SUMMARY
SETDB1 is a histone-lysine N-methyltransferase critical for germline development. However, its function in early meiotic prophase I remains unknown. Here we report that Setdb1-null spermatocytes display aberrant centromere clustering during leptotene, bouquet formation during zygotene, and subsequent failure in pairing and synapsis of homologous chromosomes as well as compromised meiotic silencing of unsynapsed chromatin, which leads to meiotic arrest before pachytene and apoptosis of spermatocytes. H3K9m3 is enriched in (peri-)centromeric regions and is present in many sites throughout the genome, with a subset changed in the Setdb1 mutant. These observations indicate that SETDB1-mediated H3K9 trimethylation is essential for the bivalent formation in early meiosis. Transcriptome analysis reveals the function of SETDB1 in repressing transposons and transposon-proximal genes and in regulating meiotic and somatic lineage genes. These findings highlight a mechanism in which SETDB1-mediated H3K9 trimethylation during early meiosis ensures the formation of homologous bivalents and survival of spermatocytes.
Keywords: SETDB1, pericentromeric heterochromatin, synapsis, homologous bivalent
INTRODUCTION
Meiosis refers to a special type of cell division through which a diploid germ cell undergoes a single round of DNA replication followed by two rounds of divisions to generate haploid gametes. To achieve faithful chromosome segregation during the first meiotic division (meiosis I), homologous chromosomes must form bivalents that interact with the meiotic spindle as a unit in a way to allow homologous centromeres to orient to opposite poles (Dunleavy and Collins, 2017; Gatti et al., 2012; Lane and Kauppi, 2019; Moore and Orr-Weaver, 1998; O'Donnell and O'Bryan, 2014; Radford et al., 2017). Defects in the establishment of homologous bivalents can lead to gametogenic arrest or aneuploidy, resulting in infertility or embryonic loss (Bolcun-Filas and Handel, 2018; Herbert et al., 2015; Jones and Lane, 2012; MacLennan et al., 2015). The prophase of meiosis I ensure the formation of homologous bivalents, including the pairing, synapsis, and recombination of homologous chromosomes (Bolcun-Filas and Handel, 2018; Bolcun-Filas and Schimenti, 2012; Hunter, 2015). Failure to complete synapsis activates the meiotic pachytene checkpoint and leads to meiotic arrest and subsequent apoptosis (Handel et al., 1999; Roeder and Bailis, 2000; Tsubouchi et al., 2018). Thus, understanding the mechanism underlying the formation of bivalents can provide insights into meiocyte loss and sterility.
The formation of bivalents is initiated by pairing homologous chromosomes with their unique homologous partners and forming a synaptonemal complex with short filaments of proteinaceous axial elements assembled along each homologous chromosome (Cahoon and Hawley, 2016; Fraune et al., 2016; Gao and Colaiacovo, 2018; Geisinger and Benavente, 2016). At the leptotene stage, recombination begins with the generation of DNA double-strand breaks (DSBs) and further recruits recombination proteins to facilitate a search for homologous sequences to repair the DSBs. During the zygotene stage, the axial element extends across the length of each chromosome axis and bring the homolog axes into proximity, stimulating synapsis. By the pachytene stage, homologous chromosomes are paired and synapsis spreads along the axis of all paired chromosomes except the X and Y chromosomes, in which only pseudoautosomal regions are paired. During the pachytene stage, recombination machinery generates crossovers by repairing a subset of DSBs, which provides a link between homologous chromosomes after the synaptonemal complex disassembles in the diplotene stage and enables homologous chromosomes to orient to opposite spindle poles (Alleva and Smolikove, 2017; Hochwagen and Amon, 2006; Kurdzo and Dawson, 2015; Obeso et al., 2014).
Increasing evidence suggests that histone modifications, which regulate the specialized architecture of meiotic chromosomes, play a fundamental role in pairing, synapsis, and recombination of homologs. Previous studies unequivocally demonstrate that methylation of histone H3 lysine 9 (H3K9me) at pericentromeric heterochromatin is required for proper chromosome interactions (Peters et al., 2001). Early in meiosis, before homologous chromosome pairing occurs, Suppressor of Variegation 3-9 Homolog Complexes (SUV39H1/2)-mediated H3K9 trimethylation at pericentromeric heterochromatin is recognized by HP1γ and further recruits G9a-mediated H3K9 dimethylation to maintain centromere clustering, which promotes homologous chromosome pairing and synapsis (Takada et al., 2011). Interestingly, in Suv39h1/2 double knockout spermatocytes, the signal of H3K9 methylation at pericentromeric heterochromatin is still observed at the zygotene stage and restored at the pachytene stage (Peters et al., 2001). Therefore, it remains elusive whether other H3K9 methyltransferases are involved in the regulation of H3K9 methylation-mediated centromere clustering, homologous pairing, and synapsis during meiosis.
SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) is a histone-lysine N-methyltransferase that mediates H3K9 trimethylation (H3K9me3) (Matsui et al.; Schultz et al., 2002; Wang et al., 2003). In addition to its known function in facilitating heterochromatin formation, it represses gene expression in euchromatin regions in diverse cell types and controls cellular identity and differentiation (Bilodeau et al., 2009; Koide et al., 2016; Lohmann et al., 2010; Yuan et al., 2009). Before birth in early fetal germline, knockout of SETDB1 in primordial germ cells (PGCs) leads to a decrease of H3K27me3 and H3K9me3 marks and DNA methylation at H3K9me3-enriched retrotransposons in gonocytes, with a concomitant derepression of endogenous retroviruses (ERVs) and ERV-proximal genes. Furthermore, SETDB1 deficiency is associated with a reduced number of male PGCs and postnatal hypogonadism (Liu et al., 2014), and defects in meiotic progression during oogenesis (Eymery et al., 2016; Kim et al., 2016). A study by Hirota et al. shows that during synapsis formation in mid-pachytene spermatocyte, meiotic DNA damage response (DDR) factors recruit SETDB1 to X and Y chromosomes and induce XY-chromatin remodeling and silencing (Hirota et al., 2018). However, the role of SETDB1 in meiotic pairing and other events prior to pachytene is not well understood.
In this study, we report that loss of SETDB1 results in early meiotic arrest at the transition from the zygotene to the pachytene stage and eventually male sterility. Knockout of SETDB1 in the germline results in abnormal distribution of H3K9me3 at pericentromeric heterochromatin, which leads to disruption of centromere clustering and bouquet formation, increased asynapsis of homologous chromosomes, and compromised meiotic silencing of unsynapsed chromatin (MSUC). Furthermore, Setdb1 knockout spermatogenic cells show inappropriate derepression of transposons, activation of transposon-proximal genes, and misregulation of meiotic and somatic genes. Our study reveals the function of SETDB1 in maintaining pericentromeric heterochromatin, establishing homologous bivalents, and ensuring proper transcript expression in early meiosis.
RESULTS
Setdb1 germline knockout male mice display spermatogenic arrest at the zygotene stage of meiosis I
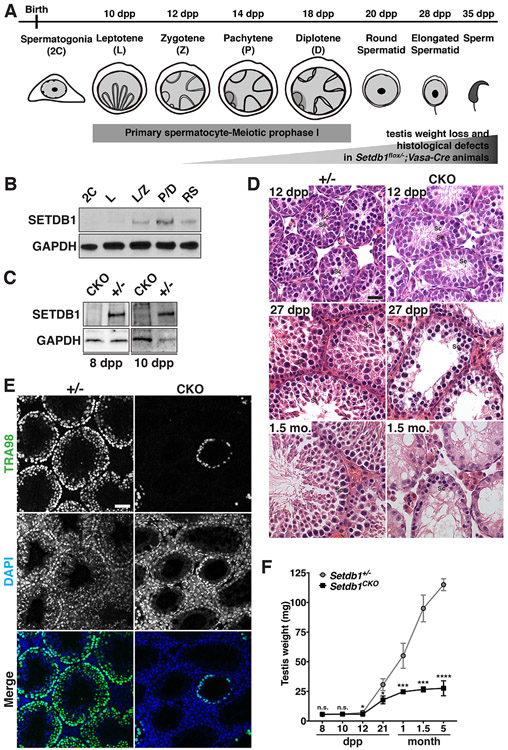
To investigate the role of SETDB1 in spermatogenesis, we first characterized its expression pattern during spermatogenesis. Figure 1A summarizes the timeline of the first wave of spermatogenesis in mice, in which leptotene, zygotene, pachytene, and diplotene cells emerge at 10 days postpartum (dpp), 12 dpp, 14 dpp, and 18 dpp, respectively. Western blot analysis of FACS-purified spermatogenic populations collected at different time points (Figure 1B) showed that the SETDB1 protein level was relatively low in somatic and spermatogonia/progenitor cells (2C), and was upregulated as spermatogenic cells entered early meiotic prophase I (leptotene and zygotene stages, L/Z), expressed at a high level during the pachytene and diplotene stages (P/D), and remained present in round spermatids (RS) after meiosis (Figure 1B), This is consistent with the profile of Setdb1 RNA expression (da Cruz et al., 2016; Hirota et al., 2018) (Figure S1). To further determine its localization in spermatocytes, we performed meiotic chromosome spreads with an anti-SCP3 antibody to recognize spermatocytes at substages of meiotic prophase I (Dobson et al., 1994) and CREST antisera to mark the centromere. Immunofluorescence analysis from 16dpp testes revealed a wide-spreading SETDB1 binding to chromosomes from early leptotene to pachytene, with lower levels of presence in the nucleoplasm (Figure S2B). This is consistent with its known function in both euchromatic regions and pericentromeric heterochromatin regions (Bilodeau et al., 2009; Koide et al., 2016; Lohmann et al., 2010; Yuan et al., 2009). These results together imply that SETDB1 may have a role in early meiosis.
