Histopathology features extracted by machine learning can identify MDS stage, prognosis, and genetic lesions, linking tissue morphology to underlying genetics and offering new tools for improved clinical evaluation for MDS.
Abstract
In myelodysplastic syndrome (MDS) and myeloproliferative neoplasm (MPN), bone marrow (BM) histopathology is assessed to identify dysplastic cellular morphology, cellularity, and blast excess. Yet, other morphologic findings may elude the human eye. We used convolutional neural networks to extract morphologic features from 236 MDS, 87 MDS/MPN, and 11 control BM biopsies. These features predicted genetic and cytogenetic aberrations, prognosis, age, and gender in multivariate regression models. Highest prediction accuracy was found for TET2 [area under the receiver operating curve (AUROC) = 0.94] and spliceosome mutations (0.89) and chromosome 7 monosomy (0.89). Mutation prediction probability correlated with variant allele frequency and number of affected genes per pathway, demonstrating the algorithms' ability to identify relevant morphologic patterns. By converting regression models to texture and cellular composition, we reproduced the classical del(5q) MDS morphology consisting of hypolobulated megakaryocytes. In summary, this study highlights the potential of linking deep BM histopathology with genetics and clinical variables.
Significance:
Histopathology is elementary in the diagnostics of patients with MDS, but its high-dimensional data are underused. By elucidating the association of morphologic features with clinical variables and molecular genetics, this study highlights the vast potential of convolutional neural networks in understanding MDS pathology and how genetics is reflected in BM morphology.
See related commentary by Elemento, p. 195.
Introduction
Current diagnosis of myelodysplastic syndrome (MDS) is based on identifying cellular dysplasia by visual inspection of bone marrow (BM) aspirate or biopsy (1). Karyotype, blast proportion, and peripheral blood (PB) cell count are assessed for disease subclassification according to World Health Organization (WHO) guidelines and for risk stratification by the Revised International Prognostic Scoring System (IPSS-R) criteria (1, 2).
Deep learning enables accurate visual pattern recognition with convolutional neural networks (CNN), where multiple processing layers detect and decode image data into numerical features (3). CNNs have recently led to significant breakthroughs in the analysis of biomedical images, facilitating diagnosis of skin tumors, retinal disease, intracranial hemorrhage, and breast cancer (3,4,5,6). In the context of routine hematoxylin and eosin (H&E) tissue stains, similar algorithms have improved Gleason scoring in prostate cancer, outcome prediction in colorectal cancer, and even discrimination of solid cancer patients by driver mutation status (4,7,8,9).
Here, we investigate the potential of CNN-based morphologic analysis in hematology. To improve our understanding of MDS histopathology and its association with clinical factors, we predict diagnosis, prognosis, IPSS-R risk score, mutated genes, cytogenetics, and patient age and gender by utilizing solely BM morphologic features (study flow presented in Supplementary Fig. S1). We demonstrate the highest detection accuracy for point mutations, such as TET2 and ASXL1. The mutation prediction probability correlates with variant allele frequency (VAF) within the sample, confirming the identification of mutation-specific features. To study interactions between disease determinants and BM histology, we introduce a novel multidimensional image analysis approach that combines information at tile, segmented nucleated hematopoietic cell (NHC), and pixel levels, ultimately facilitating the interpretation of complex BM histopathologic patterns.
Results
Unsupervised Modeling of BM Texture Recognized Morphologic Lineages and Distinct Myelodysplastic Clusters
To dissect BM morphology, we extracted ImageNet-configured visual features of H&E-stained BM biopsies from patients with MDS (236 samples from 143 subjects), patients with MDS/myeloproliferative neoplasm (MPN; 87 samples from 51 subjects), and healthy controls (11 samples from 11 subjects) using VGG16 and Xception CNNs (Fig. 1A; Supplementary Fig. S1). Images of tissue microarray (TMA) cores were grayscaled and split into 500 tiles enabling the examination of detailed tissue patterns. To investigate the spectrum of CNN texture patterns, we mapped image tiles from diagnostic BM samples of MDS, MDS/MPN, and control subjects with two-dimensional uniform manifold approximation and projection (UMAP) representation (Fig. 1B). Unsupervised image segregation was principally driven by stromal and cellular texture. Images with cellular content were observed to subcluster according to NHC and red blood cell (RBC) abundance and lipid droplet density. As up to 60% of patients with MDS may present with a hypercellular BM phenotype at diagnosis, complete discrimination of patients with MDS and MDS/MPN with an unsupervised approach is unlikely (10). Here, patients with MDS/MPN harbored an increased number of hypercellular tiles, while patients with MDS demonstrated heterogeneous histopathologic phenotypes (Fig. 1B). Notably, tiles from healthy subjects represented a balanced cellular and lipid droplet composition, with scarce proportion of stroma.
Figure 1.
Study design. A, TMAs were constructed from formalin-fixed, paraffin-embedded BM trephine biopsies and stained with H&E. Images were analyzed at tile, pixel, and NHC levels. In the tile-level analysis, TMA spot images were split into 500 small patches, and morphologic features were extracted with ImageNet-pretrained convolutional neural networks. We developed elastic net–regularized algorithms to predict multiple clinical and molecular genetic variables using only morphologic features. Tile-level feature features for each prediction model were joined into activation maps where the probability of each tile to associate with an endpoint is visualized with a heatmap color panel. A Weka pixel classifier was trained to identify NHCs, red blood cells, stroma, and lipid droplets from images. Moreover, the nuclei of NHCs were segmented, and their geometric measures were extracted. RBC, red blood cells. B, Extracted visual features from image tiles of diagnostic MDS, MDS/MPN, and healthy subjects are plotted using UMAP. We have selected 10 random tiles from each TMA sample for the UMAP visualization. Colors represent different PhenoGraph clusters (larger image) or diagnoses (top right image). Dashed circles represent morphologic subcategories where PhenoGraph clusters have been regrouped with k-means clustering. Superposed lines demonstrate morphologic trajectories generated with a slingshot analysis. Study flow is described in detail in Supplementary Fig. S1. C, UMAP projection of TMA samples. Visual features of tile images are combined at the TMA sample level by their mean value. TMA samples are then clustered with the k-means method (numbered large circles). Circle colors represent the corresponding diagnosis, tissue proportion of NHCs (D) and stroma (E), and WHO classification (F). CMML, chronic myelomonocytic leukemia; MDS-Del(5q), MDS with deletion 5q; MDS-MLD, MDS with multilineage dysplasia; MDS-RAEB-1, MDS with refractory anemia with excess blasts type 1; MDS-RAEB-2, MDS with refractory anemia with excess blasts type 2; MDS-RS, MDS with ring sideroblasts; MDS-SLD, MDS with single-lineage dysplasia; MDS/MPN-U, MDS/MPN, unclassifiable. G, Each k-means cluster has been compared with remaining clusters (Wilcoxon rank-sum test and Benjamini–Hochberg P-value correction) for segmented NHC, pixel-level image analysis parameters and clinical information. Clinical information is not reported for healthy patients (cluster 1). Variables are described in Supplementary Table S2. B-Hb, hemoglobin level; B-leuk, leucocyte count; B-neut, neutrophil count; B-PLT, platelet count; WBC, white blood cell.
