Abstract
Supervised learning method requires a large volume of annotated datasets. Collecting such datasets is time-consuming and expensive. Until now, very few annotated COVID-19 imaging datasets are available. Although self-supervised learning enables us to bootstrap the training by exploiting unlabeled data, the generic self-supervised methods for natural images do not sufficiently incorporate the context. For medical images, a desirable method should be sensitive enough to detect deviation from normal-appearing tissue of each anatomical region; here, anatomy is the context. We introduce a novel approach with two levels of self-supervised representation learning objectives: one on the regional anatomical level and another on the patient-level. We use graph neural networks to incorporate the relationship between different anatomical regions. The structure of the graph is informed by anatomical correspondences between each patient and an anatomical atlas. In addition, the graph representation has the advantage of handling any arbitrarily sized image in full resolution. Experiments on large-scale Computer Tomography (CT) datasets of lung images show that our approach compares favorably to baseline methods that do not account for the context. We use the learnt embedding to quantify the clinical progression of COVID-19 and show that our method generalizes well to COVID-19 patients from different hospitals. Qualitative results suggest that our model can identify clinically relevant regions in the images.
Introduction
While deep neural network trained by the supervised approach has made breakthroughs in many areas, its performance relies heavily on large-scale annotated datasets. Learning informative representation without human-crafted labels has achieved great success in the computer vision domain (Wu et al. 2018; Chen et al. 2020a; He et al. 2020). Importantly, the unsupervised approach has the capability of learning robust representation since the features are not optimized towards solving a single supervised task. Self-supervised learning has emerged as a powerful way of unsupervised learning. It derives input and label from an unlabeled dataset and formulates heuristics-based pre-text tasks to train a model. Contrastive learning, a more principled variant of self-supervised learning, relies on instance discrimination (Wu et al. 2018) or contrastive predictive coding (CPC) (Oord, Li, and Vinyals 2018). It has achieved state-of-the-art performance in many aspects, and can produce features that are comparable to those produced by supervised methods (He et al. 2020; Chen et al. 2020a). However, for medical images, the generic formulation of self-supervised learning doesn’t incorporate domain-specific anatomical context.
For medical imaging analysis, a large-scale annotated dataset is rarely available, especially for emerging diseases, such as COVID-19. However, there are lots of unlabeled data available. Thus, self-supervised pre-training presents an appealing solution in this domain. There are some existing works that focus on self-supervised methods for learning image-level representations. (Chen et al. 2019) proposed to learn image semantic features by restoring computerized tomography (CT) images from the corrupted input images. (Taleb et al. 2019) introduced puzzle-solving proxy tasks using multi-modal magnetic resonance images (MRI) scans for representation learning. (Bai et al. 2019) proposed to learn cardiac MR image features from anatomical positions automatically defined by cardiac chamber view planes. Despite their success, current methods suffer from two challenges: (1) These methods do not account for anatomical context. For example, the learned representation is invariant with respect to body landmarks which are highly informative for clinicians. (2) Current methods rely on fix-sized input. The dimensions of raw volumetric medical images can vary across scans due to the differences in subjects’ bodies, machine types, and operation protocols. The typical approach for pre-processing natural images is to either resize the image or crop it to the same dimensions, because the convolutional neural network (CNN) can only handle fixed dimensional input. However, both approaches can be problematic for medical images. Taking chest CT for example, reshaping voxels in a CT image may cause distortion to the lung (Singla et al. 2018), and cropping images may introduce undesired artifacts, such as discounting the lung volume.
To address the challenges discussed above, we propose a novel method for context-aware unsupervised representation learning on volumetric medical images. First, in order to incorporate context information, we represent a 3D image as a graph of patches centered at landmarks defined by an anatomical atlas. The graph structure is informed by anatomical correspondences between the subject’s image and the atlas image using registration. Second, to handle different sized images, we propose a hierarchical model which learns anatomy-specific representations at the patch level and learns subject-specific representations at the graph level. On the patch level, we use a conditional encoder to integrate the local region’s texture and the anatomical location. On the graph level, we use a graph convolutional network (GCN) to incorporate the relationship between different anatomical regions.
Experiments on a publicly available large-scale lung CT dataset of Chronic Obstructive Pulmonary Disease (COPD) show that our method compares favorably to other unsupervised baselines and outperforms supervised methods on some metrics. We also show that features learned by our proposed method outperform other baselines in staging lung tissue abnormalities related to COVID-19. Our results show that the pre-trained features on large-scale lung CT datasets are generalizable and transfer well to COVID-19 patients from different hospitals. Our code and supplementary material are available at https://github.com/batmanlab/Context_Aware_SSL
In summary, we make the following contributions:
We introduce a context-aware self-supervised representation learning method for volumetric medical images. The context is provided by both local anatomical profiles and graph-based relationship.
We introduce a hierarchical model that can learn both local textural features on patch and global contextual features on graph. The multi-scale approach enables us to handle arbitrary sized images in full resolution.
We demonstrate that features extracted from lung CT scans with our method have a superior performance in staging lung tissue abnormalities related with COVID-19 and transfer well to COVID-19 patients from different hospitals.
We propose a method that provides task-specific explanation for the predicted outcome. The heatmap results suggest that our model can identify clinically relevant regions in the images.
Method
Our method views images of every patient as a set of nodes where nodes correspond to image patches covering the lung region of a patient. Larger lung (image) results in more spread out patches. We use image registration to an anatomical atlas to maintain the anatomical correspondences between nodes. The edge connecting nodes denote neighboring patches after applying the image deformation derived from image registration. Our framework consists of two levels of self-supervised learning, one on the node level (i.e., patch level) and the second one on the graph level (i.e., subject level). In the following, we explain each component separately. The schematic is shown in Fig. 1.