Figure 1.
Defective spermatogenesis in Setdb1flox/−;Vasa-Cre mice.
(A) The timeline of the first wave of mouse spermatogenesis. Major germ cell types and stages of spermatogenesis are shown. Homologous chromosomes are illustrated by the gray lines in the nuclei of leptotene, zygotene, pachytene, and diplotene spermatocyte. The dark gray region of the nuclei in pachytene and diplotene spermatocyte indicates the XY body. (B) FACS-purified germ cells isolated from various age mice testes were prepared for western blot analysis. GAPDH serves as an internal control. 2C; diploid spermatogonia/somatic cells, L; leptotene, L/Z; leptotene and zygotene, P/D; pachytene and diplotene, RS; round spermatid. See also Figure S1 for Setdb1 RNA expression. (C) FACS-purified germ cells isolated from 8- and 10-dpp Setdb1 control (Setdb1flox/+;Vasa-Cre, +/−) and conditional knockout (Setdb1flox/−;Vasa-Cre, CKO) mice testes were prepared for western blot analysis. GAPDH serves as an internal control. See also Figure S2 for immunostaining. (D) Testicular sections of Setdb1 control (+/−) and conditional knockout (CKO) mice at 12 dpp, 27 dpp, and 1.5 month (mo.) were stained with hematoxylin and eosin. Se, Sertoli cell; Sc, spermatocyte; St, spermatid. Scale bars represent 30 μm. See also Figure S3A. (E) Histological sections of Setdb1 control (+/−) and conditional knockout (CKO) testes at 1.5 month stained with the TRA98 antibody to indicate spermatogenic cells. Blue, DAPI. Scale bars represent 30 μm. (F) Comparison of the testicular weight between Setdb1 control (+/−) and conditional knockout (CKO) mice at 8 dpp, 10 dpp, 12 dpp, 21 dpp, 1 month, 1.5 month and 5 month. See also Figure S3B. The results represent the mean ± SD of at least three independent experiments. (*P<0.05; ***P<0.001; ****P<0.0001; n.s., P>0.05)
To investigate the potential role of SETDB1 in early meiosis, we knocked out Setdb1 before germ cells reached the first wave of meiotic entry, which occurs at 8-10 dpp. To this end, we crossed Setdb1flox/flox mice to a Vasa-Cre mouse line that expresses Cre in embryonic germ cells from approximately E15-18 (Gallardo et al., 2007; Lohmann et al., 2010). The resulting mutant male mice (Setdb1flox/−;Vasa-Cre+/−) developed into adults without apparent abnormality except for being infertile. Western blot analysis of SETDB1 in the conditional knockout testes and control testes at 8 and 10 dpp confirmed that SETDB1 was absent in the knockout germ cells (Figure 1C). Immunofluorescence analysis of SETDB1 further showed that it was absent in the knockout germ cells (Figure S2). Gross examination of testes revealed that there was no difference in the histological structure of seminiferous tubules between Setdb1 conditional knockout and the control at 6, 8,10, and 12 dpp (Figure 1D, upper panel; Figure S3A). However, post-meiotic spermatids were absent from Setdb1 conditional knockout seminiferous tubules at 27 dpp (Figure 1D, middle panel), leading to a drastic germ cell atrophy in 1.5-month-old mice (Figure 1D, bottom). This defect was confirmed by immunofluorescence analysis using the TRA98 antibody that marks the all developing germ cells (Figure 1E). The characterization of testicular size throughout development showed that Setdb1 conditional knockout testes were smaller than the littermate control group from 12 dpp onward (Figure 1F and Figure S3B). These results reveal the importance of SETDB1 in regulating meiosis. Given that Setdb1 deletion in pachytene cells causes mid pachytene apoptosis (Hirota et al., 2018), it is likely that the initial defect in our model occurs during or even before the emerge of pachytene spermatocytes.
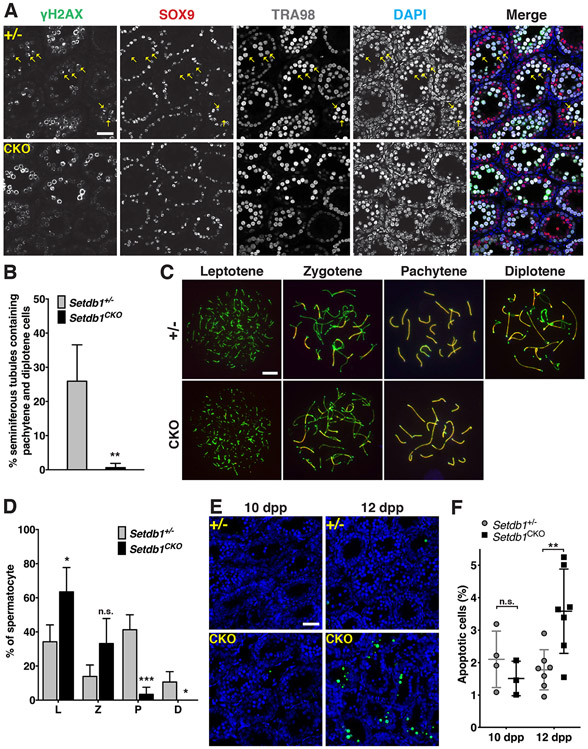
To further examine the early meiotic defect of Setdb1 knockout spermatocytes, we examined the presence of pachytene spermatocytes by using immunofluorescence analysis in testicular sections. γ-H2AX concentrates on unique foci in the XY body in pachytene and diplotene spermatocytes (Figure 2A; (Mahadevaiah et al., 2001)). Co-staining of TRA98 (marking germ cells) and SOX9 (marking somatic cells) antibodies revealed that 25.9 % of control testicular tubules contained pachytene spermatocytes, but only 0.6% of the knockout tubules contained pachytene spermatocytes (Figure 2A and B). This indicates that Setdb1 knockout spermatocytes were arrested before the pachytene stage. To more precisely determine which stage of meiosis was affected by the absence of SETDB1, we performed meiotic chromosome spreads using anti-synaptonemal complex protein 3 (SCP3) and synaptonemal complex protein 1 (SCP1) antibodies to mark the axial/lateral elements and transverse filaments, respectively (Dobson et al., 1994). At 16 dpp, control testes contained spermatocytes in all substages of meiotic prophase I, with the majority being pachytene cells (41% of total spermatocytes, Figure 2C and D). In contrast, pachytene-like cells were sporadic in Setdb1 conditional knockout testes (3% of total spermatocytes, Figure 2C and D). In addition, most zygotene and pachytene-like cells showed abnormal synapse structures (details below), with leptotene cells being the predominant meiotic cells in the knockout testes (63% of total spermatocytes). Although there was 10% diplotene cells in the control testes, it was absent in the knockout testes (Figure 2C and D). These observations indicate that the SETDB1 depletion leads to a significant defect in early meiosis. To evaluate whether the defect in early meiosis was associated with cell death, we performed TUNEL staining of the testicular sections at different postnatal ages (Figure 2E and F; Figure S3C). We found that significantly more apoptotic cells were detected in knockout tubules when zygotene cells (12 dpp) and pachytene cells first emerged (16 dpp), but not in younger (10 dpp) testes that contained only leptotene stage spermatocytes. These findings reveal a requirement of SETDB1 in the survival of early meiotic cells at zygotene and pachytene stages.
Figure 2.
Sparse presence of early- to mid-pachytene-like cells in Setdb1 conditional knockout testes.
(A) Histological sections of Setdb1 control (+/−) and conditional knockout (CKO) testes at 13 dpp were stained with anti-γH2AX (green), anti-SOX9 (red) and TRA98 (gray) antibodies. Pachytene spermatocytes were determined by the presence of the XY body, a single γH2AX foci in SOX9-negative and TRA98-positive cells (arrow). Blue, DAPI. Scale bars represent 30 μm. (B) Percentage of seminiferous tubules containing pachytene cells shown in (A). (C) Spermatocyte spreads from Setdb1 control (+/−) and conditional knockout (CKO) testes at 16 dpp. Spermatocyte nuclei were stained with anti-SCP3 (green) and SCP1 (red) antibodies, indicating the axial/lateral elements and transverse filaments of the synaptonemal complex, respectively. The leptotene, zygotene, pachytene, and diplotene stages of prophase I, respectively, are shown. No diplotene spermatocytes were found in Setdb1 CKO testes. The scale bar represents 7.5 μm. (D) Percentages of the L (leptotene), Z (zygotene), P (pachytene), and D (diplotene) spermatocytes detected from (C). (E) TUNEL assay. Comparison of apoptotic cells in Setdb1 control (+/−) and conditional knockout (CKO) testes at 10 and 12 dpp. Blue, DAPI; Green, TUNEL labeling. The scale bar represents 30 μm. See also Figure S3C. (F) Percentage of apoptotic cells shown in (E).
