Abstract
The human gene CC3 is a metastasis suppressor for small cell lung carcinoma (SCLC) in vivo. The ability of CC3 to impair the apoptotic resistance of tumor cells is likely to contribute to metastasis suppression. We describe here an alternatively spliced RNA of CC3, designated TC3, that encodes an unstable protein with antiapoptotic activity. TC3 and CC3 proteins share amino-terminal sequences, but TC3 has a unique short hydrophobic carboxyl terminus. Overexpression of CC3 results in massive death of rodent fibroblasts, but TC3 protects cells from CC3-induced death and from other death stimuli such as treatment with tumor necrosis factor or overexpression of Bax protein. The death-inducing activity of CC3 resides within its amino-terminal domain, which is conserved in TC3. The carboxyl terminus of TC3 is responsible for the antiapoptotic function of TC3; mutations in this domain abolish the ability of TC3 to protect cells from apoptosis. TC3 protein is short-lived due to its rapid degradation by proteasome, and it forms complexes with a regulatory subunit of proteasome known as s5α. The signal for the rapid degradation of TC3 resides within its carboxyl terminus, which is capable of conferring instability on a heterologous protein. The proapoptotic activity of CC3 in SCLC cells is induced by a wide variety of signals and involves disruption of the mitochondrial membrane potential (Δψm). The CC3 protein has sequence similarity to bacterial short-chain dehydrogenases/reductases and might represent a phylogenetically old effector of cell death similar to the recently identified apoptosis-inducing factor. CC3 and TC3 have opposing functions in apoptosis and represent a novel dual regulator of cell death.
Apoptosis is a genetically controlled process that is fundamental to the development and homeostasis of multicellular organisms. Aberrations in apoptosis signaling pathways result in a variety of pathological conditions and are common in cancer cells. Resistance to apoptosis is an important factor in tumor development, and the ability to inhibit programmed cell death may contribute to the emergence of aggressive and resistant phenotypes in human cancers (45). Recently, several studies uncovered a role for apoptotic resistance in metastasis, the most threatening aspect of tumor progression. These findings link development of the metastatic phenotype to the acquisition of enhanced resistance to apoptosis (13, 28, 51). Prolonged cell survival could be critical to metastasizing tumor cells at several steps in the process, such as when they are blood-borne or form a micrometastatic lesion (14). Indeed, apoptosis-related proteins have been demonstrated to modulate the metastasis of tumor cells: abrogation of p53-mediated apoptosis facilitates experimental metastasis (32), expression of the proapoptotic kinase death-associated protein (DAP) suppresses metastasis of two murine tumors (15), while elevated expression of the anti-apoptotic gene bcl-2 leads to an increase in the metastatic potential of melanoma and gastric carcinoma (44, 53). Expression of a recently identified member of the IAP (inhibitor of apoptosis) family, survivin (2), is limited to cancer cells and correlates inversely with survival rates in colorectal cancer patients (18). Thus, acquisition of apoptotic resistance might be an important and even necessary step during progression of tumors to a fully malignant metastatic phenotype.
We have previously described a new metastasis suppressor gene, CC3, whose expression is absent in highly metastatic lines of small cell lung carcinoma (SCLC). Introduction of CC3 expression into the SCLC line inhibits metastasis in vivo in SCID-hu mice (41). CC3 also suppresses metastasis of murine melanoma B16 when delivered systemically in the form of liposome-DNA complexes (24). CC3 encodes a protein whose sequence is highly conserved in evolution, with homologous genes being present in Caenorhabditis elegans, Saccharomyces cerevisiae, and even Escherichia coli. The mechanism of metastasis suppression by CC3 is not fully understood, but the proapoptotic properties of CC3 protein are likely to contribute to inhibition of metastasis. Expression of CC3 in SCLC cells increases the apoptotic responses of these cells to death signals such as growth factor withdrawal and chemotherapeutic drugs (41). Loss of CC3 in highly metastatic cells might confer resistance to death-inducing signals and thus help to ensure their survival under the unfavorable conditions encountered in the metastatic process. CC3 exemplifies the molecular link between the metastatic character of SCLC cells and their ability to ignore apoptotic signals. In this paper we describe an alternatively spliced product of CC3 locus named TC3. TC3 encodes an unstable protein that shares its N-terminal domain with CC3 but has a short unique carboxyl terminus. TC3 interacts with a regulatory subunit of proteasome; this could be related to the rapid degradation of TC3. We show that CC3 is a potent inducer of death in rodent fibroblasts; proapoptotic activities of CC3 reside within the amino-terminal domain that it shares with TC3. Unexpectedly, the short C terminus of TC3 confers upon it antiapoptotic properties capable of inhibiting apoptosis induced by CC3 and other death stimuli.
MATERIALS AND METHODS
cDNA cloning.
Cloning of CC3 and TC3 cDNA was performed with the Marathon-Ready cDNA from Clontech (Palo Alto, Calif.), using CC3 specific oligonucleotides and primer AP1 complementary to the adapter sequences attached to the ends of cDNAs. The CC3 antisense primer corresponded to positions 784 to 812 in the published sequence of CC3 RNA (41); the sense primer for amplification of 3′-end sequences was complementary to positions 90 to 116 of CC3 RNA. PCR products were cloned into TA cloning vector pCRII (InVitrogen, San Diego, Calif.).
Expression constructs.
The mammalian expression vectors used were pcDNA3neo (InVitrogen) and pcDNApuro. The latter was constructed from pPUR (Promega) by inserting the cytomegalovirus promoter and polylinker of pcDNA3. cDNA inserts were epitope tagged when necessary and subcloned into pcDNA vectors by standard techniques. All constructs were verified by sequencing and analysis of protein translation in vitro or by Western blot analysis of transfected cells. The cDNA for the C. elegans homologue of CC3 was constructed from the genomic cosmid clone C33F10. The predicted CC3 homologue C33F10.3, containing one intron, was amplified from cosmid DNA, and intron sequences were removed by standard PCR techniques. Green fluorescent protein (GFP) fusion constructs were engineered in the vector pEGFP-N1 (GFP.CC3 and GFP.TC3-N) or pEGFP-C1 (GFP.TC3 and GFP.TC3-C) and verified by sequence analysis.
Transfection of cultured cells.
For transient-transfection studies, subconfluent RAT1A and MCF7 cells were transfected in 12- or 6-well plates with 0.6 or 1.2 μg of DNA by using Lipofectamine or Cellfectin (Gibco-BRL). Plasmid pCMV-βGal DNA constituted 1/10 of the total DNA transfected; the amount of DNA per transfection was kept constant by adding required amount of pcDNA3neo. Cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) as described previously (29) 24 h after transfection. HEK293 cells were transfected in 6-cm dishes with 6 μg of DNA by using the CalPhos Maximizer kit (Clontech) and harvested for Western blot analysis 24 h later. Stably transfected N417 cell lines were generated by Lipofectamine transfection, selection in G418 (for pcDNA3neo), puromycin (for pcdnaPUR), or both for double transfectants, and single-cell cloning by limiting dilution.
Immunoblot analysis of total and fractionated lysates.
Cell lysates were prepared in a lysis buffer containing 150 mM NaCl, 50 mM Tris (pH 7.5), 1% Triton X-100, and a protease inhibitor cocktail (Boehringer Mannheim). Lysates were normalized for protein content and analyzed by Western blotting with anti-HA antibody 12CA5, anti-Au1 antibody (Babco), anti-Bcl-2 monoclonal antibody (Boehringer Mannheim), or anti-CC3 polyclonal serum generated against the hexahistidine fusion of the purified amino-terminal domain of CC3. Bound antibodies were detected using with the enhanced chemiluminescence system (Amersham Life Sciences). For cell fractionation, cells were collected; washed twice in phosphate-buffered saline (PBS); resuspended in hypotonic buffer consisting of 10 mM Tris (pH 7.4), 10 mM MgCl2, and protease inhibitors; and incubated on ice for 10 min. The cells were homogenized with 30 to 40 strokes of a tightly fitting pestle in a Dounce glass homogenizer. After restoration of tonicity, the homogenate was centrifuged at 1,000 × g for 5 min and nuclear pellets were extracted in lysis buffer. The supernatant was centrifuged at 14,000 × g for 15 min to produce a pellet of the heavy membrane fraction; the supernatant was then centrifuged at 100,000 × g for 1 h to separate a fraction enriched in light membranes from the S100 cytoplasmic fraction. For total-membrane preparation, the intermediate centrifugation at 14,000 × g was omitted. Equal amounts of protein were electrophoretically separated and analyzed by Western blotting.