Figure 1:
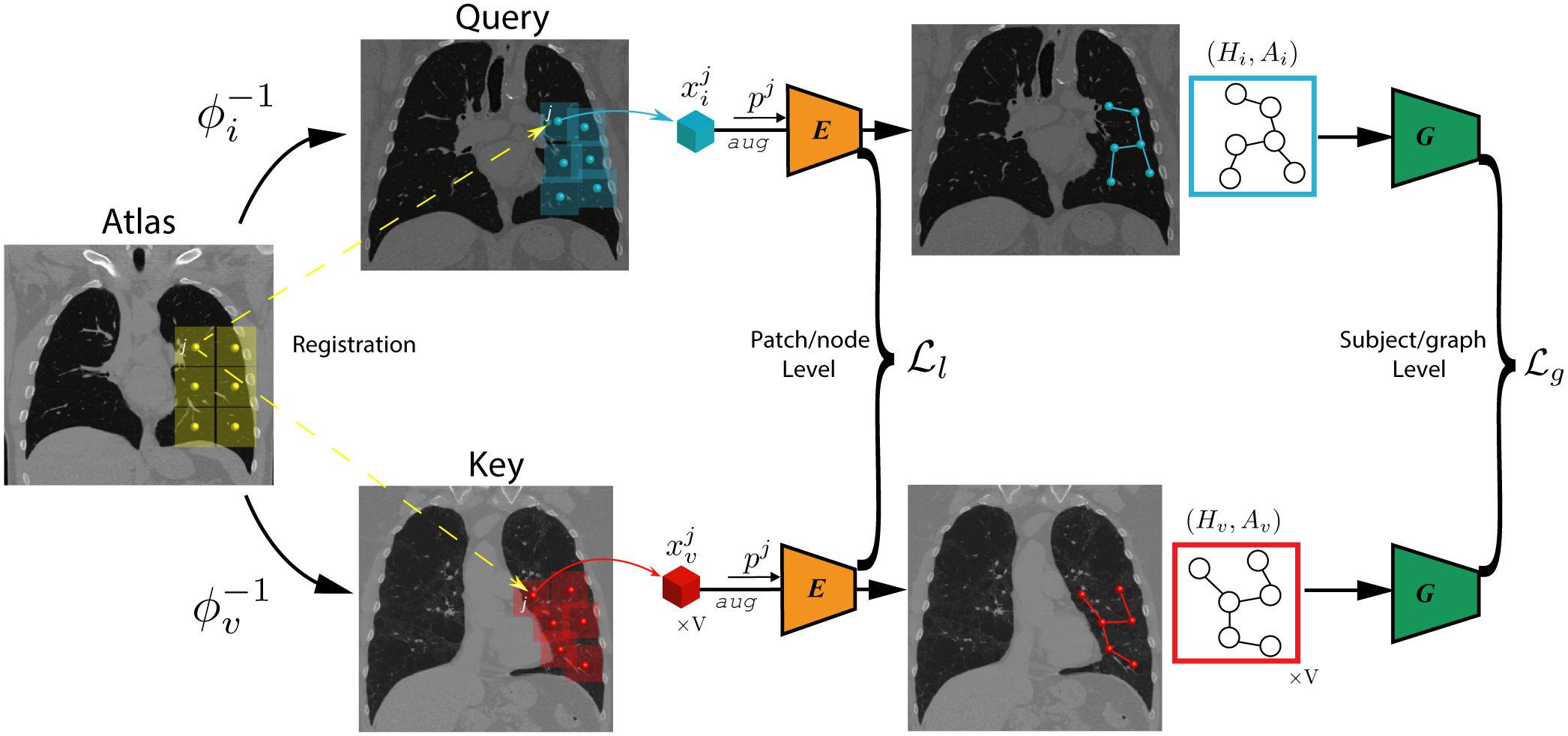
Schematic diagram of the proposed method. We represent every image as a graph of patches. The context is imposed by anatomical correspondences among patients via registration and graph-based hierarchical model used to incorporate the relationship between different anatomical regions. We use a conditional encoder E(·,·) to learn patch-level textural features and use graph convolutional network G(·,·) to learn graph-level representation through contrastive learning objectives. The detailed architecture of the networks are presented in Supplementary Material.
Constructing Anatomy-aware Graph of Patients
We use Xi to denote the image of patient i. To define a standard set of anatomical regions, we divide the atlas image into a set of N equally spaced 3D patches with some overlap. We use to denote the center coordinates of the patches in the Atlas coordinate system. We need to map to their corresponding location for each patient. This operation requires transformations that densely map every coordinate of the Atlas to the coordinate on patients. To find the transformation, we register a patient’s image to an anatomical atlas by solving the following optimization problem:
| (1) |
where Sim is a similarity metric (e.g., ℓ2 norm), ϕi(·) is the fitted subject-specific transformation, Reg(ϕi) is a regularization term to ensure the transformation is smooth enough. The ϕi maps the coordinate of the patient i to the Atlas. After solving this optimization for each patient, we can use the inverse of this transformation to map to each subject (i.e., ). We use well-established image registration software ANTs (Tustison et al. 2014) to ensure the inverse transformation exists. To avoid clutter in notation, we use as a shorthand for . In this way, patches with the same index across all subjects map to the same anatomical region on the atlas image:
| (2) |
To incorporate the relationship between different anatomical regions, we represent an image as a graph of patches (nodes), whose edge connectivity is determined by the Euclidean distance between patches’ centers. With a minor abuse of notation, we let denote the set of patches that cover the lung region of subject i. More formally the image Xi is represented as where is node (patch) information and denotes the set of edges. We use an adjacency matrix Ai ∈ N × N to represent , defined as:
| (3) |
where dist(·,·) denotes the Euclidean distance; and are the coordinates of centers in patches and , respectively; ρ is the threshold hyper-parameter that controls the density of graph.