By clustering image tiles with PhenoGraph, we observed distinct subgroups of tissue texture. In turn, we hypothesized that these patterns could be interconnected into morphologic entities (Fig. 1B). Slingshot lineage analysis is commonly used in single-cell RNA-sequencing data analysis to identify cellular development trajectories (11). The method aims to identify evolutionary branches from a common starting point using minimum spanning tree-based clustering (11). Here, we repurposed slingshot analysis for image data and demonstrate that tiles with a high proportion of either lipid droplets, hypercellularity, or RBCs were connected to tiles of hypocellular tissue (Fig. 1B). These connections suggest that adipose, hypercellular, and RBC-rich tissue areas rarely transform from one to another, but instead are more likely to arise from areas of hypocellular texture.
Tile features from MDS, MDS/MPN, and healthy subjects were averaged by adding the features of all tiles from one TMA spot and dividing by the number of tiles. Aggregated features were two-dimensionally projected with UMAP (Fig. 1C and http://hruh-20.it.helsinki.fi/mds_visualization). Unsupervised k-means clustering was used for subgrouping aggregated features, and each cluster was analyzed for associations with clinical variables. A distinct cluster representing healthy subjects (cluster 1) was clearly distinguishable and homogenous in contrast to the four myelodysplastic subgroups (Fig. 1C; Supplementary Fig. S2A). It also confirmed that the selected transfer learning approach identifies relevant biological features, even though these have been designed with the nonhistologic ImageNet image dataset (Fig. 1D). Clustering was principally driven by tissue texture content, which included the proportion of NHC, stroma, and lipid droplets and RBC and nuclear metrics of NHCs (Fig. 1D–G).
Cluster 3, defined histologically by high bone stroma content, was enriched with the WHO MDS subtypes elevated blast type 1 (EB-1; P = 0.03, χ2 test) and 2 (EB-2; P = 0.01, χ2 test), dimmer hematoxylin staining, and leukopenic PB cell count (Fig. 1F and G). Cluster 5 was characterized by high NHC proportion and hypercellular BM typical for patients with chronic myelomonocytic leukemia (CMML; P = 0.005, χ2 test) and unclassifiable MDS/MPN (P = 0.13, χ2 test). Moreover, these were also associated with darker hematoxylin staining. Cluster 4 harbored hypoplastic MDS (hMDS; P = 0.002, χ2 test) as well as low erythroid cell frequency linked with del(5q) MDS (12). Of note, the unsupervised clustering structure of MDS and MDS/MPN samples only partly overlapped with the WHO disease classification, likely as it was driven by tissue texture features, whereas the WHO classes are defined by BM cytomorphology, cytogenetics, blast proportion, and cytopenias. However, CNN-guided tissue analysis could potentially assist classification of challenging cases according to WHO criteria.
The MDS BM Morphology Is Linked to Mutation, Karyotype, Gender, and Prognostic Status
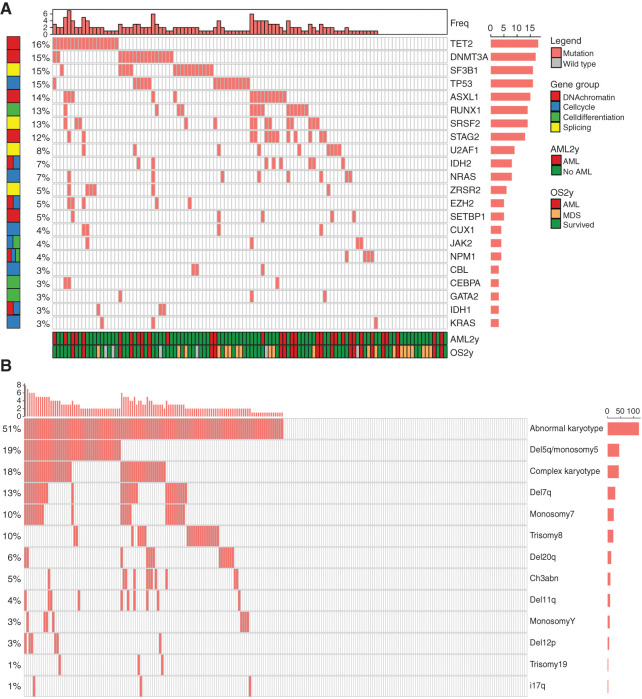
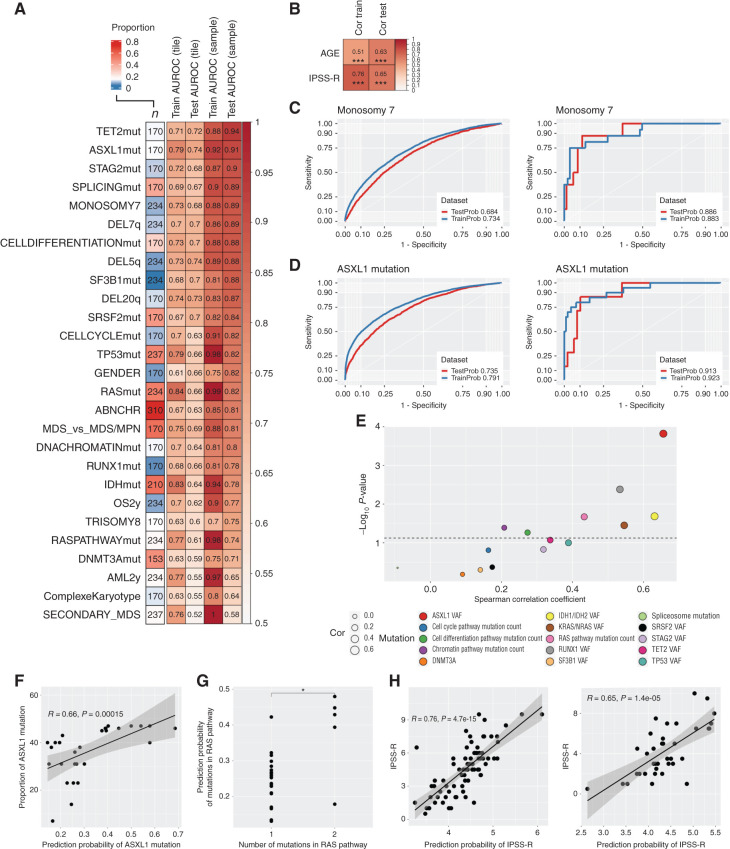
MDS is characterized by recurrent oncogenic somatic variants in driver genes and chromosomal aberrations, and the mutation profile detected in our patient cohort corresponded well to the known mutational landscape in MDS (Fig. 2A and B; Supplementary Fig. S2B and S2C; refs. 13,14,15). We adapted a transfer learning approach, where we used VGG16 and Xception network features extracted from tile-level H&E images to develop elastic net–regularized regression models (Supplementary Fig. S1). Predictions at the TMA spot level outweighed tile-level detection values due to uncertainties related to the limited image data in individual tiles. After averaging tile-level predictions at the TMA sample level, we could accurately detect notably TET2, ASXL1, and STAG2 mutations; chromosome 7 monosomy; and 7q deletion from morphologic features (Fig. 3A–D). Moreover, morphologic features were associated with point mutations in genes regulating splicing, cell differentiation, and cell cycle, which are commonly affected in MDS.