Yeast two-hybrid screening.
The “bait” construct was prepared by subcloning cDNA corresponding to the first 106 amino acids of CC3 into expression vector pAS2-1 (Clontech). Yeast strain GC1945 was transformed with the resulting construct, and expression of the fusion protein was verified by Western blot analysis with the GAL4 DNA binding domain antibody (Santa Cruz Biotechnology). Human liver cDNA library in pACTII was screened as specified by the manufacturer (Clontech). Yeast colonies growing on selective agar medium were screened for their ability to activate the lacZ promoter by a filter assay.
Analysis of protein interactions in 293 cells.
293 cells were cotransfected as described above, and 24 h later they were lysed in a buffer containing 150 mM NaCl, 50 mM Tris (pH 7.5), 5 mM EDTA, 1% Triton X-100, and protease inhibitor cocktail (Boehringer Mannheim). Lysates were subjected to immunoprecipitation with antibody to HA or Au1 (Babco) and washed four times in lysis buffer, and the immunocomplexes were analyzed by Western blotting. In some experiments, the cells were treated with 0.5 μM proteasome inhibitor MG132 to increase the expression levels of proteins.
Flow cytometry.
The DNA content of cells was quantified by propidium iodide staining of ethanol-fixed cells and flow cytometry on a FACscan apparatus (Beckton-Dickinson Immunocytometry) as described previously (7). For analysis of DNA fragmentation in transfected cells by using GFP as a marker, cells were first fixed in 3% paraformaldehyde with 300 mM sucrose for 20 min, washed in PBS, and fixed in ethanol for DNA analysis (20). CellQuest software was used to analyze the DNA content of transfected GFP-positive cells. The percentage of cells with disrupted ΔΨm was determined by staining live cells with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1; Molecular Probes, Inc.) as described previously (6) followed by flow cytometry. Briefly, cells were incubated with 0.1 μM JC-1 for 30 min in complete medium, washed with PBS, and analyzed on a FACScan apparatus for FL1 and FL2. A minimum of 10,000 events per sample were acquired and analyzed with the CellQuest software. In some experiments, cells were incubated with a different cyanine, 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3); Molecular Probes Inc.]. Accumulation of this dye into mitochondria was performed similar to JC-1; fluorescence was recorded with FL1.
Nucleotide sequence accession number.
The TC3 sequence was deposited in the GenBank database under accession no. AF092095.
RESULTS
Identification of an alternatively spliced version of CC3.
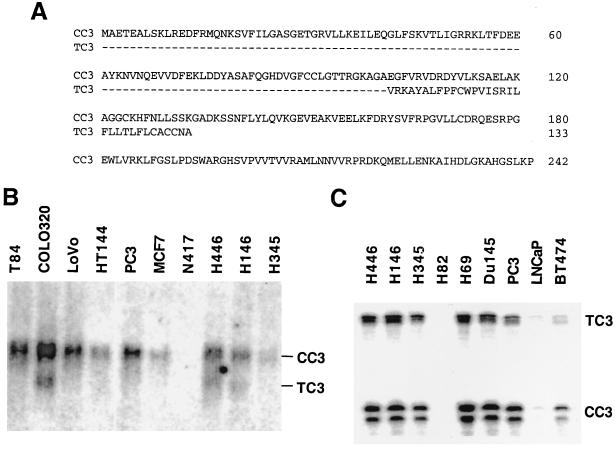
Because the previously reported cDNA of CC3 was derived from a tumor cell line (41), we have cloned CC3 cDNAs from normal human placenta through a modification of the rapid amplification of cDNA end method. To clone the CC3 cDNAs extending in the 5′ end direction, adapter-ligated cDNA was amplified with a CC3 antisense primer to a sequence at the 3′ end of CC3 open reading frame and an adapter-specific primer. We found that the protein-coding region of CC3 cDNA clones from human placenta was identical to the previously reported one (41). In the second cloning approach, we used a PCR primer corresponding to the sequence around the first methionine codon in CC3 RNA to identify all CC3 cDNAs extending in the 3′-end direction. Most of the clones obtained corresponded to the 1.6-kb RNA of CC3, but several were only about 700 bp long. Sequence analysis of these shorter cDNA clones showed their identity to CC3 in the 5′-end sequences coding for the first 101 amino acids followed by a different 3′ end and a poly(A) tail. The open reading frame present in these cDNAs was 133 amino acids long, of which the carboxyl-terminal 32 amino acid residues were unique and highly hydrophobic (Fig. 1A). This shorter form of RNA, designated TC3 (for truncated CC3), is most probably derived by alternative splicing. TC3 RNA of about 0.8 kb was detectable in some but not all human tumor cell lines by Northern blot analysis (Fig. 1B). Analysis of RNA from a number of tumor cell lines by RNase protection with a TC3-specific probe demonstrated the presence of TC3 RNA in all lines expressing CC3 RNA, although TC3 RNA was present at lower levels (Fig. 1C). Expression of TC3 RNA was also very low in most normal human tissues compared to the levels of CC3 RNA (results not shown).
FIG. 1.
Primary structure of CC3 and TC3 proteins and RNA expression analysis. (A) Comparison of the predicted amino acid sequences of CC3 and TC3. Dashes, indicate identical residues. (B) Expression of CC3 and TC3 in human tumor cell lines. Northern blot analysis of electrophoretically separated poly(A) RNA (3 μg per lane) was performed with a radioactively labeled fragment containing the entire TC3 cDNA. (C) RNase protection analysis of TC3 and CC3 RNA was performed with a riboprobe synthesized from the TC3 cDNA fragment of 300 bp between the HindIII site at position 278 and the ScaI site at position 568 in the nucleotide sequence of TC3. This fragment spans the presumed variant splice junction. A 290-nucleotide product results from protection by TC3 RNA; CC3 RNA protects only 120 nucleotides of common sequence 5′ to the variant splice junction. The relative intensity of protected riboprobe fragments does not correctly reflect the relative abundance of two RNAs due to the different sizes of the fragments and the different amounts of radioactivity contained within them.
We wished to study the apoptosis-related properties of both TC3 and CC3 in experiments with stably transfected cell lines. CC3 RNA and protein were stably expressed in several tumor cells lines (reference 41 and data not shown), but attempts to establish stable lines expressing transfected TC3 protein were unsuccessful. Transfection of TC3 or epitope-tagged TC3 expression constructs into human tumor cell lines of different origins resulted in selection of clonal populations with undetectable levels of TC3 protein, although exogenously introduced TC3 RNA could be easily detected (results not shown). We have also analyzed a number of normal tissues and cultured cells for presence of TC3 protein by Western blotting with the polyclonal antiserum raised against the N-terminal polypeptide of CC3 and TC3. Although the endogenous CC3 protein was detected, no TC3 reactivity was seen (data not shown).
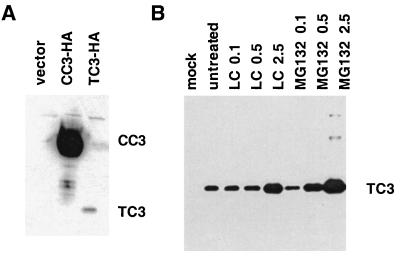
To address the potential reason for the observed lack of expression of TC3 protein, constructs expressing CC3 or TC3 identically tagged with three consecutive hemagglutinin (HA) epitopes at their amino termini were introduced into transformed human embryonic kidney 293 cells in a transient-transfection assay. Expression of tagged TC3 protein of the predicted size was detected, although at levels many times lower than in parallel cultures transfected with HA-tagged CC3 (Fig. 2A). The extremely low levels of TC3 protein were not due to its cytotoxicity, because introduction of TC3 had no effect on the viability of 293 cells within the duration of the assay (data not shown).
FIG. 2.
