Abstract
The role of membrane potential in most intracellular organelles remains unexplored because of the lack of suitable tools. Here, we describe a fluorescent DNA-nanodevice that reports absolute membrane potential and can be targeted to organelles in live cells. This nanodevice, denoted Voltair, bears a voltage-sensitive fluorophore, a reference fluorophore for ratiometry, and acts as an endocytic tracer. Using Voltair we could measure the membrane potential of different organelles in situ in live cells. Voltair can potentially guide the rational design of biocompatible electronics as well as expand our understanding of how membrane potential regulates organelle biology.
Introduction:
Membrane potential is a key property of all biological membranes and is a fundamental signaling cue in all cells 1,2. Although the membrane potential of the plasma membrane is relatively straightforward to measure, that of organelles is not. Since organelle lumens have very different ionic compositions from the cytosol, they are expected to harbor distinctive membrane potentials 3. Electrophysiology on isolated organelles reveals that membrane potential is a prime regulator of their function4. For example, the high negative membrane potential of mitochondria changes along with cellular metabolism and regulates mitochondrial fission and fusion5,6. Lysosome membrane potential regulates lysosomal functions such as the refilling of lumenal calcium and its fusion with other organelles7–8.
Importantly, the role of membrane potential in the function of many other organelles remains unaddressed as they are refractory to electrophysiology and organelle-specific probes are lacking9. The pH sensitivity of fluorescent proteins limits their applicability in organelles where lumenal pH and membrane potential are co-dependent10. Electrochromic hemicyanine dyes or photoinduced electron transfer (PeT) based voltage sensitive dyes are particularly attractive for organelles due to their low capacitive loads, high temporal resolution, photostability and response range11. However, unlike proteins, voltage sensitive dyes cannot be targeted to specific organelles other than mitochondria 12.
We have created a DNA-based nanodevice, called Voltair, that functions as a non-invasive, organelle-targetable, ratiometric reporter of absolute membrane potential of organelles in situ. DNA nanodevices can quantitatively map diverse analytes such as ions, reactive species and enzymes in organelles13–16. Using the 1:1 stoichiometry of DNA hybridization one can incorporate a reference fluorophore and any desired detection chemistry in a precise ratio to yield ratiometric probes17. By displaying molecular trafficking motifs on such DNA nanodevices one can localize them within organelles18,19.
Using Voltair, we measured the membrane potential of different organelles such as the early endosome, the late endosome and the lysosome, and mapped membrane potential changes as a function of endosomal maturation. We targeted Voltair to the recycling endosomes as well as the trans-Golgi network and quantified the membrane potentials of these organelles as well18,19. By measuring the contribution of the electrogenic Vacuolar H+ -ATPase (V-ATPase) proton pump to the membrane potentials of various organelles, we found that each organelle membrane showed different electrochemical characteristics.
Design and Characterization of Voltair
Resting membrane potential is measured by the difference in electrical potential of two electrodes, a sample electrode inserted in the biological membrane and a reference electrode located outside the biological membrane of interest (Fig. 1a)20. The DNA nanodevice Voltair, uses a similar concept, where the fluorescence of a reporter probe inserted into the biological membrane is compared to a reference probe at a different wavelength located outside the biological membrane (Fig. 1a).
Fig. 1. Design and characterization of Voltair probes:
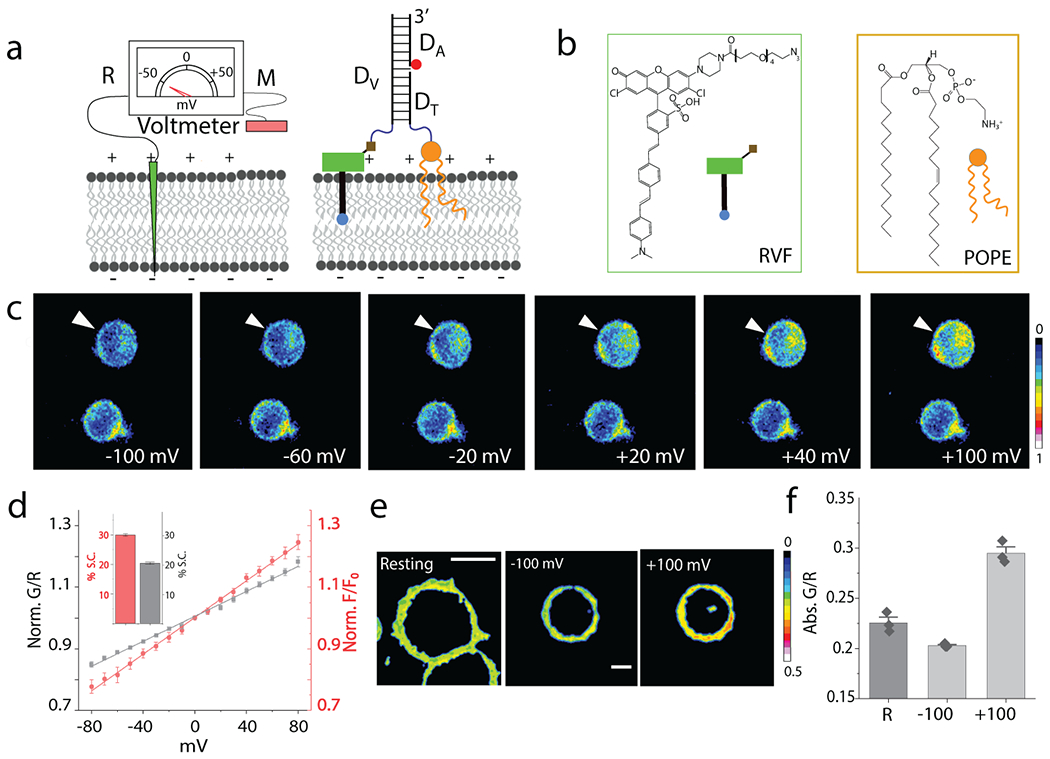
(a) Schematic of the working principle of DNA voltmeter Voltair: Measuring probe (M, Green) is a voltage sensitive dye (RVF) conjugated to a DNA duplex that is membrane-tethered by attachment to a lipid anchor (POPE). Reference probe (R, red) is DNA duplex with a reference dye (Atto647N, red sphere) that together with RVF reports membrane potential ratiometrically. (b) Structure of a conjugatable version of RVF (RVF-N3) and the lipid anchor POPE. (c) Pseudo-color images show pixel-wise ratio of RVF to Atto647N fluorescence intensities (G/R) as a function of membrane potential applied by whole cell voltage clamping. Voltage clamped cell shown by white arrow head. Scale: 10 μm. (d) Sensor response of VoltairPM (gray trace) and unmodified RVF (red trace) as a function of plasma membrane potential in HEK 293T cells. G/R values are normalized to that at 0 mV. Inset: Percentage signal change (S.C.) of VoltairPM (black) and unmodified RVF (red) from 0 to 100 mV. Error bar represents mean ± s.d. from n = 6 cells. Experiments were repeated thrice independently with similar results. (e, f) Absolute G/R values of the plasma membranes of resting cells (Res) and cells voltage clamped at −100 mV and +100 mV. Error bar represents mean ± s.e.m. of three independent trials, n = 15 cells. Scale: 10 μm.
Voltair is a 38-base pair DNA duplex with three modules (Fig. 1a and Supplementary Table S1). The first module, denoted Dv, is a 38-mer single-stranded DNA conjugated at its 3′ end to a previously characterized voltage sensing dye (RVF). The synthesis is described in Methods (Supplementary Fig. S1–S4, Supplementary Note 1)21. The fluorophore in RVF is quenched by PeT due to hyperconjugation with the lone pair of electrons on its dimethyl aminobenzyl moiety (Fig. 1b)11,21. Plasma-membrane depolarization decreases electron transfer and increases RVF fluorescence (Fig. 1a and Supplementary Movie S1). RVF has high photostability, no capacitive load, is pH insensitive from pH 4.5 – 7.5, and therefore deployable at different physiological pH (Supplementary Fig. S5, Supplementary Note 2).
The second module in Voltair is the reference probe, Atto647N, that corrects for intensity changes due to different sensor abundance arising from inhomogeneous probe distribution. Atto647N is attached to the 5′ end of DA and has high photostability, minimal spectral overlap with RVF, insensitivity to pH, voltage and other ions (red circle, Fig. 1a). The ratio of RVF and Atto647N intensities are thus proportional only to membrane potential.
The third module is a targeting moiety that differs in every Voltair variant targeted to a specific organelle. In VoltairPM, Voltair localizes to the plasma-membrane as the 5′ end of DT bears a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) moiety via a tetra-ethylene glycol linker22. When VoltairPM is added to the culture medium, the POPE moiety anchors VoltairPM to the cell surface (Supplementary Fig. S6, Supplementary Note 3). Thus, RVF is inserted in a defined orientation with respect to the membrane potential vector, and the anionic DNA duplex prevents RVF flipping within the membrane. For ~2 h post-labeling, neither RVF nor DNA conjugated to POPE are endocytosed by HEK 293T cells (Supplementary Fig. S6).
