wide association studies (GWASs) for lung function have been based on European populations, limiting the generalisability across populations. Large-scale lung function GWASs in other populations are lacking.
Methods
We included 100 285 subjects from the China Kadoorie Biobank (CKB). To identify novel loci for lung function, single-trait GWAS analyses were performed on forced expiratory volume in 1 s (FEV1), forced vital capacity (FVC) and FEV1/FVC in the CKB. We then performed genome-wide cross-trait analysis between lung function and obesity traits (body mass index (BMI), BMI-adjusted waist-to-hip ratio and BMI-adjusted waist circumference) to investigate the shared genetic effects in the CKB. Finally, polygenic risk scores (PRSs) of lung function were developed in the CKB and their interaction with BMI's association on lung function were examined. We also conducted cross-trait analysis in parallel with the CKB using up to 457 756 subjects from the UK Biobank (UKB) for replication and investigation of ancestry-specific effects.
Results
We identified nine genome-wide significant novel loci for FEV1, six for FVC and three for FEV1/FVC in the CKB. FEV1 and FVC showed significant negative genetic correlation with obesity traits in both the CKB and UKB. Genetic loci shared between lung function and obesity traits highlighted important biological pathways, including cell proliferation, embryo, skeletal and tissue development, and regulation of gene expression. Mendelian randomisation analysis suggested significant negative causal effects of BMI on FEV1 and on FVC in both the CKB and UKB. Lung function PRSs significantly modified the effect of change in BMI on change in lung function during an average follow-up of 8 years.
Conclusion
This large-scale GWAS of lung function identified novel loci and shared genetic aetiology between lung function and obesity. Change in BMI might affect change in lung function differently according to a subject's polygenic background. These findings may open new avenues for the development of molecular-targeted therapies for obesity and lung function improvement.
Short abstract
Novel loci provide additional insights into the genetic basis of lung function. Understanding the shared genetic aetiology of lung function and obesity may open new avenues for molecular-targeted therapies for obesity and lung function improvement. http://bit.ly/38oCnez
Introduction
Impaired lung function is associated with lung disease risk and mortality, such as chronic obstructive pulmonary disease (COPD) [1]. Clinical and epidemiological studies have shown many risk factors can affect lung function [2]. Among these risk factors, obesity has been one of the most rapidly growing public health issues with a nearly tripled prevalence over the past 30 years [3]. Specifically, according to a population-based study on 121 965 subjects, obesity is associated with approximately 2 times higher risk of reduced lung function (e.g. forced expiratory volume in 1 s (FEV1) and forced vital capacity (FVC)) [4]. Obesity is also associated with increased risk of respiratory diseases, such as asthma and COPD [4, 5]. However, such findings have also raised new questions about whether the genetic risk factors can contribute to the coexistence of lung function reduction and obesity.
We and others have recently identified shared genetic architecture among respiratory diseases, including asthma and COPD [6–9], indicating pleiotropic effects impacting both diseases. Lung function and obesity are both highly heritable traits, with an estimated heritability up to 70% [10–14]. The inverse association between lung function and obesity suggested potential shared genetic risk factors between these conditions [15]. However, knowledge of the shared genetic basis of lung function and obesity is limited.
To date, most lung function genome-wide association study (GWAS) participants have been of European descent [13, 14, 16, 17]; only few studies included a small number of non-European participants [18, 19]. Thus, large-scale GWASs based on non-European populations are critical to extend our understanding of the genetic heterogeneity across different populations [20, 21]. In addition, it is critical to understand the shared genetic architecture of lung function with other complex traits (e.g. obesity), which is robust to environmental confounding [22]. Thus, in the current study, we conducted a large-scale GWAS and cross-trait/cross-population analysis in the China Kadoorie Biobank (CKB) and UK Biobank (UKB) to address three aims: 1) to identify novel genetic risk loci for lung function traits that include FEV1, FVC and FEV1/FVC in the Chinese population; 2) to investigate shared genetic effects between the lung function traits and obesity traits (body mass index (BMI), BMI-adjusted waist-to-hip ratio (WHRadjBMI) and BMI-adjusted waist circumference (WCadjBMI)) in both Chinese and European populations; and 3) by using both the CKB and UKB follow-up cohorts, to investigate whether the baseline BMI and longitudinal change in BMI would affect lung function, taking into account the polygenic background of lung function.
Methods
Study design, settings and participants
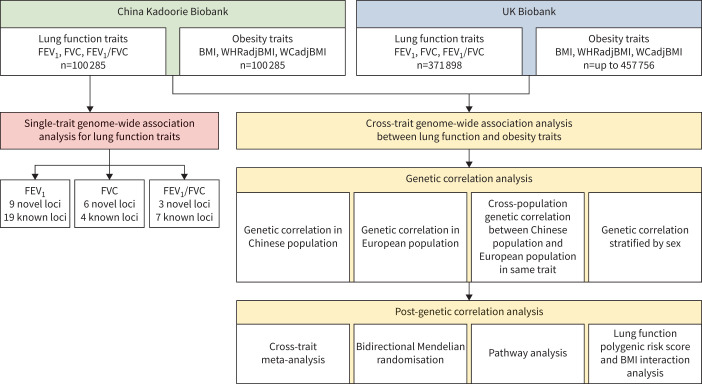
The overall study design can be found in figure 1. In brief, this study has two analytical stages. The first stage is to identify novel loci for lung function in the Chinese population by using single-trait GWAS analysis. The second stage is to investigate shared genetic effects between lung function and obesity by using cross-trait GWAS analysis in both the CKB and UKB.
FIGURE 1.
Overall study design. FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; BMI: body mass index; WHRadjBMI: BMI-adjusted waist-to-hip ratio; WCadjBMI: BMI-adjusted waist circumference.
The CKB study is a prospective cohort study of more than 500 000 participants in China. Details of the CKB have been described previously [23]. In brief, the CKB recruited 512 715 adults aged 30–79 years from 10 regions (Harbin, Qingdao, Suzhou, Liuzhou, Haikou, Henan, Gansu, Sichuan, Zhejiang and Hunan) across China. All participants gave informed written consent. Questionnaire data, physical measurements and blood samples were collected at the baseline survey during 2004–2008. Two follow-up surveys were taken in 2008 and during 2013–2014, which involved ∼5% randomly chosen surviving participants.
The UKB study has been described in detail elsewhere [7, 24]. In brief, the UKB study is a prospective cohort study of more than 500 000 participants living in the UK. In total, 503 325 participants registered in the National Health Service with ages ranging from 40 to 69 years were recruited out of 9.2 million mailed invitations. Baseline data were collected (2004–2008) using questionnaires and anthropometric assessments were performed. In the UKB, we restricted to subjects of European ancestry. All detailed genotyping, quality control and imputation procedures are described in the UKB website (http://biobank.ctsu.ox.ac.uk). All participants provided informed consent to the UKB.
Ascertainment of lung function and obesity traits
FEV1, FVC and FEV1/FVC were adjusted for age, age squared, sex, height, smoking status (ever versus never) and assessment centre in a linear regression model [14]. The resulting residuals were inverse normal transformed [14].
BMI (measured or self-reported weight in kilograms per height in metres squared) was adjusted for age, age squared, sex and assessment centre in a linear regression model [14]. Waist and hip circumferences were also measured in the CKB and UKB participants. The waist-to-hip ratio and waist circumference were adjusted for age, age squared, BMI, sex and assessment centre in a linear regression model. The resulting residuals were inverse normal transformed. Detailed physical measurement procedures can be found in the supplementary material, the previous CKB study [25] and the UKB website [26].
CKB genotyping procedure
The CKB has conducted three phases of genotyping. A custom-designed biobank array, to provide optimised genome-wide coverage for the Chinese population, was developed by the University of Oxford's Clinical Trial Service Unit and Epidemiological Studies Unit (Oxford, UK) in collaboration with the Beijing Genomics Institute (Shenzhen, China) and Affymetrix (now Thermo Fisher Scientific, Santa Clara, CA, USA). This 700K single nucleotide polymorphism (SNP) array was used to genotype ∼32 000 CKB participants in the first phase. A revised and updated version of the original array which covers ∼803K SNPs was used to genotype ∼69 000 participants in the second and third phases.
Variants with call rate >0.98, plate effect p>10−6, batch effect p>10−6, Hardy–Weinberg equilibrium (HWE) deviations p>10−6 (combined 10 degrees of freedom Chi-squared test from 10 regions) and minor allele frequency (MAF) difference from 1000 Genomes East Asian frequencies <0.2 were identified, resulting in genotypes for 532 415 biallelic variants present on both array versions. The qualified genotypes for each chromosome were phased with SHAPEIT. Then, imputation was performed for each 5-Mb interval with IMPUTE 4 based on haplotypes derived from the 1000 Genomes phase III.
