Abstract
A panel (ENVA-1) of well-defined blinded samples containing wild-type and mutant human immunodeficiency virus type 1 (HIV-1) reverse transcriptase was analyzed by automated DNA sequencing in 23 laboratories worldwide. Drug resistance mutations at codons 41, 215, and 184 were present in the panel samples at different ratios to the wild type. The presence of mutant genotypes was determined qualitatively and quantitatively. All laboratories reported the presence of sequence heterogeneities at codons 41, 215, and 184 in one or more of the panel samples, though not all reported the correct codon genotypes. Two laboratories reported a mutant genotype in samples containing only the wild type, whereas two and three laboratories failed to detect the mutant genotypes at codons 41 and 215, respectively, in a completely mutant DNA population. Mutations present at relative concentrations of 25% of the total DNA population were successfully identified by 13 of 23, 10 of 23, and 16 of 23 labs for codons 41, 215, and 184Val, respectively. For more than 80% of those laboratories that qualitatively detected the presence of a mutation correctly, the estimated wild type/mutant ratio was less than 25% different from the input ratio in those samples containing 25 to 50% or 75% mutant input. This first multicenter study on the quality of DNA sequencing approaches for identifying HIV-1 drug resistance mutations revealed large interlaboratory differences in the quality of the results. The application of these procedures in their current state would in several cases lead to inaccurate or even incorrect diagnostic results. Therefore, proper quality control and standardization are urgently needed.
The incomplete inhibition of human immunodeficiency virus type 1 (HIV-1) replication may result in the emergence of viral isolates with reduced susceptibility (resistance) to an antiviral drug. For most drugs used in the treatment of HIV-1-infected individuals to date, specific resistance-conferring mutations have been identified (16). The determination of these mutations in clinical isolates may serve as a surrogate for laborious and time-consuming classical biological drug susceptibility determinations (2, 6, 13, 17).
DNA sequencing, using various automated technologies, is widely applied to identify drug resistance mutations (17). In addition to enzymatic sequencing approaches used with most methodologies, an innovative technology involving the differential hybridization of nucleic acid sequences with short oligonucleotide arrays present on microchips is currently under evaluation (9).
The high mutation rate of HIV results in continuous changes in the composition of the viral population and the presence of sequence heterogeneities (sequence mixtures) along the viral genome (3, 11, 12). Population-based sequencing approaches enable the genotypic characterization of the predominant viral species in a patient. On the other hand, analysis of multiple (individual) cloned genes provides a detailed insight into the interactions of mutations present on the same genome and also allows for a more reliable detection of minority species. In general the sensitivities of various sequencing approaches in the detection of minority virus populations are highly variable (4).
At present, genotypic resistance determinations are performed mainly by research laboratories, without any systematic interlaboratory standardization or quality control. However, genotypic resistance determinations are of increasing diagnostic value in a number of clinical settings, including monitoring of patient resistance prior to therapy initiation or at the time of therapy failure (1, 5, 14).
This study was conducted to investigate the quality and relative sensitivity of DNA sequence analysis procedures in a large number of laboratories worldwide. Using their standard laboratory methods and technologies, participants analyzed a coded set of plasmid mixtures containing nucleotide mixtures at several drug resistance codons and reported the results of both qualitative and quantitative interpretation.
MATERIALS AND METHODS
ENVA-1 panel.
A panel of nine plasmid mixtures at a concentration of 100 pg/ml of water (approximately 20 × 106 DNA copies/ml) was distributed among participating research and commercial laboratories in Europe and the United States. Four different plasmid constructs harboring HIV-1 HxB2 reverse transcriptase (RT) genes (amino acids 1 to 563) without flanking HIV sequences were used to prepare the panel: one containing a completely wild-type RT sequence, one containing zidovudine resistance mutations at codons 41 (ATG→CTG) and 215 (ACC→TAC), and two containing either the 184Ile (ATG→ATA) or 184Val (ATG→GTG) mutation, conferring resistance to lamivudine. Mutations had been introduced into the wild-type HxB2 sequence via site-directed mutagenesis and confirmed by sequence analysis of the complete RT genes (2). The concentration of the source plasmids was determined spectrophotometrically, and subsequently mixtures containing 25, 50, and 75% of the mutant genotype were prepared. The composition of these mixtures was checked and confirmed by using a semiquantitative point mutation assay (data not shown) (7, 19). Thereafter, the source plasmids were mixed at different ratios to create a panel of nine samples with wild-type and resistant genotypes at various relative concentrations between 0 and 100% (Table 1). It should be noted that the mutations at codons 41 and 215 are present on the same source plasmid. Therefore, the amount of mutant codon 41 in a sample is always identical to the amount of mutant codon 215 in that sample.