VoltairIM labels intracellular organelles by leveraging the ability of duplex DNA to double as a ligand for scavenger receptors23. The engagement of endocytic receptors by DNA-based targeting motifs overrides the plasma-membrane residence of Voltair and targets it to specific intracellular organelles. Thus, the DNA duplex binds scavenger receptors and undergoes scavenger receptor-mediated endocytosis, trafficking to early endosomes, which mature to late endosomes, and finally to lysosomes, in a time-dependent manner18. Voltair variants are assembled by annealing equimolar amounts of the component strands to yield probes with a precise 1:1 stoichiometry of the sensing dye (RVF) and reference dye (Atto647N). The formation and integrity of Voltair variants were confirmed by gel electrophoresis (Supplementary Fig. S3, Supplementary Note 1).
We characterized the voltage-sensitive response of VoltairPM using whole-cell voltage clamping (Methods and Supplementary Fig. S7). Both VoltairPM and VoltairIM report on membrane potential by exciting RVF (G) and Atto647N (R) and monitoring their emission intensities at 540 nm and 665 nm respectively (Supplementary Fig. S4). The plasma membrane of HEK293T cells was efficiently labeled by VoltairPM, as seen by colocalization with Cellmask™ (Supplementary Fig. S6). Single VoltairPM labeled cells were voltage clamped from −100 mV to +100 mV and fluorescence images of RVF (G channel) and Atto647N (R channel) were acquired at each clamped voltage, from which the pseudo-colored G/R images were generated (Fig 1c, Methods). The intensity in the G channel of voltage-clamped cells changed as a function of the applied voltage, while the R channel stayed constant (Supplementary Fig. S8, Movie S2). The uniform 1:1 ratio of RVF: Atto647N in VoltairPM yield highly reproducible G/R values as a function of applied voltage (Fig. 1d). The fold change in G/R signal from 0 mV to +100 mV (G/R) was ~1.2 for VoltairPM which matched well with plain RVF dye (Fig. 1d, inset) 21. Electrochromic voltage sensitive dyes like di-4-ANEPPDHQ, are also sensitive to lipid composition 24. VoltairPM however, was insensitive to lipid composition, consistent with its PeT-based mechanism of voltage sensing (Supplementary Fig. S9). The G/R value of VoltairPM in resting cells showed that resting potential was −50 mV, consistent with literature (Fig. 1e,f)25.
Targeting VoltairIM to membranes of specific endocytic organelles
We then targeted the variant, VoltairIM, to organelles on the endo-lysosomal pathway using scavenger receptor mediated endocytosis (Fig. 2a). DNA-based probes can label specific endocytic organelles in diverse cell types that express scavenger receptors (Fig S10). Since HEK 293T cells do not express scavenger receptors26, we over-expressed human macrophage scavenger receptor (hMSR1) fused to CFP in HEK293T cells by transient transfection. This led to effective VoltairIM internalization (Fig. 2b,c). Colocalization between hMSR1-CFP and VoltairIM as well as competition experiments revealed that VoltairIM uptake was mediated by scavenger receptors (Fig 2b,c and Supplementary Fig. S11)14.
Fig. 2. Targeting VoltairIM to membranes of specific endocytic organelles:
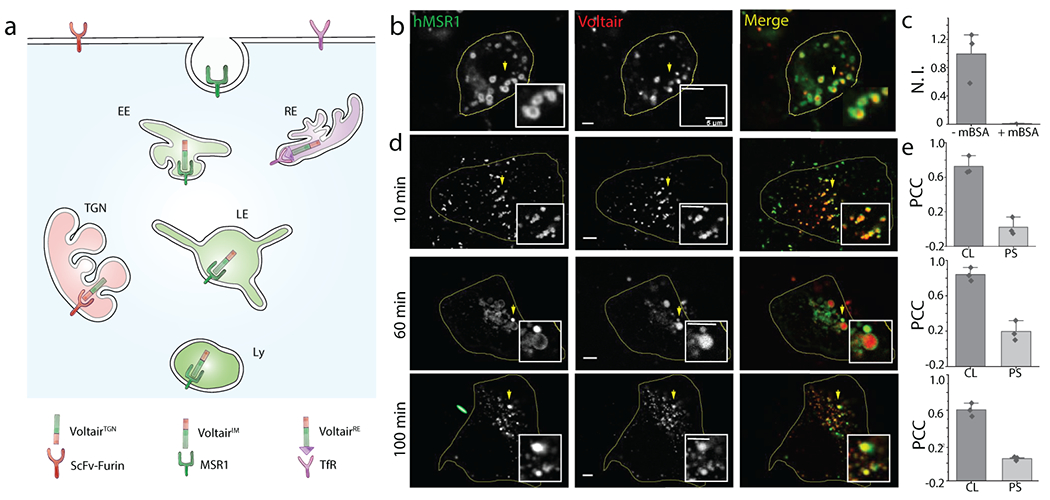
(a) Schematic of targeting strategy: VoltairIM undergoes scavenger receptor mediated endocytosis by binding scavenger receptors. Endocytosed VoltairIM traffics in a time-dependent manner from the plasma membrane to the early endosome, the late endosome and then the lysosome. (b) Representative colocalization between internalized VoltairIM (red channel) and human macrophage scavenger receptors (hMSR1-CFP, green channel) transfected in HEK 293T cells. (c) Normalized intensity of VoltairIM uptake in cells, in absence and presence of mBSA (30 eq.) Error bar represents mean ± s.e.m. of three independent trials, n =30 cells (d) Representative colocalization between various endocytic organelle markers (green channel) and VoltairIM (red channel) at the indicated chase times. Early endosomes are labelled with Transferrin-Alexa546 (Tf), late endosomes are labelled with Rab7-mRFP (Rab7) and lysosomes are labelled with TMR dextran (Dextrans). Scale bar = 5 μm. (e) Pearson’s correlation coefficient (PCC) of Voltair colocalization (CL) with corresponding endocytic markers in (d) along with pixel shift (PS). Error bar represents mean ± s.e.m. of three independent trials, n = 20 cells.
We determined the timepoints of localization of internalized VoltairIM devices at each stage of the endo-lysosomal pathway by time-dependent colocalization with various endocytic markers (Fig. 2d,e, Supplementary Fig. S12–S14, Supplementary Note 4) 27,28. We found that VoltairIM showed 70% colocalization in early endosomes at ~10 min, 85% colocalization in late endosomes at ~60 min and 60% colocalization in lysosomes at ~100 min. In VoltairIM, the RVF moiety acts both as a voltage sensitive dye and a lipid anchor that tethers VoltairIM to the lumenal face of the organelle membrane (Supplementary Fig. S15). The duplex DNA moiety in VoltairIM enables scavenger receptor binding while permitting RVF insertion into the membrane surrounding the receptor (Supplementary Fig. S16). Thus, integration onto a duplex DNA scaffold imposes the scavenger receptor-mediated endocytic program on to RVF (Supplementary Fig. S16b, S17).
To map the response of Voltair in intracellular membranes, we voltage-clamped VoltairIM-labeled lysosomes while simultaneously imaging VoltairIM ratiometrically. Treating COS-7 cells with vacuolin-1 swells up lysosomes to 1-3 μm (Methods, Supplementary Fig S18a)8. Such VoltairIM-labeled, enlarged lysosomes were isolated and voltage clamped from −100 mV to +100 mV (Methods, Supplementary Fig. S18b, Fig. 3a). Fluorescence images of RVF (G) and Atto647N (R) were acquired at different membrane potentials and an intra-organellar calibration plot of normalized G/R versus membrane potential was constructed (Fig. 3b,c). VoltairIM in lysosomes quantitatively recapitulated its voltage-sensing characteristics in the plasma membrane (Fig. 3c, inset). All further experiments used the intra-organellar calibration profile of VoltairIM to compute membrane potential.