Learning Patch-level Representation
Local anatomical variations provide valuable information about the health status of the tissue. For a given anatomical region, a desirable method should be sensitive enough to detect deviation from normal-appearing tissue. In addition, the anatomical location of lesion plays a role in patients’ survival outcomes, and the types of lesion vary across different anatomical locations in lung. In order to extract anatomy-specific features, we adopt a conditional encoder E(·,·) that takes both patch and its location index j as input. It is composed with a CNN feature extractor C(·) and a MLP head fl(·), thus we have the encoded patch-level feature:
| (4) |
where ‖ denotes concatenation,.
We adopt the InfoNCE loss (Oord, Li, and Vinyals 2018), a form of contrastive loss to train the conditional encoder on the patch level:
| (5) |
where denotes the representation of query patch , k+ and k−denotes the representation of the positive and the negative key respectively, and τ denotes the temperature hyper-parameter. We obtain a positive sample pair by generating two randomly augmented views from the same query patch , and obtain a negative sample by augmenting the patch at the same anatomical region j from a random subject v, specifically:
Learning Graph-level Representation
We adopt the Graph Convolutional Network (GCN) (Duvenaud et al. 2015) to summarize the patch-level (anatomy-specific) representation into the graph-level (subject-specific) representation. We consider each patch as one node in the graph, and the subject-specific adjacent matrix determines the connection between nodes. Specifically, the GCN model G(·,·) takes patch-level representation Hi and adjacency matrix Ai as inputs, and propagates information across the graph to update node-level features:
| (6) |
where , I is an identity matrix, is a diagonal node degree matrix of , is a N × F matrix containing F features for all N nodes in the image of the subject i, and W is a learnable projection matrix, σ is a nonlinear activation function.
We then obtain subject-level representation by global average pooling all nodes in the graph followed by a MLP head fg:
| (7) |
We adopt the InfoNCE loss to train the GCN on the graph level:
| (8) |
where denotes the representation of the entire image Xi, t+ and t− denotes the representation of the positive and the negative key respectively, and τ denotes the temperature hyper-parameter. To form a positive pair, we take two views of the same image Xi under random augmentation at patch level. We obtain a negative sample by randomly sample a different image Xv, specifically:
Overall Model
The model is trained in an end-to-end fashion by integrating the two InfoNCE losses obtained from patch level and graph level. We define the overall loss function as follows:
| (9) |
Since directly backpropagating gradients from to the parameters in the conditional encoder E is unfeasible due to the excessive memory footprint accounting for a large number of patches, we propose an interleaving algorithm that alternates the training between patch level and graph level to solve this issue. The algorithmic description of the method is shown below:
Algorithm 1.
Interleaving update algorithm
| Require: Conditional encoder E(·, ·), GCN G(·, ·). |
| Input: Image patch , anatomical landmark pj, adjacency matrix Ai. |
| for step t = 1, Tmax do |
| for step tl = 1, Tl do |
| Randomly sample a batch of Bl subjects |
| for j = 1, N do |
| Update E by backpropagating |
| end for |
| end for |
| for step tg = 1 ,Tg do |
| Randomly sample a batch of Bg subjects |
| Update G by backpropagating |
| end for |
| end for |
Model Explanation
Understanding how the model makes predictions is important to build trust in medical imaging analysis. In this section, we propose a method that provides task-specific explanation for the predicted outcome. Our method is expanded from the class activation maps (CAM) proposed by (Zhou et al. 2016). Without loss of generality, we assume a Logistic regression model is fitted for a downstream binary classification task (e.g., the presence or absence of a disease) on the extracted subject-level features . The log-odds of the target variable that Yi = 1 is:
| (10) |
where , the MLP head fg in Eq. 7 is discarded when extracting features for downstream tasks following the practice in (Chen et al. 2020a,b), β and W are the learned logistic regression weights. Then we have as the activation score of the anatomical region j to the target classification. We use a sigmoid function to normalize , and use a heatmap to show the discriminative anatomical regions in the image of subject i.
Implementation Details
We train the proposed model for 30 epochs. We set the learning rate to be 3 × 10−2. We also employed momentum = 0.9 and weight decay = 1 × 10−4 in the Adam optimizer. The patch size is set as 32 × 32 × 32. The batch size at patch level and subject level is set as 128 and 16, respectively. We let the representation dimension F be 128. The lung region is extracted using lungmask (Hofmanninger et al. 2020). Following the practice in MoCo, we maintain a queue of data samples and use a momentum update scheme to increase the number of negative samples in training; as shown in previous work, it can improve performance of downstream task (He et al. 2020). The number of negative samples during training is set as 4096. The data augmentation includes random elastic transform, adding random Gaussian noise, and random contrast adjustment. The temperature τ is chosen to be 0.2. There are 581 patches per subject/graph, this number is determined by both the atlas image size and two hyperparameters, patch size and step size. The experiments are performed on 2 GPUs, each with 16GB memory. The code is available at https://github.com/batmanlab/Context_Aware_SSL.
Related works
Unsupervised Learning
Unsupervised learning aims to learn meaningful representations without human-annotated data. Most unsupervised learning methods can be classified into generative and discriminative approaches. Generative approaches learn the distribution of data and latent representation by generation. These methods include adversarial learning and autoencoder based methods. However, generating data at pixel space can be computationally intensive, and generating fine detail may not be necessary for learning effective representation.
Discriminative approaches use pre-text tasks for representation learning. Different from supervised approaches, both the inputs and labels are derived from an unlabeled dataset. Discriminative approaches can be grouped into (1) pre-text tasks based on heuristics, including solving jigsaw puzzles (Noroozi and Favaro 2016), context predication (Doersch, Gupta, and Efros 2015), colorization (Zhang, Isola, and Efros 2016) and (2) contrastive methods. Among them, contrastive methods achieve state-of-the-art performance in many tasks. The core idea of contrastive learning is to bring different views of the same image (called ‘positive pairs’) closer, and spread representations of views from different images (called ‘negative pairs’). The similarity is measured by the dot product in feature space (Wu et al. 2018). Previous works have suggested that the performance of contrastive learning relies on large batch size (Chen et al. 2020a) and large number of negative samples (He et al. 2020).