GWAS analysis
We selected variants that did not deviate from HWE (p>1×10−12), per variant missing rates <10%, per sample missing rate <10%, MAF >1% and imputation quality score (INFO) >0.8. Detailed data summary, quality control and imputation information can be found in the supplementary material. The genotype–phenotype association test was carried out in 100 285 samples from the CKB and up to 457 756 samples from the UKB. For lung function and obesity traits, we carried out linear mixed model (LMM) association analyses and adjusted for genotyping array, 10 ancestry principal components in the CKB and 30 ancestry principal components in the UKB to assess the association between the traits’ z-scores and imputed genotype dosages under an additive genetic model by using BOLT-LMM version 2.3 [27]. After association analysis, we applied the PLINK clumping function to determine top loci that were independent to each other. Specifically, variants with p<1×10−5, r2>0.2 and <500 kb away from the peak were assigned to that peak's clump. The genes within each clump were identified by the overlap between gene regions and clump region. Novel loci were defined at two levels: clump and variant (supplementary material). In brief, if the independent clump region did not overlap with any loci in the GWAS catalogue (search date: 3 April 2020) for the same trait, we defined it as a novel locus. If there was an overlap between the clump region and GWAS catalogue, we further checked if the sentinel variant was novel, which is defined by low linkage disequilibrium (LD) r2<0.2 between the sentinel variant and any variants within the clump region from the GWAS catalogue.
Cross-trait genetic correlation
We used cross-trait LD score regression (LDSC) to estimate genetic correlation between the causal effects of two traits (ranging from −1 to 1) based on summary statistics of each trait's GWAS [28]. We specified LDSC to estimate the regression intercept to account for shared subjects between different traits’ GWAS [29]. We applied Bonferroni correction (p<0.05/9) to account for multiple testing in the LDSC analysis.
Sex-specific genetic correlation
Previous studies showed the association between lung function and obesity could differ in males and females [30, 31]. Thus, we evaluated the genetic correlation between lung function and obesity in males and females separately.
Cross-population genetic correlation
To assess the genetic heterogeneity between Chinese and European populations, we also estimated genome-wide cross-population genetic correlation for lung function and obesity traits by applying S-LDXR [32] with the baseline-LD-X model annotations. We applied Bonferroni correction (p<0.05/6) to account for multiple testing in the S-LDXR analysis.
Cross-trait meta-analysis
Cross-phenotype association (CPASSOC) combines the effect estimate and standard error of the GWAS summary statistics to test the hypothesis of association between the SNP with both traits [33]. A heterogeneous version of CPASSOC (SHet) was used in this study.
SHet is a cross-phenotype meta-analysis method based on a fixed effects model. It is more powerful when there is a heterogeneous effect present across studies, which is common when testing multiple phenotypes [34]. SHet uses the sample size of a trait as the weight instead of using the effect standard error. It can also account for effect correlation due to overlapping or related subjects within and among different studies or cohorts.
Overrepresentation enrichment analysis
In order to understand the shared biological pathways between lung function and obesity, we extracted the genes from the clumping procedure for both lung function and obesity, and used the WebGestalt tool [35] to assess the enrichment of the identified genes in Gene Ontology (GO) biological pathways. If they were significantly enriched in both lung function and obesity, we considered them as shared biological pathways. A false discovery rate (FDR) method was used to correct for multiple testing.
Mendelian randomisation analysis
We applied generalised summary data-based Mendelian randomisation (GSMR) [36] under default settings to infer putative causal relationships between BMI and lung function traits from GWAS summary statistics. To avoid overlapping subjects in the MR analysis, we used the BMI GWAS from Biobank Japan (n=158 284) [37] and the lung function GWAS from the CKB, and we used the GIANT BMI GWAS (nmax=322 154) [12] and the lung function GWAS from the UKB. A more detailed description of the Biobank Japan and GIANT GWAS data can be found in the supplementary material. Since GSMR requires a minimum of 10 LD-independent instruments with p<5×10−8, we restricted our analyses to traits that satisfy this criterion. Prior to running GSMR, we removed SNPs with strand ambiguity, INFO <0.9 and in the human leukocyte antigen (HLA) region (chr6:25–34M). We applied Bonferroni correction (0.05/6) to account for the number of trait pairs in the MR analysis.
Lung function polygenic risk score and BMI interaction (PRSlung function×BMI) analysis
We constructed the polygenic risk scores (PRSs) for three lung function traits using LDpred [38]. Details of PRS construction can be found in the supplementary material. We also constructed three additional lung function PRSs using weights of 279 SNPs reported by Shrine et al. [14] (only 275 SNPs were available in the CKB data). To investigate the interaction effect between lung function PRSs and baseline BMI or its longitudinal change (nCKB=21 791 and nUKB=12 019) on lung function, we fitted two linear regression models to test the PRSlung function×BMI effect:
Baseline model:
Change model:
where baseline (t0) is at 2004–2010 and follow-up (t1) is at 2012–2014; other covariates are principal components 1–10 for the CKB and principal components 1–30 for the UKB, age, age squared, sex, standing height, smoking status (ever/never), genotyping array, and assessment centre. For the baseline model, we set normal BMI and the deciles 2–9 group as reference; for the change model, we set BMI stable and the deciles 2–9 group as reference.
Results
GWAS and SNP-based heritability
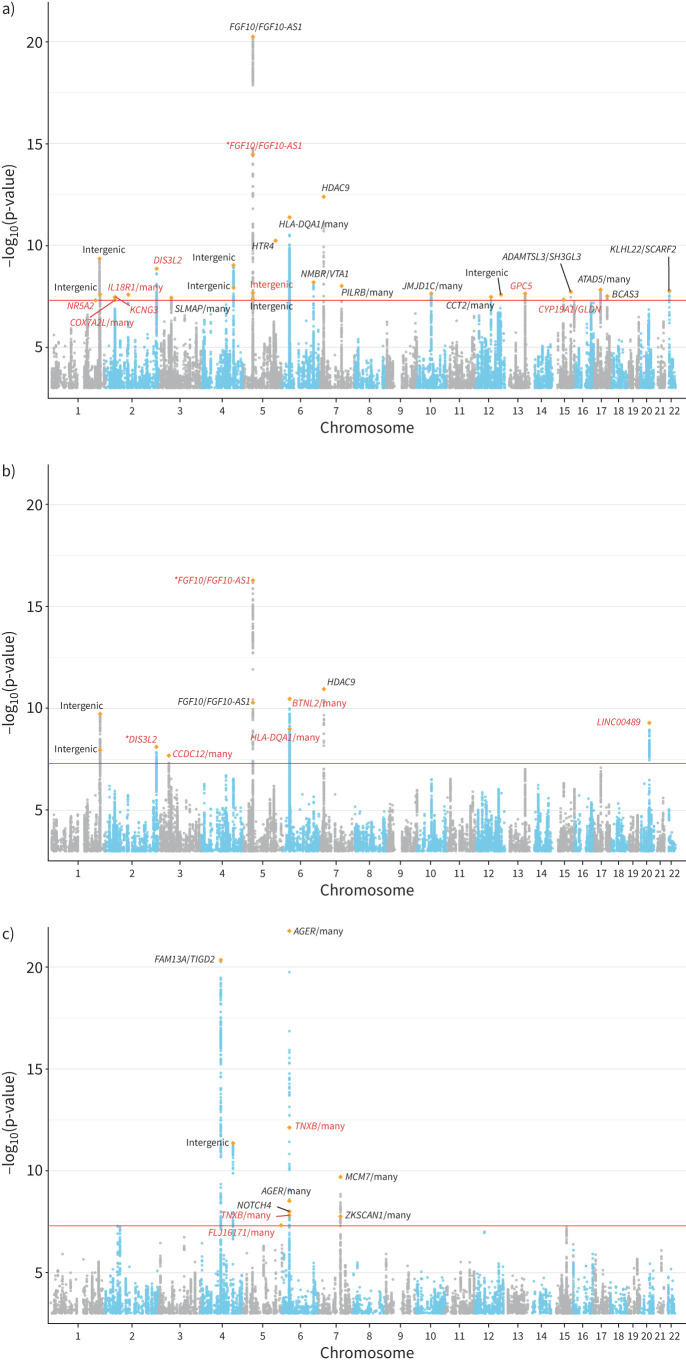
The baseline demographic characteristics of the CKB and UKB cohorts are summarised in supplementary table S1. GWAS results for all traits showed no evidence of inflation due to population stratification (supplementary figures S1 and S2). In the CKB, LDSC estimates of SNP-based heritability on the observed scale were (mean±se) 13.07±0.88% for FEV1, 11.12±0.82% for FVC, 5.12±0.67% for FEV1/FVC, 21.77±1.23% for BMI, 8.72±0.83% for WHRadjBMI and 10.65±0.95% for WCadjBMI. We identified 28 genome-wide significant (p<5×10−8) loci for FEV1, 10 for FVC and 10 for FEV1/FVC (figure 2). After comparing with GWAS catalogue results for FEV1, FVC and FEV1/FVC (supplementary tables S2–S4), we determined a total of 18 novel loci for the three lung function traits (table 1). We further conducted the replication analysis for the novel loci in a recently published large-scale lung function GWAS (supplementary table S5) by Shrine et al. [14]. A total of 11 loci were available in the Shrine et al. [14] data. Among them, we found four were significant (p<0.05/13) with consistent effect size direction. The nonreplicated loci were likely due to distinct effect allele frequencies between the CKB and Shrine et al. [14] (supplementary table S5). For example, the MAF of the sentinel SNP rs1861229 in the CKB is 0.52, but 0.17 in the Shrine et al. [14] study. Among these previously reported loci showing genome-wide significant association with lung function in Shrine et al. [14], AGER, AP4M1, DIS3L2, FAM13A, FGF10, HLA-DQA1 and HTR4 are notable genes that play important roles in lung function. In terms of novel loci, we identified GPC5 as a novel gene for FEV1 (sentinel SNP rs528366: p=2.30×10−8). In addition, we identified 20q11.23 as a novel region for FVC (sentinel SNP rs6063386: p=5.00×10−10). The sentinel SNP is mapped to a long intergenic nonprotein coding RNA (lncRNA), LINC00489. Among the novel loci for FEV1/FVC, two are within the 6p21.33 region and the sentinel SNP was mapped to TNXB, which was known for its association with lung function traits (FEV1 and FEV1/FVC) and COPD [39, 40]. Detailed summary statistics information on genome-wide significant loci for the three lung function traits can be found in supplementary tables S6–S8.