Fig. 3. VoltairIM measures lysosomal membrane potential:
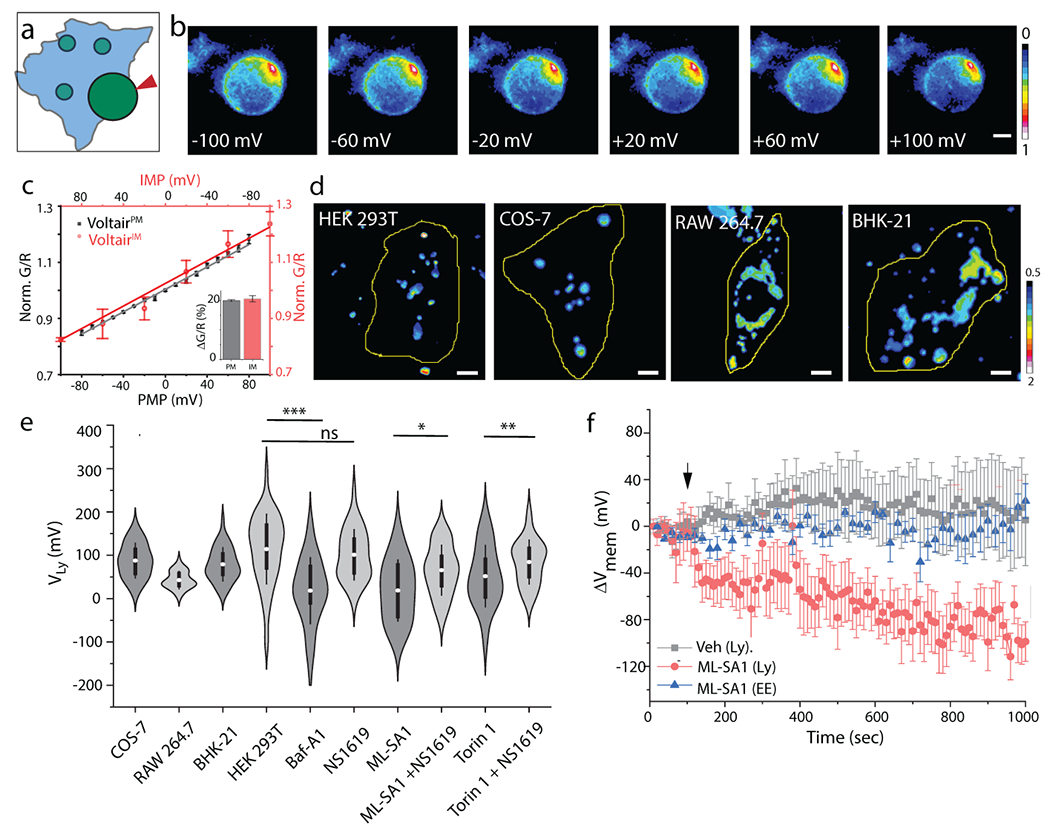
(a) Schematic showing voltage clamping of enlarged lysosome labeled with VoltairIM. Red arrowheads indicate patch pipette. (b) Pseudo-color images show pixel-wise G/R ratio of VoltairIM labelled on the inner leaflet of lysosomes as a function of membrane potential applied by whole lysosome voltage clamping. (c) Sensor response of VoltairPM (gray trace, square) and VoltairIM (red trace, circle) as a function of membrane potential (PMP: Plasma membrane potential, IMP: Intracellular membrane potential). Gray error bars indicate mean ± s.d., n = 6 cells. Red error bars indicate mean± s.d., n = 4 lysosomes. Inset: Percentage change in signal of VoltairPM (black) and VoltairIM (red) from 0 to 100 mV in (c). (d) Representative pseudocolor G/R images of the VoltairIM labeled lysosomes from different cell types. Scale bar = 5 μm. (e) Measured lysosomal resting membrane potential values (VLys) of COS-7, RAW 264.7, BHK-21 cells and HEK 293T values in presence of V-ATPase inhibitor Bafilomycin (Baf-A1, 500 nM), mTORC1 inhibitor Torin-1 (1 μM), the TRPML1 activator ML-SA1 (20 μM), and the Slo1 activator, NS1619 (15 μM). Violin plots show kernel smooth distribution, box indicates interquartile range 25-75; Error bars indicate mean ± s.d. of 100 lysosomes; ns, not significant (P > 0.05); **P < 0.01; *P < 0.05 (Unpaired two sample t-test). (f) Time-lapse plot of ΔVmem vs time upon TRPML1 activation in VoltairIM labeled lysosomes (red trace), in VoltairIM labeled Endosome (blue trace). DMSO (Veh.) treatment (black trace), Error bar represents mean ± s.d. from n = 10 cells. Experiments were repeated thrice independently with similar results.
In situ measurement of absolute lysosomal membrane potential
We first validated VoltairIM in lysosomes given the extensive prior art with electrophysiology7,8. Fluorescence images in the G and R channels of VoltairIM labeled lysosomes in HEK293T cells were converted to G/R images from which the G/R distribution could be obtained (Fig. 3d,e, Extended Data Fig. 1). Considering the lumen to be positive and the cytoplasmic face negative, the membrane potential of lysosomes (VLy) was found to be +114 mV. This is consistent with electrophysiology in different cell types9. Values of VLy in different cell lines showed slightly lower overall values of +80 mV (lumen positive) in COS-7 and BHK-21 compared to HEK 293T cells (Extended Data Figure 1). Lysosomes in RAW macrophages were even more depolarized with VLy of +40 mV (lumen positive), also consistent with literature (Fig. 3d,e)29. The variation in VLy in different cell types could arise from the differential lysosomal activity across these cell types.
VoltairIM reports absolute membrane potential up to 1h after it reaches lysosomes in HEK293T cells (Supplementary Fig. S20, Supplementary Note 5). We therefore checked whether VoltairIM could report on changes in VLy upon treatment with pharmacological modulators of lysosomal pumps, channels and transporters. The proton pump V-ATPase acidifies organelles by hydrolyzing ATP and generating membrane potential across the organelle membrane29. Inhibiting V-ATPase with bafilomycin A1 stops proton transport, which neutralizes membrane potential in purified, highly acidic synaptic vesicles 30. We found that VoltairIM labeled lysosomes showed a large increase in G/R values upon bafilomycin A1 treatment revealing that VLys was dissipated upon V-ATPase inhibition (Supplementary Fig. S21). VLys was reduced by ~100 mV from the resting value, as also seen in other V-ATPase-regulated organelles such as synaptic vesicles (Fig. 3c–e, Supplementary Fig. S21, Supplementary Note 6)30.
Next, we modulated specific lysosomal ion channels. While on the lysosome, mTORC1 inhibits both TPC2 and TRPML1 channels (Supplementary Fig. S22A) 31,32. When nutrient levels plummet, mTORC1 dissociates from the lysosome, relieving the inhibition on TPC2 and TRPML1 channels. This triggers lysosomal Ca2+ release and is expected to reduce VLys. Low VLys is expected to activate the voltage dependent channel Slo1, which would promote K+ influx into the lysosome and activate TRPML17. Cytosolic Ca2+ elevation caused by TRPML1 channel opening is expected to feedback positively to release lysosomal Ca2+ and drive further K+ influx 7.
Treating HEK 293T cells with an activator of TRPML1 (ML-SA1) gave a ΔVLys of 90 mV (Fig. 3e, Supplementary Fig S22b, d, Supplementary Movie S3). This decrease in membrane potential was specific to the lysosome, because time-lapse imaging of membrane potential in early endosomes (VEE) showed no change upon ML-SA1 treatment (Fig. 3f, Supplementary Fig. S23, Supplementary Movie S4). Treatment with ML-SA1 and the BK channel agonist, NS1619 gave ΔVLys of 50 mV (Fig. 3e, Supplementary Fig. S22d). These values agree well with electrophysiology4,7. Inhibiting mTORC1 with Torin-1 reduced VLys, by nearly 60 mV presumably due to TPC2 and TRPML1 channel activation (Supplementary Fig. S22b,c). Treatment with Torin-1 and a TPC2 channel inhibitor (trans-ned-19), gave a ΔVLys of 35 mV due to TPC2 channel inhibition (Supplementary Fig. S22b,c) 33. This agrees closely with electrophysiology on isolated lysosomes where TPC2 channel inhibition gave ΔVLys of 20 mV 31.
Membrane potential of organelles along recycling and retrograde trafficking pathways
We then sought to measure the membrane potential as a function of endosomal maturation. Although the membrane potential of isolated lysosomes is known, no prior values are known for early or late endosomes. We therefore labeled early endosomes, late endosomes and lysosomes specifically with VoltairIM as described (Fig 2d, Supplementary Fig. S24). The G/R values of ~200 organelles were computed for each endosomal stage from which the membrane potential was determined (Fig. 4a,b). The membrane potential of early endosomes (VEE) and late endosomes (VLE) was found to be +153 mV and +46 mV respectively (Supplementary Table S2). We observed a spread in values of membrane potential in endocytic organelles (Fig. 4b). At least for endo-lysosomes, we know that they are heterogenous and comprise subpopulations that enclose different ionic concentrations14. This could in part explain the observed spread in organellar membrane potential.