TC3 protein is subject to degradation by proteasome. (A) Western blot analysis of CC3 and TC3 identically tagged at the N termini with three consecutive HA epitopes transiently expressed in 293 cells. Cell lysates were prepared 24 h after transfection and analyzed by blotting with anti-HA antibody. (B) 293 cells were transiently transfected with TC3-HA expression construct or empty vector (mock) and treated with the proteasome inhibitor lactacystin (LC) or MG132 at the indicated micromolar concentrations for 18 h posttransfection, at which time cell lysates were prepared and analyzed as for panel A.
The low expression levels of TC3 protein in transfected cells could be due to rapid degradation of the newly translated TC3 protein. To address the possibility that TC3 protein is degraded by proteasome, TC3-transfected 293 cells were treated with the specific proteasome inhibitor lactacystin (8) or MG132 (39). As shown in Fig. 2B, treatment with inhibitors increased the levels of TC3 protein in a dose-dependent manner. More slowly migrating bands, possibly corresponding to ubiquitinated forms of TC3, could be seen at the highest dose of inhibitors (Fig. 2B). We conclude that TC3 is an unstable protein subject to rapid degradation by proteasome.
The C terminus of TC3 confers instability on a heterologous protein.
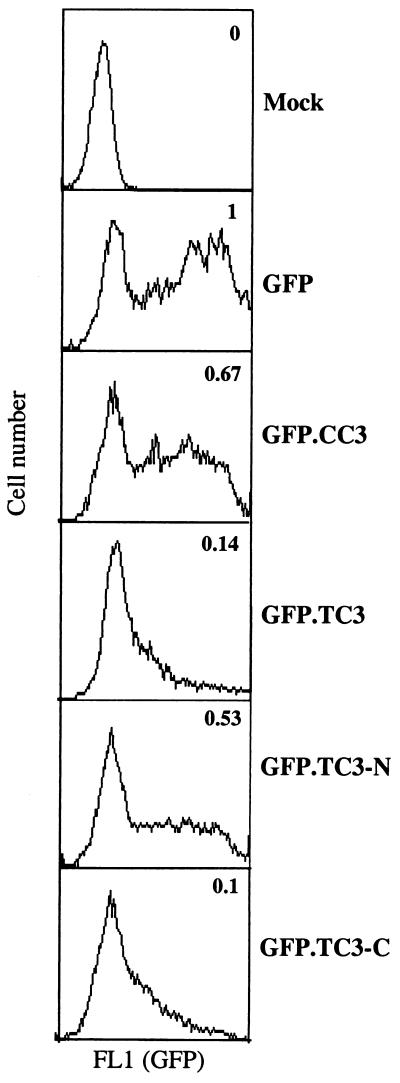
To define the regions of TC3 protein that are responsible for its rapid degradation by proteasome, we have constructed fusion constructs of CC3 or TC3 with GFP. The open reading frame of GFP was fused with full length CC3 (GFP.CC3), TC3 (GFP.TC3), the N-terminal domain common to both CC3 and TC3 (GFP.TC3-N), and the C-terminal domain of TC3 (GFP.TC3-C). The resulting plasmids were transfected into 293 cells, and flow-cytometric analysis was performed 24 h later. As expected, the expression levels of GFP.TC3 as measured by FL1 fluorescence of live cells were dramatically lower than those of GFP alone and GFP.CC3 (Fig. 3). The shared N-terminal domain of TC3 and CC3 had only a slight effect on the levels of expression of GFP, while the C terminus of TC3 reduced it to the levels seen with GFP.TC3 construct. Therefore, the C terminus of TC3 acts independently as a signal for rapid protein degradation.
FIG. 3.
Mapping of the domain of the TC3 protein responsible for the protein instability. HEK293 cells were transfected with the GFP fusion constructs indicated, and live cells were analyzed 24 h later by flow cytometry for FL1 corresponding to the expression levels of GFP. The median fluorescence of transfected populations (GFP-positive cells only) was calculated from the histograms. Numbers indicate the relative fluorescence produced by various fusions compared to the fluorescence of unmodified GFP protein designated as 1 (arbitrary units).
TC3 protein interacts with a regulatory subunit of proteasome s5α.
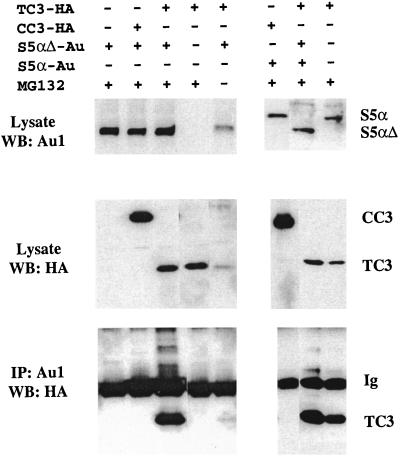
To identify candidate proteins that interact with TC3 or CC3, we have performed a yeast two-hybrid screen of the human liver cDNA two-hybrid library in the yeast expression vector pACTII. The bait consisted of the first 106 amino acids of CC3 (of which 101 are identical in CC3 and TC3) fused in frame to the GAL4 DNA binding domain in vector pAS2-1. Stable transformation of yeast strain GC1945 with the resulting construct lead to a retardation of the yeast growth rate (data not shown). Screening of a small number of transformants (2 × 105) for histidine prototrophy produced only three colonies. All three were found to contain cDNA of the same gene with sequence identity to the human proteasomal regulatory subunit s5α (9). The three clones contained inserts of 1.2 kbp corresponding to the full-length s5α cDNA with the exception of an internal in-frame deletion of sequence encoding 47 amino acids between positions 10 and 56 in the published sequence (9). The interaction of this variant s5α (s5αΔ) with the N-terminal region of CC3 was confirmed to be specific within the yeast two-hybrid system by examination of the induction of β-galactosidase activity. However, we could detect no interaction between s5αΔ and the full-length CC3 subcloned into yeast two-hybrid vectors (results not shown).
We tested the interaction of TC3 and s5αΔ in vivo. 293 cells were transiently transfected with amino-terminally HA-tagged TC3 or CC3 expression vectors and Au1 epitope-tagged s5αΔ. We found that the levels of s5αΔ protein transiently expressed in 293 cells were very low (Fig. 4) but were substantially increased when proteasome activity is inhibited. Transfected cells were thus treated with a relatively low dose of the proteasomal inhibitor MG132 to stabilize both TC3 and s5αΔ proteins. Coimmunoprecipitation analysis revealed complex formation between TC3 and s5αΔ but not between CC3 and s5αΔ (Fig. 4), confirming the results obtained in yeast protein interaction tests. To exclude the possibility that interaction of CC3 and s5αΔ could not be detected for technical reasons, we performed coimmunoprecipitation experiments with a CC3 construct where the HA tag was placed at the carboxyl terminus and also with the anti-CC3 polyclonal serum rather than anti-HA antibody. The negative results of these experiments further confirmed the lack of specific interaction between CC3 and s5αΔ.
FIG. 4.
TC3 interacts with s5α. 293 cells were transiently transfected with the indicated expression constructs and vector DNA to keep the amount of DNA per transfection constant. Where indicated, the cells were treated with 0.5 μM MG132 for the last 16 h of transfection. Cell lysates were prepared 24 h after transfection and subjected to Western blotting (WB) or immunoprecipitation followed by Western blotting with the indicated antibodies. Ig, immunoglobulin.
Higher-molecular-weight forms of TC3 protein were detected in complexes with s5αΔ, indicating that both unmodified and ubiquitinated forms of TC3 are capable of binding to s5αΔ. However, the levels of TC3 protein were not affected by coexpression of s5αΔ (Fig. 4). The internal deletion in s5αΔ does not involve its polyubiquitin-binding domains (11, 54) but removes the region that was shown to be indispensable for the ultimate degradation of at least some ubiquitinated proteins in yeast (11). We examined the interaction of TC3 and CC3 with full-length s5αΔ protein and the potential effect of fully functional s5α on TC3 protein levels. As shown in Fig. 4, full-length s5α also interacted specifically with TC3 but not with CC3. Smaller amounts of TC3 protein were consistently expressed and coprecipitated with full-length s5α than with s5αΔ (Fig. 4). Apparently, even under condition of partial inhibition of the proteolytic function of proteasome, introduction of s5α leads to increased degradation of TC3.
Subcellular localization of CC3 and TC3 proteins.