Figure 2.
Distribution of genomic alterations. A, Oncoprint visualization of mutation pattern and gene groups in patients with MDS. The top bars represent the frequency of mutations per patient. The right bars represent the frequency of patients with a given mutated gene. The bottom bars represent the survival status of each patient. AML, acute myeloid leukemia. B, Oncoprint visualization of cytogenetics pattern in samples from MDS patients. The top bars represent the frequency of the described karyotype aberrations per patient. The right bars represent the frequency of patients with a given karyotype aberration.
Figure 3.
Supervised learning on BM morphologic features. A, Heatmap displaying AUROC values of elastic net–regularized logistic regression models. The leftmost column shows the number of samples included in the analysis, and the cell color represents the distribution of the binary variables to be predicted. The following two columns inform the AUROC values of the models in the training (2/3) and test (1/3) dataset in the tile-level images and the last two columns at the TMA spot level. Abbreviations used for predicted variables: ABNCHR, presence of any abnormal chromosome; AML2y, progression to acute myeloid leukemia (AML) within 2 years of follow-up; CELLCYCLEmut, mutation in genes regulating cell cycle; CELLDIFFERENTIATIONmut, mutation in genes regulating cell differentiation; DNACHROMATINmut, mutation in genes regulating DNA chromatin structure; IDHmut, mutation in IDH1 or IDH2; OS2y, overall survival event within 2 years of follow-up; RASmut, mutation in NRAS or KRAS; RASPATHWAYmut, mutation in genes regulating the RAS pathway; SPLICINGmut, spliceosome mutations. B, Similar plot for elastic net–regularized linear regression models. The leftmost column shows the Spearman correlation value in the training dataset and rightmost in the test dataset. ***, P < 0.001. Tile-level (left) and TMA spot–level (right) AUROC for the logistic regression of monosomy 7 (C) and ASXL1 mutation status (D). The analysis shows the consistency of the predicted and true occurrence of an aberration. E, Scatter plot for the Spearman correlation (x-axis) between logistic regression predicting mutation status and the observed gene VAF for individual genes or number of genes mutated for functional pathways. F, Linear regression (R represents Spearman correlation) between prediction probability of ASXL1 mutation and its detected VAF. G, Wilcoxon comparison for predicted mutation probability and detected frequency of altered genes in the RAS pathway. *, P < 0.05. H, Linear regression (R represents Spearman correlation) between the predicted and observed IPSS-R score in the training (left) and test (right) datasets.
Secondary MDS and complex karyotype were often observed in the same patients (P = 0.008, χ2 test), and their morphologic features were most challenging to learn possibly due to higher variability in tissue texture. Features extracted with the Xception CNN model were more generalizable than those extracted with VGG16, concomitant with higher feature heterogeneity observed in the Xception correlation matrix (Supplementary Figs. S3 and S4). In addition, Lasso and elastic net penalization provided more generalizable models than ridge regression (Supplementary Fig. S4).
Next, we investigated the validity of models predicting mutated genes and dysregulated pathways. The detection probability of a distinct mutation correlated significantly with its VAF within the sample for ASXL1, KRAS/NRAS, IDH1/IDH2, and RUNX1 genes, and showed a trend for TET2 and TP53 genes (Fig. 3E and F; Supplementary Fig. S5). Moreover, the predicted likelihood of RAS, cell differentiation, and chromatin structure regulating gene pathway dysregulation correlated with the number of genes mutated in the respective pathways (Fig. 3G). This signifies that the prediction probability is higher when the sample contains more mutated cells, represented as higher VAF, or the sample has a higher proportion of mutated genes in the same pathway. This implies that prominent genetic changes translate to distinct morphologic image features. Of note, the initial models were developed using data from all patients with available myeloid mutation panel analysis, while the correlation analysis was restricted to samples with known mutation, indicating BM tissue morphology to be impacted by molecular genetics and emphasizing the algorithms' ability to identify variant-related histopathologic patterns in an unprecedented fashion.
Contrary to the histopathologic evaluation of solid tumors, tissue morphology is not factored in risk stratification of patients with MDS. Instead, the IPSS-R score accounting for PB cell count, BM blast burden, and cytogenetics is the most established stratification tool, and was associated in this dataset with risk for acute myeloid leukemia (AML) and overall survival (OS; Supplementary Fig. S6A and S6B; ref. 2). We could predict IPSS-R score, OS, and progression to AML by solely employing H&E-stained slides (Fig. 3A, B, and H). When evaluating progression to AML, BM morphology was associated with inferior prediction capability compared to the IPSS-R score but performed slightly better than IPSS-R when predicting OS within 2 years (Supplementary Fig. S6C and S6D). The best model was achieved by combining both deep histopathology and IPSS-R scores (Supplementary Fig. S6D). These results might be impacted by differences in patient selection, as our cohort represented an unbiased real-world cohort, while patients treated with disease-modifying therapies were excluded from the landmark prognostic study by Greenberg and colleagues (2). However, patient prognostication and treatment stratification could be significantly improved by including morphologic features.