FIGURE 2.
Manhattan plots for genome-wide association analysis of 100 285 Chinese subjects in the China Kadoorie Biobank cohort for three lung function traits: a) forced expiratory volume in 1 s (FEV1), b) forced vital capacity (FVC) and c) FEV1/FVC. The x-axis denotes the genomic position (chromosomes 1–22); the y-axis denotes the –log10(p-value) of association test and starts at –log10(p-value)=3. The most significant novel variant in each independent clump is highlighted by an orange diamond symbol. Genes in black were previously reported and genes in red are novel. An asterisk on some genes indicates a novel variant. The genome-wide significance level (p=5×10−8) is denoted by the red line.
TABLE 1.
18 novel loci associated with forced expiratory volume in 1 s (FEV1), forced vital capacity (FVC) and FEV1/FVC in the China Kadoorie Biobank
| Trait | Sentinel SNP | Clump region | N | A1 | A2 | A1 FREQ | BETA | se | p-value | Genes within clump region |
| FEV1 | rs145972739 | chr1:200031115 –200031115 |
1 | A | G | 0.97 | −0.083 | 0.015 | 4.90×10−8 | NR5A2 |
| rs1861229 | chr2:102992079 –103208610 |
28 | A | G | 0.52 | 0.026 | 0.005 | 2.60×10−8 | IL18R1, IL18RAP, MIR4772, SLC9A4 | |
| rs28695435 | chr2:232797462 –233092939 |
181 | G | A | 0.31 | −0.029 | 0.005 | 1.40×10−9 | DIS3L2 | |
| rs222482 | chr2:42391012 –42703861 |
188 | C | T | 0.27 | −0.028 | 0.005 | 4.70×10−8 | COX7A2L, EML4, KCNG3, LOC102723824 | |
| rs112952987 | chr2:42638788 –42703942 |
21 | G | A | 0.94 | 0.050 | 0.009 | 3.40×10−8 | KCNG3 | |
| rs117331805 | chr5:43813683 –44687091 |
96 | A | G | 0.94 | −0.076 | 0.010 | 3.50×10−15 | FGF10, FGF10-AS1 | |
| rs117675260 | chr5:44474070 –44623745 |
28 | G | A | 0.96 | −0.069 | 0.012 | 2.10×10−8 | Intergenic region | |
| rs528366 | chr13:92381450 –92572381 |
224 | T | C | 0.78 | −0.030 | 0.005 | 2.30×10−8 | GPC5 | |
| rs77578670 | chr15:51607186 –51645049 |
23 | C | T | 0.75 | −0.028 | 0.005 | 4.60×10−8 | CYP19A1, GLDN | |
| FVC | rs143944819 | chr2:232797462 –233101499 |
185 | A | G | 0.77 | 0.033 | 0.006 | 7.50×10−9 | DIS3L2 |
| rs6442039 | chr3:46902129 –47410564 |
158 | C | G | 0.52 | −0.025 | 0.004 | 2.00×10−8 | CCDC12, KIF9, KIF9-AS1, KLHL18, MYL3, NBEAL2, NRADDP, PTH1R, SETD2 | |
| rs78732306 | chr5:44048662 –44583962 |
177 | T | C | 0.91 | −0.068 | 0.008 | 5.10×10−17 | FGF10, FGF10-AS1 | |
| rs28366282 | chr6:32196697 –32713674 |
1767 | C | T | 0.76 | 0.035 | 0.005 | 3.30×10−11 | BTNL2, C6orf10, HCG23, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DRB6 | |
| rs139447342 | chr6:32396905 –32636434 |
661 | C | T | 0.3 | 0.032 | 0.005 | 1.00×10−9 | HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DRB6 | |
| rs6063386 | chr20:36206453 –36273380 |
127 | C | T | 0.33 | 0.030 | 0.005 | 5.00×10−10 | LINC00489 | |
| FEV1/FVC | rs186784089 | chr5:174393318 –174393318 |
1 | G | A | 0.99 | 0.120 | 0.022 | 4.70×10−8 | FLJ16171 |
| rs149101418 | chr6:31944375 –32113312 |
17 | T | G | 0.55 | 0.035 | 0.005 | 7.50×10−13 | ATF6B,C4A, C4B, C4B_2, CYP21A1P, CYP21A2, FKBPL, STK19, TNXA, TNXB | |
| rs200214283 | chr6:31976290 –32133380 |
4 | C | T | 0.9 | −0.045 | 0.008 | 1.50×10−8 | ATF6B, C4A, C4B, C4B_2, CYP21A1P, CYP21A2, EGFL8, FKBPL, LOC100507547, PPT2, PPT2-EGFL8, PRRT1, STK19, TNXA, TNXB |
SNP: single nucleotide polymorphism; N: number of variants meeting the criteria of p<1×10−5 and r2>0.2 within the clump region; A1: effect allele; A2: noneffect allele; BETA: BOLT-LMM regression effect size.
In the UKB, LDSC estimates of SNP-based heritability on the observed scale were (mean±se) 20.13±0.76% for FEV1, 20.26±0.74% for FVC, 23.95±1.43% for FEV1/FVC, 27.41±1.07% for BMI, 13.88±0.94% for WHRadjBMI and 16.58±0.83% for WCadjBMI. The single-trait GWAS results are consistent with the Shrine et al. [14] study.
Genetic correlation between lung function and obesity traits
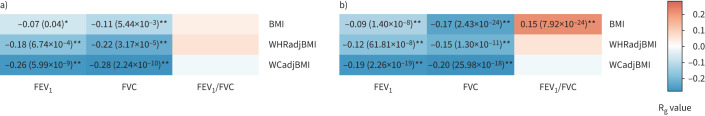
We investigated the genetic correlation between lung function and obesity traits in both the CKB and UKB. As shown in figure 3, we found that two lung function traits have significant negative genetic correlation with obesity traits in the CKB (e.g. Rg= −0.26, p=5.99×10−9 for FEV1–WCadjBMI, Rg= −0.11, p=5.44×10−3 for FVC–BMI and Rg= −0.28, p=2.24×10−10 for FVC–WCadjBMI). We found that the genetic correlation is generally stronger between lung function and central obesity traits than BMI. The UKB genetic correlations also showed consistent findings with the CKB in most of the trait pairs, although different for one trait pair (e.g. Rg=0.15, p=7.92×10−24 for FEV1/FVC–BMI in the UKB, but not significant in the CKB) (figure 3). Sex-specific analyses found stronger genetic correlation between lung function and obesity traits in females than in males (supplementary tables S9 and S10). In addition, the cross-population genetic correlation analysis showed that one of these traits had an estimated cross-population Rg significantly <1 (Rgcross-population=0.86, p=3.76×10−6 for BMI) (supplementary table S11).