The amino acid sequence of CC3 has no identifiable protein motifs that might provide a clue to its subcellular localization. We have examined localization of CC3 protein in the stably transfected SCLC line N417 (Fig. 5A). CC3 protein was found predominantly in the membrane and nuclear fractions but not in the particulate free cytoplasmic fraction. Identical pattern of subcellular distribution of endogenous CC3 protein was found in HeLa cells (results not shown).
FIG. 5.
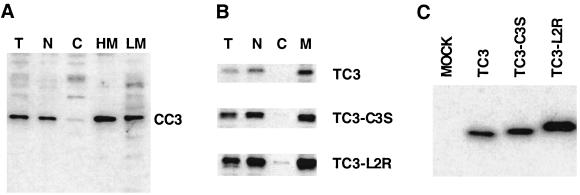
Subcellular localization of the CC3 and TC3 proteins. (A). Western blot analysis of subcellular fractions from N417cc3 cells (T, total; N, nuclear; C, S100 fraction; HM and LM, heavy and light membrane fractions, respectively). Equal amounts of protein extracts from the fractions indicated were electrophoresed and blotted with polyclonal anti-CC3 serum. (B) Analysis of TC3 protein and its mutant forms transiently expressed in 293 cells. Fractions were analyzed as in panel A with anti-HA antibody. M, total membrane fraction. (C) Expression levels of TC3 and mutant proteins in transiently transfected 293 cells analyzed by Western blotting with anti-HA antibody.
The subcellular distribution of TC3 protein was addressed in experiments with transiently transfected cells. We have found that most of the transiently expressed TC3 protein cofractionates with the membrane fraction and some cofractionates with the nuclear fraction of 293 cells (Fig. 5B). We hypothesized that membrane localization of TC3 might be determined by its hydrophobic carboxyl terminus, which scores highly when analyzed for the likelihood of the presence of a transmembrane helix. In addition, the C-terminal amino acids of TC3 are CACCNA (Fig. 1A), raising the possibility that one of the cysteine residues serves as a substrate for prenylation (56) and subsequently directs the protein to cellular membranes. To address the latter possibility, we treated 293 cells transiently expressing TC3 protein with the prenylation inhibitor lovostatin, but that treatment did not result in dislocation of TC3 protein from the membranes (data not shown). In addition, we did not detect incorporation of 3H from radioactive mevalonolactone into TC3 protein, which also argued against the possibility of prenylation of TC3. Finally, all three cysteine residues in the carboxyl terminus of TC3 were changed to serines (TC3-C3S), but that did not result in dislocation of the protein from membranes (Fig. 5B). Since these data indicated the lack of prenylation of TC3 protein, we examined the possibility that the hydrophobicity of the C-terminus of TC3 itself is responsible for its membrane localization. We have used a computer program predicting the likelihood of membrane localization (TMPred) to determine the hydrophobic residues within the TC3 C terminus that, when mutated, will greatly reduce the probability of membrane localization. Mutations of two leucine residues (positions 120 and 123) to arginine residues were introduced into TC3 expression vector (TC3-L2R) because they were predicted to abolish the overall hydrophobic character of the sequence and its potential to serve as a membrane-spanning domain. However, the subcellular localization of TC3-L2R was essentially unchanged compared to that of the TC3 protein, with only a very minor fraction of the mutated form found in the cytoplasmic fraction (Fig. 5B). Surprisingly, there was an appreciable effect of mutations in the carboxyl terminus of TC3 on protein stability, which was more pronounced in TC3-L2R than the TC3-C3S mutant (Fig. 5C). However, the levels of expression achieved by transfection of TC3-L2R were still much lower than those observed for identically epitope-tagged transfected CC3 construct (results not shown). These results indicate that the carboxyl-terminal sequences of TC3 protein are not responsible for the membrane localization of TC3 but contribute to its rapid degradation. The identities of the nuclear and membrane localization signals in CC3 and TC3 proteins remain to be determined, but they are likely to reside within the shared amino terminal.
Overexpression of CC3 kills cells, while TC3 has anti-death activity.
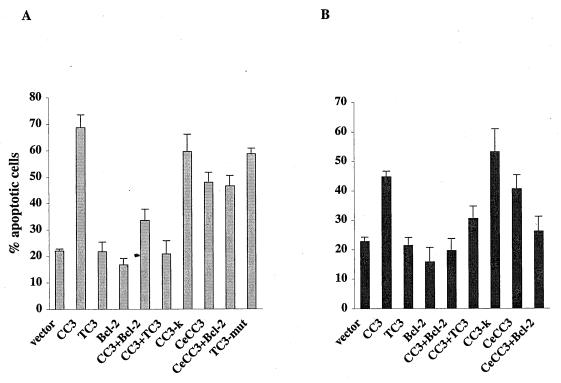
Transient-expression assays (29) were used to evaluate the effect of CC3 and TC3 overexpression on the viability of RAT1a rodent fibroblasts. Short-term expression of CC3 in RAT1a cells induced the death of about 70% of the total transfected cells (Fig. 6A). Dying cells exhibited a rounded condensed morphology and cytoplasmic blebbing and detached from the dish. Introduction of TC3 did not induce a loss of viability of transfected cells. Cotransfection of a twofold excess of Bcl-2 vector with CC3 suppressed most but not all deaths induced by CC3. However, cotransfection of a twofold excess of TC3 with CC3 completely inhibited death induction by CC3, indicating that TC3 actually has a death-protective function. By using a construct that encoded only the amino-terminal 94 amino acids of CC3 (CC3-K), we have localized the cytotoxic activity of CC3 to its amino-terminal sequences shared with TC3. Transfection of TC3-L2R expression vector induces the death of RAT1a cells with an efficiency similar to that for CC3, indicating that the death-protective function of TC3 resides in its unique C terminus. The TC3-C3S mutant construct also showed a similar death activity (data not shown). Mutations in the unique C terminus of TC3 sequence abolish its anti-death function and allow the expression of the death-promoting activity of the shared N terminus. The cDNA of the conserved homologue of CC3 from C. elegans (CeCC3) was also tested in transient-transfection assays with RAT1a cells and was found to induce cell death, albeit with a frequency somewhat lower than that for CC3 (Fig. 6A). Cotransfection of a twofold excess of Bcl-2 did not inhibit death induced by the C. elegans homologue of human CC3.
FIG. 6.
Induction of cell death by CC3 and its inhibition by TC3. (A) Quantitation of cell death in RAT1a cells. The indicated expression constructs were cotransfected in excess with pCMV-β-gal. At 20 to 24 h after transfection, the cells were stained with X-Gal and examined microscopically. Data represent the percentage of round apoptotic cells as a function of total X-Gal-positive cells and are expressed as the mean and standard deviation of at least four independent experiments. (B) Quantitation of cell death in MCF7 cells. Experiments and data presentation are as for panel A.
The death-inducing activity of CC3 and its inhibition by TC3 were also examined in transient-transfection experiments with the human breast carcinoma cell line MCF7.CC3 was significantly less potent in killing MCF7 cells than RAT1a cells (Fig. 6B). Killing of MCF7 cells by CC3 differed from that of RAT1a cells in yet another way: it was more efficiently inhibited by Bcl-2 than by TC3 (Fig. 6B). CeCC3-induced death of MCF7 cells could also be suppressed by Bcl-2, unlike CeCC3-induced death of RAT1a cells (Fig. 6). Apparently, the death-inducing activity of CC3 is significantly attenuated in MCF7 cells and is inhibited by Bcl-2.
To examine if CC3-induced death is accompanied by DNA fragmentation, we performed experiments with RAT1a cells in which an excess of CC3 or TC3 expression constructs was cotransfected with an expression plasmid for GFP. Thus, transfected cells could be individually analyzed for DNA content by propidium iodide staining and flow cytometry (20). By this method, the percentage of cells with reduced DNA content among the transfected population was estimated as 13% for control empty vector, 39% for CC3-transfected cells, and 12% for cells transfected with both CC3 and TC3 (data are the average of two experiments). Apparently, DNA fragmentation occurs in a significant proportion of RAT1a cells that are forced to overexpress CC3 protein. We conclude that CC3 is a potent inducer of apoptosis in RAT1a cells but less so in MCF7 cells and that CC3-induced death is inhibited by alternatively spliced version, TC3.
CC3-induced death is caspase dependent in tumor cells but not in nontransformed cells.