Fig. 4. Membrane potential of organelles along endocytic, recycling and retrograde trafficking pathway:
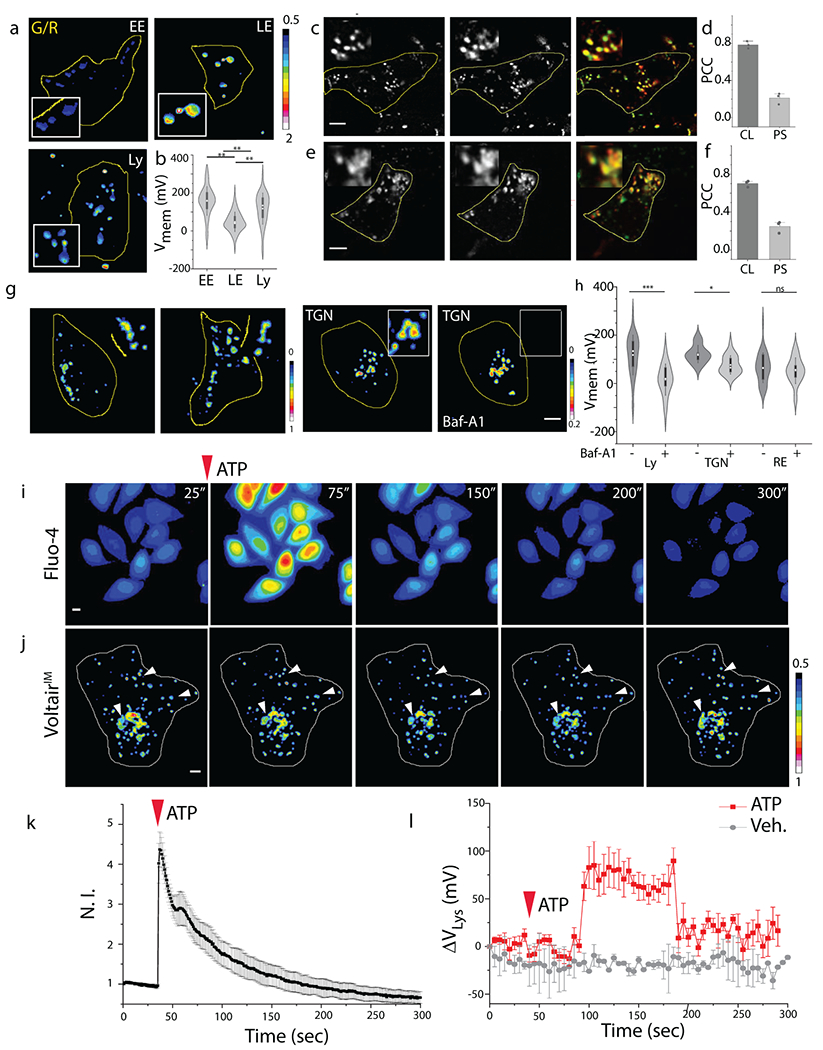
(a) Representative pseudocolor G/R images of the indicated organelles labeled with VoltairIM. Scale: 5 μm. (b) Measured membrane potentials of early endosomes (EE), late endosomes (LE) and lysosomes (Ly). Violin plots show kernel smooth distribution, box indicates interquartile range 25-75; Error bars indicate mean ± s.d. of 200 vesicles; **P < 0.01 (one-way ANOVA with Tukey post hoc test). (c) Colocalization between internalized VoltairRE (red channel) and recycling endosome marker (Transferrin-Alexa546, green channel) in HEK 293T cells. (d) Pearson’s correlation coefficient (PCC) of colocalization (CL) and pixel shift (PS) in (c). Error bar represents mean ± s.e.m. of three independent trials, n = 20 cells. (e) Colocalization between internalized VoltairTGN (red channel) and trans-Golgi network marker (TGN46-mCherry, green channel) in ScFv-Furin transfected HEK 293T cells. Scale = 5 μm, inset = 5 μm. (f) Pearson’s correlation coefficient (PCC) of colocalization (CL) and pixel shift (PS) in (e). Error bar represents mean ± s.e.m. of three independent trials, n = 20 cells. (g) Representative pseudo-color G/R images of RE and TGN of HEK 293T cells in absence and presence of bafilomycin A1 (500 nM). Scale = 5 μm. (h) Violin plot of resting membrane potential of organelles and changes upon inhibition of V-ATPase. Box indicates interquartile range 25-75; Error bars indicate mean ± s.d; ns, not significant (P > 0.05); ***P < 0.001, *P < 0.05 (one-way ANOVA with Tukey post hoc test). (i) Representative pseudo-color time lapse images of ATP induced cytosolic calcium increase imaged using Fluo-4 AM dye. Scale = 10 μm. (j) Representative pseudo-color time lapse images of G/R ratio from VoltairIM in presence of 100 μM ATP. White arrowheads highlight the lysosomes undergoing ATP induced hyperpolarization. (k) Quantification of (i) as a fluorescent intensity vs time plot, error bar represents standard deviation from n = 10 cells; Scale = 10 μm. (l) Quantification of (j) as a normalized G/R ratio from whole cell vs time plot, 100 μM ATP treated (Black trace), Untreated (red trace). Decrease in G/R (observed in movie) represents increase in positive membrane potential. Error bar represents standard deviation from n = 6 cells; Scale = 10 μm. Experiments were repeated thrice independently with similar results.
Surprisingly the gradient of membrane potential accompanying endosomal maturation did not reflect that of ion gradients (Supplementary Fig. S25). Protons, chloride and calcium levels increase progressively during endosomal maturation (Supplementary Fig. S25). In contrast, membrane potential is highest in the early endosome, drops ~3 fold in the late endosome and increases again in lysosomes. With respect to cytoplasmic concentrations, the levels of lumenal [Ca2+] and [Cl−] for the early endosome or the lysosome, are similar i.e., ~102-103 fold higher [Ca2+], 1-2-fold higher [Cl−] (Supplementary Fig. S25)34–36. However, the lysosome lumen has ~103 fold higher [H+], while the early endosome lumen has only ~10 fold higher [H+] than the cytosol37 . Thus, the membrane potential for the early endosome should be much lower than what we have observed (VEE = +153 mV; VLY = +114 mV). Considering the low abundances of other free ions, this provides a compelling case for Na+ and K+ transporters or exchangers in maintaining the high positive membrane potential of the early endosome. In fact, late endosomes are posited to have high K+, which is consistent with our observations38. Our hypothesis is supported by observations at the plasma membrane. The differences in [Ca2+] and [Cl−] across the plasma membrane are comparable, yet its membrane potential is set by the differences in [Na+] and [K+] across the membrane39.
We re-designed Voltair to give VoltairRE and VoltairTGN to measure the membrane potential of recycling endosomes and the trans-Golgi network. Both organelles have been hypothesized to have negligible membrane potential40. VoltairRE, displays an RNA aptamer against the human transferrin receptor (Supplementary Fig. S26), was uptaken by the transferrin receptor pathway and trafficked to recycling endosomes as evidenced by 80% colocalization with Alexa 546 labelled transferrin (Fig. 4c,d)18. VoltairTGN was targeted to the trans-Golgi network in HEK 293 cells expressing furin fused to a single-chain variable fragment recombinant antibody, scFv 19. The scFv domain binds d(AT)4 sequence that is included in VoltairTGN. Thus, VoltairTGN is trafficked retrogradely to the trans Golgi network evidenced by 70% colocalization with TGN46-mCherry and no colocalization with endocytic organelles (Fig. 4e,f, Supplementary Fig. S27).
We then measured the resting membrane potential of the recycling endosome (VRE) and trans-Golgi network (VTGN), neither of which has been previously possible, and evaluated the contribution of V-ATPase to membrane potential in each organelle. We found that VRE was +65 mV (lumen positive), similar to plasma membrane (cytosol negative). When V-ATPase activity was inhibited with bafilomycin A1, VRE showed no change revealing a negligible contribution of V-ATPase to the membrane potential41. The magnitude and the V-ATPase dependence of VRE mirrors that of the plasma membrane, indicating that both membranes share similar electrical characteristics.
VoltairTGN labelled cells were similarly imaged and G/R values of the TGN in ~30 cells revealed that VTGN was +121 mV (lumen positive) (Fig 4g,h). Such high membrane potential is surprising as the Golgi has been posited by others to have negligible membrane potential40. In addition, V-ATPase inhibition substantially reduced VTGN. However, unlike in lysosomes, the membrane potential of the TGN could not be completely neutralized and still showed +75 mV across the membrane. This suggests that other electrogenic transporters at the TGN, possibly Na+/K+ ATPases could significantly contribute to the VTGN42. It also reveals that different organelle membranes have distinct electrochemical behaviors.
Time lapse imaging of membrane potential changes in lysosomes:
Finally, to test whether Voltair could provide temporal information, we mapped VLY of large numbers of intact lysosomes in situ in response to acute pharmacological triggers. Cytosolic Ca2+ levels are stringently regulated by various organelles such as endoplasmic reticulum, mitochondria and plasma membrane 43. Lysosomes are also hypothesized to buffer cytosolic Ca2+, which, when elevated is expected to perturb the electrochemical homeostasis of lysosomes44. We therefore elevated cytosolic Ca2+ using ATP and measured VLY along with cytosolic Ca2+ levels in COS-7 cells. Extracellular ATP elevates cytosolic Ca2+ by interacting with P2 purinergic receptors on the plasma membrane, generating Ins(1,4,5)P3 to activate IP3 receptors on the endoplasmic reticulum which releases Ca2+ 45.