FIGURE 3.
Genome-wide genetic correlation between three lung function traits and three obesity traits in the a) China Kadoorie Biobank (CKB) and b) UK Biobank (UKB) cohorts. FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; BMI: body mass index; WHRadjBMI: BMI-adjusted waist-to-hip ratio; WCadjBMI: BMI-adjusted waist circumference. The colour of each box scales with the magnitude of the genetic correlation (Rg). *: pairs of traits with nominal significant genetic correlation (p<0.05); **: pairs of traits with significant genetic correlation after correcting for multiple testing (p<0.05/9). Boxes without labelling are trait pairs with nonsignificant genetic correlation.
Cross-trait meta-analysis
For the trait pairs that showed significant genetic correlation after Bonferroni correction (we also included the BMI–FEV1 trait pair despite p=0.038), we applied CPASSOC for genome-wide cross-trait meta-analysis to identify shared genetic variants among each of the trait pairs (pmeta<5×10−8; single-trait p<1×10−5). A total of six trait pairs in the CKB and seven trait pairs in the UKB were included for the cross-trait meta-analysis. In the CKB, after pruning, we found seven loci significantly associated with BMI and FEV1, five loci with WHRadjBMI and FEV1, seven loci with WCadjBMI and FEV1, four loci with BMI and FVC, one locus with WHRadjBMI and FVC, and one locus with WCadjBMI and FVC. Among these loci, we highlighted three shared loci since they were shared loci in multiple pairs of lung function and obesity traits. The first locus is DIS3L2 on 2q37.1 (BMI–FEV1, WCadjBMI–FEV1, BMI–FVC and WCadjBMI–FVC). The second locus is HLA-DQA1 (BMI–FEV1 and BMI–FVC). The third locus consists of several sentinel SNPs that were all mapped within 12p13.2, with genes including ATXN2 and ACAD10 (table 2). Out of the 25 shared loci identified in the CKB, five of them were also identified in the same trait pairs in the UKB (table 2 and supplementary tables S12–S18).
TABLE 2.
25 shared genetic loci between lung function (forced expiratory volume in 1 s (FEV1) and forced vital capacity (FVC)) and obesity (body mass index (BMI), BMI-adjusted waist-to-hip ratio (WHRadjBMI) and BMI-adjusted waist circumference (WCadjBMI)) traits in the China Kadoorie Biobank
| Trait pair | Sentinel SNP | Clump region | N | A1 | A2 | BETA1 | P1 | BETA2 | P2 | P | Genes within clump region | Overlap with UKB# |
| FEV1 and BMI | rs73995038 | chr2:232797462 –233165478 |
193 | A | G | −0.027 | 2.40×10−9 | −0.024 | 1.30×10−7 | 1.15×10−15 | DIS3L2 | No |
| rs801170 | chr5:139973696 –140230371 |
303 | C | T | 0.023 | 3.70×10−7 | 0.022 | 7.20×10−7 | 9.11×10−13 | CD14, DND1, HARS, HARS2, IK, MIR3655, NDUFA2, PCDHA1, PCDHA2, PCDHA3, PCDHA4, PCDHA5, PCDHA6, PCDHA7, PCDHA8, PCDHA9, TMCO6, VTRNA1-1, VTRNA1-2, VTRNA1-3, WDR55, ZMAT2 | No | |
| rs9271730 | chr6:32397794 –32667412 |
1728 | G | A | 0.022 | 3.10×10−6 | 0.029 | 3.30×10−10 | 7.72×10−15 | HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DRB6 | No | |
| rs11066001 | chr12:111629389 –112119171 |
19 | T | C | 0.024 | 7.60×10−6 | 0.028 | 3.40×10−7 | 1.99×10−11 | ATXN2, BRAP, CUX2, FAM109A, MIR6760, SH2B3 | No | |
| rs144504271 | chr12:112140669 –113117897 |
16 | G | A | 0.028 | 1.00×10−6 | 0.029 | 3.20×10−7 | 1.88×10−12 | ACAD10, ADAM1A, ALDH2, ERP29, HECTD4, MAPKAPK5, MAPKAPK5-AS1, MIR6761, MIR6861, NAA25, PTPN11, RPL6, TMEM116, TRAFD1 | No | |
| rs2078863 | chr12:111846028 –112355472 |
137 | T | C | 0.020 | 5.60×10−6 | 0.020 | 9.50×10−6 | 5.20×10−10 | ACAD10, ADAM1A, ALDH2, ATXN2, BRAP, MAPKAPK5, MAPKAPK5-AS1, MIR6761, SH2B3 | No | |
| rs5742653 | chr12:102397730 –102910374 |
339 | C | T | 0.021 | 4.50×10−6 | 0.022 | 1.10×10−6 | 2.03×10−11 | CCDC53, IGF1, NUP37, PARPBP, PMCH | No | |
| FEV1 and WHRadjBMI | rs11066325 | chr12:112834586 –113150735 |
7 | T | C | 0.036 | 2.00×10−9 | 0.031 | 2.30×10−7 | 5.44×10−16 | PTPN11, RPL6 | No |
| rs3809297 | chr12:111293470 –111718231 |
88 | G | T | 0.029 | 8.80×10−7 | 0.026 | 6.80×10−6 | 2.53×10−11 | CCDC63, CUX2, LOC100131138, MYL2 | No | |
| rs4646776 | chr12:111886967 –112678697 |
19 | G | C | 0.037 | 5.60×10−11 | 0.029 | 2.20×10−7 | 1.51×10−17 | ACAD10, ADAM1A, ALDH2, ATXN2, BRAP, ERP29, HECTD4, MAPKAPK5, MAPKAPK5-AS1, MIR6761, MIR6861, NAA25, SH2B3, TMEM116, TRAFD1 | No | |
| rs7175531 | chr15:51415799 –51556959 |
49 | T | C | 0.027 | 1.60×10−7 | 0.022 | 7.80×10−6 | 2.01×10−12 | CYP19A1, MIR4713 | No | |
| rs6142351 | chr20:33864484 –34336720 |
302 | G | A | −0.032 | 5.20×10−11 | −0.024 | 1.20×10−6 | 5.03×10−16 | C20orf173, CEP250, CPNE1, EIF6, ERGIC3, FAM83C, FAM83C-AS1, FER1L4, GDF5, MMP24, MMP24-AS1, NFS1, RBM12, RBM39, ROMO1, SPAG4, UQCC1 | Yes | |
| FEV1 and WCadjBMI | rs12048493 | chr1:149922960 –149995265 |
4 | A | C | −0.023 | 1.00×10−6 | 0.022 | 6.10×10−6 | 1.92×10−10 | OTUD7B | Yes |
| rs6604614 | chr1:218568359 –218690948 |
71 | C | G | −0.030 | 5.10×10−9 | −0.027 | 1.90×10−7 | 1.06×10−16 | TGFB2 | Yes | |
| rs16828537 | chr2:232797462 –233211117 |
264 | A | G | 0.033 | 3.90×10−13 | −0.025 | 3.30×10−8 | 3.91×10−19 | DIS3L2 | No | |
| rs11066065 | chr12:111846028 –112824473 |
707 | C | G | 0.025 | 2.20×10−8 | 0.024 | 1.20×10−7 | 1.92×10−16 | ACAD10, ADAM1A, ALDH2, ATXN2, BRAP, ERP29, HECTD4, MAPKAPK5, MAPKAPK5-AS1, MIR6761, MIR6861, NAA25, SH2B3, TMEM116, TRAFD1 | No | |
| rs11066325 | chr12:112834586 –113150735 |
7 | T | C | 0.053 | 6.70×10−19 | 0.031 | 2.30×10−7 | 5.08×10−27 | PTPN11, RPL6 | No | |
| rs4646776 | chr12:111827203 –112678697 |
31 | G | C | 0.055 | 4.90×10−23 | 0.029 | 2.20×10−7 | 7.56×10−31 | ACAD10, ADAM1A, ALDH2, ATXN2, BRAP, ERP29, HECTD4, MAPKAPK5, MAPKAPK5-AS1, MIR6761, MIR6861, NAA25, SH2B3, TMEM116, TRAFD1 | No | |
| rs78572043 | chr12:111293470 –111718231 |