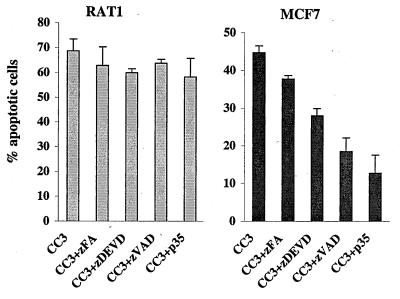
We have examined if caspases play a role in CC3-induced death, by interfering with caspase activity in transiently transfected cells. Transfected cultures were treated with the specific peptide inhibitor of CPP32-like caspases zDEVD-fmk (31), with the broad-spectrum caspase inhibitor zVAD-fmk (57), or with the unrelated protease inhibitor zFA-fmk. The baculovirus protein caspase inhibitor p35 (10) was cotransfected at a twofold excess with CC3-expressing plasmid. None of the above caspase inhibitors had a discernible effect on the frequency of generation of apoptotic morphology among transfected RAT1a cells (Fig. 7A). This indicates that death of RAT1a cells triggered by CC3 proceeds in spite of the inhibition of caspase function. However, death induced by CC3 in MCF7 cells was completely inhibited by p35 and zVAD-fmk and partially inhibited by zDEVD-fmk, while zFA-fmk had no protective effect (Fig. 7B). Apparently, the death of tumor cells induced by overexpression of CC3 proceeds in a manner that could be defined as classical apoptosis in that it is caspase dependent. However, the CC3-induced death of RAT1 cells involves a caspase-independent pathway.
FIG. 7.
Effect of caspase inhibitors on cell death induction by CC3. (A) RAT1a cells were transiently transfected and analyzed as described in the legend to Fig. 4. The protease inhibitors were added at a final concentration of 25 μM immediately after transfection; pcDNA35 was cotransfected with pcDNACC3 in a twofold excess. (B) MCF7 cells were transfected and analyzed as for panel A.
TC3 provides partial protection from diverse death stimuli.
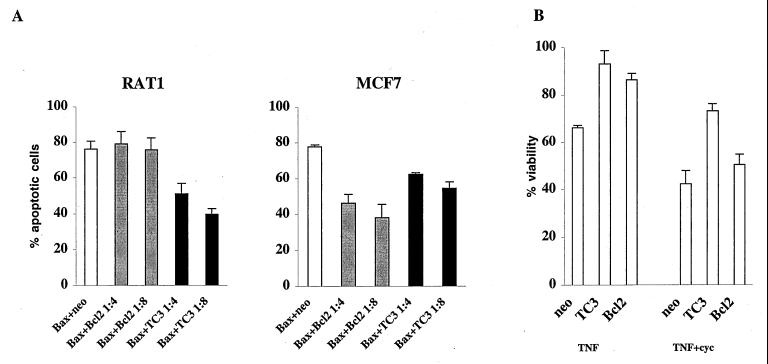
We next examined if overexpression of TC3 is capable of protecting cells from apoptosis induced by agents other than CC3. RAT1a and MCF7 cells were transiently cotransfected with the expression construct for murine bax (33) and a four- or eightfold excess of TC3, Bcl-2, or empty vector. Again, differences emerged between responses of the RAT1a and the MCF7 cells. Although bax was similarly efficient in killing both cell types, RAT1a cells were better protected by TC3 then by Bcl-2, while MCF7 cells were more efficiently protected by Bcl-2 (Fig. 8A).
FIG. 8.
TC3 offers partial protection against cell killing by bax and TNF-α. (A) TC3 but not Bcl-2 partially blocks mBax-induced apoptosis in RAT1a cells. The cells were transiently transfected and analyzed as described in legend to Fig. 4. mbax was cotransfected with a four- or eightfold excess of empty vector (neo), Bcl-2, or TC3 vector as indicated. The results are the mean and standard deviation of three experiments. However, TC3 protection from mBax-induced death is not efficient in MCF7 cells. (B) Protection of MCF7 cells from TNF-α-induced apoptosis by TC3. MCF cells were cotransfected with pCMV-β-gal and indicated vectors. TNF-α (40 ng/ml) treatment was initiated 20 h after transfection, and cells were stained with X-Gal 24 h later. TNF-α (40 ng/ml) and cycloheximide (cyc) (2 μg/ml) treatments were initiated 30 h after transfection, and cells were stained 14 h later. Results (mean and standard deviation of three experiments) are expressed as the percent viability over control transfected untreated cultures.
The antiapoptotic properties of TC3 were further examined in MCF7 cells treated with tumor necrosis factor alpha (TNF-α). As shown in Fig. 8B, MCF7 cells transiently transfected with TC3 or Bcl-2 became resistant to TNF-α-induced death. When transfected cells were treated with a combination of TNF-α and cycloheximide, expression of TC3 still offered protection against TNF-α-induced apoptosis, and this protection was somewhat higher than that offered by Bcl-2 (Fig. 7C). This indicates that the death-inhibitory function of TC3 is less dependent on de novo protein synthesis than is death inhibition by Bcl-2. We conclude that the antiapoptotic activity of TC3 protein is not limited to protection from CC3-induced death but is at least partially effective against other death stimuli.
The proapoptotic activity of CC3 is induced by a wide variety of signals.
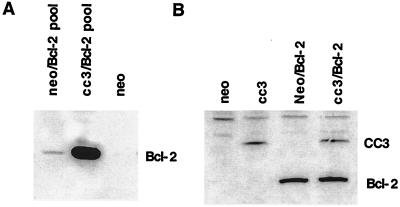
To study the proapoptotic properties of CC3 protein in a stable expression system, we used the previously described SCLC line N417 stably transfected with CC3. We examined if CC3-induced apoptotic responses are dependent on the nature of the signal used and if they are inhibitable by the antiapoptotic protein Bcl-2, whose expression is not detectable in N417 cells. Bcl-2 expression vector conferring puromycin resistance was introduced into a CC3-expressing clone of N417 cells, N417cc3.2 (41), and into the control clone of N417 cells, N417neo. Examination of puromycin-resistant pooled transfected cells showed that N417cc3.2 cells expressed very high levels of Bcl-2 protein while N417neo cells contained much lower levels of Bcl-2 (Fig. 9A). A number of single-cell clones were selected from both transfected populations and examined for levels of Bcl-2 protein expression. All N417cc3/bcl-2 clones expressed relatively high levels of Bcl-2 protein, but N417 clones that expressed similarly high levels of Bcl-2 exhibited a pronounced growth retardation (unpublished observation), in agreement with previous reports on the growth-inhibitory properties of Bcl-2 in tumor cell lines (5, 26, 35). This negative effect on growth rate probably accounts for selection against high levels of Bcl-2 in pooled transfected N417neo cells. The N417cc3 cells, which are predisposed to apoptosis (41), apparently tolerate well the high levels of the anti-apoptotic Bcl-2 protein. Clones with similar levels of Bcl-2 protein with or without CC3 protein were chosen for further analysis (see example of the clones in Fig. 9B).
FIG. 9.
Introduction of Bcl-2 protein expression into N417 cells and N417cc3 cells. (A) Western blot analysis of Bcl-2 protein levels in total transfected populations of N417neo and N417cc2 control cells. (B) Expression of CC3 and Bcl-2 in stably transfected N417 clones. Total-cell extracts from clones stably transfected with the indicated expression constructs were analyzed by Western blotting with polyclonal anti-CC3 antibody to detect CC3 protein and with the anti-Bcl-2 monoclonal antibody (Dako).