Representation Learning for Graph
Graphs are a powerful way of representing entities with arbitrary relational structure (Battaglia et al. 2018). Several algorithms proposed to use random walk-based methods for unsupervised representation learning on the graph (Grover and Leskovec 2016; Perozzi, Al-Rfou, and Skiena 2014; Hamilton, Ying, and Leskovec 2017). These methods are powerful but rely more on local neighbors than structural information (Ribeiro, Saverese, and Figueiredo 2017). Graph convolutional network (GCN) (Duvenaud et al. 2015; Kipf and Welling 2016) was proposed to generalize convolutional neural networks to work on the graphs. Recently, Deep Graph Infomax (Velickovic et al. 2019) was proposed to learn node-level representation by maximizing mutual information between patch representations and corresponding high-level summaries of graphs.
Experiments
We evaluate the performance of the proposed model on two large-scale datasets of 3D medical images. We compare our model with various baseline methods, including both supervised approaches and unsupervised approaches.
Datasets
The experiments are conducted on three volumetric medical imaging datasets, including the COPDGene dataset (Regan et al. 2011), the MosMed dataset (Morozov et al. 2020) and the COVID-19 CT dataset. All images are re-sampled to isotropic 1mm3 resolution. The Hounsfield Units (HU) are mapped to the intensity window of [−1024, 240] and then normalized to [−1, 1].
COPDGene Dataset
COPD is a lung disease that makes it difficult to breathe. The COPDGene Study (Regan et al. 2011) is a multi-center observational study designed to identify the underlying genetic factors of COPD. We use a large set of 3D thorax computerized tomography (CT) images of 9,180 subjects from the COPDGene dataset in our study.
MosMed Dataset
We use 3D CT scans of 1,110 subjects from the MosMed dataset (Morozov et al. 2020) provided by municipal hospitals in Moscow, Russia. Based on the severity of lung tissues abnormalities related with COVID-19, the images are classified into five severity categories associated with different triage decisions. For example, the patients in the mild category are followed up at home with telemedicine monitoring, while the patients in the critical category are immediately transferred to the intensive care unit.
COVID-19 CT Dataset
To verify whether the learned representation can be transferred to COVID-19 patients from other sites, we collect a multi-hospital 3D thorax CT images of COVID-19. The combined dataset has 80 subjects, in which 35 positive subjects are from multiple publicly available COVID-19 datasets (Jun et al. 2020; Bell 2020; Zhou et al. 2020), and 45 healthy subjects randomly sampled from the LIDC-IDRI dataset (Armato III et al. 2011) as negative samples.
Quantitative Evaluation
We evaluate the performance of proposed method by using extracted representations of subjects to predict clinically relevant variables.
COPDGene dataset
We first perform self-supervised pre-training with our method on the COPDGene dataset. Then we freeze the extracted subject-level features and use them to train a linear regression model to predict two continuous clinical variables, percent predicted values of Forced Expiratory Volume in one second (FEV1pp) and its ratio with Forced vital capacity (FVC) (FEV1/FVC), on the the log scale. We report average R2 scores with standard deviations in five-fold cross-validation. We also train a logistic regression model for each of the six categorical variables, including (1) Global Initiative for Chronic Obstructive Lung Disease (GOLD) score, which is a four-grade categorical value indicating the severity of airflow limitation, (2) Centrilobular emphysema (CLE) visual score, which is a six-grade categorical value indicating the severity of emphysema in centrilobular, (3) Paraseptal emphysema (Para-septal) visual score, which is a three-grade categorical value indicating the severity of paraseptal emphysema, (4) Acute Exacerbation history (AE history), which is a binary variable indicating whether the subject has experienced at least one exacerbation before enrolling in the study, (5) Future Acute Exacerbation (Future AE), which is a binary variable indicating whether the subject has reported experiencing at least one exacerbation at the 5-year longitudinal follow up, (6) Medical Research Council Dyspnea Score (mMRC), which is a five-grade categorical value indicating dyspnea symptom.
We compare the performance of our method against: (1) supervised approaches, including Subject2Vec (Singla et al. 2018), Slice-based CNN (González et al. 2018) and (2) unsupervised approaches, including Models Genesis (Zhou et al. 2019), MedicalNet (Chen, Ma, and Zheng 2019), MoCo (3D implementation) (He et al. 2020), Divergence-based feature extractor (Schabdach et al. 2017), K-means algorithm applied to image features extracted from local lung regions (Schabdach et al. 2017), and Low Attenuation Area (LAA), which is a clinical descriptor. The evaluation results are shown in Table 1. For all results, we report average test accuracy in five-fold cross-validation.