The results represent the mean ± SD of at least three independent experiments. (*P<0.05; **P<0.01; ***P<0.001; n.s., P>0.05)
SETDB1 deficiency perturbs distribution of histone H3K9me3 on chromosomes during prophase I of meiosis
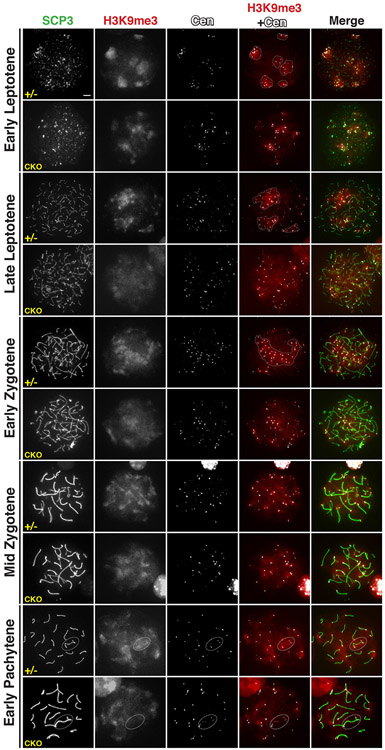
Given the role of SETDB1 in H3K9 methylation, we investigated H3K9me3 patterns in Setdb1 conditional knockout spermatocytes by meiotic chromosome spreads using an anti-SCP3 antibody to recognize spermatocytes at substages of meiotic prophase I and CREST antisera to mark the centromere. In the control spermatocytes at all substages of meiotic prophase I, H3K9me3 signal, a hallmark of pericentromeric heterochromatin, was enriched around the outer edge of centromere (Figure 3), as previously reported (Peters et al., 2001; Tachibana et al., 2007; Takada et al., 2011; Visnes et al., 2014). However, in all Setdb1 conditional knockout spermatocytes,H3K9me3 surrounding centromeres became less enriched, starting from the early leptotene stage. The diffused pattern of H3K9me3 distribution at pericentromeric areas continued throughout the pachytene stage. These observations implicate the essential function of SETDB1 in maintaining the trimethylated state of H3K9 at pericentromeric heterochromatin in early meiosis.
Figure 3.
H3K9 trimethylation in Setdb1 mutant spermatocytes.
Spermatocyte spreads from Setdb1 control (+/−) and conditional knockout (CKO) testes at 16 dpp. Spermatocyte nuclei were stained with anti-SCP3 (green), H3K9me3 (red), and centromere protein (CREST, gray) antibodies. The leptotene, zygotene, and pachytene stages of prophase I, respectively, are shown. Dotted line in leptotene cells indicates the clustered centromeres. Dotted line in the zygotene cell indicates the bouquet. Dotted line oval in early-pachytene cells indicates the XY body. The scale bar represents 5 μm.
Strikingly, in all Setdb-deficient leptotene cells, centromere clustering was compromised as compared to controls (Figure 3). This is in line with the previous observation that H3K9me3 at pericentromeric heterochromatin is required for proper centromere clustering at leptotene stage (Takada et al., 2011). The defect in centromere clustering apparently further affected the formation of the bouquet that initiates homologous pairing in early zygotene cells (Figure 3; (Scherthan, 2007). This implies that homologous pairing in the knockout cells may be affected. Moreover, in the Setdb1 knockout pachytene cells that rarely appeared, H3K9me3 was absent in the X and Y chromosomes (Figure 3), as previously reported (Hirota et al., 2018). These results reflect that the abolition of SETDB1-mediated H3K9 trimethylation may have an impact on homologous pairing/meiotic synapsis, due to changes in the overall configuration of centromere clustering and the bouquet formation.
SETDB1 is required for meiotic chromosome synapsis
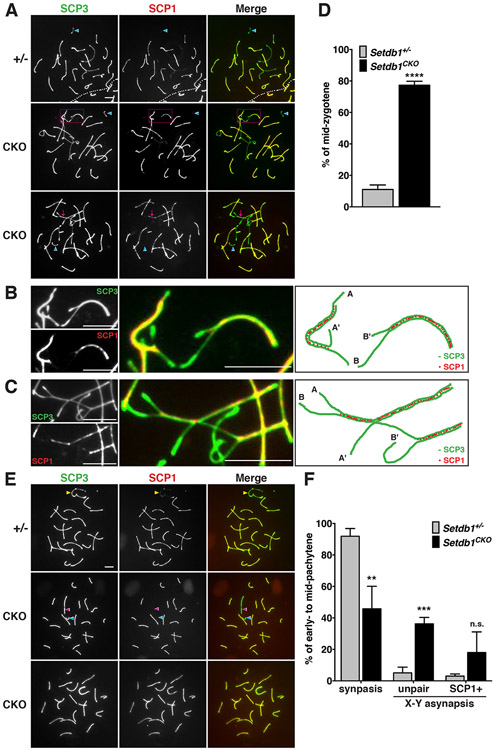
To investigate whether SETDB1-mediated H3K9 trimethylation is critical for homologous chromosome pairing, we examined the configuration of the synaptonemal complex, which stabilizes the initial association of homologous chromosomes. We performed meiotic chromosome spreads with anti-SCP1 and SCP3 antibodies. In normal mid-zygotene cells, all homologous chromosomes were paired and formed synapses except X and Y chromosomes (Figure 4A, top panel). However, abnormal pairing configurations of chromosomes were detected in Setdb1 knockout mid-zygotene cells, including heterologous associations between non-homologous chromosomes (Figure 4A, middle panel, and Figure 4B) and the formation of trivalent (Figure 4A, bottom panel, and Figure 4C). As compared to controls, the frequency of abnormal homologous chromosome pairings was significantly higher in the absence of SETDB1 (Figure 4D), affirming the importance of SETDB1-mediated H3K9 trimethylation in preventing homologous pairing aberration. Consistently, we observed that in control early- to mid-pachytene cells, the autosomal pairing was complete, and the synapse had formed at the pseudoautosomal region of the X and Y chromosome (Figure 4E, top panel). In addition, the majority of control pachytene cells displayed complete chromosome synapsis in controls (91.9% of early- to mid-pachytene cells; Figure 4F). However, in the pachytene-like-cells in Setdb1 knockout that appeared rarely, the percentage of complete chromosome synapsis was significantly reduced (45.8% of early- to mid-pachytene cells). The knockout cells presented fully synapsed autosomes, but the X and Y chromosomes were not paired (Figure 4E, middle panel). The frequency of the unpaired X and Y chromosomes was significantly higher in Setdb1 knockout pachytene cells than in controls (36.2% vs 5.1% of early- to mid-pachytene cells; Figure 4F). In some of the pachytene-like mutant cells, both X and Y chromosomes were abnormally paired and co-stained with both SCP3 and SCP1 (Figure 4E, bottom panel). The frequency of abnormal X-Y synapsis was significantly increased in Setdb1 knockout pachytene cells (from 3% of control to 18% of mutant early- to mid-pachytene cells; Figure 4F). These observations indicate that SETDB1-mediated H3K9 trimethylation is critical for meiotic chromosomal synapsis of both autosomes and the X and Y chromosomes.
Figure 4.
SETDB1 deletion caused defects in homologous pairing.
Spermatocyte spreads from Setdb1 control (+/−) and conditional knockout (CKO) testes at 16 dpp. The nuclei were stained with anti-SCP3 (green) and SCP1 (red) antibodies, indicating the axial/lateral elements and transverse filaments of the synaptonemal complex, respectively. (A) Formation of the synaptonemal complex in mid-zygotene. Y chromosome indicates by the blue arrowhead. Magenta dotted line rectangle in the middle panel indicates the asynapsed chromosomes. Magenta arrow in the bottom panel indicates the unsynapsed trivalent. (B) Higher-magnification of the middle panel in (A). The right painting illustrates asynapsed chromosomes. The A-A’ and B-B’ represent the pairing of non-homologous chromosomes that are different in length. (C) Higher-magnification of the bottom panel in (A). The right painting illustrates the unsynapsed trivalent labeled by partial synapsed A-A’ and A-B chromosomes. (D) Percentages of the abnormal mid-zygotene nuclei with synaptic defects shown in (A). (E) Formation of the synaptonemal complex in early- to mid-pachytene cells. Yellow arrowhead in the top panel indicates X and Y chromosomes synapse in the pseudoautosomal regions. Unpaired X and Y chromosomes in the middle panel indicate by the pink and blue arrowhead, respectively. The bottom panel shows both XY chromosomes or autosomes present the SCP1-positive signals. (F) Percentages of nuclei with synapsed or asynapsed X-Y chromosomes at the early- to mid-pachytene stage demonstrated in (E).
The results represent the mean ± SD of three independent experiments. (**P<0.01; ***P<0.001; ****P<0.0001; n.s., P>0.05) All scale bars represent 5 μm.