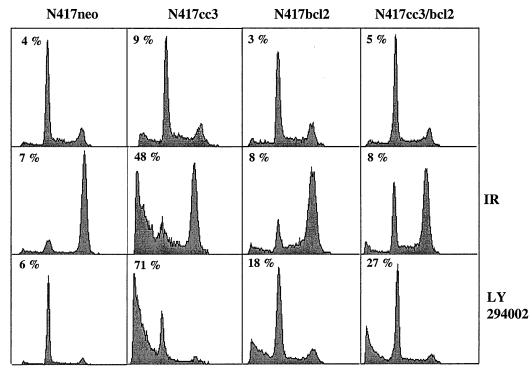
N417 clones were subjected to treatment with a variety of apoptotic inducers: the anticancer drug etoposide, gamma irradiation, the broad-spectrum protein kinase inhibitor staurosporine (STS) or the specific inhibitor of phosphatidylinositol 3-kinase (PI-3 kinase) LY294002. The last of these was chosen because N417 cells contain a constitutively active PI-3 kinase pathway (data not shown). The extent of apoptosis was measured by quantifying the proportion of cells undergoing DNA fragmentation (7) and (for etoposide and STS treatments) loss of mitochondrial membrane potential (Δψm) through opening of the permeability transition pore complex (55). The latter was measured by uptake of the J-aggregate-forming cation JC-1 (6). Representative results on the extent of DNA fragmentation induced by irradiation and treatment with LY-94002 are shown in Fig. 10; data on treatment with etoposide and STS are summarized in Table 1. Dissipation of Δψm and DNA fragmentation in significant numbers of cells were observed specifically in N417cc3 clones. None of the treatments, with the exception of STS, induced substantial DNA fragmentation in N417neo. Bcl-2 inhibited DNA fragmentation induced by the presence of CC3 protein in N417 cells subjected to all apoptotic treatments, although inhibition was incomplete in the cells treated with the PI-3 kinase inhibitor (Fig. 10). However, a lower concentration of LY294002 or shorter treatment times induced DNA fragmentation in CC3-expressing clones only (results not shown). A reduction in Δψm following treatment was also observed in N417cc3 cells only and was largely suppressed by Bcl-2 (Table 1). Similar results were obtained when changes in Δψm were measured by examination of incorporation of a different cyanine dye, DiOC6(3). We conclude that CC3 induces the apoptosis of SCLC cells in response to a variety of different apoptotic signals and that CC3-mediated apoptotic responses are largely inhibitable by Bcl-2.
FIG. 10.
DNA fragmentation analysis of apoptosis in N417 clones treated with 8 Gy of gamma irradiation (IR) or 30 μM of LY294002. Cells were analyzed 24 h after irradiation and 24 h after continuous treatment with LY294003. The top row shows results for untreated cells. Numbers indicate the percentage of cells with DNA content less than 2n.
TABLE 1.
Effects of CC3 and Bcl-2 on apoptotic responses of N417 cellsa
| Cell line | % of cells with reduced DNA content
|
% of cells with disrupted Δψm
|
||||
|---|---|---|---|---|---|---|
| No drug | Etoposide | STS | No drug | Etoposide | STS | |
| N417neo | 3.6 ± 1.6 | 8.2 ± 1.7 | 20.3 ± 4.4 | 4.8 ± 1.1 | 4.8 ± 1.8 | 8.7 ± 2.8 |
| N417cc3 | 9.0 ± 0.7 | 29.2 ± 0.6 | 54.6 ± 4.2 | 8.9 ± 0.8 | 18.1 ± 4.8 | 31.6 ± 7.2 |
| N417bcl-2 | 3.3 ± 1.3 | 4.8 ± 0.4 | 7.3 ± 1.8 | 5.6 ± 2.6 | 8.0 ± 2.5 | 8.7 ± 2.7 |
| N417cc3/bcl-2 | 4.1 ± 2.4 | 6.8 ± 0.8 | 5.6 ± 1.8 | 7.1 ± 2.0 | 8.5 ± 4.9 | 16.8 ± 3.8 |
Cells were treated with 100 μg of etoposide per ml or 50 nM STS for 24 h and analyzed as described in Materials and Methods. Results are presented as the mean of three to four determinations ± standard deviation. Similar results were obtained with at least one more independently derived clone expressing the indicated proteins.
We have addressed the potential mechanism of induction of apoptosis by CC3 by examining several effectors and executors of the apoptotic response in N417 and N417cc3 cells. We have found that introduction of CC3 expression had no effect on the proapoptotic proteins Bax and Bad, caspase 3 or 9, or the cdk inhibitors p21WAF and p27KIP (data not shown). We have also examined if CC3 or TC3 expression have an effect on the levels of Bcl-2 and Bax proteins in a transient-cotransfection assay of 293 cells and found no changes in the expression levels of these proteins in presence of either CC3 or TC3 (data not shown).
DISCUSSION
In this paper we have described the alternatively spliced products CC3 and TC3, which have opposing effects on the induction of apoptosis. Transient expression of CC3 in nontransformed RAT1a fibroblasts causes the death of the majority of transfected cells. Attempts to achieve stable expression of CC3 in immortalized rodent fibroblasts resulted in reduced colony numbers after transfection and selection, and the surviving colonies had extremely low protein levels (data not shown). However, the death-inducing activity of CC3 is significantly attenuated in MCF7 tumor cells. CC3 protein can be stably expressed at significant levels in a variety of tumor cell lines derived from diverse tumors such as SCLC, breast cancer, colon cancer, neuroblastoma, and melanoma (unpublished data). As shown here for MCF7 cells (Fig. 6B) and in unpublished data for other tumor cell lines, only a relatively small percentage of tumor cells directly undergo apoptosis is in response to CC3 overexpression.
The limited killing of MCF7 cells by overexpressed CC3 could be prevented by caspase inhibitors, indicating that activation of caspases is the critical event in the CC3-induced death of transformed cells. However, RAT1a cells die by a process that is caspase independent, although it shows other hallmarks of apoptosis such as cell shrinkage, membrane blebbing, and DNA fragmentation. Our unpublished results show that NIH 3T3 mouse fibroblasts are also killed by CC3 overexpression in a similar manner. The death pathway triggered by CC3 in RAT1a cells and leading to caspase-independent death is apparently not functional in MCF7 cells. The death-inducing activity of CC3 in nontransformed cells is similar to the death-inducing effects of overexpression of Bax protein (49) and of some other death signals and proteins (17, 22, 27, 30, 37, 46).
The mechanism of CC3-induced or CC3-mediated apoptosis remains to be elucidated. We show here that introduction of CC3 protein expression into SCLC cells forces the expression of apoptotic responses, including a drop in Δψm. Mitochondrial dysfunction is a feature of apoptosis induced by a variety of death signals in different cells and is likely to play a key role in the process of commitment to cell death that develops independently of caspase activation (19). The absence of p53 protein in N417 cells (43) is probably significant for the lack of mitochondrial response, particularly in view of the data indicating a role for p53 in the formation of reactive oxygen species and induction of mitochondrial permeability transition (36). Induction of apoptotic responses by CC3 in N417 cells apparently proceeds in a p53-independent manner. Because CC3 induces apoptosis irrespective of the nature of the signals used, it is likely to activate the cell death program at a point where different signal converge on common effectors or executors. At the same time, because CC3-mediated apoptotic responses are largely inhibitable by Bcl-2, the point of action of CC3 in death signal transduction is still upstream of Bcl-2. However, Bcl-2 protein is capable of intercepting and blocking death signal pathways at multiple junctures (38), making it difficult to predict the mechanism by which it might prevent CC3-induced apoptosis.
CC3-mediated apoptosis of SCLC cells appears not to involve downstream effects on several known apoptotic regulators. One tangible clue to the mechanism of CC3-induced apoptosis comes from the recently published finding that CC3 has homology to the bacterial short-chain dehydrogenase/reductase family (3). This finding is reminiscent of the recently identified apoptosis-inducing factor (AIF), a flavoprotein with homology to bacterial oxidoreductases (42). CC3 appears to be similar to AIF in more than one way: both have a higher degree of homology to prokaryotic rather than eukaryotic oxidoreductases, and both apparently can induce apoptosis in a caspase-independent manner (25, 42). This indicates that CC3 might be a phylogenetically old caspase-independent effector of cell death, like AIF.