Table 1:
Evaluation on COPD dataset
| Method | Supervised | logFEV1pp | logFEV1/FVC | GOLD | CLE | Para-septal | AE History | Future AE | mMRC |
|---|---|---|---|---|---|---|---|---|---|
| Metric | R-Square | % Accuracy | |||||||
| LAA-950 | ✗ | 0.44±.02 | 0.60±.01 | 55.8 | 32.9 | 33.3 | 73.8 | 73.8 | 41.6 |
| K-Means | ✗ | 0.55±.03 | 0.68±.02 | 57.3 | - | - | - | - | - |
| Divergence-based | ✗ | 0.58±.03 | 0.70±.02 | 58.9 | - | - | - | - | - |
| MedicalNet | ✗ | 0.47±.10 | 0.59±.06 | 57.0±1.3 | 40.3±1.9 | 53.1±0.7 | 78.7±1.3 | 81.4±1.7 | 47.9±1.2 |
| ModelsGenesis | ✗ | 0.58±.01 | 0.64±.01 | 59.5±2.3 | 41.8±1.4 | 52.7±0.5 | 77.8±0.8 | 76.7±1.2 | 46.0±12 |
| MoCo | ✗ | 0.40±.02 | 0.49±.02 | 52.7±1.1 | 36.5±0.7 | 52.5±1.4 | 78.6±0.9 | 82.0 ±1.2 | 46.4±1.7 |
| 2D CNN | ✓ | 0.53 | - | 51.1 | - | - | 60.4 | - | - |
| Subject2Vec | ✓ | 0.67 ±.03 | 0.74 ±.01 | 65.4 | 40.6 | 52.8 | 76.9 | 68.3 | 43.6 |
| Ours w/o CE | ✗ | 0.56±.03 | 0.65±.03 | 61.6±1.2 | 48.1±0.4 | 55.5±0.8 | 78.8 ±1.2 | 80.8±1.7 | 50.4±1.0 |
| Ours w/o Graph | ✗ | 0.60±.01 | 0.69±.01 | 62.5±1.0 | 49.2±1.1 | 55.8±1.3 | 78.7±1.5 | 80.7±1.7 | 50.6±0.8 |
| Ours | ✗ | 0.62±.01 | 0.70±.01 | 63.2±1.1 | 50.4 ±1.3 | 56.2 ±1.1 | 78.8 ±1.3 | 81.1±1.6 | 51.0 ±1.0 |
- indicates not reported.
The results show that our proposed model outperforms unsupervised baselines in all metrics except for Future AE. While MoCo is also a contrastive learning based method, we believe that our proposed method achieves better performance for three reasons: (1) Our method incorporates anatomical context. (2) Since MoCo can only accept fixed-size input, we resize all volumetric images into 256×256× 256. In this way, lung shapes may be distorted in the CT images, and fine-details are lost due to down-sampling. In comparison, our model supports images with arbitrary sizes in full resolution by design. (3) Since training CNN model with volumetric images is extremely memory-intensive, we can only train the MoCo model with limited batch size. The small batch size may lead to unstable gradients. In comparison, the interleaving training scheme reduces the usage of memory footprint, thus it allows us to train our model with a much larger batch size.
Our method also outperforms supervised methods, including Subject2Vec and 2D CNN, in terms of CLE, Paraseptal, AE History, Future AE and mMRC; for the rest clinical variables, the performance gap of our method is smaller than other unsupervised methods. We believe that the improvement is mainly from the richer context information incorporated by our method. Subject2Vec uses an unordered set-based representation which does not account for spatial locations of the patches. 2D CNN only uses 2D slices which does not leverage 3D structure. Overall, the results suggest that representation extracted by our model preserves richer information about the disease severity than baselines.
Ablation study:
We perform two ablation studies to validate the importance of context provided by anatomy and the relational structure of anatomical regions: (1) Removing conditional encoding (CE). In this setting, we replace the proposed conditional encoder with a conventional encoder which only takes images as input. (2) Removing graph. In this setting, we remove GCN in the model and obtain subject-level representation by average pooling of all patch/node level representations without propagating information between nodes. As shown in Table 1, both types of context contribute significantly to the performance of the final model.
MosMed dataset
We first perform self-supervised pre-training with our method on the MosMed dataset. Then we freeze the extracted patient-level features and train a logistic regression classifier to predict the severity of lung tissue abnormalities related with COVID-19, a five-grade categorical variable based on the on CT findings and other clinical measures. We compare the proposed method with benchmark unsupervised methods, including MedicalNet, ModelsGenesis, MoCo, and one supervised model, 3D CNN model. We use the average test accuracy in five-fold cross-validation as the metric for quantifying prediction performance.
Table 2 shows that our proposed model outperforms both the unsupervised and supervised baselines. The supervised 3D CNN model performed worse than the other unsupervised methods, suggesting that it might not converge well or become overfitted since the size of the training set is limited. The features extracted by the proposed method show superior performance in staging lung tissue abnormalities related with COVID-19 than those extracted by other unsupervised benchmark models. We believe that the graph-based feature extractor provides additional gains by utilizing the full-resolution CT images than CNN-based feature extractor, which may lose information after resizing or downsampling the raw images. The results of ablation studies support that counting local anatomy and relational structure of different anatomical regions is useful for learning more informative representations for COVID-19 patients.
Table 2:
Evaluation on MosMed dataset
| Method | Supervised | % Accuracy |
|---|---|---|
| MedicalNet | ✗ | 62.1 |
| ModelsGenesis | ✗ | 62.0 |
| MoCo | ✗ | 62.1 |
| 3D CNN | ✓ | 61.2 |
| Ours w/o CE | ✗ | 63.3 |
| Ours w/o Graph | ✗ | 64.3 |
| Ours | ✗ | 65.3 |
COVID-19 CT Dataset
Since the size of COVID-19 CT Dataset is very small (only 80 images are available), we don’t train the networks from scratch with this dataset. Instead, we use models pre-trained on the COPDGene dataset and the MosMed dataset to extract patient-level features from the images in the COVID-19 CT Dataset, and train a logistic regression model on top of it to classify COVID-19 patients. We compare the features extracted by the proposed method to the baselines including MedicalNet, ModelsGenesis, MoCo (unsupervised), and 3D CNN (supervised). We report the average test accuracy in five-fold cross-validation.
Table 3 shows that the features extracted by the proposed model pre-trained on the MosMed dataset perform the best for COVID-19 patient classification. They outperform the features extracted by the same model pre-trained on the COPDGene dataset. We hypothesize that this is because the MosMed dataset contains subjects with COVID-19 related pathological tissue, such as ground glass opacities and mixed attenuation. However, the COPDGene dataset also shows great performance for transfer learning with both the ModelsGenesis model and our model, which shed light on the utility of unlabeled data for COVID-19 analysis.