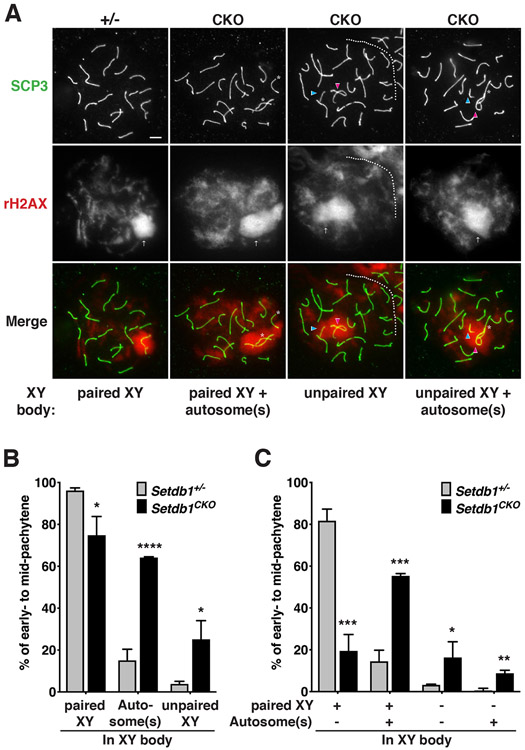
We then examined if autosomal asynapsis in Setdb1-knockout spermatocytes leads to MSUC in the early-pachytene stage. Normally, asynapsed chromosomes or chromosome segments are transcriptionally silenced, and accumulate BRCA1 and ATR on asynapsed axes and the spreading of γH2AX into the associated chromatin loops (Manterola et al., 2009; Turner et al., 2005). It then leads to spermatocyte loss due to the activation of checkpoint and apoptosis during the pachytene stage, and ultimately leads to sterility (de Rooij and de Boer, 2003). To investigate the MSUC response in Setdb1 knockout spermatocytes, we performed meiotic chromosome spreads using anti-SCP3 and γH2AX antibodies to recognize pachytene cells and mark the transcriptionally silenced XY chromatin domain and asynapsed autosomes. In both genotypes, the γH2AX signal was present at the XY body in the early-pachytene cells (Figure 5A), and the majority of X and Y chromosomes were paired (Figure 5B, columns 1 and 2). However, in most control spermatocytes, the XY body (defined by γH2AX signal covering the X and Y chromosomes) does not contain autosomes, but 64% of Setdb1 mutant cells the XY body included autosomes (Figure 5B, columns 3 and 4). The XY body with paired X and Y chromosomes in 55% of Setdb1 mutant cells also contained autosomes (Figure 5C, columns 3 and 4). In addition, the percentage of unpaired and silenced X and Y chromosome in the XY body was significantly increased from only 4% in the control to 25% in the mutant spermatocytes (Figure 5B, columns 5 and 6 and 5C, columns 5-8), The frequency of autosome presenting in the XY body containing unpaired X and Y chromosomes are also significantly increased (Figure 5C, columns 7 and 8). These defects implicate that abolition of SETDB1 leads to autosomal asynapsis and further actives MSUC response in early-pachytene cells. These observations were consistent with an increase in apoptosis at the pachytene stage in the absence of early meiotic SETDB1 (Figure S3C). Altogether, these results demonstrate that SETDB1-mediated H3K9 trimethylation is required for homologous chromosome pairing and meiotic synapsis to prevent spermatocyte loss and male sterility.
Figure 5.
The zygotene wave of γH2AX asynapsis signaling is preserved in SETDB1-deficient pachytene.
(A) Spermatocyte spread from Setdb1 control (+/−) and conditional knockout (CKO) testes at 16 dpp. Spermatocyte nuclei were stained with anti-SCP3 (green) and γH2AX (red) antibodies, indicating the axial/lateral elements of the synaptonemal complex and XY body, respectively. The occurrence of asynaptic X-Y and autosomes in early-to mid-pachytene. Unpaired X and Y chromosomes indicate by the pink and blue arrowhead, respectively. Arrow indicates the XY body. Asterisk indicates the autosome presents in the XY body. The scale bar represents 5 μm. (B-C) Frequency of asynaptic X-Y and autosomes in early-to mid-pachytene detected from A. The results represent the mean ± SD of three independent experiments. (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001)
SETDB1 deficiency perturbs H3K9me3 in a limited number of sites throughout the genome during meiosis
To further investigate the above-observed roles of SETDB1 in the H3K9 methylation of meiotic chromosomes and in their pairing, we conducted chromatin immunoprecipitation followed by sequencing (ChIP-Seq) to map H3K9me3 (Figure 6 and Figure S4) and H3K9me2 (Figure S5) changes in the genome of Setdb1 conditional knockout germ cells that were purified for the “epithelial cell adhesion molecule” protein (EpCAM+) that marks differentiating spermatogonia and early spermatocytes (Kanatsu-Shinohara et al., 2011). Juvenile testes at 10 dpp were used to enrich leptotene spermatocytes (Goetz et al., 1984; Margolin et al., 2014) and to avoid the possibility of analyzing a different cell composition due to apoptosis and germ cell loss from older testes (Figure 1 and 2). We also mapped the changes in a SETDB1-independent mark, H3K27me3, for comparison (Figure S6). All samples were analyzed in biological duplicates. Genomic binning analysis with non-overlapping 1-kb windows showed that all ChIP-Seq replicates had high correlations (Figures S4A, S5A, and S6A).
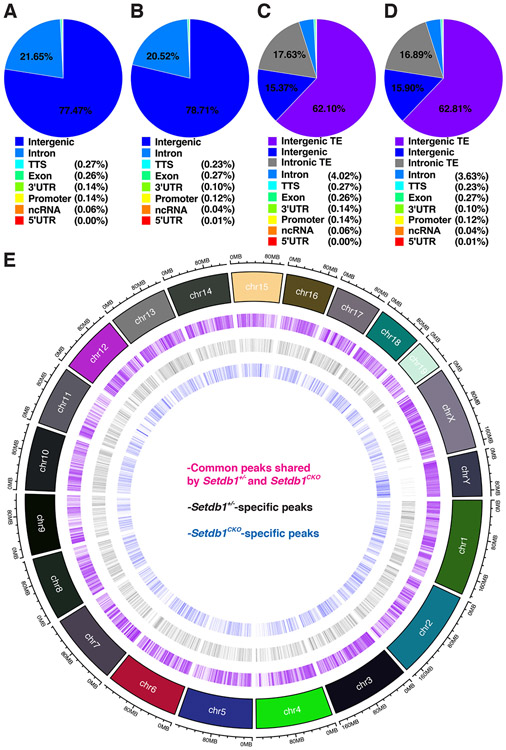
Figure 6.
ChIP-Seq analysis reveals the disruption of global H3K9me3 deposition in Setdb1 knockout. See also Figure S4-7.
(A-D) Distribution of H3K9me3 in control (A and C) and Setdb1 mutant (B and D) peaks in spermatocytes with regard to genes vs intergenic regions (A and B) and transposons (TE) vs genes (C and D).
(E) A genome-wide map of H3K9me3 peaks that are common or unique to the control and Setdb1 mutant spermatocytes.
In control spermatocytes, 33,407 H3K9me3 peaks were identified (Figure S4B). Among them, 77.47% were in the intergenic regions while 22.53% of the peaks were in protein-coding genes, of which 96% of the peaks were in introns and only 4% were in the rest of the genes (Figure 6A). In Setdb1-deficient spermatocytes, 34,429 peaks were identified (Figure S4B), with a distribution between intergenic regions and genes similar to that of the control spermatocytes (Figure 6B). When we further classified peaks into genes, transposons (including those embedded in genes), and non-transposon intergenic regions, 62.1% of the peaks were in transposons and only 15.37% were in transposon-free intergenic regions (Figure 6C). Likewise, most of the intronic H3K9me3 methylation were in transposon sequences in these introns. The mutant cells showed again similar distribution (Figure 6D). Finally, when we compared the control peaks with mutant peaks, only 4,579 peaks disappeared in the mutant and 5,486 new peaks gained in the mutant (Figure 6E). Expectedly, most of these control- and mutant-specific H3K9me3 peaks are in transposons and introns (Figure S7). This indicates that SETDB1 selectively contribute to only a small fraction of H3K9me3 in the spermatocyte epigenome with transposons and protein coding genes as its main targets.
SETDB1 deficiency mildly de-silences transposon expression
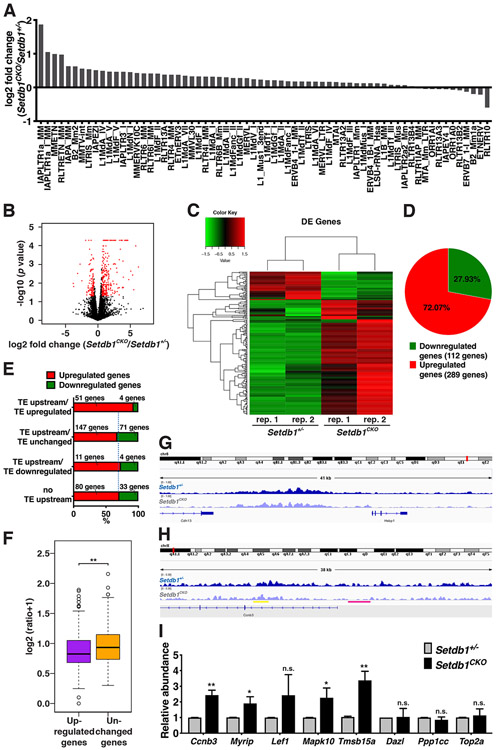
Given the abundant presence of SETDB1 Peaks in transposons and the reported role of SETDB1 in regulating transposons in PGCs (Liu et al., 2014) and pachytene spermatocytes (Hirota et al., 2018), we investigated if the expression of transposon during early meiosis is disturbed in Setdb1 knockout germ cells. For this purpose, we performed RNA-Seq analysis of spermatocytes isolated the same way as for ChIP-Seq analysis. From RNA-Seq analysis, each replicate generated more than 30 million paired-end reads that were mapped to the mouse genome (see Methods Details). Analysis of transposon expression in Setdb1 conditional knockout germ cells showed that only 55 transposons were mildly upregulated within a few fold and 11 transposons were slightly downregulated also within a few fold as compared to control spermatocytes (Figure 7A and Table S1), consistent with the mild effect of SETDB1 in regulating ERVs in pachytene spermatocyte (Hirota et al., 2018). This implies that SETDB1-mediated H3K9me3 only has a slight effect in the expression of TEs at the early stage of meiotic prophase I.
Figure 7.
RNA-Seq analysis reveals the derepression of genes in response to Setdb1 knockout.