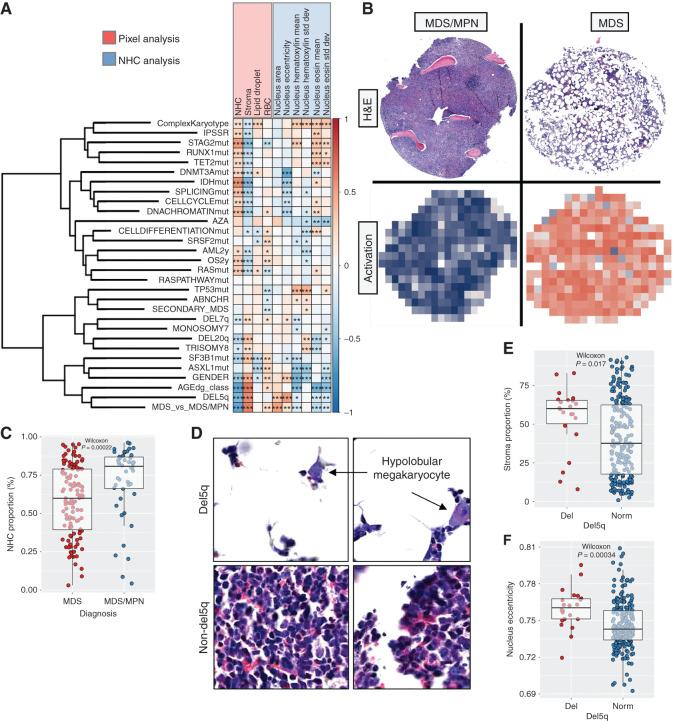
Interpretation of Supervised Prediction Models with Multilevel Image Analysis
MDS is differentially diagnosed from MDS/MPN by cytomorphologic and histopathologic examination, evaluation of PB counts, flow cytometry, karyotype, and increasingly, molecular genetics. In our study, tissue samples were partitioned into smaller TMA cores, possibly limiting effective diagnostic segregation. Yet, we could discern patients with MDS from patients with MDS/MPN with an area under the receiver operating curve (AUROC) validation accuracy of 0.81 (Fig. 3A). To help understand which texture patterns are associated with various clinical variables, we interpreted regression models by correlating tile-level tissue texture predictions with pixel- and NHC-level metrics (Fig. 4A). In addition, we present both tile- and TMA spot–level images with the highest and lowest association with the predicted endpoint as well as TMA-level activation maps (Supplementary Fig. S7). Pixel classification was trained to identify NHCs, RBCs, stroma including fibrotic stroma and bone trabeculae, and lipid droplets, constituting the major tissue elements in standard H&E staining. In addition, NHCs were segmented to extract nuclear measurements such as size, circularity, and H&E dye variations. As expected, MDS likelihood increased if the sample represented hypoplastic texture (Fig. 4B and C). MDS morphology was also associated with dimmer H&E staining and larger nuclear size, likely due to a technically larger cell segmentation area in hypocellular BM (Fig. 4A; Supplementary Fig. S7A).
Figure 4.
Interpretation of supervised prediction models. A, Correlation matrix for logistic and linear regression model predictions (rows) and pixel-level and segmented NHC–level image analysis metrics aggregated per sample. The color of individual matrix cells represents the Spearman correlation and the asterisks the Benjamini–Hochberg–adjusted significance values: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Variables are described in Supplementary Table S2. Abbreviations used for predicted variables: ABNCHR, presence of any abnormal chromosome; AML2y, progression to acute myeloid leukemia (AML) within 2 years of follow-up; AZA, azacytidine treatment response; CELLCYCLEmut, mutation in genes regulating cell cycle; CELLDIFFERENTIATIONmut, mutation in genes regulating cell differentiation; DNACHROMATINmut, mutation in genes regulating DNA chromatin structure; IDHmut, mutation in IDH1 or IDH2; OS2y, overall survival event within 2 years of follow-up; RASmut, mutation in NRAS or KRAS; RASPATHWAYmut, mutation in genes regulating the RAS pathway; SPLICINGmut, spliceosome mutations. B, H&E-stained TMA spots and corresponding activation maps for the prediction of MDS or MDS/MPN. For the activation map, tile-level predictions have been color-scaled (blue, high probability for MDS/MPN; red, high probability for MDS). C, Scatter plot and Wilcoxon rank-sum test to compare NHC proportion by MDS and MDS/MPN diagnoses. Boxplots define the interquartile ranges and median values by diagnoses. D, Image tiles representing highest and lowest computed probability of chromosome 5q deletion. Scatter plot and Wilcoxon rank-sum test to compare the proportion of stroma (E) and nuclei eccentricity (opposite of circularity; F) by del(5q) status. Boxplots define the interquartile ranges and median values by diagnoses.
Chromosome 5q deletion is associated with a decrease in erythroid precursor cells and an increase in hypolobular megakaryocytes (12). When inspecting tiles associated with 5q deletion, we discovered enrichment of megakaryocytes with abnormally circular nuclei, in line with previous findings (Fig. 4D). While hMDS was not associated with 5q deletion syndrome in our cohort (7/51 vs. 14/183 samples had 5q deletion syndrome in hMDS vs. non-hMDS, P = 0.29, χ2 test), del(5q) was identified more often in hypocellular samples and samples with higher stroma content (Fig. 4E). These findings could be due to prior treatment with lenalidomide indicated for MDS with del(5q). Moreover, del(5q) MDS samples displayed lower nucleus circularity. This could be due to different white blood cell content rather than dysplasia, as this pattern was absent in samples with complex karyotype (Fig. 4F).
Discussion
Here, we demonstrate how the intricate and heterogeneous BM morphologic landscape can be dissected and associated with clinical data using multilevel computer vision.
While the presented analytic platform is not accurate and extensive enough to replace sequencing technologies in identifying genetic lesions, deep BM morphology predicted mutations and cytogenetic aberrations with accuracy exceeding that of similar applications in solid tumors (4, 7, 16, 17). We suspect that the homogeneous BM tissue consistency and lower mutation burden in MDS may account for the results. To increase reliability, we employed transfer learning and separated the training and validation datasets at the TMA sample level. However, in the absence of an independent test cohort, the results should be interpreted with caution.