Table 3:
Evaluation on COVID-19 CT dataset
| Method | Supervised | % Accuracy |
|---|---|---|
| MedicalNet | ✗ | 85.0 |
| ModelsGenesis (Pretrained on COPDGene) | ✗ | 92.5 |
| ModelsGenesis (Pretrained on MosMed) | ✗ | 88.7 |
| MoCo (Pretrained on COPDGene) | ✗ | 75.0 |
| MoCo (Pretrained on MosMed) | ✗ | 86.3 |
| 3D CNN | ✓ | 77.5 |
| Ours (Pretrained on COPDGene) | ✗ | 90.0 |
| Ours (Pretrained on MosMed) | ✗ | 96.3 |
Model Visualization
To visualize the learned embedding and understand the model’s behavior, we use two methods to visualize the model. The first one is embedding visualization, we use UMAP (McInnes, Healy, and Melville 2018) to visualize the patient-level features extracted on the COPDGene dataset in two dimensions. Figure 2 shows a trend, from lower-left to upper-right, along which the value of FEV1pp decreases or the severity of disease increases.
Figure 2:
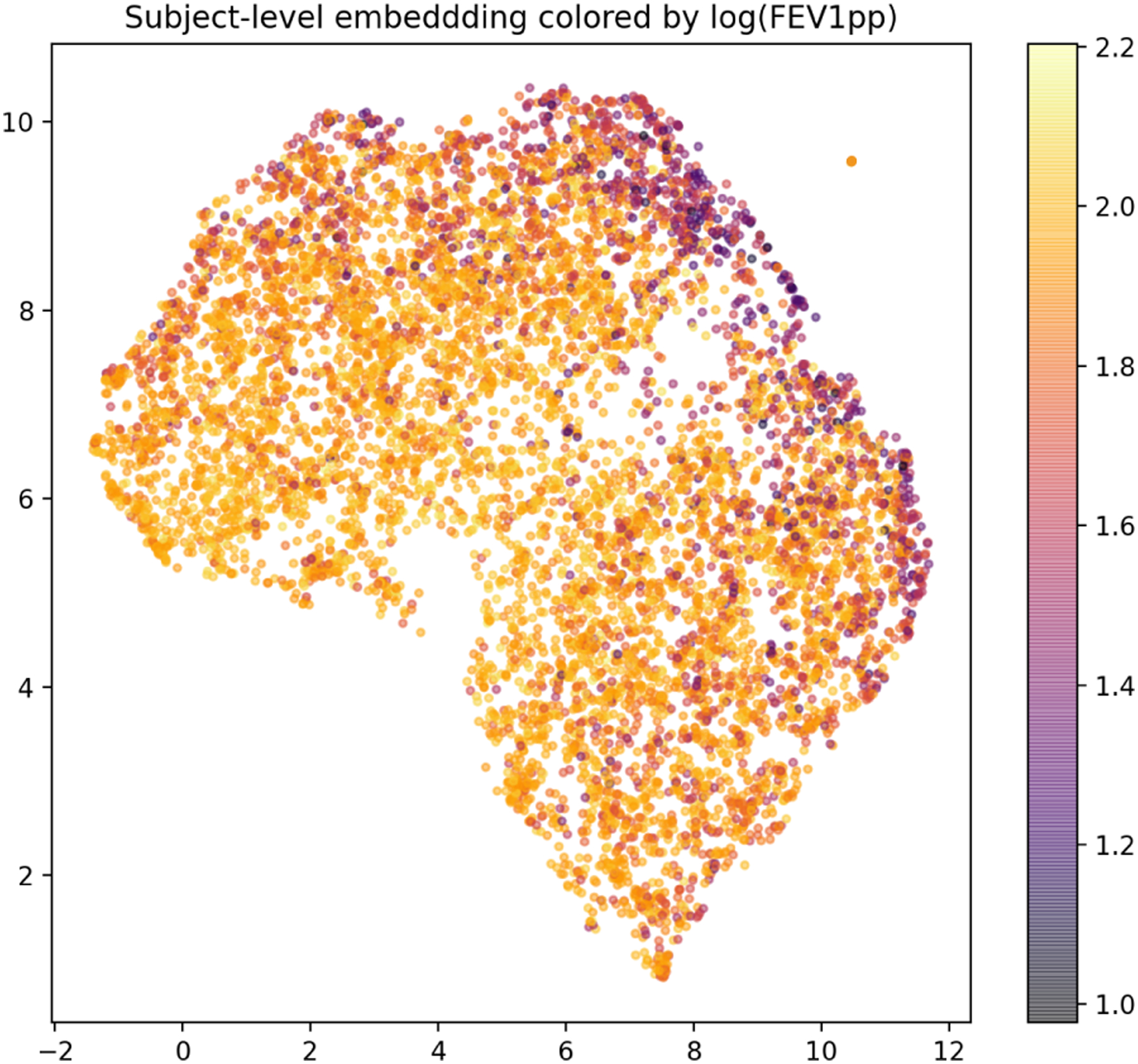
Embedding of subjects in 2D using UMAP. Each dot represents one subject colored by log FEV1pp. Note that lower FEV1pp value indicates more severe disease.
In addition, we use the model explanation method introduced before to obtain the activation heatmap relevant to the downstream task, COVID-19 classification. Figure 3 (left) shows the axial view of the CT image of a COVID-19 positive patient, and Figure 3 (right) shows the corresponding activation map. The anatomical regions received high activation scores overlap with the peripheral ground glass opacities on the CT image, which is a known indicator of COVID-19. We also found that activation maps of non-COVID-19 patients usually have no obvious signal, which is expected. This result suggests that our model can highlight the regions that are clinically relevant to the prediction. More examples can be found in Supplementary Material.