We monitored cytosolic Ca2+ and VLY as a function of time by continuously imaging both Fluo-4 and VoltairIM labeled cells from whole cell intensity images (Fig. 4i,j). Cytosolic Ca2+ peaked ~5 seconds after ATP addition (Fig 4k, Supplementary Movie S5). Interestingly, ~80 seconds after cytosolic Ca2+ peaked, we observed an acute hyperpolarization of the lysosomal membrane by +75 mV. This hyperpolarization lasted for ~100 seconds, and then resting VLY was restored (Fig 4j,l, Supplementary Movie S6). Notably, VLY restoration coincided with cytosolic Ca2+ restoration. The correlation between the cytosolic Ca2+ surge and the abrupt lysosomal hyperpolarization suggests the activation of lysosome-resident transporters that contribute to restoring cytosolic Ca2+, along with other Ca2+ buffering organelles of the cell. For example, the activity of a lysosomal Ca2+/H+ exchanger (CAX) could elevate VLY because it would accumulate positive charge within the lysosome due to dicationic Ca2+ influx versus monocationic H+ efflux46. Membrane potential variations are ~103-fold faster than Ca2+ transients and the small size of lysosomes compared to the plasma membrane are possible reasons for the sharp changes in VLY.
Conclusion:
Our studies provide evidence for either Na+ or K+ in establishing the high membrane potential of the early endosome. The recycling endosomal membrane and plasma membrane show very similar electrochemical characteristics. The membrane potential of the trans Golgi network, previously hypothesized to be negligible, is actually as high as the lysosome40. Yet, unlike the lysosome, it is not completely driven by V-ATPase. Non-invasively interrogating membrane potential in organelles that have proved previously impossible to address, offers the capacity to uncover how membrane potential regulates functionality.
Protein-based voltage indicators while powerful, have limited applicability to acidic organelles, as they are pH sensitive and cannot yet provide absolute membrane potential. Voltage sensitive dyes are attractive due to their low capacitive loads and pH insensitivity, but, on their own, cannot be targeted to organelles. Voltair unites the advantages of voltage sensitive dyes with the organelle-targetability of proteins to non-invasively measure the membrane potential of organelles.
A key requirement for voltage reporters is robust membrane association. Genetically encoded voltage reporters access the membrane of interest from the cytosolic face, while voltage-sensitive dyes such as RVF bind the outer leaflet of the plasma membrane12,47. Voltair probes bind the lumenal leaflet of the organelle. Note that the vector direction of membrane potential as measured by Voltair in organelles is inverted with respect to lysosomal electrophysiology7,8. We purposely keep our sign convention consistent that of with electrophysiology, to also convey the inverted topology of organelle membranes and plasma membrane.
Currently, Voltair can probe only endocytic organelles and the trans Golgi network (TGN). Although one can potentially target synaptic vesicles and secretory vesicles, their small size may limit probe abundance and therefore require more sophisticated imaging48. To apply Voltair to organelles on the secretory pathway, mitochondria, nucleus and peroxisomes, new knowledge of proteins that retrogradely traffic from the plasma membrane to the organelle is required49.
While Voltair is not two-photon compatible, it can be redesigned to display two-photon compatible dyes for deep tissue imaging. Unlike genetically encoded voltage reporters, Voltair cannot yet be targeted tissue specifically. Thus, mechanisms to target DNA nanodevices to excitable tissues would significantly expand its applicability. However, DNA nanodevices are preferentially internalized by immune cells in vivo; hence one can readily image organelles in transparent systems such as coelomocytes in nematodes13,36 or microglia in zebrafish brains 16.
When an organelle membrane breaks, the membrane potential is expected to fall to zero. Thus, Voltair could also act as a real-time reporter of organelle damage and membrane repair. As Voltair is a quantitative reporter at the biotic-abiotic interface, it can be used to rationally guide the design of biocompatible electronics50. Knowledge of absolute membrane potential will enable us to model cellular responses to electrical stimuli at the level of organelles.
Online Methods:
Reagents
Modified oligonucleotides (Supplementary Table S1) were purchased from IDT (USA), subjected to ethanol precipitation and quantified using UV absorbance. CellMask™ reagents and TMR-Dextran were purchased from molecular probes/Life Technologies (USA). Maleylated BSA (mBSA) and fluorescent transferrin (Tf-Alexa546) were conjugated according to previously published protocols18,37,51. Pharmacological Lysosomal modulators were purchased from Cayman Chemical (USA). Phenyl triflimide, N-Boc-piperazine, Azido-PEG4-NHS and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine lipid were purchased from TCI America (USA), Oakwood chemicals (USA), Click Chemistry tools (USA) and Avanti lipids (USA) respectively. All other reagents were purchased from Sigma-Aldrich (USA) unless otherwise specified.
Synthesis of RVF
Synthesis scheme is shown in Supplementary Fig. S1. Compound 1a (0.2 g, 0.36 mmol)11 was suspended in N,N-dimethylformamide (DMF) (1.4 mL) and cooled to 0°C . Triethylamine (2.7 mL) and Phenyl triflimide (0.25 g, 0.72 mmol, 2 eq.) were added dropwise and the reaction was stirred at room temperature for 2 hours. The reaction mixture was then diluted in water and extracted with dichloromethane (DCM) twice. Organic extracts were washed three times with brine and 1M HCl. The product was concentrated via rotor evaporation and diluted in dry DMSO (2mL). N-boc piperazine (3.72g, 20 mmol, 100eq.) was added and the reaction mixture was kept at 100°C overnight. The reaction mixture was then diluted in water, extracted in DCM twice then washed with brine twice. The mixture was dried over anhydrous Na2SO4 and concentrated under reduced pressure. Silica gel column chromatography was performed in 5% methanol in chloroform to get 1b (0.08g, 28%). ESI-MS (-) Expected mass = 728.98, found = 729.0.
A reaction tube was charged with 1b (70 mg, 0.0956 mmol), Pd(OAc)2 (6.87 mg, 0.0306 mmol, 0.32 eq.), tri-o-tolylphosphine (20.4 mg, 0.067 mmol, 0.7 eq.) and (E)-N,N-dimethyl-4-(4-vinylstyryl)aniline (26.23mg, 1.052 mmol, 1.1 eq.) which was previously synthesized according to literature 11. The tube was evacuated and backfilled with N2 three times. 1 mL of dry DMF and 500 μL dry triethylamine were added via syringe, and the reaction was stirred at 110°C overnight. The reaction mixture was diluted in water and extracted with DCM twice. The organic extract was washed with brine twice and concentrated under reduce pressure. Silica gel column chromatography was performed in 5% methanol in DCM to get orangish brown solid 1c (20 mg, 25%). ESI-MS (+), Expected mass = 851.29, found = 851.2.
A reaction vial with 1c (5 mg, 6.7 nmol) was placed in 5% TFA in DCM overnight for deprotection. TFA was removed under reduced pressure and Azido-PEG4-NHS ester (26mg, 6.7 nmol, 10.0 eq.), 300 μL dry DMF, and 200 μL triethylamine were added. The mixture was stirred for 4 hours at room temperature. The reaction mixture was then diluted in water, extracted to DCM twice, washed with brine three times, dried over anhydrous Na2SO4 and concentrated under reduced pressure. Silica gel column chromatography was performed with 2% methanol in DCM, slowly increasing the gradient to 10%, to get reddish brown solid 1d (2mg, 30%). ESI- MS(+), Expected mass = 1025.23, found = 1025.3.
Sample preparation
Sensing domain (DV, DVRE):
3’- DBCO modified 38 base strand (10 μM) was coupled to the azide containing RVF (50 μM, 5 eq.) in 20 mM sodium phosphate buffer, pH 7.4, and incubated at RT for 4 hrs35,52,53. Upon completion, unconjugated fluorophores were removed by ethanol precipitation54.
Targeting domain (DT):
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) was conjugated to NHS-PEG4-Azide using an established protocol 22. 20 μM of the 5’-DBCO modified 22 base strand (IDT, USA) was coupled to azido-POPE (40 μM, 2eq.) in 20 mM sodium phosphate buffer, pH 7.4 and stirred for 4 hours at RT (Supplementary Fig. S2).
Construction of VoltairPM, VoltairIM and VoltairRE:
Stock solution of VoltairPM was prepared at a final concentration of 10 μM by mixing DV, DT and DA (Atto647N – 5’ modified strand) at an equimolar ratio in 10 mM sodium phosphate buffer, pH 7.4 (Supplementary Fig. S2, S3). For VoltairIM samples, DV (Voltage sensing strand of VoltairPM) and DA’ (Atto647N – 3’ modified 38 mer strand) were mixed at an equimolar ratio with a final concentration of 10 μM (Supplementary Table S2). For VoltairRE samples, 10 μM of DVRE, DTf and DARE were mixed at an equimolar ratio. For all samples, annealing and gel characterization were done according to previous established protocol35,36.