94 | A | G | 0.046 | 1.10×10−13 | 0.029 | 2.70×10−6 | 4.58×10−20 | CCDC63, CUX2, LOC100131138, MYL2 | No | |
| FVC and BMI | rs6730783 | chr2:219890663 –220051676 |
63 | A | G | −0.023 | 2.00×10−7 | −0.021 | 4.00×10−6 | 2.73×10−12 | CCDC108, CNPPD1, FAM134A, IHH, MIR3131, NHEJ1, SLC23A3 | No |
| rs73995038 | chr2:232797462 –233201328 |
207 | A | G | −0.027 | 2.40×10−9 | −0.024 | 7.10×10−8 | 4.13×10−16 | DIS3L2 | No | |
| rs801170 | chr5:139791506 –140230371 |
327 | C | T | 0.023 | 3.70×10−7 | 0.022 | 1.30×10−6 | 8.74×10−13 | ANKHD1, ANKHD1-EIF4EBP3, APBB3, CD14, DND1, EIF4EBP3, HARS, HARS2, IK, MIR3655, MIR6831, NDUFA2, PCDHA1, PCDHA2, PCDHA3, PCDHA4, PCDHA5, PCDHA6, PCDHA7, PCDHA8, PCDHA9, SLC35A4, SRA1, TMCO6, VTRNA1-1, VTRNA1-2, VTRNA1-3, WDR55, ZMAT2 | No | |
| rs9271730 | chr6:32397794 –32667412 |
2271 | G | A | 0.022 | 3.10×10−6 | 0.028 | 1.30×10−9 | 2.21×10−14 | HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DRB6 | No | |
| FVC and WHRadjBMI | rs2425059 | chr20:33847253 –34412049 |
326 | T | C | 0.033 | 3.70×10−11 | 0.022 | 9.10×10−6 | 1.61×10−14 | C20orf173, CEP250, CPNE1, EIF6, ERGIC3, FAM83C, FAM83C-AS1, FER1L4, GDF5, MMP24, MMP24-AS1, NFS1, PHF20, RBM12, RBM39, ROMO1, SPAG4, UQCC1 | Yes |
| FVC and WCadjBMI | rs16828537 | chr2:232797462 –233211117 |
257 | A | G | 0.033 | 3.90×10−13 | −0.025 | 1.40×10−8 | 5.78×10−17 | DIS3L2 | Yes |
SNP: single nucleotide polymorphism; N: number of variants meeting the criteria of p<1×10−5 and r2>0.2 within the clump region; A1: effect allele; A2: noneffect allele; BETA1: lung function trait effect size; P1: lung function trait p-value; BETA2: obesity trait effect size; P2: obesity trait p-value; P: cross-trait meta-analysis p-value; UKB: UK Biobank. #: if the cross-trait meta-analysis clump region is overlapped with the same trait pair results in the UKB.
Pathway analysis
To gain biological insights into the shared genes, we assessed the enrichment of the independent loci for each trait and the identified set of shared genes between lung function and obesity traits in GO biological process categories, and observed many significant enrichments in the UKB results (FDR: q<0.05 for both traits) (supplementary table S19). Consistent with the gene function of shared loci, GO biological process highlighted several common biological pathways for lung function and obesity traits, such as cell proliferation, embryo, skeletal and tissue development, and regulation of gene expression. However, we did not observe any significant enrichment in the CKB results.
Mendelian randomisation
We applied GSMR to perform causal inference between BMI and lung function traits. In the East Asian population analysis, we observed a significant negative causal effect of BMI (per standard deviation) on FEV1 (bxy= −0.08, p=2.46×10−4) and FVC (bxy= −0.11, p=3.44×10−7) (table 3). We did not observe a significant causal effect of BMI on FEV1/FVC. In the reverse direction, we observed either a small magnitude or nonsignificant causal effect.
TABLE 3.
Estimates of causal effect size for body mass index (BMI) and lung function traits (forced expiratory volume in 1 s (FEV1), forced vital capacity (FVC) and FEV1/FVC)
| Population | Trait 1 | Trait 2 | Direction | Causal effect size# | se | p-value | nSNP |
| East Asian | BMI | FEV1 | → | −0.0773 | 0.021 | 2.46×10−4 | 68 |
| ← | 0.0884 | 0.033 | 6.53×10−3 | 17 | |||
| FVC | → | −0.1084 | 0.021 | 3.44×10−7 | 67 | ||
| ←¶ | 6 | ||||||
| FEV1/FVC | → | 0.0332 | 0.021 | 0.115732 | 69 | ||
| ←¶ | 3 | ||||||
| European | BMI | FEV1 | → | −0.1057 | 0.012 | 4.65×10−18 | 50 |
| ← | 0.0250 | 0.012 | 0.0308 | 411 | |||
| FVC | → | −0.1564 | 0.012 | 1.23×10−37 | 50 | ||
| ← | −0.0180 | 0.013 | 0.154475 | 379 | |||
| FEV1/FVC | → | 0.1622 | 0.014 | 8.01×10−33 | 48 | ||
| ← | 0.0393 | 0.009 | 4.75×10−6 | 599 |
nSNP: number of single nucleotide polymorphisms in the instrumental variable; →: trait 1→trait 2 causal direction; ←: trait 2→trait 1 causal direction. #: causal effect sizes are in units of per standard deviation increase in exposure; ¶: the FVC and FEV1/FVC genome-wide association studies do not have enough SNPs at the genome-wide significance level to construct the instrument variable.
PRSlung function×BMI analysis
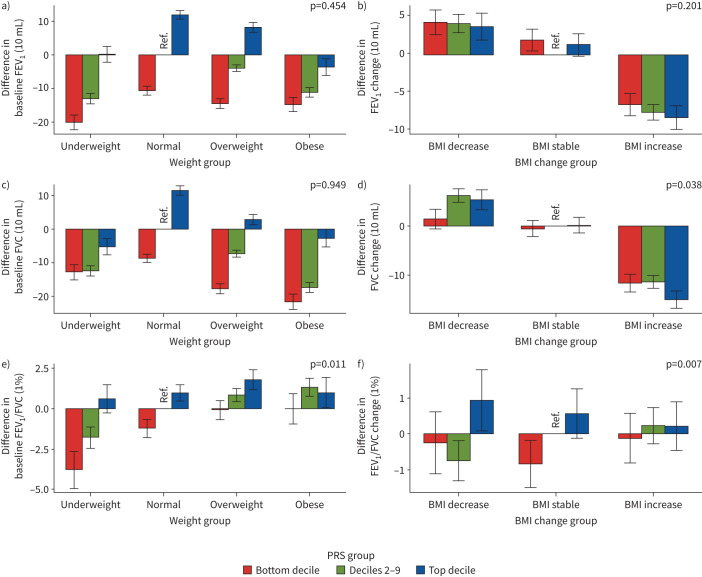
We constructed seven PRS models from LDpred for each lung function trait and selected the PRS model with the highest discriminatory performance (R2) for interaction analysis (supplementary table S20). We found a significant interaction between baseline BMI and PRS of FEV1/FVC on FEV1/FVC (p=0.011) (figure 4a and supplementary table S21). Generally, compared with the reference group, the bottom PRS decile plus underweight group has the lowest FEV1. For FVC, the largest reduction was found in the bottom PRS decile plus obese group. For FEV1/FVC, we observed that the bottom PRS decile plus underweight group had the lowest FEV1/FVC, and overweight and obese groups had increases. In the change model, we found a significant interaction between change in BMI and PRS of FVC (p=0.038) on change in FVC or between change in BMI and PRS of FEV1/FVC (p=0.007) on change in FEV1/FVC (figure 4b and supplementary table S21). Overall, compared with the reference group, the BMI increase group had reduced FEV1 and FVC, and the effect was largest in the top PRS decile. For the UKB, we did not find a significant interaction effect in the baseline model. However, we found a borderline significant interaction between change in BMI and PRS of FVC (p=0.068) on change in FVC (supplementary table S22 and supplementary figure S3). Finally, we constructed additional lung function PRSs using weights of 275 SNPs from the Shrine et al. [14] study and did not find any significant PRSlung function×BMI effect (supplementary table S23 and supplementary figure S4).
FIGURE 4.