TABLE 1.
Composition of ENVA-1 panel
| Panel sample | % of codon(s)a
|
||||
|---|---|---|---|---|---|
| 41 + 215 WT | 41 + 215 MT | 184 Met WT | 184 Ile MT | 184 Val MT | |
| S-1 | 100 | 0 | 50b | 25 | 25 |
| S-2 | 95 | 5 | 95 | 0 | 5 |
| S-3 | 90 | 10 | 10 | 0 | 90 |
| S-4 | 75 | 25 | 50 | 50 | 0 |
| S-5 | 50 | 50 | 50 | 0 | 50 |
| S-6 | 25 | 75 | 75 | 0 | 25 |
| S-7 | 10 | 90 | 90 | 0 | 10 |
| S-8 | 5 | 95 | 95 | 0 | 5 |
| S-9 | 0 | 100 | 100 | 0 | 0 |
WT, wild type; MT, mutant.
A mixture of 50% wild-type codon 184Met (ATG) plus 25% 184Val (GTG) plus 25% 184Ile (ATA) results in a net 75% wild-type nucleotide at both the first and last nucleotides of the codon.
Genotypic analysis of the panel samples.
The panel samples (ENVA-1 panel) were coded and sent to the participants at room temperature. Participants treated the material as purified DNA and performed the genotypic analysis starting from amplification, by using their standard in-house sequencing procedures. In addition, participants filled out a questionnaire requesting information on the technologies and analysis procedures applied.
Panel samples consisted of complete wild-type or mutant genotypes or a mixture of these. In all samples containing a mixed genotype the minority genotype was always present at a concentration of least 5% (Table 1). Participants were requested to perform sequence analysis on all the samples and report the interpreted nucleotide sequence between nucleotides 30 and 800 of RT (amino acids 10 to 265) for each of the samples. In addition, laboratories were requested to provide semiquantitative information (percentages) on the heterogeneic positions, based on the sequencing results for each of the mixed nucleotide positions.
Data analysis.
Data were collected on standard report forms and entered into a central database at the European Network for the Evaluation of New Antiviral Treatments (ENVA) headquarters. Information was collected on the sequence analysis procedures and technology, i.e., sequencing hardware, DNA labelling technology, and analysis of one or both strands. In addition, the obtained qualitative and quantitative results for each of the heterogeneous positions in the panel were collected, as well as all additional mutations reported by the participants. After being entered into the database, the blinded results were reviewed for data entry errors by each of the participants.
For each of the participants we calculated the so-called mutation score (M). M is the total number of positions at which the presence of a mutation was reported, irrespective of its concentration relative to that of the wild type. The total (cumulative) number of mutations present in the panel (i.e., codons harboring between 5 and 100% of the mutations) was 25, indicating that the maximal M that could be achieved was 25.
Subsequent analyses focused on the determination of the sensitivities and accuracies of the methodologies in relation to the input mutant or wild-type concentration.
RESULTS
Participating laboratories and technologies used. Sequence analysis.
A total of 23 laboratories participated on a voluntary basis in the study: 11 European and 12 U.S. labs. The European labs were ENVA collaborating laboratories studying HIV-1 drug resistance (eight), independent research labs (two), or commercial DNA sequencing laboratories (one). The U.S.-based laboratories were ACTG laboratories (ten), a biotechnology company laboratory (one), and a laboratory that performed sequence analysis on a contract basis for clinical trials (one).
As shown in Table 2, none of the labs used manual sequencing technologies for the analysis of the panel. Seventeen of 23 labs (74%) used Applied Biosystems International (ABI) (Foster City, Calif.) automated fluorescent sequencing technology (ABI models 310, 373, and 377). The remaining six laboratories used one of the other automated platforms, i.e., ALF or ALF-express (Pharmacia Upjohn, Uppsala, Sweden), Amersham Vistra (Amersham, Little Chalfont, United Kingdom), or Affymetrix gene chip technology (Santa Clara, Calif.). This distribution of sequencing hardware and sequencing technologies allowed a comparison of individual laboratory performance but did not provide sufficient data to assess the relative performance of the different technologies.