(A) Analysis of transposon expression change in Setdb1 knockout condition in RNA-Seq data. See also Table S1 for the details of transposons. (B) Volcano plot of differential expressed genes with log2 fold change (x-axis) against - log10 (P-value) (y-axis) in Setdb1 knockout and control cells. DEGs (fold change ≥ 1.5, P ≤ 0.05) are in red. (C) A heatmap representing clusters of differential expressed genes in Setdb1 knockout and control cells. See also Table S2-3 for details of DEGs. (D) A pie chart indicating the percentage of up-and downregulated genes identified by the gene expression analysis. (E) Analysis of DEGs containing TEs within 2 kb upstream of the transcription start site and oriented in the same direction (TE upstream). DEGs are divided into four categories; DEGs that contain upregulated TEs in the upstream (TE upstream/TE upregulated), DEGs that contain unchanged TEs in the upstream (TE upstream/TE unchanged), DEGs that contain downregulated TEs in the upstream (TE upstream/TE downregulated), and DEGs that do not contain TEs upstream (no TE upstream). The percentages of genes that are upregulated or downregulated in each category are shown in red or green, respectively. (F) H3K9me3 ChIP signal within 1.5 kb upstream of upregulated DEGs and unchanged genes are shown. (G-H) Genome browser views showing H3K9me3 coverage of the (G) Cdh13 and (H) Ccnb3 gene loci in Setdb1 control (+/−) and conditional knockout (CKO) germ cells. Yellow and pink lines indicate the H3K9me3 unchanges and changes in Ccnb3 gene loci, respectively. (I) qRT-PCR validation for RNA-Seq. Equal amounts of total RNA isolated from Setdb1 control (+/−) and conditional knockout (CKO) germ cells are subjected to qRT-PCR with primers specific to Ccnb3, Myrip, Lef1, Mapk10, Tmsb15a, Dazl, Ppp1cc, Top2a, and Gapdh. Data presented were normalized to Gapdh. The results represent the mean ± SD of three independent experiments. (*P<0.05; **P<0.01; n.s., P>0.05)
SETDB1 directly target many genes to regulate their expression in early meiosis
We then examined the gene expression in Setdb1 knockout spermatocytes. With a cutoff of 1.5-fold change, we identified 401 differentially expressed genes (DEGs) containing mRNAs, lncRNAs, and pseudogenes (Figure 7B). Among them, 289 genes were upregulated and 112 genes were downregulated in the conditional knockout as compared to controls (Figure 7C and D; Tables S2 and S3), consistent with the function of SETDB1 in H3K9me3-mediated gene silencing.
To investigate if aberrantly expressed transposons affect the expression of nearby genes, we classified all DEGs based on the absence (Figure 7E, column 4) or presence of an annotated transposons within 2 kb upstream of the transcription start site (TSS) that oriented in the same directions. Among 55 DEGs with an upstream transposon upregulated in the mutant, 51 (93%) genes were also upregulated (Figure 7E). Among 218 DEGs with an upstream transposon unchanged in the mutant, 147 (67%) genes were upregulated. Among 15 DEGs with an upstream transposon downregulated in the mutant, 11 (73%) genes were upregulated. Among remaining 113 DEGs without a transposon with 2 kb upstream, 80 (73%) genes were upregulated. This indicates that upregulation of transposons influences certain downstream genes in a SETDB1-depedent manner during early stages of meiotic prophase.
We next sought to examine if SETDB1 suppressed gene expression through the trimethylation of H3K9. We found that in Setdb1 conditional knockout spermatocytes, a significantly weaker signal of H3K9me3 in the promoter regions (±1.5 kb at TSS) of upregulated genes was observed as compared to the promoter regions of unchanged genes (Figure 7F), indicating that SETDB1 represses a subset of gene expression through establishing H3K9 trimethylation at their promoter regions.
To investigate if SETDB1 may affect genes involved in meiotic progression, we further turned our attention to the 289 upregulated genes. Interestingly, we identified two upregulated genes, Ccnb3 and Myrip, which had been shown to cause meiotic defects while overexpressed (Eggers et al., 2015; Nguyen et al., 2002; Refik-Rogers et al., 2006). Ccnb3 encodes CYCLIN B3, a pre-pachytene cyclin which is expressed only during leptotene and zygotene phases. Forced-expression of CYCLIN B3 after the zygotene stage led to defective spermatogenesis and apoptosis in transgenic mice (Nguyen et al., 2002; Refik-Rogers et al., 2006). MYRIP encodes Myosin VIIA And Rab Interacting Protein. A duplication of MYRIP has been detected in male patients with meiotic arrest (Eggers et al., 2015). To verify the expression of Ccnb3 and Myrip mRNAs, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed to measure the relative abundance of their transcripts in Setdb1 knockout and control germ cells. The RNA levels of both genes had a significant two-to-three fold increase in Setdb1 knockout germ cells as compared to controls (Figure 7I), implying that the abolition of SETDB1 may lead to the gain-of-function effects of CCNB3 and MYRIP during meiotic prophase. There is no transposon within the 2 kb upstream of their TSS, inferring SETDB1 represses their gene expression not through a concomitant influence on transposons. However, the promoter region of the Ccnb3 gene is under-methylated at H3K9 in the mutant cells (Figure 7H), suggesting that SETDB1-mediated methylation directly regulates its expression. This correlation also holds true of most other genes whose expression is upregulated in the mutant cells. In contrast, for genes that are not upregulated in the mutant, their H3K9me3 level are not changed, as illustrated by the Cdh13 gene (Figure 7G).
Gene ontology (GO) analysis further revealed that somatic related processes were enriched among the top GO terms associated with the upregulated genes (Tables S4 and S5). To confirm these somatic gene expressions, we chose the 3 upregulated DEGs–Lef1, Mapk10, and Tmsb15a, which are without transposons within the 2 kb upstream of their TSS, and verify their RNA levels in Setdb1 knockout and control germ cells by qRT-PCR. Three genes, Dazl, Ppp1cc, and Top2a, whose expression was not changed in Setdb1 knockout spermatocytes served as controls. We observed a consistent increase in the RNA levels of Lef1, Mapk10, and Tmsb15a but not the three control genes, in the Setdb1 knockout spermatocytes (Figure 7I). This indicates that SETDB1 functions in repressing somatic genes in early meiotic cells. These results reveal that SETDB1 not only plays a critical role in centromere clustering and homologous pairing, but also in preventing ectopic expression of somatic and meiotic genes in early meiosis.
DISCUSSION
The structural integrity of bivalents is a prerequisite for proper chromosome segregation in meiosis I. During the formation of bivalents, histone modification constitutes a primary regulatory mechanism contributing to the dynamics of chromatin structure and timely expression of lineage-specific genes. In this study, our findings reveal a potential role of SETDB1 in (peri-)centromeric heterochromatin-mediated centromere clustering for proper progression of homologous chromosome synapsis. Furthermore, we showed the function of SETDB1 in repressing transposons and specific meiotic and somatic genes to ensure the proper expression of genes for the progression of early meiotic prophase I.
One of the critical findings of this study is the function of SETDB1-mediated H3K9 trimethylation in establishing centromere clustering at the leptotene stage and in bouquet formation at the zygotene stage. These early functions of SETDB1 in leptotene and zygotene stages explain the origin of apoptosis at the pachytene stage as reported recently by Hirota et al. (2018). We are able to discover the earlier function of SETDB1 during meiosis possibly because we used the Vasa-Cre driver that is expressed from E15-18, much earlier than Ngn3-Cre used by Hirtato et al. that starts expression at 7 dpp. Thus it might have depleted the SETDB1 protein more by the onset of meiosis.
Although previous studies have revealed SUV39H1/2 as an enzyme generates H3K9me3 at pericentromeric regions in early meiotic prophase I (Peters et al., 2001; Takada et al., 2011), our results demonstrate the essential role of SETDB1 in H3K9me3 distribution at the same stage (Figure 3), firmly indicating that both SETDB1 and SUV39H1/2 are required for establishing pericentromeric heterochromatin and for HP1γ/G9a-related centromere clustering. This is consistent with a report by Fritsch et al. that several histone methyltransferases, including G9A, SETDB1, SUV39H1, and GLP, coexist in the same complex and interdependently stabilize each other in HeLa cells and mouse embryonic fibroblasts (Fritsch et al., 2010). Specifically, these four histone methyltransferases functionally cooperate and not only to establish heterochromatin but also regulate euchromatic genes. Given the known link of H3K9me3 to HP1γ/G9a-mediated centromere clustering, our studies thus extend the potential roles for SETDB1-SUV39H1/2 functional cooperation in H3K9 trimethylation during early meiosis.
Although we have identified a regulatory function of SETDB1-mediated H3K9 trimethylation in pairing and synapsis of homologous chromosomes, its role in homologous recombination is still unresolved. Several studies have reported that a spatial organization of active histone markers such as H3K4me3, H2Bub, or H4K5ac, are displayed at the meiotic recombination hotspots (Hernandez-Hernandez et al., 2009; Ivanovska and Orr-Weaver, 2006). Open chromatin then allows the PRDM9 zinc-finger protein to bind its cognate binding sequences followed by endonuclease SPO1-mediated DSB cleavage at the leptotene stage (Getun et al., 2017; Getun et al., 2012; Parvanov et al., 2010). Given the important role of SETDB1 in repressive histone H3K9me3 mark deposition, it is conceivable that loss of SETDB1 leads to an open chromatin conformation that is accessible by SPO11, which may then mis-regulate the hotspot distribution and create unexpected DSBs. Therefore, although we have successfully demonstrated the role of SETDB1 in centromere clustering and homologous pairing, it is likely that the absence of SETDB1-mediated H3K9 trimethylation also affects homologous recombination in early meiosis.