Relationship of the distribution of three lung function polygenic risk scores (PRSs) with body mass index (BMI) in the China Kadoorie Biobank for a, c, e) the baseline model and b, d, f) the change model: a, b) forced expiratory volume in 1 s (FEV1), c, d) forced vital capacity (FVC) and e, f) FEV1/FVC. For the baseline model, we set normal BMI and the deciles 2–9 group as reference; for the change model, we set BMI stable and the deciles 2–9 group as reference. For the baseline model, the x-axis denotes different BMI categories by the following definitions: underweight BMI <18.5 kg·m−2, normal BMI 18.5–24.9 kg·m−2, overweight BMI 25.0–29.9 kg·m−2 and obese BMI ≥30.0 kg·m−2. The y-axis denotes the difference between lung function measurements for each group and the reference group. For the change model, the x-axis denotes different BMI change categories: BMI decrease is defined as BMIt1−BMIt0≤ −1 kg·m−2, BMI stable is defined as −1 kg·m−2<BMIt1−BMIt0≤1 kg·m−2 and BMI increase is defined as BMIt1−BMIt0>1 kg·m−2. The y-axis denotes the difference between lung function measurements change (lung functiont1−lung functiont0) for each group and the reference group. The PRS groups were defined as: bottom decile, deciles 2–9 and top decile. The p-value on each plot represents the lung function and baseline BMI or BMI change interaction p-value from baseline or change models.
Discussion
To the best of our knowledge, the current study is the largest GWAS of lung function in the Chinese population. We found a strong genetic correlation and shared genetic loci between lung function and obesity traits. We replicated these Chinese findings in the UKB and also identified population-specific genetic effects. We also found shared biological pathways between lung function and obesity traits, such as cell proliferation, embryo, skeletal and tissue development, and regulation of gene expression.
In this study, we identified nine new loci for FEV1, six for FVC and three for FEV1/FVC. Of these, we highlighted a novel gene associated with FEV1, GPC5 on 13q31.3. GPC5 is a member of the glypican gene family. Evidence to date suggests that the main function of the glypicans is to regulate the signalling pathway of bone morphogenetic proteins, Wnt, Hedgehog and fibroblast growth factors [41], which are involved in modulation of lung function [42], pulmonary fibrogenesis [43] and COPD pathobiology [44]. GPC5 was also found to contribute to an increased risk of lung cancer in never-smokers [45]. For FVC, we also found a novel independent region, 20q11.23, where the sentinel SNP is mapped to a lncRNA, LINC00489, although the function of this region needs to be further investigated. For FEV1/FVC, we note that several independent loci were within the 6p21 region, which was known for its association with lung function traits (FEV1 and FEV1/FVC) and COPD [39, 40]. This region contains genes such as AGER, ATF6B, NOTCH4 and TNXB, of which AGER has been reported to play a potential functional role in lung function [46]. AGER protein, a receptor for advanced glycation end-products (RAGE), is a multiligand receptor of the immunoglobulin superfamily and interacts with distinct molecules implicated in homeostasis, development, inflammation, diabetes and neurodegeneration [46]. RAGE signals depend on cell type and context. RAGE expression increases following cigarette smoke exposure and is partially responsible for inducing the pro-inflammatory signalling pathways (e.g. NF-κB) [47]. In addition, we noticed two novel loci (mapped genes DIS3L2 and FGF10/FGF10-AS1) that are significant in both FEV1 and FVC, indicating their pleotropic effect between different lung function traits.
Our LDSC analysis showed a strong genome-wide genetic correlation between lung function and obesity traits in both Chinese and European populations. We observed a strong negative genetic correlation of obesity traits with FEV1 and FVC in both populations, but a nonsignificant genetic correlation between obesity traits and FEV1/FVC in the Chinese population. The genetic correlation results in the European population were consistent with those from the Chinese population, except for BMI–FEV1/FVC, which was also highly significant in the European population (Rg=0.15, p=7.92×10−24). Previous studies showed that FEV1 and FVC are reduced in the presence of obesity [48], but the FEV1/FVC ratio is usually unaffected [49]. There are several biological mechanisms that could potentially explain how lung function impairment and obesity are associated. First, the mechanical effects of obesity produce airway narrowing and closure and increased respiratory system resistance. Compared with healthy weight individuals, airway narrowing in obesity correlates with airway closure and airway hyperresponsiveness [50]. Airway narrowing and closure lead to air trapping and ventilation inhomogeneity [51]. In addition, we found that the genetic correlation is stronger between lung function and central obesity than with global obesity. Compared with global obesity, which does not take account of fat distribution, abdominal and thoracic fat are more likely to play a role in lung function impairment. This is because they have direct mechanical effects on the diaphragm and chest wall expansion during forced inspiration [52, 53], a typical symptom of restrictive lung disease [54].
Cross-trait meta-analysis identified significantly independent loci shared between lung function and obesity traits. In the Chinese population, the locus DIS3L2 on 2q37.1 was found to be shared between multiple lung function and obesity traits (BMI–FEV1, WCadjBMI–FEV1, BMI–FVC and WCadjBMI–FVC in the Chinese population; WC–FVC in the European population). A previous study found DIS3L2 to be a gene that contributes to an overgrowth syndrome (e.g. Perlman syndrome [55]), suggesting its critical role in the regulation of cell growth and division. Such a function is also consistent with the findings from the pathway analysis, where the shared genes are mainly enriched in pathways related to cell proliferation and embryo, skeletal and tissue development. Notably, these shared pathways show the important role of growth for both lung function and obesity, and are partially distinct from the pathways identified in a recent lung function GWAS study [14]. Unsurprisingly, we also found many loci in the HLA region that were shared by obesity traits and lung function in both populations. HLA is a gene complex that contains abundant pleiotropy for many complex diseases [6, 7, 9, 56] and is especially involved in immune-related process [57]. In the European population, we also identified many shared loci between lung function and obesity traits. However, we found that most of the shared loci were distinct between the two populations.
Although the relationship between lung function and obesity was established in epidemiological studies [5, 15, 31, 58], it remains unclear whether obesity is a driving component in lung function or a comorbidity of its presence. The MR estimates in the current study suggested a negative causal effect of BMI to FEV1 and FVC and a positive causal effect to FEV1/FVC. These estimates provide evidence that BMI might reduce lung function in both East Asian and European populations, although the causal relationship between BMI and FEV1/FVC can still be bidirectional. The results of causal association from BMI to lung function traits are consistent with Wielscher et al. [59], showing negative causal associations of BMI with FEV1 and FVC and a positive causal association of BMI with FEV1/FVC.
In this study, we found evidence of genetic heterogeneity in Chinese and European populations. At the genome-wide level, the cross-population genetic correlation analysis showed that BMI had an estimated cross-population Rg statistically significantly <1, indicating heterogeneity in genetic regulation of BMI across Chinese and European populations. At the variant level, the cross-trait meta-analysis showed the majority of the shared variants are different in Chinese and European populations, which could be due to the distinct genetic background and sample size in the two populations, and also gene×environment interaction [32].
Several recent lung function PRS studies have focused on the association between lung function PRS and COPD risk, and interaction with smoking status [14, 60]. Our study investigated PRSlung function×BMI in both cross-sectional and longitudinal settings. Our interaction analysis showed that maintaining a normal BMI improved lung function and the beneficial effect is more profound in subjects with a high lung function genetic profile. The results suggest that BMI might be a mediator of genetic effects on lung function, which is consistent with the MR results showing that BMI is a causal risk factor of lung function. We also observed the top PRS group showed the most beneficial effect by BMI change. These findings have important implications for lung function improvement because they can provide potential intervention on BMI to individuals at risk before lung function reduction based on more precise risk stratification by using PRSs.