Figure 3:
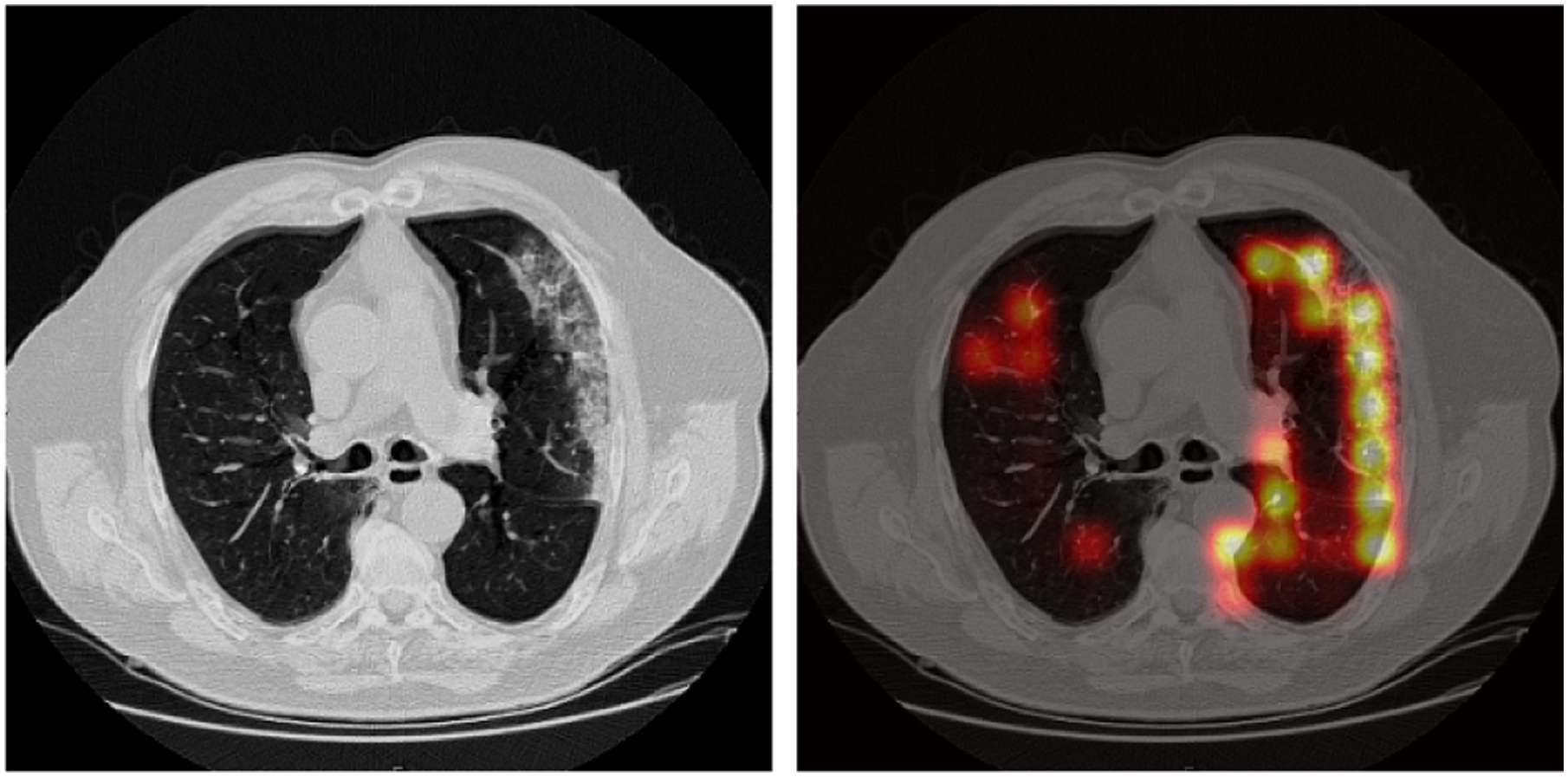
An axial view of the activation heatmap on a COVID-19 positive subject in the COVID-19 CT dataset. Brighter color indicate higher relevance to the disease severity. The figure illustrates that high activation region overlaps with the ground glass opacities.
Conclusion
In this paper, we introduce a novel method for context-aware unsupervised representation learning on volumetric medical images. We represent a 3D image as a graph of patches with anatomical correspondences between each patient, and incorporate the relationship between anatomical regions. In addition, we introduced a multi-scale model which includes a conditional encoder for local textural feature extraction and a graph convolutional network for global contextual feature extraction. Moreover, we propose a task-specific method for model explanation. The experiments on multiple datasets demonstrate that our proposed method is effective, generalizable and interpretable.
Supplementary Material
References
- Armato SG III; McLennan G; Bidaut L; McNitt-Gray MF; Meyer CR; Reeves AP; Zhao B; Aberle DR; Henschke CI; Hoffman EA; et al. 2011. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Medical physics 38(2): 915–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai W; Chen C; Tarroni G; Duan J; Guitton F; Petersen SE; Guo Y; Matthews PM; and Rueckert D 2019. Self-supervised learning for cardiac mr image segmentation by anatomical position prediction. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 541–549. Springer. [Google Scholar]
- Battaglia PW; Hamrick JB; Bapst V; Sanchez-Gonzalez A; Zambaldi V; Malinowski M; Tacchetti A; Raposo D; Santoro A; Faulkner R; et al. 2018. Relational inductive biases, deep learning, and graph networks. arXiv preprint arXiv:1806.01261. [Google Scholar]
- Bell DJ 2020. COVID-19 CT segmentation dataset. URL https://radiopaedia.org/articles/covid-19-4?lang=us.