TABLE 2.
Technologies used and M for ENVA-1a
| Technology | Total no. of labs | Strands analyzed | M (per lab) | Mean M |
|---|---|---|---|---|
| ABI dye primer | 11 | Single | 18 | 15.6 |
| Both | 19, 19, 18, 18, 16, 16, 15, 14, 12, 7 | |||
| ABY dye terminator | 6 | Single | 16, 13, 0 | 14.4 (excluding 0 score) |
| Both | 17, 17, 9 | 12.0 (including 0 score) | ||
| ALF | 3 | Single | 19 | 13.7 |
| Both | 12, 10 | |||
| Affymetrix | 2 | Single | ND | 20 |
| Both | 22, 18 | |||
| Vistra | 1 | Single | 13 | ND |
| Both | ND |
ND, not determined.
Detection of resistance mutations.
The drug resistance-conferring mutations were located at three amino acid and four independent nucleotide positions, i.e., codon 41 (ATG→CTG), 184Val (ATG→GTG), 184Ile (ATG→ATA), and 215 (ACC→TAC). The nine panel samples encoded a complete wild-type genotype at 11 of these nucleotide positions and a total of 25 positions at which a mutant (drug-resistant) genotype was present in at least 5% of the RT population. For each of the participating laboratories the total number of positions was scored at which a mutant (resistant) genotype was reported irrespective of its relative concentration (M) (Table 2). The maximum M was 25. The highest score obtained was 22, whereas one of the participants failed to detect a correct mutant genotype at any of the 25 positions. No differences in M were observed between labs that based the results on sequence analysis of one strand compared to those which analyzed both strands.
A summary of the qualitative results for each of the panel samples is shown in Tables 3 and 4. Twenty of 23 laboratories reported the presence of mutant codons 41 and 215 in the sample containing a complete mutant population. Seventeen and 18 of these 20 labs reported the presence of only the mutant genotype at codons 41 and 215, respectively. The remaining three and two laboratories reported the presence of wild-type and mutant mixtures. In addition, two laboratories reported the presence of only wild-type codons 41 and 215 in these completely mutant-encoding samples. A third participant reported the mutant codon 41 but not 215 in this sample. In two cases at two different laboratories, a mixture of wild-type and mutant genotypes at codon 41 or 215 was reported in samples containing only wild-type DNA.
TABLE 3.
Frequency of mutant detection in relation to sample composition
| % Mutant input | No. of laboratories that reported for:
|
Concordance (%)a | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Codon 41
|
Codon 215
|
||||||||
| Mutant | No mutant | Incorrect codon call | Codon not determined | Mutant | No mutant | Incorrect codon call | Codon not determined | ||
| 0 | 1 | 20 | 1 | 1 | 1 | 22 | 0 | 0 | 19/21 (90) |
| 5 | 1 | 20 | 0 | 2 | 0 | 22 | 0 | 1 | 20/21 (95) |
| 10 | 4 | 16 | 2 | 1 | 4 | 19 | 0 | 0 | 18/20 (90) |
| 25 | 13 | 8 | 1 | 1 | 10 | 11 | 2 | 0 | 15/19 (79) |
| 50 | 18 | 3 | 1 | 1 | 19 | 3 | 0 | 1 | 18/21 (86) |
| 75 | 20 | 0 | 1 | 2 | 21 | 2 | 0 | 0 | 18/20 (90) |
| 90 | 20 | 2 | 0 | 1 | 20 | 2 | 1 | 0 | 19/21 (90) |
| 95 | 20 | 1 | 1 | 1 | 20 | 2 | 0 | 1 | 19/21 (90) |
| 100 | 20 | 2 | 0 | 1 | 20 | 3 | 0 | 0 | 21/22 (95) |
Results are given as a ratio of the number of laboratories with concordant results for both codon 41 and 215 to the total number of laboratories without incorrect codon calls and with both codons analyzed.
TABLE 4.