On the basis of our findings, transfer learning of H&E images can shed light on disease pathology and gene targets by linking molecular genetics with tissue dysplasia. It would be interesting to compare the texture of MDS BM samples collected in clinical trials or derived from animal models to images in our interactive platform (http://hruh-20.it.helsinki.fi/mds_visualization). The mechanism of action of any drug could thus be examined with respect to the signaling pathways or molecular genetic aberrations described in this study. The potential of this approach could further expand if explored by combining genetic variants with protein production quantitated with immunohistochemistry to comprehensively observe the effects of transcriptional, spliceosomal, and translational regulation on eventual protein production.
The analysis was conducted for images digitized with 20× objective, as we initially aimed to combine cellular-level morphologic patterns with clinical variables. However, the resolution remains suboptimal for the distinction of subcellular organelles, and quantification of chromatin or nucleus-to-cytoplasm ratio. Addressing these questions would likely require analysis of May-Grunwald-Giemsa (MGG)–stained BM aspirate samples with better morphologic quality using high-magnification (100×) microscopes and oil immersion.
While presence of bone trabeculae could be related to sampling artifacts, the BM microenvironment and erythropoietin are known to regulate the osteoblast–osteoclast balance, and ultimately bone remodeling (18, 19). Moreover, both cellular and acellular components contribute to the BM niche (20). Therefore, bone trabeculae were not excluded from image analysis, and further examination of the BM microenvironment is needed to validate associations with molecular genetics.
Digital pathology and adaptation of CNNs have had substantial influence in the field of solid tumors (21). Standardized clinical application would require multicenter collaboration both to collect sufficient training data and to impact clinical routine. Our current results suggest that image analysis of H&E-stained BM samples is insufficient alone to reliably differentiate MDS and MDS/MPN subtypes. However, especially the trained eye of the pathologist may benefit by learning novel associations between morphology and molecular genetics. Moreover, CNN-based analytics could reduce intra- and interobserver variability of histopathologic analysis, and incorporating additional clinical information could assist in automating and objectively classifying patients with MDS. A recent study took a slightly different approach and used hematopathologic reports instead of image analysis to link BM morphology and clinical variables (22). Cytomorphologic findings such as dysplasia, monocytosis, and elevated megakaryocytes from MGG-stained BM smears and presence of myelofibrosis from H&E-stained BM biopsies were shown to associate with cytopenia and mutations, emphasizing that machine learning–based platforms could support MDS diagnosis.
The black box dilemma hinders clinical translation of deep learning algorithms (23). To increase model transparency, we decoded CNN-extracted morphologic patterns associated with molecular and clinical determinants with a holistic methodology linking image analysis at the tile, pixel, and cellular levels. CNN-based feature extraction, pixel classification, and NHC segmentation required little parameter optimization to increase reproducibility and scalability to any image analysis study employing deep learning. Image features associated with MDS versus MDS/MPN diagnosis and del(5q) MDS were affirmed by a hematopathologist and were consistent with previous knowledge (1, 12). Dissection of complex computer vision models could be achieved with similar approaches based on neural network–based semantic and instance segmentation methods.
Taken together, deep mining of the BM tissue texture on a larger scale could assist pathologists by revealing intricate morphologic patterns defining disease subtypes and eventually improving clinical stratification of patients with MDS.
Methods
Patients
The study population comprised patients with MDS (n = 143) and MDS/MPN (n = 51) and control subjects (n = 11) treated from 2000 to 2018 at the Department of Hematology in the Helsinki University Hospital (HUH; Helsinki, Finland; Supplementary Fig. S1; Table 1). The formal MDS or MDS/MPN diagnosis and classification adhered to WHO guidelines and were assigned by the treating clinical hematologist on the basis of patient history, PB cell counts, BM cytomorphology evaluated by two laboratory hematology specialists, and cytogenetics according to WHO guidelines. If needed, flow cytometry results were included in the evaluation. The diagnostic conclusion, especially if challenging, was confirmed by a regular tumor board of clinical hematologists and a laboratory hematologist at our institution (24, 25).
Table 1.
Patient characteristics at diagnosis
| MDS (n = 143) | MDS/MPN (n = 51) | |
|---|---|---|
| Gender, male | 73 (51.0%) | 36 (70.6%) |
| Age, mean (range) | 64.8 [15.5–88.6] | 67.5 (36.4–87.0) |
| Etiology | ||
| De novo | 118 (82.5%) | 47 (92.2%) |
| Secondary (treatment-related) | 25 (17.5%) | 4 (7.8%) |
| WHO MDS classification | ||
| MDS-SLD | 20 (14.0%) | |
| MDS-MLD | 33 (23.1%) | |
| MDS-RS | 16 (11.2%) | |
| MDS-EB-1 | 25 (17.5%) | |
| MDS-EB-2 | 35 (24.5%) | |
| MDS with isolated del(5q) | 10 (7.0%) | |
| MDS-U | 4 (2.8%) | |
| WHO MDS/MPN classification | ||
| CMML | 35 (68.6%) | |
| MDS/MPN-U | 16 (31.4%) | |
| Survival status 2 years after diagnosis | ||
| Alive | 60 (42.0%) | 32 (62.7%) |
| Deceased | 70 (49.0%) | 19 (37.3%) |
| Not defined or censored due to alloHSCT | 13 (9.1%) | 0 (0.0%) |
| Progression to AML within 2 years of diagnosis | ||
| No | 51 (35.7%) | 28 (54.9%) |
| Yes | 42 (29.4%) | 11 (21.6%) |
| Not defined or censored due to alloHSCT | 50 (35.0%) | 12 (23.5%) |
| Azacytidine-treated within 1 year of diagnosis | ||
| No | 78 (54.5%) | 31 (60.8%) |
| Yes | 65 (45.5%) | 20 (39.2%) |
| IPSS risk class | ||
| Low | 24 (16.8%) | |
| Intermediate-1 | 57 (39.9%) | |
| Intermediate-2 | 26 (18.2%) | |
| High | 24 (16.8%) | |
| Not defined | 12 (8.4%) | |
| IPSS-R risk class | ||
| Very low | 17 (11.9%) | |
| Low | 33 (23.1%) | |
| Intermediate | 27 (18.9%) | |
| High | 26 (18.2%) | |
| Very high | 28 (19.6%) | |
| Not defined | 12 (8.4%) | |
| AlloHSCT | ||
| No | 117 (81.8%) | 43 (84.3%) |
| Yes | 24 (16.8%) | 8 (15.7%) |
| Not defined | 2 (1.4%) | 0 (0.0%) |
Abbreviations: alloHSCT, allogeneic hematopoietic stem cell transplantation; MDS-EB-1, MDS with excess blasts 1; MDS-EB-2, MDS with excess blasts 2; MDS-MLD, MDS with multilineage dysplasia; MDS/MPN-U, MDS/MPN, unclassifiable; MDS-RS, MDS with ring sideroblasts; MDS-SLD, MDS with single-lineage dysplasia; MDS-U, MDS, unclassifiable.