We also acknowledge the limitations in the current study. First, the CKB GWAS sample size is only a quarter the size of the UKB's. This leads to some findings that may not be directly comparable between the two cohorts. Second, the FVC and FEV1/FVC measurements for the Haikou and Qingdao regions (n=14 000) have may be biased in comparison with the other eight regions in the CKB. Thus, we further conducted the sensitivity analysis removing the two regions. The sensitivity analysis (supplementary figures S5 and S6) showed the effect sizes of FVC and FEV1/FVC novel loci were highly consistent with the primary analysis (using the full GWAS cohort), although the p-values modestly increased after removing the two regions due to less power. In addition, the results of the sensitivity analysis for genetic correlation are consistent with the primary analysis (supplementary table S24). The CKB FEV1/FVC PRS has also been used in a recent study and showed consistent results compared with other independent ancestry groups [14]. Third, we chose to use the lung function spirometry measures with two time-points in the UKB (data fields 3062 and 3063), thus the most cleaned lung function spirometry measures (data fields 20 150 and 20 151) could not be used. However, we showed our GWAS results are consistent with the results of the Shrine et al. [14] study, which used the most cleaned lung function spirometry measures. Fourth, although the use of PRSs allows researchers to effectively capture a useful fraction of genetic effects, the PRSlung function×BMI analysis remains susceptible to confounding or bias due to LD between SNP markers in the PRS model and the causal variants of BMI [61, 62]. However, our sensitivity analyses showed that this potentially has little impact on the PRSlung function×BMI analyses and adjusting height is not likely to introduce collider bias between the BMI and lung function association (supplementary tables S25–S27). Finally, the LD patterns in the HLA region are highly complex and the signals we reported are likely tagging the causal variants instead of actually being the causal variants due to the limitation of imputation data. Sequencing data is recommended to identify causal variants in the HLA region.
In conclusion, the current study is the first large-scale GWAS analysis of lung function in the Chinese population. Our study extends existing knowledge of the genetic landscape of lung function traits by leveraging large-scale Chinese and European genetic cohorts. We applied single- and cross-trait analyses, and identified novel loci for lung function traits in the Chinese population, shared genetic effects between lung function and obesity traits, genetic heterogeneity in Chinese and European populations, and a PRSlung function×BMI effect. These new findings provide greater knowledge of the genetic basis of lung function in the Chinese population, and shared genetics between lung function and obesity, which will foster subsequent translational, clinical and public health research.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary appendix and figures ERJ-00199-2021.Supplement (1.2MB, pdf)
Supplementary tables ERJ-00199-2021.Tables (205.4KB, xlsx)
Shareable PDF
Acknowledgements
This research has been conducted using the China Kadoorie Biobank (CKB) and UK Biobank (UKB) (application numbers 16549 and 45052). We greatly thank the participants in the study and the members of the research teams at both the CKB and UKB. We thank GIANT and the Biobank Japan consortium for providing genome-wide association study summary statistics data. We also thank Zachary Schwartz (Harvard T.H. Chan School of Public Health, Boston, MA, USA) for language editing assistance.
Footnotes
This article has supplementary material available from erj.ersjournals.com
This article has an editorial commentary: https://doi.org/10.1183/13993003.01615-2021
Author contributions: Z. Zhu, C. Yu and L. Liang the designed the study. All authors acquired or interpreted the data. Z. Zhu, J. Li, B. Ma and H. Shi did the statistical analysis. L. Li, W. Cao, C. Yu, J. Lv and Y. Guo obtained funding. All authors reviewed and approved the manuscript.
Conflict of interest: Z. Zhu has nothing to disclose.
Conflict of interest: J. Li has nothing to disclose.
Conflict of interest: J. Si has nothing to disclose.
Conflict of interest: B. Ma has nothing to disclose.
Conflict of interest: H. Shi has nothing to disclose.
Conflict of interest: J. Lv has nothing to disclose.
Conflict of interest: W. Cao has nothing to disclose.
Conflict of interest: Y. Guo has nothing to disclose.
Conflict of interest: I.Y. Millwood has nothing to disclose.
Conflict of interest: R.G. Walters has nothing to disclose.
Conflict of interest: K. Lin has nothing to disclose.
Conflict of interest: L. Yang has nothing to disclose.
Conflict of interest: Y. Chen has nothing to disclose.
Conflict of interest: H. Du has nothing to disclose.
Conflict of interest: B. Yu has nothing to disclose.
Conflict of interest: K. Hasegawa reports grants from the NIH and Novartis, outside the submitted work.
Conflict of interest: C.A. Camargo Jr has nothing to disclose.
Conflict of interest: M.F. Moffatt has nothing to disclose.
Conflict of interest: W.O.C. Cookson has nothing to disclose.
Conflict of interest: J. Chen has nothing to disclose.
Conflict of interest: Z. Chen has nothing to disclose.
Conflict of interest: L. Li has nothing to disclose.
Conflict of interest: C. Yu has nothing to disclose.
Conflict of interest: L. Liang has nothing to disclose.
Support statement: This work was supported by the National Key R&D Program of China (2016YFC1303904, 2016YFC0900500, 2016YFC0900501 and 2016YFC0900504) and National Natural Science Foundation of China (81941018, 91846303 and 91843302). The China Kadoorie Biobank baseline survey and the first re-survey were supported by a grant from the Kadoorie Charitable Foundation in Hong Kong. The long-term follow-up is supported by grants from the UK Wellcome Trust (212946/Z/18/Z, 202922/Z/16/Z, 104085/Z/14/Z and 088158/Z/09/Z), National Natural Science Foundation of China (81390540, 81390541 and 81390544), and Chinese Ministry of Science and Technology (2011BAI09B01). The funders had no role in the study design, data collection, data analysis and interpretation, writing of the report, or the decision to submit the article for publication. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Garcia-Aymerich J, Serra Pons I, Mannino DM, et al. Lung function impairment, COPD hospitalisations and subsequent mortality. Thorax 2011; 66: 585–590. doi: 10.1136/thx.2010.152876 [DOI] [PubMed] [Google Scholar]
- 2.Ostrowski S, Barud W. Factors influencing lung function: are the predicted values for spirometry reliable enough? J Physiol Pharmacol 2006; 57: Suppl. 4, 263–271. [PubMed] [Google Scholar]
- 3.World Health Organization . Obesity and overweight. 2020. www.who.int/news-room/fact-sheets/detail/obesity-and-overweight Date last accessed: 1 July 2020.
- 4.Leone N, Courbon D, Thomas F, et al. Lung function impairment and metabolic syndrome: the critical role of abdominal obesity. Am J Respir Crit Care Med 2009; 179: 509–516. doi: 10.1164/rccm.200807-1195OC [DOI] [PubMed] [Google Scholar]
- 5.Li J, Zhu L, Wei Y, et al. Association between adiposity measures and COPD risk in Chinese adults. Eur Respir J 2020; 55:1901899. doi: 10.1183/13993003.01899-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Guo Y, Shi H, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK biobank. J Allergy Clin Immunol 2020; 145: 537–549. doi: 10.1016/j.jaci.2019.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Z, Lee PH, Chaffin MD, et al. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet 2018; 50: 857–864. doi: 10.1038/s41588-018-0121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z, Wang X, Li X, et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir Res 2019; 20: 64. doi: 10.1186/s12931-019-1036-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Z, Zhu X, Liu CL, et al. Shared genetics of asthma and mental health disorders: a large-scale genome-wide cross-trait analysis. Eur Respir J 2019; 54: 1901507, doi: 10.1183/13993003.01 [DOI] [PubMed] [Google Scholar]
- 10.Palmer LJ, Knuiman MW, Divitini ML, et al. Familial aggregation and heritability of adult lung function: results from the Busselton Health Study. Eur Respir J 2001; 17: 696–702. doi: 10.1183/09031936.01.17406960 [DOI] [PubMed] [Google Scholar]
- 11.Herrera BM, Lindgren CM. The genetics of obesity. Curr Diab Rep 2010; 10: 498–505. doi: 10.1007/s11892-010-0153-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015; 518: 197–206. doi: 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wain LV, Shrine N, Artigas MS, et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet 2017; 49: 416–425. doi: 10.1038/ng.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrine N, Guyatt AL, Erzurumluoglu AM, et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet 2019; 51: 481–493. doi: 10.1038/s41588-018-0321-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon AE, Peters U. The effect of obesity on lung function. Expert Rev Respir Med 2018; 12: 755–767. doi: 10.1080/17476348.2018.1506331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loth DW, Soler Artigas M, Gharib SA, et al. Genome-wide association analysis identifies six new loci associated with forced vital capacity. Nat Genet 2014; 46: 669–677. doi: 10.1038/ng.3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soler Artigas M, Wain LV, Miller S, et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Commun 2015; 6: 8658. doi: 10.1038/ncomms9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyss AB, Sofer T, Lee MK, et al. Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nat Commun 2018; 9: 2976. doi: 10.1038/s41467-018-05369-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Ortega VE, Ampleford EJ, et al. Genome-wide association study of lung function and clinical implication in heavy smokers. BMC Med Genet 2018; 19: 134. doi: 10.1186/s12881-018-0656-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakaue S, Kanai M, Karjalainen J, et al. Trans-biobank analysis with 676 000 individuals elucidates the association of polygenic risk scores of complex traits with human lifespan. Nat Med 2020; 26: 542–548. doi: 10.1038/s41591-020-0785-8 [DOI] [PubMed] [Google Scholar]
- 21.Shi H, Burch KS, Johnson R, et al. Localising components of shared transethnic genetic architecture of complex traits from GWAS summary data. Am J Hum Genet 2020; 106: 805–817. doi: 10.1016/j.ajhg.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z, Hasegawa K, Camargo CA Jr, et al. Investigating asthma heterogeneity through shared and distinct genetics: insights from genome-wide cross-trait analysis. J Allergy Clin Immunol 2021; 147: 796–807. doi: 10.1016/j.jaci.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Chen J, Collins R, et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol 2011; 40: 1652–1666. doi: 10.1093/ije/dyr120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12: e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Lee L, Chen J, et al. Cohort profile: the Kadoorie Study of Chronic Disease in China (KSCDC). Int J Epidemiol 2005; 34: 1243–1249. doi: 10.1093/ije/dyi174 [DOI] [PubMed] [Google Scholar]
- 26.UK Biobank . Body composition measurement. Version 1.0. 2020. https://biobank.ctsu.ox.ac.uk/crystal/crystal/docs/body_composition.pdf Date last accessed: 5 August 2021.