Frequency of mutant detection in relation to sample composition
| % Mutant input | No. of laboratories that reported for codon 184
|
|||
|---|---|---|---|---|
| Mutant | No mutant | Incorrect codon call | Codon not determined | |
| 184 Val | ||||
| 0 | 0 | 21 | 2 | 0 |
| 5 | 3 | 19 | 1 | 0 |
| 10 | 5 | 15 | 3 | 0 |
| 25 | 16 | 5 | 2 | 0 |
| 50 | 20 | 2 | 1 | 0 |
| 75 | ||||
| 90 | 22 | 0 | 1 | 0 |
| 95 | ||||
| 100 | ||||
| 50 (184 Met/Ile) | 19a | 3 | 1 | 0 |
| 25 (Met/Ile) + 25 (Met/Val) | 15b | 5 | 2 | 1 |
In all 19 cases 184Ile reported.
In six cases (ATG→GTG), only first nucleotide change detected. In eight cases (ATG→GTA), both first and third nucleotide changes detected. In one case (ATG→ATA), only third nucleotide change detected.
Two of the participants reported incorrect codon calls, codons not reflecting the wild-type or mutant genotype, in several samples at both resistance codons (Table 3). The incorrect mutant genotypes reported by these laboratories were for codon 41 (AAG, GTG, or TTG instead of CTG) and codon 215 (TCC instead of TAC). In eleven cases from three laboratories and three cases from two laboratories, respectively, codons 41 and 215 were not determined, i.e., no sequence results were reported for these codons.
For codon 184 the frequency of incorrect codon calls, i.e., reported codons that did not reflect the wild type (ATG) or mutants (ATA and GTG), was higher than for codons 41 and 215 (Tables 3 and 4). All codon 184 incorrect calls reported in the samples originated from three participants. The incorrect mutant genotypes reported by these laboratories for codon 184 were ACG, ATC, or GTC instead of GTG.
In sample S-1 containing the 184 wild-type codon (ATG) in combination with both codon 184 mutants (ATA and GTG), population sequencing could not distinguish between the mutant nucleotides being present on one codon (resulting in GTA) or two codons (resulting in ATA and GTG). Therefore, the detection of GTA alone or the combination of ATA and GTG together in sample S-1 were both considered correct.
Only a minority of laboratories reported the presence of a mutant when it was present in less than 25% of the population. About half of the laboratories reported a mutation at codon 41 or 215 when the mutant genotype was present in at least 25% of the DNA population (Table 3). The proportion of labs detecting the mutant genotypes steadily increased with a further increase in the relative concentration of the mutant genotype. The presence of 5% mutant codon 41 was detected by only a single participant. However, this was the same participant who incorrectly reported the presence of a mutant genotype in the sample containing a complete wild-type DNA population.
When the first nucleotide of codon 184 encoded the mutant valine genotype (ATG→GTG) in 25% of the DNA population, its presence was reported by 16 of the laboratories when in a background of 75% wild-type methionine (ATG) and by 14 of the laboratories when present in a background of 50% wild-type (ATG) and 25% codon 184 isoleucine mutants (ATA; Table 4). The presence of a third nucleotide change (ATG→ATA) in the sample harboring the three codon 184 genotypes (sample S-1) was detected by only nine laboratories, suggesting that the sensitivities for the detection of different mutations may not be equal for each of the mutations.
Mutations present in the panel at codons 41 and 215 originated from a single plasmid. Therefore, by definition the input concentrations for both mutations within a sample were identical. Concordance of the results for these codons was checked for each of the participants that had analyzed both codons and reported the correct wild-type and mutant codons. For each panel sample, concordant results were obtained by 74 to 95% of the laboratories, with the lowest level of concordance (79%) in the panel sample containing 25% mutant genotypes (Table 3). Even in the samples containing a complete wild-type or mutant DNA population, an incomplete concordance was observed at 90 and 95%, respectively.
Quantitative detection of sequence heterogeneities.