In vitro spectroscopic measurements:
Fluorescence spectra were recorded on a FluoroMax-4 scanning Spectro-fluorometer (Horiba Scientific, Edison, NJ, USA). For recording spectra, 100 nM VoltairIM in UB4 buffer (20 mM HEPES, MES and sodium acetate, 150 mM KCl, 5 mM NaCl, 1 mM CaCl2 and MgCl2) of desired pH, were excited at 520 nm and 650 nm and emission spectra were collected between 525 – 600 nm and 655-750 nm respectively (Supplementary Fig. S4).
Cell culture, plasmids and transfection:
Human embryonic kidney cells (HEK 293T), BHK-21 cells, human dermal fibroblasts (HDF), RAW 264.7, THP-1 and T-47D cells were a kind gift from Dr. Bryan Dickinson ( University of Chicago), Dr. M. Gack (University of Chicago), J. Rowley’s lab (University of Chicago), Dr. Christine A. Petersen (University of Iowa), Dr. D. Nelson (University of Chicago) and G. Greene (University of Chicago), respectively. COS-7 cells were purchased from ATCC. Cell lines were purchased from ATCC prior to receiving them as gifts and were authenticated by short tandem repeats (STR). All cell lines were checked for mycoplasma contamination using Hoechst-33342 staining. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen Corporation, USA) containing 10% heat inactivated Fetal Bovine Serum (FBS) (Invitrogen Corporation, USA), 100 U/mL penicillin and 100 μg/mL streptomycin and maintained at 37°C under 5% CO2. HEK 293T cells were passaged and plated at a confluency of 20 – 30% for electrophysiology experiments, and 50 – 70% for transfection and intracellular measurements.
The hMSR1 sequence was cloned into the PCS2NXE vector (4,103 bp) containing the CMV promoter for overexpression in mammalian cell lines. The hMSR1-CFP plasmid (5973 bp) was constructed by cloning hMSR1 sequence into pECFP-C1 plasmid (4731 bp). The identity of each construct was confirmed by sequencing, using forward primer (5’ to 3’) GGGACATGGGAATGCAATAG and reverse primer (5’ to 3’) CTCAAGGTCTGAGAATGTTCCC.
The mCherry-TGNP-N-10 was a gift from Michael Davidson (Addgene plasmid #55145) and Rab7-RFP was a gift from Ari Helenius (Addgene plasmid #14436, 55). Construction of scFv-furin construct is reported previously 19. HEK 293T cells were transiently transfected with respective plasmids using TransIT®-293 transfection reagent (MIRUS). After a 4-hour incubation the transfected medium was replaced with fresh medium. Labeling experiments were performed on cells 48 hours post transfection.
Electrophysiology:
A schematic of the electrophysiology equipment used for whole cell patch clamp recording is shown in Supplementary Fig. S7. Recordings were performed with an Axopatch 200A amplifier (Molecular Devices), digitized using an NI-6251 DAQ (National Instruments). The amplifier and digitizer were controlled using WinWCP software (Strathclyde Electrophysiology Software). Borosilicate glass capillaries (Sutter) of dimension 1.5 mm x 0.86 mm (OD/ID) were pulled using a Sutter P-97 Micropipette puller (program: Heat – Ramp, Pull – 0, Vel – 21, Time – 1(Delay), Loops – 5). Patch pipettes with resistances between 5-10 MOhm were used in voltage clamping experiments. The patch pipette was positioned using an MP325 motorized manipulator (Sutter). Image Acquisition software Metamorph premier Ver. 7.8.12.0 was linked to an NI-6501 DAQ to enable voltage triggered image acquisition. For all measurements the extracellular solution composition was (in mM) 145 NaCl, 20 glucose, 10 HEPES, pH 7.4, 3 KCl, 2 CaCl2, 1 MgCl2 (310 mOsm) and the intracellular solution composition was (in mM) 115 potassium gluconate, 10 EGTA, 10 HEPES, pH 7.2, 5 NaCl, 10 KCl, 2 ATP disodium salt, 0.3 GTP trisodium salt (290 mOsm).
For plasma membrane voltage clamping experiments, 1 μM RVF or 500 nM VoltairPM was incubated with HEK 293T cells for 30 mins in Hank’s Balanced Salt Solution (Thermofisher) at RT. Labelled cells were washed three times with PBS and incubated in extracellular solution for whole cell voltage clamping. Whole cell voltage clamping was performed according to an established protocol 56. For background subtraction, bleaching correction and lamp fluctuation compensation, imaging field was chosen with at least one more HEK 293T cell that is not clamped. Once clamped, membrane potential is changed from −100 to +100 mV in 10 mV increments at 1000 ms intervals. Around 200 ms after the voltage is changed three images are taken in quick succession. Voltage clamp experiments were also performed with extracellular solutions of different pH, to study the effect of pH on voltage sensitivity of RVF (Supplementary Fig. S5).
Endo-lysosomal electrophysiology:
COS-7 cells were treated with 1 μM vacuolin-1 overnight to increase the size of lysosomes to 1-3 μm 57. Enlarged lysosomes of COS-7 cells were labeled with 500 nM VoltairIM, by 30 mins pulse in HBSS followed by 2 hr. chase in complete media containing 1 μM vacuolin-1 (Supplementary Fig S18). Enlarged endolysosome containing VoltairIM was pushed out of the ruptured cell (ref) (Supplementary Fig. S16). Borosilicate glass capillaries (Sutter) of dimension 1.5 mm x 0.86 mm (OD/ID) were pulled using the program: Heat – 520, Pull – 0, Vel – 20, Time – 200, Loops – 4. Fire polished patch pipettes with resistances between 15-20 MOhm were used in voltage clamping experiments. After giga-ohm seal formation, break in was performed by a zap protocol (5V: 0.5-5s) till appearance of capacitance transients. In order to minimize fluorescence interference from other lysosomes, patched lysosome is moved away from the cell prior to imaging. For all measurements the cytoplasmic solution composition was (in mM) 140 K-gluconate, 4 NaCl, 1 EGTA, 20 HEPES, pH 7.2, 0.39 CaCl2, 2 MgCl2, 2 ATP disodium salt, 0.3 mM GTP and the composition of pipette solution was 145 NaCl, 2 CaCl2, 1 MgCl2, 10 HEPES, 10 MES pH 4.6, 10 glucose, 5 KCl. The voltage clamping and imaging protocol was followed as discussed in electrophysiology section above.
Microscopy:
Wide field microscopy was carried out on an IX83 inverted microscope (Olympus Corporation of the Americas, Center Valley, PA, USA) using either a 100X or 60X, 1.4 NA, DIC oil immersion objective (PLAPON) and Evolve Delta 512 EMCCD camera (Photometrics, USA) and controlled using Metamorph premier Ver 7.8.12.0 (Molecular Devices, LLC, USA). Images were acquired with exposure 100 ms and EM gain at 100 for Atto647N, exposure 200 ms and EM gain 300 for RVF. RVF channel images were obtained using 500/20 band pass excitation filter, 535/30 band pas emission filter and 89016 dichroic. For Atto647N, images were obtained using the 640/30 band pass excitation filter, 705/72 band pass emission filter and 89016 dichroic. For cytosolic calcium recording, fluorescence of Fluo-4 was recorded by exciting at 480 nm and collecting emission at 520 nm. Images were acquired using a 480/20 band pass excitation filter, 520/40 band pass emission filter and 89016 dichroic.
Intracellular membrane potential measurements were performed by recording images in the sensing channel (G) and the normalizing channel (R) in cells where each specific compartment was labeled. After acquisition of G and R images, intracellular membrane potential was neutralized (~ 0 mV) by adding 50 μM valinomycin and monensin in high K+ buffer, for 20 mins at room temperature. A set of G and R images of same cells were acquired after valinomycin and monensin or pharmacological treatments as shown in Supplementary Fig. S17. These images of neutralized endosomes were used as a baseline measurement to correct for variations in autofluorescence. To record all endo-lysosomal compartments in the cell, Z-stacks (30 planes, Z distance = 0.8 μm) were captured and a maximum intensity projection was used to produce a single image for analysis.
Confocal images were captured with Leica TCS SP5 II STED laser scanning confocal microscope (Leica Microsystems, Inc. Buffalo Grove, IL, USA) equipped with a 63X, 1.4 NA, Oil immersion objective. RVF was excited using an argon laser with 514 nm wavelength, CFP by 458 nm and Atto647N using a He-Ne laser with 633 nm wavelength. CellMask orange stain was excited by 543 nm and all emissions were filtered using Acousto Optical Beam Splitter (AOBS) with settings suitable for each fluorophore and recorded using hybrid detectors (HyD).