Our current RNA-Seq datasets reveal only slight desilencing of transposons and relatively small changes in gene expression in Setdb1 knockout germ cells, despite that transposons are major targets of SETDB1. This is different from previous studies that report the function of SETDB1 in regulating a significant transcriptome/euchromatic genes in other cell types (Bilodeau et al., 2009; Koide et al., 2016; Lohmann et al., 2010; Yuan et al., 2009). This difference may be due to the nature of a low complexity of gene expression in leptotene and zygotene cells, in accord with a transcriptionally quiescent state during early meiosis (Ernst et al., 2019; Kierszenbaum and Tres, 1974; Monesi, 1964; Morgan et al., 2019; Page et al., 2012). Alternatively, other epigenetic factors (Peters et al., 2001; Takada et al., 2011), in addition to SETDB1, might mediate transcriptome silencing at this stage. Despite the difference, we found SETDB1 suppresses the expression of somatic genes, such as hematopoietic and neuronal lineage genes, in spermatocytes. This is consistent with the findings that SETDB1 controls cellular identity of embryonic stem cells and hematopoietic cells (Bilodeau et al., 2009; Koide et al., 2016; Lohmann et al., 2010; Yuan et al., 2009), and, in our case, of germ cells in the testis.
There is a positive correlation between upregulated DEGs containing upstream transposon sequences that are also upregulated, indicating that SETDB1 may sometimes control gene expression through repressing their upstream transposons. This is also consistent with the findings in PGCs and ESCs, where aberrant ERV transcription in the SETDB1 knockout leads to expression of chimeric genic transcripts (Liu et al., 2014; Karimi et al., 2011).
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Haifan Lin (Haifan.lin@yale.edu), Yale Stem Cell Center and Department of Cell Biology, Yale University School of Medicine, New Haven, CT 06520 USA.
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
ChIP-seq and RNA-seq data are available at the NCBI SRA under the accession number PRJNA674950. This study did not generate any unique code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Setdb1flox/flox mice were generated as reported (Lohmann et al., 2010). Vasa (or Ddx4)-Cre mice (Tg(Ddx4-Cre)1Dcas) were generated by Dr. Diego H. Castrillon and obtained from The Jackson Laboratory (Bar Harbor, ME). To achieve efficient inactivation of Setdb1 in early spermatocytes, we generated Setdb1+/−;Vasa-Cre/Vasa-Cre males that were mated to Setdb1flox/flox females to produce Setdb1 conditional knockout (Setdb1flox/−;Vasa-Cre+/−) animals and control littermates (Setdb1flox/+;Vasa-Cre+/−). The conditional knockout animals and control littermates were on a mixed C57BL/6 and FVB background. In this study, only male animals were used due to our focus on testis development. All animal procedures were approved by the Animal Care and Use Committee of Yale University.
METHOD DETAILS
Western blot analysis of testicular germ cells
For western blot analysis, testes of different ages were used to enrich specific germ cell types. Somatic cells and spermatogonia (2C), and leptotene (L) cells were isolated from 10 dpp testes. Leptotene/zygotene (L/Z) cells, pachytene/diplotene (P/D) cells, and round spermatid (RS) cells were isolated from 20 dpp testes. Decapsulated testes at various ages were treated with 0.5 mg/ml collagenase type IV (Thermo Fisher, Waltham, MA) and 100 μg/ml DNase I (MilliporeSigma, Burlington, MA) in Accutase solution (MilliporeSigma, Burlington, MA) at 35°C for 10 min. Cells were then triturated and pass through a 16-gauge needle to remove large cell aggregates. After the addition of 10% volume of fetal bovine serum (FBS), cell suspensions were filtered through a 70-μm cell strainer (Thermo Fisher, Waltham, MA) and washed once with DMEM containing 10% FBS (Thermo Fisher, Waltham, MA). Single-cell suspensions were then incubated with 0.5% FBS, 5 μg/ml Hoechst 33342, 1 μg/ml PI in DMEM at 35°C for 20 min. After washing, the cells were FACS-sorted based on DNA content and the efficiency of Hoechst 33342 staining, which varies with cell type (Getun et al., 2011). To examine SETDB1 levels in Setdb1 conditional knockout and control testes, germ cells were purified using the anti-EpCAM antibody (Thermo Fisher, clone G8.8, Waltham, MA) with Dynabeads Biotin Binder (Thermo Fisher, Waltham, MA) and DynaMag-2 Magnet (Thermo Fisher, Waltham, MA). The purity of cells was further validated with FACS, and only cells with purity above 90% were used for experiments. For western blot analysis, anti-SETDB1 antibody (Cell Applications Inc., CP10377; dilution: 1:1,000, San Diego, CA) and anti-GAPDH antibody (Cell Signaling Technology, #5174; dilution: 1:1,000, Danvers, MA) were used.
Spermatocyte Spreads
Spermatocyte spreads were prepared as follows: Briefly, the enriched germ cells isolated from adult mouse testes were suspended in 0.1 M sucrose supplied with protease inhibitor. The cells were applied to the micro slides with a fixation solution (0.1% Triton x-100 in 1% PFA). The slides were in a humid chamber for 3 hrs prior to air-drying. After blocking with 10% antibody dilution buffer stock (ADB) (3% BSA, 10% normal goat serum, 0.05% Triton X-100, 1xPBS), the slides were incubated with the mouse monoclonal SCP-3 antibody (Santa Cruz Biotechnology, Tx, sc-74569), SCP-1 antibody (Abcam, Cambridge, MA, ab15090), γH2AX antibody (MilliporeSigma, Burlington, MA, #05-636), SETDB1 antibody (Abcam, Cambridge, MA, ab107225), centromere antibody (Antibodies Incorporated, 15-235-0001), and/or H3K9me3 antibody (Abcam, Cambridge, MA, Ab8898) overnight at 4°C followed by the secondary antibody for 1 hr at RT. Cells were counterstained with DAPI and washed with PBS. The slides were mounted using Vectashield mounting medium.
Apoptosis analysis
Analysis of cell death was performed on testicular tissue sections using ApopTag Fluorescein In Situ kit (Chemicon International, Temecula, CA) according to manufacturer’s directions. Sections were counterstained with DAPI to visualize nuclei.
Immunohistochemistry
For histological sections, testes were fixed in 4% paraformaldehyde (PFA) overnight and embedded in wax before cutting 5-μm sections by the Research Histology Facility at Yale School of Medicine. Immunofluorescence was performed using 1:100 mouse anti-SETDB1 antibody (Abcam, Cambridge, MA, ab107225), 1:500 mouse anti-γH2AX (MilliporeSigma, Burlington, MA, #05-636), 1:500 rabbit anti-SOX9 (MilliporeSigma, Burlington, MA, AB5535), and/or 1:500 rat TRA98 (Abcam, Cambridge, MA, Ab82527) primary antibodies. Fluorescent staining was imaged by Confocal Leica TCS SP5. To evaluate seminiferous tubule containing pachytene cells, testicular tubules with the presence of TRA98-positive germ cells containing γH2AX-foci were scored on the random field of testicular tubule cross-sections. Tubules with γH2AX- and SOX9- double-positive cells representing apoptotic somatic cells were excluded from scoring.
Isolation of testicular germ cells
To prepare germ cells for RNA-Seq and ChIP-Seq, 10-dpp decapsulated testes were treated with 0.5 mg/ml collagenase type IV (Thermo Fisher, Waltham, MA) and 100 λg/ml DNase I (MilliporeSigma, Burlington, MA) in Accutase solution (MilliporeSigma, Burlington, MA) at 35°C for 10 min. Cells were then triturated and pass through a 16-gauge needle to remove large cell aggregates. After the addition of 10% volume of fetal bovine serum (FBS), cell suspensions were filtered through a 70-μm cell strainer (Thermo Fisher, Waltham, MA) and washed once with DMEM containing 10% FBS (Thermo Fisher, Waltham, MA). On average, each mouse yielded 1.5 million cells. Cells were then incubated with a 1:50 biotin-conjugated anti-EpCAM antibody (Thermo Fisher, Waltham, MA, clone G8.8) at 100 μl for 45 min at 4°C. Cells were then washed with DMEM containing 10% FBS. Cells were then resuspended in 400ul DMEM containing 10% FBS, and 20 μl of prewashed Dynabeads Biotin Binder (Thermo Fisher, Waltham, MA) was added. Cells were incubated with beads for 20 min at 4°C with rotation. EpCAM positive cells were then separated from the rest of the sample on a DynaMag-2 Magnet (Thermo Fisher, Waltham, MA) and washed with DMEM containing 10% FBS for three times. The purity of cells was further validated with FACS, and only cells with purity above 90% were used for experiments.
RNA-Seq
RNA-Seq libraries were constructed using TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA). Libraries were sequenced for 100 cycle paired-end using HiSeq 2000 (Illumina, San Diego, CA). All the raw paired-end sequences were mapped to mouse genome mm10 and annotated transcripts GRCm38.81 by using tophat2 (v2.1.0) with a default setting. Next, Cuffdiff was executed to analyze changes in the RNA expression level and its significance for each gene based on two replicates of each condition with the upper-quartile-norm option. Differentially expressed genes (DEGs) were selected using the following criteria: 1) at least 1.5-fold change; and 2) p-value less than 0.05. Heatmap plots of DEGs were produced by the R package. GO and KEGG analyses were applied by the DAVID package (https://david.ncifcrf.gov). The annotation of DEGs was based on Ensembl annotation GRCm38.81.
For transposon expression change in the knockout condition, mRNA sequences were mapped to Repbase transposons with default option by bowtie2-2.8.2. To study the transposon insertion in upstream of DEGs, upstream 2 kb windows were scanned for any presence of transposons. All transposons located within a 2 kb window of identified DEGs and oriented in the same transcription direction were examined for their expression level change in control and in the knockout condition by Rsubread with the default option. Read counts were normalized to FPKM based on all mapped reads. For the transposon expression level calculation, multiple mapped reads were excluded.