We have demonstrated that TC3 protein produced from alternatively spliced CC3 RNA has anti-apoptotic activity in transient-transfection assays. TC3 protects cells from apoptosis induced not only by CC3 but also partially by Bax and TNF-α. This indicates that TC3 expression targets a common step that is shared in apoptotic processes induced by divergent death stimuli. Although TC3 RNA is detectable in many of cell lines and tissues examined, TC3 protein could not be detected after attempts to express it stably in a number of cell types. TC3 protein levels are tightly regulated by rapid degradation that involves proteasome. Two lines of evidence point to proteasome involvement in TC3 degradation: first, the levels of TC3 protein could be elevated by inhibiting the proteolytic activity of proteasome, and second, TC3 protein interacts with a regulatory subunit of proteasome s5α in vivo. Moreover, transient coexpression of TC3 and s5α results in reduction of TC3 levels. s5α is the only subunit of proteasome with the demonstrated ability to bind polyubiquitin chains (47), although it is not essential for degradation of all polyubiquitinated substrates in vivo (11). The N-terminal region absent in s5α cDNA cloned here is most critical for the function of s5α in proteasome (11). In accordance with these data, cotransfection of the “deletion” variant s5α with TC3 did not affect TC3 levels in our experiments (data not shown). However, cotransfection of the full-length s5α resulted in an increased degradation rate of TC3, even in the presence of partial inhibition of the proteasome proteolytic function. This indicated that TC3 protein levels in cells might be kept disappearingly low through its interaction with the full-length s5α and subsequent proteolysis by the catalytic 20S proteasome. CC3 protein does not interact with s5α, and its levels are not tightly regulated by proteasome. It is plausible that the shared N terminus of CC3 is prevented from interaction with s5α either by an intermolecular interaction with the relatively long C terminus of CC3 or by binding of other proteins.
We have localized the signal that triggers the degradation of TC3 protein to its C-terminal domain. The sequence of this polypeptide is highly hydrophobic, and mutations in it result in increased stability of TC3. These data are reminiscent of the degradation signal Deg1 found in the S. cerevisiae mating locus transcription factor MATα2 (16). A hydrophobic sequence within MATα2 that is critical for its rapid degradation was predicted to form and amphipathic helix. Helical-wheel analysis of the carboxyl terminus of TC3 predicts a possible α-helical segment (results not shown). In addition, a screen for random polypeptides that act as ubiquitination-dependent degradation signals in yeast produced a number of sequences that are highly hydrophobic (12, 21) and are rich in leucine and phenylalanine residues, similar to the TC3 carboxyl terminus. MATα2 protein is stabilized in diploid yeast by interaction with MATa1 protein, which masks the Deg1 signal. The degradation signal in the carboxyl terminus of TC3 could be masked by an unknown protein interaction under certain conditions or in certain tissues that have yet to be identified.
The death-inducing activity of CC3 is contained within its amino-terminal domain, which is common with the anti-apoptotic TC3 protein. The protective antiapoptotic effect of TC3 resides in the unique carboxyl terminus of this protein, because mutations in this region abolish the protective function of TC3 and convert it to a proapoptotic protein. It is unknown how that short sequence reverses the proapoptotic function of the shared amino-terminal domain. Preliminary results indicate that the C-terminal portion of TC3 linked to a heterologous protein (GFP) is not sufficient for suppression of cell death. Conceivably, specific interactions of TC3 with cellular proteins or its localization to cellular compartments rely on its carboxyl terminus, but the shared amino terminus is still necessary for antiapoptotic effects. Our results show that the subcellular localizations of CC3 and TC3 proteins are quite similar, at least as determined by crude fractionation. The bulk of both proteins is distributed between the nuclear and membrane fractions, although there are no discernible nuclear localization signals in the sequences of the two proteins. Considering the small size of the proteins (28 kDa for CC3 and 15 kDa for TC3), simple diffusion through nuclear pore could account for their nuclear localization. The membrane localization is likely to be determined by the shared amino-terminal domains of the two proteins, because abolishing the hydrophobic character of the C terminus of TC3 did not change its localization.
Recently CC3 was independently identified as a protein named TIP30, which is capable of binding to the human immunodeficiency virus type 1 Tat protein, by Xiao et al. (50), who at the time were unaware of the identity of TIP30 to CC3. TIP30 apparently acts as a cofactor that enhances Tat-activated transcriptional elongation. The role of Tat in elongation is believed now to rely mainly on the ability of Tat to recruit two cellular Cdk-cyclin complexes that stimulate RNA polymerase II processivity (reviewed in reference 52). Thus, the significance of the physical interaction between Tat and CC3 remains to be explored, although it might indicate a potential role for CC3 in transcription through interaction with cellular transcription factors. Alternatively, this interaction might be relevant to the fact that Tat protein is capable of inducing apoptosis in certain cell types (23).
Our results show that the CC3 genomic locus encodes two opposing functions that are active in the apoptotic pathway. The role of alternative splicing in generating cell death-related proteins with different specificities was shown for several genes including those encoding members of Bcl-2, caspase, and death receptor families (1, 4, 34, 40, 48). Similar to CC3/TC3 locus, alternative splicing results in proteins with opposing functions in the case of Bcl-x (4), the caspase Ich-1 (48), and the C. elegans protein ced-4 (40). CC3 and TC3 represent a novel dual regulator of cell death. The relative ratio between CC3 and TC3 proteins might influence the critical decision of a cell to live or die when faced with a death signal.
ACKNOWLEDGMENTS
We thank S. Chissoe for the cosmid clone C33F10, A. Gross and S. Korsmeeyer for mbax plasmid, J. Wilson and L. K. Miller for p35 plasmid, I. Lonnroth and M. Rechsteiner for full-length s5α cdna clones, and A. Ballmain, F. McCormick, and K. Smith-McCune for critical reading of the manuscript.
This work was supported by Public Health Service grant RO1 CA71422 from the National Cancer Institute and by institutional funds from the Cancer Research Institute, UCSF.
REFERENCES
- 1.Alnemri E S, Fernandes-Alnemri T, Litwack G. Expression of four novel isoforms of human interleukin-1converting enzyme with different apoptotic activities. J Biol Chem. 1995;270:4312–4317. doi: 10.1074/jbc.270.9.4312. [DOI] [PubMed] [Google Scholar]
- 2.Ambrosini G, Adida C, Altieri D C. A novel anti-apoptosis gene, surviving, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 3.Baker M E. TIP30, a cofactor for HIV-1 Tat-activated transcription, is homologous to short-chain dehydrogenases/reductases. Curr Biol. 1999;9:R471. doi: 10.1016/s0960-9822(99)80297-8. [DOI] [PubMed] [Google Scholar]
- 4.Boise L H, Gonzalez-Garcia M, Postema C E, Ding L, Lindsten T, Turk L A, Mao X, Nunez G, Thompson C B. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 5.Borner C. Diminished cell proliferation associated with the death-protective activity of Bcl-2. J Biol Chem. 1996;271:12695–12698. doi: 10.1074/jbc.271.22.12695. [DOI] [PubMed] [Google Scholar]
- 6.Cossarizza A, Baccarini-Contri M, Kalshnikova G, Franceschi C. A new method for the cytofluorimetric analysis of mitochondrial membrane potential using the J-aggregate forming lipophilic cation 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1) Biochem Biophys Res Commun. 1993;197:40–45. doi: 10.1006/bbrc.1993.2438. [DOI] [PubMed] [Google Scholar]
- 7.Darzynkiewicz Z, Li X, Gong J. Assays of cell viability: discrimination of cells dying by apoptosis. Methods Cell Biol. 1994;41:15–38. doi: 10.1016/s0091-679x(08)61707-0. [DOI] [PubMed] [Google Scholar]
- 8.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 9.Ferrell K, Deveraux Q, van Nocker S, Rechsteiner M. Molecular cloning and expression of a multiubiquitin chain binding subunit of the human 26S protease. FEBS Lett. 1996;381:143–148. doi: 10.1016/0014-5793(96)00101-9. [DOI] [PubMed] [Google Scholar]