Time lapse Imaging:
Lysosomes of COS-7 cells labeled with VoltairIM (Supplementary Fig. S20b,c) were sequentially imaged on a widefield microscope with 60X magnification in G and R channel at 10-20s interval for a duration of 20 mins. Image acquisition was briefly paused at indicated time for addition of indicated chemical to cells. To minimize bleaching, time-lapse imaging was done at single focal plane. Z-drift compensator module from IX83 was applied to minimize out of focus drift during time lapse imaging. The extracellular ATP induced cytosolic calcium increase was imaged by labeling COS-7 cells with cell permeable calcium sensitive dye Fluo4-AM ester 45. Time-lapse imaging of Fluo-4 labeled COS-7 cells were acquired at 1s interval for 20 mins.
Competition assay:
Maleylated BSA based competition assay was performed according to previously established protocols37.
Co-localization and labeling experiments:
To find out the trafficking time of DNA device in specifically labeling EE, LE or Ly of HEK 293T cells transfected with hMSR1, time dependent colocalization experiments were performed as reported previously19,58. Fluorescent Transferrin was used to specifically label EE by pulsing it for 10 minute prior to imaging and to label RE with an additional chase time of 30 mins (Supplementary Fig S26) 28. Rab7-RFP is a well-established late endosome (LE) marker, and was transiently expressed in HEK 293T cells to label LE 27. Transient expression of TGN46-mCherry specifically labels trans Golgi network (TGN) 59. Finally, TMR-Dextran a specific marker for lysosome (Ly) after previously reported chase times, was pulsed for 1 hour, followed by 16 hours chase in complete media to label lysosomes 27. The trafficking time of DNA device with transferrin aptamer and d(AT)4 tag in labeling RE and TGN, respectively, have been established previously 18,19,60. Briefly, recycling endosomes are targeted by pulsing VoltairRE in 1X HBSS, for 10 mins at 37°C, followed by 30 mins of chase in complete media at 37°C. Trans Golgi network is targeted by pulsing VoltairIM to ScFv-Furin transfect HEK 293T cells, in complete media containing cycloheximide (CHX) for 90 mins at 37°C, followed by 90 mins chase.
Pharmacological drug treatments:
VoltairIM labelled cells were treated with ML-SA1 (20 μM), NS1619 (15 μM) or trans-ned-19 (1 μM) for 15 mins in HBSS solution at room temperature. Torin-1 (1 μM) was treated to labelled cells during the chase period (50 mins), to inhibit mTOR. Bafilomycin-A1 (500 nM) was added to cells for 30 mins in 1X HBSS and incubated at 37°C prior organellar voltage measurements. After acquisition of G and R images of drug treated cells, intracellular membrane potential was neutralized (~ 0 mV) by adding 50 μM valinomycin and monensin in high K+ buffer, for 20 mins at room temperature (Extended Data Figure 1).
Image analysis:
Images were analyzed with Fiji (NIH, USA). For organellar voltage measurements, regions of cells containing single isolated endosomes/lysosomes in each Atto647N (R) image were manually selected and the coordinates saved in the ROI plugin. Similarly, for background computation, a nearby region outside endosomes/lysosomes were manually selected and saved as an ROI. The same regions were selected in the RVF (G) image by recalling the ROIs. After background subtraction, mean intensity for each endosome (G and R) was measured and exported to OriginPro (OriginLab, USA). A ratio of G to R intensities (G/R) was obtained from these values by dividing the mean intensity of a given endosome in the G image with the corresponding intensity in the R image. To minimize the measurement error due to low fold change of Voltair probes, the same endosomes or lysosomes are measured post addition of valinomycin and monensin which neutralizes the membrane potential in presence of 150 mM KCl. For TGN voltage measurements, total cell intensity was recorded and background subtraction was performed by manually selecting a region outside the cell. For a given experiment, membrane potential of an organelle population was determined by converting the mean [G/R]V - [G/R]O value of the distribution to voltage values according to intracellular voltage calibration profile (Fig. 3c). The mean value of each organelle population across three trials on different days is determined and the final data is presented as mean ± S.E.M. Representative images are shown in pseudo-color images, where G and R images were modified by thresholding in ImageJ to get G’ and R’ images. Using ImageJ’s Image calculator module, G’ images were divided by R’ images to generate an image where each pixel represents [G/R]V.
For whole cell patch clamp, image analysis was performed using custom Matlab code. A series of images corresponding to a voltage sweep from −100 to +100 mV was collected and input into the program. By identifying changes in intensity from the first to last image a region of interest corresponding to the clamped cell was selected. Other cells present in the image were also selected based on an intensity threshold. The region containing no cells was used to subtract background noise from the detector from all regions. Intensity from unclamped cells was measured in each image and used to correct for photobleaching or fluctuation in lamp intensity. After background corrections the average intensity of the patch clamped cell was then measured for each individual image in the series and normalized against the value at −60 mV.
Plasma membrane Cholesterol modulation:
HeLa cells were incubated with either 5mM Methyl-β-cyclodextrin (MβCD) in HBSS alone or 4.5mM MβCD complexed with 0.5mM cholesterol for 1 h. The former treatment depletes cholesterol levels in the plasma membrane while the latter treatment increases cholesterol levels in the plasma membrane 61. HeLa cells treated thus, were then labeled with VoltairPM in HBSS for 20 mins and voltage clamped from −100 to +100 mV and simultaneously imaged in the red and green channels.
Statistical analysis
For statistical analysis between two samples, two-sample two tailed test assuming unequal variance were used. For comparison of multiple samples, one-way ANOVA with a post hoc Tukey test or Fischer test was used. All statistical analysis was performed in Origin (Student version). Violin plots show the Kernel smooth distribution of data points and embedded box plots indicate 25-75% percentile, with median shown as white circle and error bars represents standard deviation.
Extended Data
Extended Data Fig. 1.
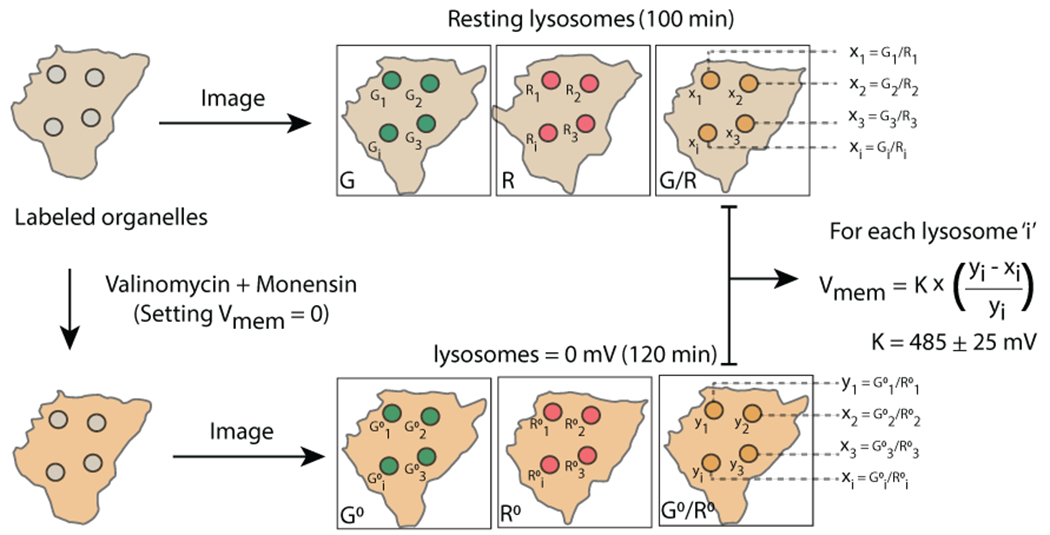
Schematic of Lysosomal Vmem measurement, Imaging protocol of organelles labeled with VoltairIM. resting organelles are imaged in G and R channels, neutralized with valinomycin-monensin and second set of images are acquired in G and R channels. Resting membrane potential of the organelle is calculated by normalizing G/R values from untreated lysosomes (xi) to neutralized lysosomes (yi).
Supplementary Material
Acknowledgments:
We thank Professors John Kuriyan, Jack W. Szostak, Haoxing Xu and Francisco Bezanilla for critical comments; Kasturi Chakraborty, Xiaoli Zhang, Joao L. C. Souza, Jary Delgado and Yuanwen Jiang for discussions; the Integrated Light Microscopy and mass spectrometry facilities at the University of Chicago.
Funding: This work was supported by Women’s Board of the University of Chicago; FA9550-19-0003 from AFOSR, NIH grants R21NS114428, 1R01NS112139-01A1, Pilot and Feasibility award from an NIH-NIDDK center grant P30DK42086 to University of Chicago’s Digestive Diseases Research Center; Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust, C-084. M.S. was a Heisenberg fellow supported by the Deutsche Forschungsgemeinschaft.
Footnotes
Competing interests: Authors declare no competing interests.
Data availability: The data that support the plots within this paper and other finding of this study are available from the corresponding author upon reasonable request.
Code availability: The MATLAB code written for voltage clamping experiments are available from the corresponding author upon reasonable request.
Additional Information: Supplementary information is available in the online version of the paper. Reprints and permission information is available online at www.nature.com/reprints. Correspondence and requests for materials should be addressed to Y. Krishnan.