BM trephine biopsies are used in standard diagnostic procedures and occasionally for assessing treatment response and disease progression. As all available samples were obtained, no selection bias in sample collection occurred. Diagnostic (MDS n = 143, MDS/MPN n = 51) and follow-up specimens (MDS n = 93, MDS/MPN n = 36) were collected prior to and after initiating a disease-modifying treatment, respectively. According to ethics board guidelines, control BM trephine samples were collected from subjects without diagnosis of hematologic malignancy, chronic infection, or autoimmune disorder in the 6-year follow-up (Supplementary Table S1). Of the 11 control subjects, six were males and their median age was 57.0 (range, 40.0–82.0) years at the time of BM sampling. Patients with MDS and MDS/MPN were slightly older than control subjects (P = 0.02 and P = 0.002, respectively) but did not differ significantly by sex (Table 1).
The study complied with the Declaration of Helsinki and the HUS institutional ethics committee. All study patients gave written informed consent. All clinical data were collected from the HUS datalake, a General Data Protection Regulation (GDPR)–compliant database.
Sequencing
Genomic DNA was isolated from diagnostic BM aspirates (n = 108) using the QIAsymphony DSP DNA kit. According to the manufacturer's protocols, 20 ng of DNA for each sample was used for library preparation with the Ion AmpliSeq Library Kit 2.0 (Thermo Fisher Scientific) and the AmpliSeq primers (Thermo Fisher Scientific) for the in-house myeloid cancer gene panel. This panel covers all exons in ASXL1, BCOR, CDKN2A, CEBPA, CREBBP, CUX1, DNMT3A, EP300, ETV6, EZH2, GATA2, KDM6A, NF1, PHF6, RAD21, SETD2, STAG2, TET2, TP53, and ZRSR2 genes and hotspot exons in BRAF, CALR, CBL, CSF3R, FLT3, GATA1, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, PDGFRA, PTPN11, RUNX1, SETBP1, SF3B1, SMC1A, SMC3, SRSF2, U2AF1, and WT1 genes. Libraries were barcoded with the Ion Xpress Barcode Adapters (Thermo Fisher Scientific) and quantified with the Ion Library TaqMan Quantitation Kit. Template preparation and chip loading were performed on the Ion Chef system (Thermo Fisher Scientific). Sequencing was carried out on the Ion Proton or Ion GeneStudio S5 system (Thermo Fisher Scientific). Data analysis was carried out with Torrent Suite Software v.5.8 (Life Technologies). The Ion Reporter software v4.6 (Thermo Fisher Scientific) was used to filter out noncoding and polymorphic variants. All variants listed after filtering were visualized in the Integrative Genomics Viewer (IGV) to manually discard alterations generated by incorrect calling.
Driver gene mutations were defined using a clinical-grade, myeloid amplicon sequencing panel capable of identifying mutations with VAF >2% (mean sequencing depth 6,000×; Supplementary Fig. S2B and S2C). In addition, data from an additional 40 samples analyzed with the Illumina TruSight myeloid amplicon sequencing panel were included (mean sequencing depth 100×). Sequencing panels produced similar results for 305/315 genes (96.8%) by analyzing nine samples in parallel with both assays.
Tissue Microarrays
Upon sampling, fresh BM biopsies were subjected to routine EDTA-based decalcification, softening the tissue and FFPE. TMAs were cast by a single 2-mm core (MDS and MDS/MPN) or double 1-mm cores (controls) per sample from representative BM biopsy areas (Fig. 1A). Duplicate control cores did not differ by their morphologic patterns (Supplementary Fig. S2A). Tissue blocks were cut into 4-μm–thick sections and stained with H&E. Slides were digitized at 0.22 μm/pixel (20× objective magnification) with the whole-slide scanner Pannoramic 250 FLASH (3DHISTECH Ltd.).
Image Analysis
Images were analyzed with three methods—(i) transfer learning, (ii) pixel classification, and (iii) NHC analysis—to capture information from different representation levels (Supplementary Fig. S1).
Transfer Learning
Image Preprocessing.
As sample processing and slide digitization affect color distribution, H&E-stained RGB images were converted into gray scale with the OpenCV Python module to reduce possible technical noise. Moreover, we standardized background pixels that did not represent BM tissue by converting them into the pixel value 255 (white). H&E images were split into 500 equal-sized tiles (small subimages of the original H&E image), in average 470 × 470 pixels. Tile size was optimized to outsize the BM lipid droplets to avoid their classification as nontissue background. Tiles with mean pixel intensity over 240 represented nontissue background and were excluded from the analysis, resulting in 73,531 tiles.
Feature Extraction.
In transfer learning, a prior algorithm or its configurations are reused to develop a new machine learning algorithm. Visual features that were previously defined in a larger training dataset can be repurposed for new image analysis tasks, reducing the need for training models from scratch (26). We adapted a transfer learning approach where image tile features were extracted with pretrained Xception and VGG16 convolutional networks that have achieved high accuracy in classifying the ImageNet dataset commonly used to develop and benchmark computer vision algorithms (27, 28). Individual tiles were resized into equal sizes (224 × 224 pixels for VGG16 and 299 × 299 pixels for Xception). Colors were rescaled between values [0,1]. Images were reformatted as three-channel gray scale where each pixel was assigned an identical value for each channel to ensure compatibility with pretrained CNNs, as ImageNet configurations have been developed for color images. For each tile image, a 2.048-bin feature vector was extracted at the second-to-last Xception network layer. As VGG16 and Xception networks do not possess a similar number of parameters, we exported features from the last layer of the VGG16 network (n = 25.088) and retained only 2.048 features with the highest variance-to-mean ratio (Supplementary Fig. S3A and S3B).
Regression Models.