- Chen L; Bentley P; Mori K; Misawa K; Fujiwara M; and Rueckert D 2019. Self-supervised learning for medical image analysis using image context restoration. Medical image analysis 58: 101539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S; Ma K; and Zheng Y 2019. Med3d: Transfer learning for 3d medical image analysis. arXiv preprint arXiv:1904.00625. [Google Scholar]
- Chen T; Kornblith S; Norouzi M; and Hinton G 2020a. A simple framework for contrastive learning of visual representations. arXiv preprint arXiv:2002.05709. [Google Scholar]
- Chen X; Fan H; Girshick R; and He K 2020b. Improved baselines with momentum contrastive learning. arXiv preprint arXiv:2003.04297. [Google Scholar]
- Doersch C; Gupta A; and Efros AA 2015. Unsupervised visual representation learning by context prediction. In Proceedings of the IEEE international conference on computer vision, 1422–1430. [Google Scholar]
- Duvenaud DK; Maclaurin D; Iparraguirre J; Bombarell R; Hirzel T; Aspuru-Guzik A; and Adams RP 2015. Convolutional networks on graphs for learning molecular fingerprints. In Advances in neural information processing systems, 2224–2232. [Google Scholar]
- González G; Ash SY; Vegas-Sánchez-Ferrero G; Onieva Onieva J; Rahaghi FN; Ross JC; Díaz A; San José Estépar R; and Washko GR 2018. Disease staging and prognosis in smokers using deep learning in chest computed tomography. American journal of respiratory and critical care medicine 197(2): 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A; and Leskovec J 2016. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W; Ying Z; and Leskovec J 2017. Inductive representation learning on large graphs. In Advances in neural information processing systems, 1024–1034. [Google Scholar]
- He K; Fan H; Wu Y; Xie S; and Girshick R 2020. Momentum contrast for unsupervised visual representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 9729–9738. [Google Scholar]
- Hofmanninger J; Prayer F; Pan J; Rohrich S; Prosch H; and Langs G 2020. Automatic lung segmentation in routine imaging is a data diversity problem, not a methodology problem. arXiv preprint arXiv:2001.11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun M; Yixin W; Xingle A; Cheng G; Ziqi Y; Jianan C; Qiongjie Z; Guoqiang D; Jian H; Zhiqiang H; Ziwei N; and Xiaoping Y 2020. Towards Efficient COVID-19 CT Annotation: A Benchmark for Lung and Infection Segmentation. arXiv preprint arXiv:2004.12537. [Google Scholar]
- Kipf TN; and Welling M 2016. Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907. [Google Scholar]
- McInnes L; Healy J; and Melville J 2018. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426. [Google Scholar]
- Morozov S; Andreychenko A; Pavlov N; Vladzymyrskyy A; Ledikhova N; Gombolevskiy V; Blokhin IA; Gelezhe P; Gonchar A; and Chernina VY 2020. MosMedData: Chest CT Scans With COVID-19 Related Findings Dataset. arXiv preprint arXiv:2005.06465. [Google Scholar]
- Noroozi M; and Favaro P 2016. Unsupervised learning of visual representations by solving jigsaw puzzles. In European Conference on Computer Vision, 69–84. Springer. [Google Scholar]
- Oord A. v. d.; Li Y; and Vinyals O 2018. Representation learning with contrastive predictive coding. arXiv preprint arXiv:1807.03748. [Google Scholar]
- Perozzi B; Al-Rfou R; and Skiena S 2014. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining, 701–710. [Google Scholar]
- Regan EA; Hokanson JE; Murphy JR; Make B; Lynch DA; Beaty TH; Curran-Everett D; Silverman EK; and Crapo JD 2011. Genetic epidemiology of COPD (COPDGene) study design. COPD: Journal of Chronic Obstructive Pulmonary Disease 7(1): 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro LF; Saverese PH; and Figueiredo DR 2017. struc2vec: Learning node representations from structural identity. In Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, 385–394. [Google Scholar]
- Schabdach J; Wells WM; Cho M; and Batmanghelich KN 2017. A likelihood-free approach for characterizing heterogeneous diseases in large-scale studies. In International Conference on Information Processing in Medical Imaging, 170–183. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla S; Gong M; Ravanbakhsh S; Sciurba F; Poczos B; and Batmanghelich KN 2018. Subject2Vec: generative-discriminative approach from a set of image patches to a vector. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 502–510. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleb A; Lippert C; Klein T; and Nabi M 2019. Multimodal self-supervised learning for medical image analysis. arXiv preprint arXiv:1912.05396. [Google Scholar]
- Tustison NJ; Cook PA; Klein A; Song G; Das SR; Duda JT; Kandel BM; van Strien N; Stone JR; Gee JC; et al. 2014. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage 99: 166–179. [DOI] [PubMed] [Google Scholar]
- Velickovic P; Fedus W; Hamilton WL; Liò P; Bengio Y; and Hjelm RD 2019. Deep Graph Infomax. In ICLR (Poster). [Google Scholar]
- Wu Z; Xiong Y; Yu SX; and Lin D 2018. Unsupervised feature learning via non-parametric instance discrimination. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 3733–3742. [Google Scholar]
- Zhang R; Isola P; and Efros AA 2016. Colorful image colorization. In European conference on computer vision, 649–666. Springer. [Google Scholar]
- Zhou B; Khosla A; Lapedriza A; Oliva A; and Torralba A 2016. Learning deep features for discriminative localization. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2921–2929. [Google Scholar]
- Zhou L; Li Z; Zhou J; Li H; Chen Y; Huang Y; Xie D; Zhao L; Fan M; Hashmi S; et al. 2020. A Rapid, Accurate and Machine-Agnostic Segmentation and Quantification Method for CT-Based COVID-19 Diagnosis. IEEE Transactions on Medical Imaging 39(8): 2638–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z; Sodha V; Siddiquee MMR; Feng R; Tajbakhsh N; Gotway MB; and Liang J 2019. Models genesis: Generic autodidactic models for 3d medical image analysis. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 384–393. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