- 27.Loh PR, Tucker G, Bulik-Sullivan BK, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 2015; 47: 284–290. doi: 10.1038/ng.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47: 1236–1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forno E, Han YY, Mullen J, et al. Overweight, obesity, and lung function in children and adults – a meta-analysis. J Allergy Clin Immunol Pract 2018; 6: 570–581. doi: 10.1016/j.jaip.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutherland TJ, McLachlan CR, Sears MR, et al. The relationship between body fat and respiratory function in young adults. Eur Respir J 2016; 48: 734–747. doi: 10.1183/13993003.02216-2015 [DOI] [PubMed] [Google Scholar]
- 32.Shi H, Gazal S, Kanai M, et al. Population-specific causal disease effect sizes in functionally important regions impacted by selection. Nat Commun 2021; 12: 1098. doi: 10.1038/s41467-021-21286-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X, Feng T, Tayo BO, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am J Hum Genet 2015; 96: 21–36. doi: 10.1016/j.ajhg.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z, Anttila V, Smoller JW, et al. Statistical power and utility of meta-analysis methods for cross-phenotype genome-wide association studies. PLoS One 2018; 13: e0193256. doi: 10.1371/journal.pone.0193256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y, Wang J, Jaehnig EJ, et al. Webgestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019; 47: W199–W205. doi: 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 2018; 9: 224. doi: 10.1038/s41467-017-02317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akiyama M, Okada Y, Kanai M, et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet 2017; 49: 1458–1467. doi: 10.1038/ng.3951 [DOI] [PubMed] [Google Scholar]
- 38.Vilhjalmsson BJ, Yang J, Finucane HK, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet 2015; 97: 576–592. doi: 10.1016/j.ajhg.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim WJ, Lee MK, Shin C, et al. Genome-wide association studies identify locus on 6p21 influencing lung function in the Korean population. Respirology 2014; 19: 360–368. doi: 10.1111/resp.12230 [DOI] [PubMed] [Google Scholar]
- 40.Artigas MS, Wain LV, Shrine N, et al. Targeted sequencing of lung function loci in chronic obstructive pulmonary disease cases and controls. PLoS One 2017; 12: e0170222. doi: 10.1371/journal.pone.0170222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol 2008; 9: 224. doi: 10.1186/gb-2008-9-5-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhamme FM, De Smet EG, Van Hooste W, et al. Bone morphogenetic protein 6 (BMP-6) modulates lung function, pulmonary iron levels and cigarette smoke-induced inflammation. Mucosal Immunol 2019; 12: 340–351. doi: 10.1038/s41385-018-0116-2 [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Shi C, Cao H, et al. The hedgehog and Wnt/beta-catenin system machinery mediate myofibroblast differentiation of LR-MSCs in pulmonary fibrogenesis. Cell Death Dis 2018; 9: 639. doi: 10.1038/s41419-018-0692-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YS, Hong G, Kim DH, et al. The role of FGF-2 in smoke-induced emphysema and the therapeutic potential of recombinant FGF-2 in patients with COPD. Exp Mol Med 2018; 50: 1–10. doi: 10.1038/s12276-018-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Sheu CC, Ye Y, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol 2010; 11: 321–330. doi: 10.1016/S1470-2045(10)70042-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller S, Henry AP, Hodge E, et al. The Ser82 RAGE variant affects lung function and serum rage in smokers and sRAGE production in vitro. PLoS One 2016; 11: e0164041. doi: 10.1371/journal.pone.0164041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson AB, Stogsdill JA, Lewis JB, et al. RAGE and tobacco smoke: insights into modeling chronic obstructive pulmonary disease. Front Physiol 2012; 3: 301. doi: 10.3389/fphys.2012.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerah F, Harf A, Perlemuter L, et al. Effects of obesity on respiratory resistance. Chest 1993; 103: 1470–1476. doi: 10.1378/chest.103.5.1470 [DOI] [PubMed] [Google Scholar]
- 49.Sin DD, Jones RL, Man SF. Obesity is a risk factor for dyspnea but not for airflow obstruction. Arch Intern Med 2002; 162: 1477–1481. doi: 10.1001/archinte.162.13.1477 [DOI] [PubMed] [Google Scholar]
- 50.Chapman DG, Berend N, King GG, et al. Increased airway closure is a determinant of airway hyperresponsiveness. Eur Respir J 2008; 32: 1563–1569. doi: 10.1183/09031936.00114007 [DOI] [PubMed] [Google Scholar]
- 51.Pellegrino R, Gobbi A, Antonelli A, et al. Ventilation heterogeneity in obesity. J Appl Physiol 2014; 116: 1175–1181. doi: 10.1152/japplphysiol.01339.2013 [DOI] [PubMed] [Google Scholar]
- 52.Canoy D, Luben R, Welch A, et al. Abdominal obesity and respiratory function in men and women in the EPIC-Norfolk Study, United Kingdom. Am J Epidemiol 2004; 159: 1140–1149. doi: 10.1093/aje/kwh155 [DOI] [PubMed] [Google Scholar]
- 53.Salome CM, King GG, Berend N. Physiology of obesity and effects on lung function. J Appl Physiol 2010; 108: 206–211. doi: 10.1152/japplphysiol.00694.2009 [DOI] [PubMed] [Google Scholar]
- 54.Melo LC, Silva MA, Calles AC. Obesity and lung function: a systematic review. Einstein 2014; 12: 120–125. doi: 10.1590/S1679-45082014RW2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Astuti D, Morris MR, Cooper WN, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 2012; 44: 277–284. doi: 10.1038/ng.1071 [DOI] [PubMed] [Google Scholar]
- 56.Sivakumaran S, Agakov F, Theodoratou E, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet 2011; 89: 607–618. doi: 10.1016/j.ajhg.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dendrou CA, Petersen J, Rossjohn J, et al. HLA variation and disease. Nat Rev Immunol 2018; 18: 325–339. doi: 10.1038/nri.2017.143 [DOI] [PubMed] [Google Scholar]
- 58.Vogelmeier CF, Criner GJ, Martinez FJ, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease 2017 Report: GOLD Executive Summary. Eur Respir J 2017; 49: 1700214. doi: 10.1183/13993003.00214-2017 [DOI] [PubMed] [Google Scholar]
- 59.Wielscher M, Minelli C, Amaral A, et al. Cardio metabolic traits and lung function: a Mendelian randomisation study. Eur Respir J 2018; 52: Suppl. 62, PA1277. doi: 10.1183/13993003.congress-2018.PA1277 [DOI] [Google Scholar]
- 60.Moll M, Sakornsakolpat P, Shrine N, et al. Chronic obstructive pulmonary disease and related phenotypes: polygenic risk scores in population-based and case-control cohorts. Lancet Respir Med 2020; 8: 696–708. doi: 10.1016/S2213-2600(20)30101-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aschard H, Vilhjalmsson BJ, Joshi AD, et al. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet 2015; 96: 329–339. doi: 10.1016/j.ajhg.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dudbridge F, Fletcher O. Gene-environment dependence creates spurious gene-environment interaction. Am J Hum Genet 2014; 95: 301–307. doi: 10.1016/j.ajhg.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary appendix and figures ERJ-00199-2021.Supplement (1.2MB, pdf)
Supplementary tables ERJ-00199-2021.Tables (205.4KB, xlsx)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00199-2021.Shareable (386.7KB, pdf)