ChIP-Seq
We performed ChIP based on a published protocol for ultra-low-input micrococcal nuclease-based native ChIP (ULI-NChIP) (Brind'Amour et al., 2015) with modifications. For chromatin preparation, 150,000 EpCAM+ germ cells were resuspended in Nuclei EZ Lysis Buffer (MilliporeSigma, Burlington, MA) containing cOmplete Protease Inhibitor Cocktail (MilliporeSigma, Burlington, MA) and 0.1mM Phenylmethylsulfonyl fluoride (MilliporeSigma, Burlington, MA). Chromatin was fragmented for 5 min using 50 U/ul MNase (NEB, Ipswich, MA) at 35 °C, and then the reaction was quenched by 0.1M EGTA. A final of 0.1% Triton and DOC was added to the reaction and rest on ice for 15 min. For immunoprecipitation, the digested chromatin was diluted at least four times in ChIP buffer (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 1 × EDTA-free complete Protease Inhibitor Cocktail and 0.1 mM phenylmethanesulfonyl fluoride). Chromatin was pre-cleared with Dynabeads (Thermo Fisher, Waltham, MA) for 1 hr at 4 °C and immunoprecipitated with 2 μg of H3K9me3 (Abcam, Ab8898, Cambridge, MA), H3K9me2 (Abcam, Ab1220, Cambridge, MA), or H3K27me3 (Diagenode, C15410069, Denville, NJ) antibody and 12.5 μl anti-mouse/anti-rabbit Dynabead complexes overnight at 4 °C. IPed complexes were washed three times with 1 ml of ChIP buffer. Protein–DNA complexes were eluted in 30 μl of ChIP elution buffer (100 mM NaHCO3 and 1% SDS) for 30min at 65 °C. IPed material was then diluted with water to a final of 150 μl, and digested with 1ul of RNAse A (Thermo Fisher, Waltham, MA) containing 200 mM NaCl for 30min at 37 °C. DNA was dissociated from the protein complex with proteinase K (MilliporeSigma, Burlington, MA) for 30min at 65 °C, and purified with The MinElute PCR Purification Kit (Qiagen, Germantown, MD).
ChIP-Seq libraries were constructed using Rubicon ThruPLEX-FD Prep kit (Takara Bio USA, Mountain View, CA). Libraries were sequenced using HiSeq 2000 (Illumina, San Diego, CA) for 50 cycle single-read. Sequences were mapped to the mouse mm10 genome using bowtie2-2.8.2 with the default option. The reproducibility was represented by the Spearman correlation between replicates based on non-overlapping 1 kb window. Since the correlation values were all >0.8, we pulled all biological replicates together for downstream analysis. Rseg was used to call peaks for each condition against the input. ChIP-Seq peaks were compared and annotated by HOMER v4.11. The ChIP signal change within 1.5 kb upstream of DEGs was examined by Rsubread. The signal was normalized to sequencing depth.
Real-time PCR
Total RNA from cells purified through FACS was isolated using the High Pure RNA Isolation Kit according to the manufacturer’s instructions. cDNA was synthesized by Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) with random hexamer. Primers for real-time PCR assays are listed in Table S6. Quantitative determination of the cDNA levels was done by real-time PCR using the Bio-Rad iTaq Universal SYBR Green Supermix system. All results represent the mean ± SD of at least three independent experiments.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was calculated by unpaired two-tailed t test using GraphPad Prism 8. For all spermatocyte spread experiments, we counted cells from at least three independent experiments. Biological duplicates were pooled from at least seven mice per replicate for RNA-seq and ChIP-seq experiments. Details are described in the Figure Legends and Method Details.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human anti-Centromere | Antibodies Incorporated | Cat# 15-234-0001, RRID:AB_2687472 |
| Rat anti-EpCAM [G8.8], Biotin | Thermo Fisher Scientific | Cat# 13-5791-82, RRID:AB_1659713 |
| Rat anti-GCNA1 [TRA98] | Abcam | Cat# ab82527, RRID:AB_1659152 |
| Rabbit anti-GAPDH [D16H11] | Cell Signaling Technology | Cat# 5174, RRID:AB_10622025 |
| Mouse anti-Histone H3K9me2 | Abcam | Cat# ab1220, RRID:AB_449854 |
| Rabbit anti-Histone H3K9me3 | Abcam | Cat# ab8898, RRID:AB_306848 |
| Rabbit anti-H3K27me3 | Diagenode | Cat# C15410069, RRID:AB_2814977 |
| Mouse anti-phospho-Histone H2A.X (Ser139) [JBW301] | MilliporeSigma | Cat# 05-636, RRID:AB_309864 |
| Mouse anti-SETDB1 antibody [5H6A12] | Abcam | Cat# ab107225, RRID:AB_10861045 |
| Mouse anti-SETDB1 | Cell Applications | Cat# CP10377 |
| Rabbit anti-SCP1 | Abcam | Cat# ab15090, RRID:AB_301636 |
| Mouse anti-SCP-3 [D-1] | Santa Cruz Biotechnology | Cat# sc-74569, RRID:AB_2197353 |
| Rabbit anti-SOX9 | MilliporeSigma | Cat# AB5535, RRID:AB_2239761 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Accutase cell detachment solution | MilliporeSigma | Cat# SCR005 |
| Ambion RNase A | Thermo Fisher Scientific | Cat# AM2270 |
| Collagenase, Type IV, powder | Thermo Fisher Scientific | Cat# 17104019 |
| cOmplete Protease Inhibitor Cocktail | MilliporeSigma | Cat# CO-RO |
| DAPI-(4,6-Diamido-2-Phenylindole, dihydrochloride) | Thermo Fisher Scientific | Cat# D1306; RRID:AB_2629482 |
| Deoxyribonuclease I from bovine pancreas | MilliporeSigma | Cat# DN25 |
| Dynabeads Biotin Binder | Thermo Fisher Scientific | Cat# 11047 |
| Eosin | MilliporeSigma | Cat# HT1101128 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10428026 |
| Hematoxylin | MilliporeSigma | Cat# H3136 |
| Phosphate Buffer Saline | MilliporeSigma | Cat# P5368 |
| Phenylmethanesulfonyl fluoride (PMSF) | AmericanBio | Cat# AB01620 |
| Proteinase K, recombinant, PCR Grade | MilliporeSigma | Cat# RPROTKSOL-RO |
| SuperScript II Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18064014 |
| Triton x-100 | MilliporeSigma | Cat# X100 |
| Tween-20 | MilliporeSigma | Cat# 113322465001 |
| Critical Commercial Assays | ||
| ApopTag Fluorescein In Situ Apoptosis Detection Kit | MilliporeSigma | Cat# S7110 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725120 |
| Micrococcal Nuclease | NEB | Cat# M0247S |
| MinElute PCR Purification Kit | Qiagen | Cat# 28004 |
| Nuclei ez lysis buffer | MilliporeSigma | Cat# N3408 |
| ThruPLEX -FD Prep Kit | Rubicon Genomics | Cat# R40012 |
| TruSeq Stranded mRNA Library Prep | Illumina | Cat# 20020594 |
| Deposited Data | ||
| RNA Sequencing Data | this manuscript | PRJNA674950 |
| ChIP Sequencing Data | this manuscript | PRJNA674950 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Setdb1flox/flox | Taiping Chen (Lohmann et al., 2010) | N/A |
| Mouse: FVB-Tg(Ddx4-cre)1Dcas/J | The Jackson Laboratory | Cat# JAX:006954, RRID:IMSR_JAX:006954 |
| Oligonucleotides | ||
| Primers for quantitative real-time PCR (qPCR), see Table S6. | this manuscript | N/A |
| Software and Algorithms | ||
| Adobe Illustrator CC | Adobe | RRID: SCR_010279 |
| Adobe Photoshop CC | Adobe | RRID:SCR_014199 |
| Bowtie | Bowtie | RRID:SCR_005476 |
| Cuffdiff | Cuffdiff | RRID:SCR_001647 |
| DAVID | DAVID | RRID:SCR_001881 |
| HOMER | HOMER | RRID:SCR_010881 |
| ImageJ | National Institutes of Health | RRID: SCR_003070 |
| Prism | Graph Pad | RRID:SCR_002798 |
| RSEG | RSEG | RRID:SCR_007695 |
| Rsubread | Rsubread | RRID:SCR_016945 |
| TopHat | TopHat | RRID:SCR_013035 |
ACKNOWLEDGMENTS
The sequencing services were conducted at Yale Stem Cell Center Genomics Core supported by the Connecticut Regenerative Medicine Research Fund and the Li Ka Shing Foundation. We are grateful to members of the Lin lab members for discussions. This work was supported by NIH R37HD42012 to H.L. and NIH R01DK106418 to T.C.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alleva B and Smolikove S (2017). Moving and stopping: Regulation of chromosome movement to promote meiotic chromosome pairing and synapsis. Nucleus 8, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB and Young RA (2009). SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes & development 23, 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E and Handel MA (2018). Meiosis: the chromosomal foundation of reproduction. Biol Reprod 99, 112–126. [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E and Schimenti JC (2012). Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol 298, 179–227. [DOI] [PubMed] [Google Scholar]
- Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM and Lorincz MC (2015). An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 6, 6033. [DOI] [PubMed] [Google Scholar]
- Cahoon CK and Hawley RS (2016). Regulating the construction and demolition of the synaptonemal complex. Nat Struct Mol Biol 23, 369–377. [DOI] [PubMed] [Google Scholar]
- da Cruz I, Rodriguez-Casuriaga R, Santinaque FF, Farias J, Curti G, Capoano CA, Folle GA, Benavente R, Sotelo-Silveira JR and Geisinger A (2016). Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics 17, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG and de Boer P (2003). Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet Genome Res 103, 267–276. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B and Moens PB (1994). Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. Journal of cell science 107 ( Pt 10), 2749–2760. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM and Collins CM (2017). Centromere Dynamics in Male and Female Germ Cells. Prog Mol Subcell Biol 56, 357–375. [DOI] [PubMed] [Google Scholar]
- Eggers S, DeBoer KD, van den Bergen J, Gordon L, White SJ, Jamsai D, McLachlan RI, Sinclair AH and O'Bryan MK (2015). Copy number variation associated with meiotic arrest in idiopathic male infertility. Fertil Steril 103, 214–219. [DOI] [PubMed] [Google Scholar]
- Ernst C, Eling N, Martinez-Jimenez CP, Marioni JC and Odom DT (2019). Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat Commun 10, 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymery A, Liu Z, Ozonov EA, Stadler MB and Peters AH (2016). The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development 143, 2767–2779. [DOI] [PubMed] [Google Scholar]