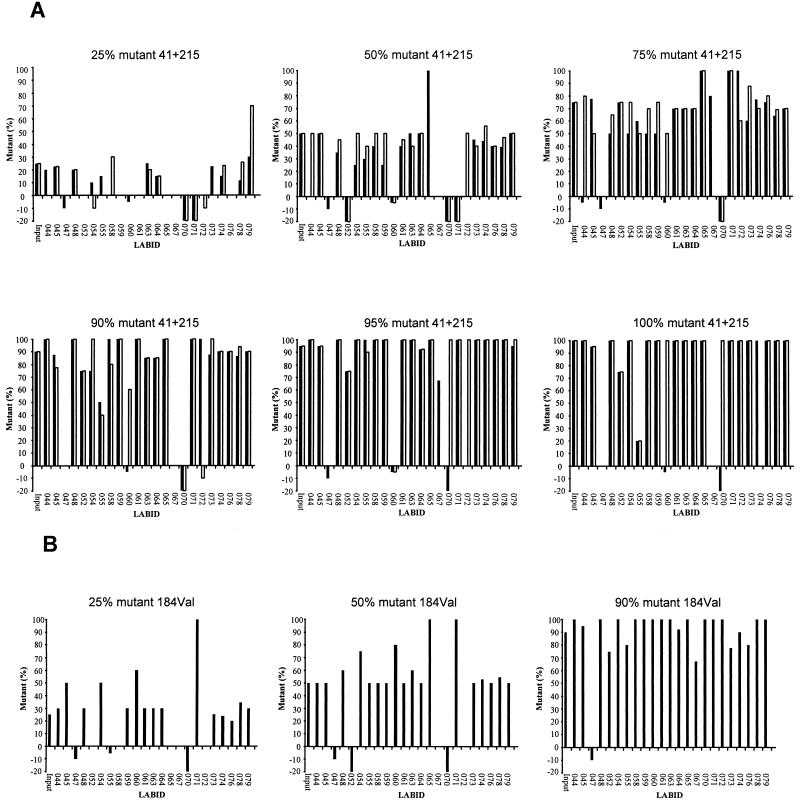
In addition to the qualitative detection of the mutations in the panel samples, laboratories also reported their best estimates for the relative proportions of wild-type and mutant genotypes at the four mutation positions in each panel sample. The majority of laboratories that reported the presence of a sequence heterogeneity also estimated its proportion, in most cases based on the double-peak spectra at these nucleotide positions in the electropherograms. These results, as shown in Fig. 1, demonstrated extensive interlaboratory differences in the estimated proportions. For those laboratories that did detect the presence of the mutation and reported a quantitative estimate, we analyzed the variation in estimated output concentrations for samples containing mixed RT populations at an input of 25 to 75% mutant genotype (Table 5). The estimated proportions were less than 25% different from the input concentration for 83 to 100% of the laboratories. For 47 to 100% of the laboratories the accuracy of the estimated proportion mutant genotype in the samples was less than 10% different from the input concentration (Table 5).
FIG. 1.
Reported percentage of mutant per panel sample and per individual laboratory. The percentage mutant input is indicated above each graph and also is shown graphically on the far left end of each graph. Individual laboratories are indicated by numbers along the horizontal axes. The bars indicate the reported percentage mutant genotype estimated upon sequence analysis. Shown are the results for codon 41 (black bars) and 215 (white bars) (A) and for codon 184-valine (B). Negative percentages indicate that a codon was not determined (−5%), an incorrect mutant codon was reported (−10%), and a mutant was detected but no quantitative information was reported (−20%).
TABLE 5.
Accuracy of estimated mutant concentrations
| % Mutant input | Laboratories reporting estimates different from input concn by:
|
|||
|---|---|---|---|---|
| <25%
|
<10%
|
|||
| Ratioa | % | Ratio | % | |
| Codon 41 | ||||
| 25 | 11/11 | 100 | 9/11 | 82 |
| 50 | 14/15 | 93 | 9/15 | 60 |
| 75 | 19/19 | 100 | 9/19 | 47 |
| Codon 215 | ||||
| 25 | 7/8 | 88 | 7/8 | 88 |
| 50 | 16/16 | 100 | 16/16 | 100 |
| 75 | 20/20 | 100 | 13/20 | 65 |
| Codon 184 | ||||
| 25 | 13/15 | 100 | 11/15 | 73 |
| 50 | 15/18 | 83 | 14/18 | 78 |
| 75 | ||||
Number of labs for which the estimated mutant concentration was less than 25% or less than 10% different from the input percentage (numerator), as a function of the total number of labs that reported a quantitative estimate (denominator).
DISCUSSION
This is the first large-scale study evaluating the quality of sequence analysis approaches for the identification of drug resistance mutations in the HIV-1 RT gene. Recent developments and improvements in genotyping technologies (9, 15, 17), a rapidly evolving understanding of the effects of resistance mutations on viral drug susceptibility, and the fact that biological drug susceptibility assays are costly, difficult to standardize, and extremely time-consuming (2, 6, 13) have led to an increasing use of genotypic drug resistance methods for HIV. At present these methods are mainly applied by research laboratories but are beginning to be implemented diagnostically. Most importantly, genotyping to determine the resistance profile in patients virologically showing treatment failure has recently been shown to be beneficial in selecting a salvage regimen (1, 5), and as such, sequencing results become of increasing importance in the clinical care of HIV-1-infected patients.