Visual features extracted from image tiles with CNNs contain numerical values. Therefore, these can be introduced as covariates to develop regression models. Image tiles were first split at the TMA sample level into training (2/3) and test (1/3) datasets. In average, 213 (range 70–336) tiles per sample were available. Models were trained with L1 (alpha = 1), L2 (alpha = 0), or elastic net–regularized (alpha = 0.5) regression models. Here, multivariate regression models are optimized for both regularizing a factor alpha and penalization rate lambda using 5-fold cross-validation. Lambda values were optimized for each fixed parameter alpha to reach minimum cross-validation error (lambda.min) and the lambda at one SE of this minimum (lambda.1se). Training of the regression models occurred at the tile level to highlight individual tissue texture and cell morphology features. The prediction results were assessed both at tile level and TMA sample level. Prediction probabilities for TMA samples were calculated by summing probabilities of each tile of the same TMA sample and dividing the total by the number of tiles. In summary, each predicted variable was estimated based on 12 algorithms (feature matrices extracted with two CNN models and three alpha and two lambda elastic net regularization parameter values).
Follow-up for survival analyses was restricted to 2 years to identify morphologic patterns associated with aggressive disease and to ensure concordance between AUROC model evaluation and the predicted variable. Patients with shorter follow-up time or receiving allogenic hematopoietic stem cell transplant before the occurrence of an event were censored. Azacytidine was administered 75 mg/m2 per day, with a median interval of 29 days (27–33 days, 25th–75th percentiles) and a median of six treatment cycles (3–11 cycles, 25%–75%). Positive treatment response was defined as continuation of treatment over 6 months or complete remission prior to 6 months. Disease progression was determined to represent treatment failure. Patients treated for less than 6 months due to short follow-up or therapy-related side effects were censored.
As longitudinal samples were not characterized with lower variance in morphologic features compared with samples from other patients, we included samples from multiple time points of the same patient for model training (Supplementary Fig. S8A and S8B). Prediction models were trained with MDS BM images only except for differential diagnosis, where both MDS and MDS/MPN samples were used. Separate prediction models for IPSS-R score and age at diagnosis were developed with linear regression and using only diagnostic MDS samples. Age was transformed into categories of <50.0, 50.0–59.9, 60.0–69.9, 70.0–79.9, and >80.0 years age categories, which ameliorated the accuracy and interpretation of results. Gender, mutations, cytogenetic aberrations, and MDS etiology were predicted with logistic regression using both diagnostic and follow-up samples. Prognosis was measured by predicting OS and AML progression using both diagnostic and follow-up samples. Only mutations and chromosomal aberrancies present in over 9% of the samples were selected. Disease etiology was assigned as either “de novo” or “secondary MDS.” Azacytidine response was predicted with logistic regression using samples taken 0 to 365 days before treatment start with an AUROC accuracy of 0.80 at tile level and 0.98 at TMA sample level. Due to the low number of available samples prior to azacytidine initiation, we did not evaluate treatment response in a separate test cohort. Samples taken during hypomethylating agent (HMA) treatment did not differ by their morphologic profile from other samples, suggesting that HMA does not alter the BM morphology in a distinct fashion (Supplementary Fig. S8C and S8D).
Pixel Classification.
Each RGB TMA spot image was analyzed with the Trainable Weka Segmentation module of the Fiji software using default parameters (29). With the assistance of hematopathologic expertise, we manually delineated areas representing NHCs, RBCs, stroma, or lipid droplet/background from 10 images and trained a tissue pixel classifier. NHCs represented nucleated cells consisting primarily of leukocytes and occasional erythroblasts and megakaryocytes. Stroma included fibrotic stroma and bone trabeculae, as both are associated with MDS pathology (30, 31). We manually annotated additional 10 test images to evaluate the accuracy of the classifier. Each pixel was classified with global 97% accuracy composed of NHC (99% accuracy), RBC (84% accuracy), stroma (80% accuracy), or lipid droplet/background (100% accuracy; Supplementary Fig. S9A and S9B). The relative area of individual classes was calculated as their proportion to a binary tissue mask. The tissue mask was created by converting H&E images into binary format, where white areas represent tissue and black areas adipose tissue and empty background. Then, we performed mask dilatation, empty hole fill, and mask erosion steps to differ adipose tissue from the background (Supplementary Fig. S9C). Lipid droplets were defined as filled image holes from the initial tissue mask.
NHC Analysis.
Each RGB TMA spot image was analyzed with the open-source software QuPath (v0.2.0; ref. 32). NHCs were segmented with the watershed cell detection and background radius 30 px, median filter radius 0 px, sigma 6 px, minimum area 10 px2, maximum area 10.000 px2, threshold 0.1, maximum background intensity 2, and cell expansion 5 px (Supplementary Fig. S10). Nucleus H&E staining intensity was defined as their optical density after color deconvolution. Nucleus size was calculated on the basis of area and eccentricity based on the deviance from a perfect circle. These metrics were extracted for individual NHCs and averaged at the TMA spot level (Supplementary Table S2).
Statistical Analysis
Continuous variables were compared with Wilcoxon rank-sum test (unpaired, two-tailed) and correlated with Spearman rank correlation coefficient. Categorical variables were compared with χ2 test. P values were adjusted with Benjamini–Hochberg correction when necessary. The log-rank test for Kaplan–Meier analysis was used for survival analysis. Model fitness was assessed by calculating statistical significance of the AUROC. AUROC values for predicting AML progression, OS, and IPSS-R were further compared with DeLong test.
For unsupervised analysis, we selected a UMAP method (33). Pheno-Graph is a graph-based community detection method designed for high-resolution single-cell data analogous to visual features (34). Single tiles were clustered with PhenoGraph with default settings to attain higher granularity to distinguish morphologic features. K-means clustering was selected for sample grouping to simplify interpretation of TMA spot grouping. The k parameter was harmonized using the consensus of 30 indices based on Euclidean distance and Ward agglomeration metrics (35). Feature extraction, regression models, and statistical analysis were performed with R 3.5.1 (Supplementary Table S3).
Code and Data Availability
Codes used for data analysis are available at https://github.com/obruck/MDS_HE_IA/. Image data and activation maps are available at http://hruh-20.it.helsinki.fi/mds_visualization.