- Fraune J, Brochier-Armanet C, Alsheimer M, Volff JN, Schucker K and Benavente R (2016). Evolutionary history of the mammalian synaptonemal complex. Chromosoma 125, 355–360. [DOI] [PubMed] [Google Scholar]
- Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M and Ait-Si-Ali S (2010). A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37, 46–56. [DOI] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB and Castrillon DH (2007). Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J and Colaiacovo MP (2018). Zipping and Unzipping: Protein Modifications Regulating Synaptonemal Complex Dynamics. Trends Genet 34, 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Bucciarelli E, Lattao R, Pellacani C, Mottier-Pavie V, Giansanti MG, Somma MP and Bonaccorsi S (2012). The relative roles of centrosomal and kinetochore-driven microtubules in Drosophila spindle formation. Exp Cell Res 318, 1375–1380. [DOI] [PubMed] [Google Scholar]
- Geisinger A and Benavente R (2016). Mutations in Genes Coding for Synaptonemal Complex Proteins and Their Impact on Human Fertility. Cytogenet Genome Res 150, 77–85. [DOI] [PubMed] [Google Scholar]
- Getun IV, Torres B and Bois PR (2011). Flow cytometry purification of mouse meiotic cells. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getun IV, Wu Z, Fallahi M, Ouizem S, Liu Q, Li W, Costi R, Roush WR, Cleveland JL and Bois PRJ (2017). Functional Roles of Acetylated Histone Marks at Mouse Meiotic Recombination Hot Spots. Mol Cell Biol 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getun IV, Wu ZK and Bois PR (2012). Organization and roles of nucleosomes at mouse meiotic recombination hotspots. Nucleus 3, 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz P, Chandley AC and Speed RM (1984). Morphological and temporal sequence of meiotic prophase development at puberty in the male mouse. Journal of cell science 65, 249–263. [DOI] [PubMed] [Google Scholar]
- Handel MA, Cobb J and Eaker S (1999). What are the spermatocyte's requirements for successful meiotic division? J Exp Zool 285, 243–250. [PubMed] [Google Scholar]
- Herbert M, Kalleas D, Cooney D, Lamb M and Lister L (2015). Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb Perspect Biol 7, a017970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hernandez A, Vazquez-Nin GH, Echeverria OM and Recillas-Targa F (2009). Chromatin structure contribution to the synaptonemal complex formation. Cell Mol Life Sci 66, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Blakeley P, Sangrithi MN, Mahadevaiah SK, Encheva V, Snijders AP, ElInati E, Ojarikre OA, de Rooij DG, Niakan KK, et al. (2018). SETDB1 Links the Meiotic DNA Damage Response to Sex Chromosome Silencing in Mice. Dev Cell 47, 645–659 e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A and Amon A (2006). Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol 16, R217–228. [DOI] [PubMed] [Google Scholar]
- Hunter N (2015). Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I and Orr-Weaver TL (2006). Histone modifications and the chromatin scaffold for meiotic chromosome architecture. Cell Cycle 5, 2064–2071. [DOI] [PubMed] [Google Scholar]
- Jones KT and Lane SI (2012). Chromosomal, metabolic, environmental, and hormonal origins of aneuploidy in mammalian oocytes. Exp Cell Res 318, 1394–1399. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Takashima S, Ishii K and Shinohara T (2011). Dynamic changes in EPCAM expression during spermatogonial stem cell differentiation in the mouse testis. PLoS One 6, e23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL and Tres LL (1974). Nucleolar and perichromosomal RNA synthesis during meiotic prophase in the mouse testis. J Cell Biol 60, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Zhao H, Dan J, Kim S, Hardikar S, Hollowell D, Lin K, Lu Y, Takata Y, Shen J, et al. (2016). Maternal Setdb1 Is Required for Meiotic Progression and Preimplantation Development in Mouse. PLoS Genet 12, e1005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide S, Oshima M, Takubo K, Yamazaki S, Nitta E, Saraya A, Aoyama K, Kato Y, Miyagi S, Nakajima-Takagi Y, et al. (2016). Setdb1 maintains hematopoietic stem and progenitor cells by restricting the ectopic activation of nonhematopoietic genes. Blood 128, 638–649. [DOI] [PubMed] [Google Scholar]
- Kurdzo EL and Dawson DS (2015). Centromere pairing--tethering partner chromosomes in meiosis I. FEBS J 282, 2458–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S and Kauppi L (2019). Meiotic spindle assembly checkpoint and aneuploidy in males versus females. Cell Mol Life Sci 76, 1135–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brind'Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y and Lorincz MC (2014). Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes & development 28, 2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann F, Loureiro J, Su H, Fang Q, Lei H, Lewis T, Yang Y, Labow M, Li E, Chen T, et al. (2010). KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cells 28, 201–212. [DOI] [PubMed] [Google Scholar]
- MacLennan M, Crichton JH, Playfoot CJ and Adams IR (2015). Oocyte development, meiosis and aneuploidy. Semin Cell Dev Biol 45, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM and Burgoyne PS (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27, 271–276. [DOI] [PubMed] [Google Scholar]
- Manterola M, Page J, Vasco C, Berrios S, Parra MT, Viera A, Rufas JS, Zuccotti M, Garagna S and Fernandez-Donoso R (2009). A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple simple robertsonian translocations. PLoS Genet 5, e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin G, Khil PP, Kim J, Bellani MA and Camerini-Otero RD (2014). Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 15, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC and Shinkai Y Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931. [DOI] [PubMed] [Google Scholar]
- Monesi V (1964). Ribonucleic Acid Synthesis during Mitosis and Meiosis in the Mouse Testis. J Cell Biol 22, 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DP and Orr-Weaver TL (1998). Chromosome segregation during meiosis: building an unambivalent bivalent. Curr Top Dev Biol 37, 263–299. [DOI] [PubMed] [Google Scholar]
- Morgan M, Kabayama Y, Much C, Ivanova I, Di Giacomo M, Auchynnikava T, Monahan JM, Vitsios DM, Vasiliauskaite L, Comazzetto S, et al. (2019). A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis. Cell Res 29, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TB, Manova K, Capodieci P, Lindon C, Bottega S, Wang XY, Refik-Rogers J, Pines J, Wolgemuth DJ and Koff A (2002). Characterization and expression of mammalian cyclin b3, a prepachytene meiotic cyclin. J Biol Chem 277, 41960–41969. [DOI] [PubMed] [Google Scholar]
- O'Donnell L and O'Bryan MK (2014). Microtubules and spermatogenesis. Semin Cell Dev Biol 30, 45–54. [DOI] [PubMed] [Google Scholar]
- Obeso D, Pezza RJ and Dawson D (2014). Couples, pairs, and clusters: mechanisms and implications of centromere associations in meiosis. Chromosoma 123, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page J, de la Fuente R, Manterola M, Parra MT, Viera A, Berrios S, Fernandez-Donoso R and Rufas JS (2012). Inactivation or non-reactivation: what accounts better for the silence of sex chromosomes during mammalian male meiosis? Chromosoma 121, 307–326. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM and Paigen K (2010). Prdm9 controls activation of mammalian recombination hotspots. Science 327, 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337. [DOI] [PubMed] [Google Scholar]
- Radford SJ, Nguyen AL, Schindler K and McKim KS (2017). The chromosomal basis of meiotic acentrosomal spindle assembly and function in oocytes. Chromosoma 126, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refik-Rogers J, Manova K and Koff A (2006). Misexpression of cyclin B3 leads to aberrant spermatogenesis. Cell Cycle 5, 1966–1973. [DOI] [PubMed] [Google Scholar]
- Roeder GS and Bailis JM (2000). The pachytene checkpoint. Trends Genet 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Scherthan H (2007). Telomere attachment and clustering during meiosis. Cell Mol Life Sci 64, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG and Rauscher FJ 3rd, (2002). SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & development 16, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Nozaki M, Takeda N and Shinkai Y (2007). Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J 26, 3346–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Naruse C, Costa Y, Shirakawa T, Tachibana M, Sharif J, Kezuka-Shiotani F, Kakiuchi D, Masumoto H, Shinkai Y, et al. (2011). HP1gamma links histone methylation marks to meiotic synapsis in mice. Development 138, 4207–4217. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Argunhan B and Tsubouchi T (2018). Exiting prophase I: no clear boundary. Curr Genet 64, 423–427. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX and Burgoyne PS (2005). Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37, 41–47. [DOI] [PubMed] [Google Scholar]
- Visnes T, Giordano F, Kuznetsova A, Suja JA, Lander AD, Calof AL and Strom L (2014). Localisation of the SMC loading complex Nipbl/Mau2 during mammalian meiotic prophase I. Chromosoma 123, 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG and Zhang Y (2003). mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell 12, 475–487. [DOI] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B and Ng HH (2009). Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes & development 23, 2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq data are available at the NCBI SRA under the accession number PRJNA674950. This study did not generate any unique code.