This first study evaluating the quality of HIV drug resistance genotyping protocols was based on the use of recombinant RT genes derived from a reference virus, into which mutations had been introduced via site-directed mutagenesis. This approach enabled a direct comparison of the reliability of DNA sequencing protocols in the absence of factors such as variation introduced by viral quasispecies, the nucleic acid extraction procedure, or the cDNA synthesis reactions. Participants used their standard sequencing hardware, procedures, and protocols to analyze the panel samples. Most of the participants used an ABI sequencer (17 of 23 labs); the remaining six labs used the additional available sequencing technologies. This distribution of technologies therefore did not allow the comparison of the results for each hardware technology. Moreover, the qualitative interpretation of the results demonstrated highly variable scores even between laboratories using the same sequencing technology, indicating that interlaboratory differences are extensive and may affect the results more than the differences between the technologies. The striking differences in the M values show that large differences exist in the quality of DNA sequencing results of laboratories dedicated to performing these types of analyses routinely on a research or even diagnostic basis.
The qualitative interpretation of the laboratory results for each of the panel samples demonstrated that none of the panel samples, including those containing only wild-type or mutant genotypes, was scored correctly by all of the laboratories. Incorrect codon calls, i.e., the detection of a codon sequence that did not reflect the wild-type or the mutant sequence, were reported for several of the panel samples, in particular for codons 41 and 184, independent of the mutant concentration. However, all these calls originated from three (13%) of the participants. No incorrect codons at the mutation sites were reported by any of the other participants.
In addition, the results for codons 41 and 215 demonstrated that some laboratories missed the presence of a resistance mutation, even in samples with a homogeneous genetic makeup, or in the opposite situation, the presence of a mutation was reported in samples containing a complete wild-type input. It is of note that the participant detecting the codon 41 mutation, when it was present at the level of 5%, also reported the presence of this mutation in the purely wild-type specimen. This suggests that the reported detection of the 5% minority species might result from a nonspecific sequencing reaction or sample contamination and does not necessarily reflect a highly sensitive and specific procedure.
Upon entry of the data into the database all participants reviewed the blinded results for data entry errors. It cannot be excluded that some errors may have been introduced somewhere in the entire multistep laboratory process or at the steps of data analysis, data reporting, and data entry into the central database. Since, except for data entry into a central database, all these procedures are part of the standard sample manipulation process in HIV-1 genotyping, the results may be a good reflection of the actual overall quality of these procedures in daily practice.
The analysis of the results did not include an evaluation of the quality of the sequence reaction, e.g., signal strength or peak heights, and its effect on the sequence interpretation. Since the decision to use the sequence results for interpretation was in the hands of the participants, without any predefined quality criteria, differences in the quality of the sequence reactions itself may partially explain the observed interlaboratory differences.
The results of this study indicate that before applying DNA sequencing diagnostically for the detection of drug resistance mutations, considerable improvements need to be made in the quality of the results and procedures. In order to achieve this and monitor the quality of these procedures on a continuous basis, the installation of proper quality control programs and the standardization of protocols are essential. A significant improvement in the overall quality may also come from the use of dedicated kits and procedures to perform HIV-1 drug resistance genotyping, as recently introduced by ABI and Visible Genetics (Toronto, Canada).
The sensitivity of detecting a mutant genotype when this was present at a relative concentration of 25% was comparable for the mutations at codons 41 and 215 and for the 184-valine mutation. Interestingly, a much lower frequency was observed for the 184-isoleucine mutation. This might be due to differences in the sensitivities of the sequencing procedures to detect each of the nucleotide changes. The 184-isoleucine mutation is the result of a G→A mutation, whereas for all three other codons at least one of the base changes is due to a mutation in which the wild-type A nucleotide is replaced (codon 41, A→G; codon 215, A→T; and C→A, codon 184Val A→G). The relatively low sensitivity for codon 184-isoleucine was specifically observed in a combination with both the wild type and valine variant, indicating that the high level of variation in the codon might have affected the interpretation of the analyzed nucleotide sequence. Another explanation might be that the codon 184-isoleucine mutation was unexpected by the participants, since the mutation is rather infrequent in clinical isolates from extensively lamivudine-treated patients (8, 10, 18).