Authors' Disclosures
O.E. Brück reports grants from Novartis International AG during the conduct of the study, as well as personal fees from Novartis International AG and Sanofi S.A. outside the submitted work. H.R. Hohtari reports grants from Incyte outside the submitted work. P.M. Ramos reports other support from Novartis during the conduct of the study, as well as other support from Novartis outside the submitted work. K.V. Porkka reports grants and personal fees from Novartis and Bristol-Myers Squibb/Celgene during the conduct of the study. S.M. Mustjoki reports grants from Cancer Foundation Finland, Gyllenberg Foundation, Sigrid Juselius Foundation, Relander Foundation, and Novartis during the conduct of the study, as well as grants and personal fees from Bristol-Myers Squibb and Novartis and grants from Pfizer outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors are grateful to the members of the Hematology Research Unit Helsinki for discussions and technical help. They thank the Helsinki Biobank for sample collection and the Digital and Molecular Pathology Unit supported by the University of Helsinki and Biocenter Finland for digital microscopy services. The authors thank the University of Helsinki language center for editing and proofreading services. This study was supported by the University of Helsinki, the Doctoral Programme in Biomedicine (DPBM), personal grants (to O.E. Brück) from Biomedicum Helsinki Foundation, Finska Läkaresällskapet, research grants (to S. Mustjoki) from the Cancer Foundation Finland, Sigrid Juselius Foundation, Signe and Ane Gyllenberg Foundation, Relander Foundation, state funding for university-level health research in Finland, a Helsinki Institute of Life Sciences Fellow grant, and an investigator-initiated research grant from Novartis.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
#S.E. Lallukka-Brück, H.R. Hohtari, and A. Ianevski contributed equally to this article.
Blood Cancer Discov 2021;2:238–49
Authors' Contributions
O.E. Brück: Conceptualization, software, formal analysis, funding acquisition, investigation, visualization, methodology, writing–original draft. S.E. Lallukka-Brück: Data curation, visualization, writing–review and editing. H.R. Hohtari: Data curation, investigation, writing–review and editing. A. Ianevski: Software, visualization, writing–review and editing. F.T. Ebeling: Investigation, writing–review and editing.P.E. Kovanen: Investigation, writing–review and editing. S.I. Kytölä: Resources, formal analysis, writing–review and editing. T.A. Aittokallio: Visualization, writing–review and editing. P.M. Ramos: Formal analysis, funding acquisition, writing–review and editing. K.V. Porkka: Formal analysis, investigation, writing–review and editing. S.M. Mustjoki: Conceptualization, resources, supervision, funding acquisition, investigation, project administration, writing–review and editing.
References
- 1.Montalban-Bravo G, Garcia-Manero G. Myelodysplastic syndromes: 2018 update on diagnosis, risk-stratification and management. Am J Hematol 2018;93:129–47. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Solé F, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012;120:2454–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lecun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- 4.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Titano JJ, Badgeley M, Schefflein J, Pain M, Su A, Cai M, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med 2018;24:1337–41. [DOI] [PubMed] [Google Scholar]
- 6.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ström P, Kartasalo K, Olsson H, Solorzano L, Delahunt B, Berney DM, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol 2020;21:222–32. [DOI] [PubMed] [Google Scholar]
- 9.Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep 2018;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schemenau J, Baldus S, Anlauf M, Reinecke P, Braunstein S, Blum S, et al. Cellularity, characteristics of hematopoietic parameters and prognosis in myelodysplastic syndromes. Eur J Haematol 2015;95:181–9. [DOI] [PubMed] [Google Scholar]
- 11.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 2018;19:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van den Berghe H, Cassiman JJ, David G, Fryns JP, Michaux JL, Sokal G. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature 1974;251:437–8. [DOI] [PubMed] [Google Scholar]
- 13.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2013;28:241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013;122:3616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason CC, Khorashad JS, Tantravahi SK, Kelley TW, Zabriskie MS, Yan D, et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia 2016;30:906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer 2020;2:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, Jung AW, Torne RV, Gonzalez S, Vöhringer H, Shmatko A, et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer 2020;2:800–10. [DOI] [PubMed] [Google Scholar]
- 18.Mellibovsky L, Diez A, Serrano S, Aubia J, Pérez-Vila E, Mariñoso ML, et al. Bone remodeling alterations in myelodysplastic syndrome. Bone 1996;19:401–5. [DOI] [PubMed] [Google Scholar]
- 19.Balaian E, Wobus M, Weidner H, Baschant U, Stiehler M, Ehninger G, et al. Erythropoietin inhibits osteoblast function in myelodysplastic syndromes via the canonical Wnt pathway. Haematologica 2018;103:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pronk E, Raaijmakers MHGP. The mesenchymal niche in MDS. Blood 2019;133:1031–8. [DOI] [PubMed] [Google Scholar]
- 21.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019;25:1301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagata Y, Zhao R, Awada H, Kerr CM, Mirzaev I, Kongkiatkamon S, et al. Machine learning demonstrates that somatic mutations imprint invariant morphologic features in myelodysplastic syndromes. Blood 2020;136:2249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opening the black box of machine learning. Lancet Respir Med 2018;6:801. [DOI] [PubMed] [Google Scholar]
- 24.MDS and CMML Guidelines: guidelines for the diagnosis and treatment of myelodysplastic syndrome and chronic myelomonocytic leukemia. 9th update. Nordic MDS Group; 2019. Available from: https://www.nmds.org/attachments/article/121/200120_Guidelines%20for%20the%20diagnosis%20and%20treatment%20of%20MDS%20and%20CMML.pdf.
- 25.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 26.Shin H, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition.arXiv:1409.1556 [Preprint]. 2014. Available from:https://arxiv.org/abs/1409.1556.
- 28.Chollet F. Xception: deep learning with depthwise separable convolutions. arXiv:1610.02357 [Preprint]. 2016. Available from:https://arxiv.org/abs/1610.02357.
- 29.Arganda-Carreras I, Kaynig V, Rueden C, Eliceiri KW, Schindelin J, Cardona A, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 2017;33:2424–6. [DOI] [PubMed] [Google Scholar]
- 30.Della Porta MG, Malcovati L. Myelodysplastic syndromes with bone marrow fibrosis. Haematologica 2011;96:180–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weidner H, Rauner M, Trautmann F, Schmitt J, Balaian E, Mies A, et al. Myelodysplastic syndromes and bone loss in mice and men. Leukemia 2017;31:1003–7. [DOI] [PubMed] [Google Scholar]
- 32.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 [Preprint]. 2018. Available from: https://arxiv.org/abs/1802.03426.
- 34.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir EAD, Tadmor MD, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015;162:184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw 2014;61. Available from:http://www.jstatsoft.org/v61/i06/paper. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Codes used for data analysis are available at https://github.com/obruck/MDS_HE_IA/. Image data and activation maps are available at http://hruh-20.it.helsinki.fi/mds_visualization.