- 10.Friesen P D, Miller L K. Divergent transcription of early 35- and 94-kilodalton protein genes encoded by the HindIII K genome fragment of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1987;61:2264–2272. doi: 10.1128/jvi.61.7.2264-2272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu H, Sadis S, Rubin D M, Glickman M, van Nocker S, Finley D, Vierstra R D. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- 12.Gilon T, Chomsky O, Kulka R G. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glinsky G V, Glinsky V V. Apoptosis and metastasis: a superior resistance of metastatic cancer cells to programmed cell death. Cancer Lett. 1996;101:43–51. doi: 10.1016/0304-3835(96)04112-2. [DOI] [PubMed] [Google Scholar]
- 14.Holmgren L, O'Reilly M S, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med. 1995;1:149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- 15.Inbal B, Cohen O, Polak-Charcon S, Kopolovic J, Vadai E, Eisenbach L, Kimchi A. DAP kinase links the control of apoptosis to metastasis. Nature. 1997;390:180–184. doi: 10.1038/36599. [DOI] [PubMed] [Google Scholar]
- 16.Johnson P R, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- 17.Johnson M D, Xiang H, London S, Kinoshita Y, Knudson M, Mayberg M, Korsmeyer S J, Morrison R S. Evidence for involvement of Bax and p53, but not caspases, in radiation-induced cell death of cultured postnatal hippocampal neurons. J Neurosci Res. 1998;54:721–33. doi: 10.1002/(SICI)1097-4547(19981215)54:6<721::AID-JNR1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Kawasaki H, Altieri D C, Lu C D, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by surviving predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58:5071–5074. [PubMed] [Google Scholar]
- 19.Kroemer G, Zamzami N, Susin S A. Mitochondrial control of apoptosis. Immunol Today. 1997;18:45–51. doi: 10.1016/s0167-5699(97)80014-x. [DOI] [PubMed] [Google Scholar]
- 20.Lamm G M, Steinlein P, Cotton M, Christofori G. A rapid, quantitative and inexpensive method for detecting apoptosis by flow cytometry in transiently transfected cells. Nucleic Acids Res. 1997;25:4855–4857. doi: 10.1093/nar/25.23.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laney J D, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 22.Lavoie J N, Nguyen M, Marcellus R C, Branton P E, Shore G C. E4orf4, a novel adenovirus death factor that induces p53-independent apoptosis by a pathway that is not inhibited by zVAD-fmk. J Cell Biol. 1998;140:637–645. doi: 10.1083/jcb.140.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J L, Friedman D J, Wang C, Metelev V, Pardi A. Induction of apoptosis in uninfected lymphocytes by HIV-1 Tat protein. Science. 1995;268:429–431. doi: 10.1126/science.7716549. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Thor A, Shtivelman E, Cao Y, Heath T D, Debs R J. Systemic gene delivery expands the repertoire of effective anti-angiogenic agents. J Biol Chem. 1999;274:13338–13344. doi: 10.1074/jbc.274.19.13338. [DOI] [PubMed] [Google Scholar]
- 25.Lorenzo H K, Susin S A, Penninger J, Kroemer G. Apoptosis inducing factor (AIF): a phylogenetically old, caspase-independent effector of cell death. Cell Death Differ. 1999;6:516–524. doi: 10.1038/sj.cdd.4400527. [DOI] [PubMed] [Google Scholar]
- 26.Mazel S, Burtrum D, Petrie H T. Regulation of cell division cycle progression by bcl-2 expression: a potential mechanism for inhibition of programmed cell death. J Exp Med. 1996;183:2219–2226. doi: 10.1084/jem.183.5.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy N J, Whyte M K, Gilbert C S, Evan G I. Inhibition of Ced-3/ICE-related proteases does not prevent cell death induced by oncogenes, DNA damage, or the Bcl-2 homologue Bak. J Cell Biol. 1997;136:215–227. doi: 10.1083/jcb.136.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McConkey D J, Greene G, Pettaway C A. Apoptosis resistance increases with metastatic potential in cells of the human LNCaP prostate carcinoma line. Cancer Res. 1996;56:5594–5599. [PubMed] [Google Scholar]
- 29.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 30.Monney L, Otter I, Olivier R, Ozer H L, Haas A L, Omura S, Borner C. Defects in the ubiquitin pathway induce caspase-independent apoptosis blocked by Bcl-2. J Biol Chem. 1998;273:6121–6131. doi: 10.1074/jbc.273.11.6121. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 32.Nikiforov M A, Hagen K, Ossovskaya V S, Connor T M F, Lowe S W, Deichman G I, Gudkov A V. p53 modulation of anchorage independent growth and experimental metastasis. Oncogene. 1996;13:1709–1719. [PubMed] [Google Scholar]
- 33.Oltvai Z N, Milliman C L, Korsmeyer S J. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 34.Papoff G, Cascino I, Eramo A, Starace G, Lynch D H, Ruberti G. An N-terminal domain shared by Fas/Apo-1 (CD95) soluble variants prevents cell death in vitro. J Immunol. 1996;156:4622–4630. [PubMed] [Google Scholar]
- 35.Pietenpol J A, Papadopoulos N, Markowitz S, Willson J K, Kinzler K W, Vogelstein B. Paradoxical inhibition of solid tumor cell growth by bcl2. Cancer Res. 1994;54:3714–3717. [PubMed] [Google Scholar]
- 36.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 37.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J C, de The H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 38.Reed J C. Bcl-2 family proteins. Oncogene. 1998;17:3225–3236. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- 39.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 40.Shaham S, Horvitz H R. An alternatively spliced C. elegans ced-4 RNA encodes a novel cell death inhibitor. Cell. 1996;86:201–208. doi: 10.1016/s0092-8674(00)80092-6. [DOI] [PubMed] [Google Scholar]
- 41.Shtivelman E. A link between metastasis and resistance to apoptosis of variant small cell lung carcinoma. Oncogene. 1997;14:2167–2173. doi: 10.1038/sj.onc.1201059. [DOI] [PubMed] [Google Scholar]
- 42.Susin S A, Lorenzo H K, Zamzami N, Marzo I, Snow B E, Brothers G M, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett D R, Aebersold R, Siderovski D P, Penninger J M, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi T, Nau M M, Chiba I, Birrer M J, Rosenberg R K, Vinocour M, Levitt M, Pass H, Gazdar A F, Minna J D. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 44.Takaoka A, Adachi M, Okuda H, Sato S, Yawata A, Hinoda Y, Takayama S, Reed J C, Imai K. Anti-cell death activity promotes pulmonary metastasis of melanoma cells. Oncogene. 1997;14:2971–2977. doi: 10.1038/sj.onc.1201147. [DOI] [PubMed] [Google Scholar]
- 45.Thompson C B. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 46.Toyoshima F, Moriguchi T, Nishida E. Fas induces cytoplasmic apoptotic responses and activation of the MKK7-JNK/SAPK and MKK6-p38 pathways independent of CPP32-like proteases. J Cell Biol. 1997;139:1005–1015. doi: 10.1083/jcb.139.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Nocker S, Deveraux Q, Rechsteiner M, Vierstra R D. Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc Natl Acad Sci USA. 1996;93:856–860. doi: 10.1073/pnas.93.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 49.Xiang J, Chao D T, Korsmeyer S J. BAX-induced cell death may not require interleukin 1b-converting enzyme-like proteases. Proc Natl Acad Sci USA. 1996;93:14559–14563. doi: 10.1073/pnas.93.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao H, Tao Y, Greenblatt J, Roeder R G. A cofactor, TIP30, specifically enhances HIV-1 Tat activated transcription. Proc Natl Acad Sci USA. 1998;95:2146–2151. doi: 10.1073/pnas.95.5.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie K, Huang S, Dong Z, Juang S H, Gutman M, Xie Q W, Nathan C, Fidler I J. Transfection with the inducible nitric oxide synthase gene suppresses tumorigenicity and abrogates metastasis by K-1735 murine melanoma cells. J Exp Med. 1995;181:1333–1343. doi: 10.1084/jem.181.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yankulov K, Bentley D. Transcriptional control: Tat cofactors and transcriptional elongation. Curr Biol. 1998;8:R447–R449. doi: 10.1016/s0960-9822(98)70289-1. [DOI] [PubMed] [Google Scholar]
- 53.Yawata A, Adachi M, Okuda H, Naishiro Y, Takamura T, Hareyama M, Takayama S, Reed J C, Imai K. Prolonged cell survival enhances peritoneal dissemination of gastric cancer cells. Oncogene. 1998;16:2681–2686. doi: 10.1038/sj.onc.1201792. [DOI] [PubMed] [Google Scholar]
- 54.Young P, Deveraux Q, Beal R E, Pickart C M, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26S protease subunit s5a. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 55.Zamzami N, Marchetti P, Castedo M, Zanin C, Vayssiere J L, Petit P X, Kroemer G. Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J Exp Med. 1995;181:1661–1672. doi: 10.1084/jem.181.5.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang F L, Casey P J. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 57.Zhivotovsky B, Gahm A, Ankarcrona M, Nicotera P, Orrenius S. Multiple proteases are involved in thymocyte apoptosis. Exp Cell Res. 1995;221:404–412. doi: 10.1006/excr.1995.1391. [DOI] [PubMed] [Google Scholar]