References:
- 1.Alberts B et al. Ion Channels and the Electrical Properties of Membranes. (2002). [Google Scholar]
- 2.Mousavi SAR, Chauvin A, Pascaud F, Kellenberger S & Farmer EE GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Grabe M & Oster G Regulation of organelle acidity. J Gen Physiol 117, 329–344 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong XZ & Dong X-P Lysosome electrophysiology. Methods Cell Biol 126, 197–215 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Meeusen S, McCaffery JM & Nunnari J Mitochondrial fusion intermediates revealed in vitro. Science 305, 1747–1752 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Ramzan R, Staniek K, Kadenbach B & Vogt S Mitochondrial respiration and membrane potential are regulated by the allosteric ATP-inhibition of cytochrome c oxidase. Biochim Biophys Acta 1797, 1672–1680 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Wang W et al. A voltage-dependent K+ channel in the lysosome is required for refilling lysosomal Ca2+ stores. J Cell Biol 216, 1715–1730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X et al. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong X-P, Wang X & Xu H TRP channels of intracellular membranes. J Neurochem 113, 313–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava J, Barber DL & Jacobson MP Intracellular pH sensors: design principles and functional significance. Physiology (Bethesda) 22, 30–39 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Miller EW et al. Optically monitoring voltage in neurons by photo-induced electron transfer through molecular wires. Proc Natl Acad Sci U S A 109, 2114–2119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller EW Small molecule fluorescent voltage indicators for studying membrane potential. Curr Opin Chem Biol 33, 74–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dan K, Veetil AT, Chakraborty K & Krishnan Y DNA nanodevices map enzymatic activity in organelles. Nat Nanotechnol 14, 252–259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung K, Chakraborty K, Saminathan A & Krishnan Y A DNA nanomachine chemically resolves lysosomes in live cells. Nat Nanotechnol 14, 176–183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B et al. Visualizing Intercellular Tensile Forces by DNA-Based Membrane Molecular Probes. J Am Chem Soc 139, 18182–18185 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Veetil AT et al. DNA-based fluorescent probes of NOS2 activity in live brains. Proc Natl Acad Sci U S A 117, 14694–14702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Famulok M, Hartig JS & Mayer G Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 107, 3715–3743 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Saha S, Prakash V, Halder S, Chakraborty K & Krishnan Y A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat Nanotechnol 10, 645–651 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Modi S, Nizak C, Surana S, Halder S & Krishnan Y Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat Nanotechnol 8, 459–467 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Rubaiy HN A short guide to electrophysiology and ion channels. J Pharm Pharm Sci 20, 48–67 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni RU et al. Voltage-sensitive rhodol with enhanced two-photon brightness. Proc Natl Acad Sci U S A 114, 2813–2818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Lengerich B, Rawle RJ & Boxer SG Covalent attachment of lipid vesicles to a fluid-supported bilayer allows observation of DNA-mediated vesicle interactions. Langmuir 26, 8666–8672 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty K, Veetil AT, Jaffrey SR & Krishnan Y Nucleic Acid-Based Nanodevices in Biological Imaging. Annu Rev Biochem 85, 349–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin L et al. Characterization and application of a new optical probe for membrane lipid domains. Biophys J 90, 2563–2575 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fliegert R et al. Modulation of Ca2+ entry and plasma membrane potential by human TRPM4b. FEBS J 274, 704–713 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Post SR et al. Class A scavenger receptors mediate cell adhesion via activation of G(i/o) and formation of focal adhesion complexes. J Lipid Res 43, 1829–1836 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Rosenfeld JL et al. Lysosome proteins are redistributed during expression of a GTP-hydrolysis-defective rab5a. J Cell Sci 114, 4499–4508 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Magadán JG, Barbieri MA, Mesa R, Stahl PD & Mayorga LS Rab22a regulates the sorting of transferrin to recycling endosomes. Mol Cell Biol 26, 2595–2614 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koivusalo M, Steinberg BE, Mason D & Grinstein S In situ measurement of the electrical potential across the lysosomal membrane using FRET. Traffic 12, 972–982 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Farsi Z et al. Single-vesicle imaging reveals different transport mechanisms between glutamatergic and GABAergic vesicles. Science 351, 981–984 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Cang C et al. mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell 152, 778–790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun X et al. A negative feedback regulation of MTORC1 activity by the lysosomal Ca2+ channel MCOLN1 (mucolipin 1) using a CALM (calmodulin)-dependent mechanism. Autophagy 14, 38–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hockey LN et al. Dysregulation of lysosomal morphology by pathogenic LRRK2 is corrected by TPC2 inhibition. J Cell Sci 128, 232–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd-Evans E, Waller-Evans H, Peterneva K & Platt FM Endolysosomal calcium regulation and disease. Biochem Soc Trans 38, 1458–1464 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Narayanaswamy N et al. A pH-correctable, DNA-based fluorescent reporter for organellar calcium. Nat Methods 16, 95–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakraborty K, Leung K & Krishnan Y High lumenal chloride in the lysosome is critical for lysosome function. elife 6, e28862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modi S et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat Nanotechnol 4, 325–330 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Hover S et al. Bunyavirus requirement for endosomal K+ reveals new roles of cellular ion channels during infection. PLoS Pathog 14, e1006845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cain CC, Sipe DM & Murphy RF Regulation of endocytic pH by the Na+,K+-ATPase in living cells. Proc Natl Acad Sci U S A 86, 544–548 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schapiro FB & Grinstein S Determinants of the pH of the Golgi complex. J Biol Chem 275, 21025–21032 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Gagescu R et al. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell 11, 2775–2791 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Numata M & Orlowski J Molecular cloning and characterization of a novel (Na+,K+)/H+ exchanger localized to the trans-Golgi network. J Biol Chem 276, 17387–17394 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Raffaello A, Mammucari C, Gherardi G & Rizzuto R Calcium at the Center of Cell Signaling: Interplay between Endoplasmic Reticulum, Mitochondria, and Lysosomes. Trends Biochem Sci 41, 1035–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atakpa P, Thillaiappan NB, Mataragka S, Prole DL & Taylor CW IP3 Receptors Preferentially Associate with ER-Lysosome Contact Sites and Selectively Deliver Ca2+ to Lysosomes. Cell Rep 25, 3180–3193. e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russa AD et al. Microtubule remodeling mediates the inhibition of store-operated calcium entry (SOCE) during mitosis in COS-7 cells. Arch Histol Cytol 71, 249–263 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Lloyd-Evans E On the move, lysosomal CAX drives Ca2+ transport and motility. J Cell Biol 212, 755–757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang HH & St-Pierre F Genetically encoded voltage indicators: opportunities and challenges. J Neurosci 36, 9977–9989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaithianathan T, Henry D, Akmentin W & Matthews G Nanoscale dynamics of synaptic vesicle trafficking and fusion at the presynaptic active zone. elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jani MS, Zou J, Veetil AT & Krishnan Y A DNA-based fluorescent probe maps NOS3 activity with subcellular spatial resolution. Nat Chem Biol 16, 660–666 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y et al. A mechanically driven form of Kirigami as a route to 3D mesostructures in micro/nanomembranes. Proc Natl Acad Sci U S A 112, 11757–11764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prakash V et al. Quantitative mapping of endosomal DNA processing by single molecule counting. Angew. Chem. Int. Ed. Engl 58, 3073–3076 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Click chemistry for biotechnology and materials science. (John Wiley & Sons, Ltd, 2009). doi: 10.1002/9780470748862 [DOI] [Google Scholar]
- 53.Thekkan S et al. A DNA-based fluorescent reporter maps HOCl production in the maturing phagosome. Nat. Chem. Biol 15, 1165–1172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore D & Dowhan D Purification and concentration of DNA from aqueous solutions. Curr. Protoc. Mol. Biol Chapter 2, Unit 2.1A (2002). [DOI] [PubMed] [Google Scholar]
- 55.Vonderheit A & Helenius A Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 3, e233 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grimm C, Vierock J, Hegemann P & Wietek J Whole-cell Patch-clamp Recordings for Electrophysiological Determination of Ion Selectivity in Channelrhodopsins. J. Vis. Exp (2017). doi: 10.3791/55497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong X-P et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature 455, 992–996 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Surana S, Bhat JM, Koushika SP & Krishnan Y An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat Commun 2, 340 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Van Galen J et al. Sphingomyelin homeostasis is required to form functional enzymatic domains at the trans-Golgi network. J. Cell Biol 206, 609–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Modi S, Halder S, Nizak C & Krishnan Y Recombinant antibody mediated delivery of organelle-specific DNA pH sensors along endocytic pathways. Nanoscale 6, 1144–1152 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Zhang Q et al. Functional relevance of Golgi- and plasma membrane-localized endothelial NO synthase in reconstituted endothelial cells. Arterioscler. Thromb. Vasc. Biol 26, 1015–1021 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