As mentioned before, the inputs for codons 41 and 215 were identical, as the mutations were coupled on the source plasmid. The concordance of results for codons 41 and 215, based on the qualitative results, was high, though it was complete in none of the samples. The maximum variation, i.e., a 50% wild-type and mutant mixture, resulted in only 79% concordance, again suggesting mutation-specific differences in mutation detection sensitivities. Apart from possible explanations mentioned before, this difference might also be due to other factors such as the distance of the mutation from the sequencing primer, resulting in divergent signal strengths at the location of the mutations.
The quantitative determination of the mutations demonstrated that laboratories capable of detecting a mutation qualitatively generally estimated its relative concentration with 25% accuracy from the input concentration. This indicates that laboratories performing good quality sequencing should be able to differentiate relative mutant concentrations in strata of 25%.
This study demonstrates that extensive differences exist in the quality of DNA sequence analysis for the identification of HIV-1 drug resistance mutations. Although the capacity to determine mutations was analyzed only for the HIV-1 RT gene, there is no reason to assume that the quality of results would be different for the protease gene or other target genes and organisms. In summary, this first multicenter evaluation of DNA sequence analysis procedures for HIV-1 drug resistance demonstrates large differences in the overall quality of the results. Many of the laboratories generated moderate to good results, while none of the labs were perfect. A small number of laboratories demonstrated poor performance. Therefore, the clinical application of DNA sequencing results should be considered with care, and clinicians should be clearly educated about the current limitations of sequencing technology for HIV-1 drug resistance genotyping.
As an important step towards the improvement of the quality of the results, the development of quality control programs for genotyping is essential.
ACKNOWLEDGMENTS
We thank all who contributed to the study: M. Arens, J. Albert, B. Clotet, R. Colgrove, D. Descamps, E. Fisher, D. Huang, V. Johnson, L. Johnston-Dow, D. de Jong, E. Lorenzo, S. Kaye, D. Kuritzkes, B. Masquelier, C. Nielsen, L. Perrin, S. Rasheed, R. Respess, R. Shafer, G. Sitbon, A. M. Vandamme, O. Weislow, and Y. Zhao. Furthermore, we thank A. M. van Loon for critically reading the manuscript. R. Tedder and S. Kaye are thanked for their help in the initial quality control of the source plasmids.
This study was supported by the Biomed-2 program of the European Commission (grant BMH4-CT96-0409).
REFERENCES
- 1.Baxter J D, Mayers D L, Wentworth D N, Neaton J D, Merigan T C the CPCRA Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. Abstracts of the 6th Conference on Retroviruses and Opportunistic Infections. 1999. A pilot study of the short-term effects of antiretroviral management based on plasma genotypic antiretroviral resistance testing (GART) in patients failing antiretorviral therapy, abstr. LB 8. [Google Scholar]
- 2.Boucher C A B, Keulen W, van Bommel T, Nijhuis M, de Jong D, de Jong M D, Schipper P, Back N K T. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob Agents Chemother. 1996;40:2404–2409. doi: 10.1128/aac.40.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 4.Demeter L M, D’Aquila R, Weislow O, Lorenzo E, Erice A, Fitzgibbon J, Shafer R, Richman D, Howard T M, Zhao Y, Fisher E, Huang D, Mayers D, Sylvester S, Arens M, Sannerud K, Rasheed S, Johnson V, Kuritzkes D, Reichelderfer P, Japour A ACTG Sequencing Working Group; AIDS Clinical Trials Group. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. J Virol Methods. 1998;75:93–104. doi: 10.1016/s0166-0934(98)00100-1. [DOI] [PubMed] [Google Scholar]
- 5.Durant, J., P. Clevenbergh, P. Halfon, P. Delguidice, S. Porsin, P. Simonet, N. Montagne, E. Dohin, J. M. Schapiro, C. Boucher, and P. Dellamonica. Improving HIV therapy with drug resistance genotyping: The Viradapt Study. Lancet, in press. [DOI] [PubMed]
- 6.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Becket L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D’Aquila R T, Crumpacker Clyde S The RV-43 Study Group; The AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaye S, Loveday C, Tedder R S. A microtitre format point mutation assay: application to the detection of drug resistance in human immunodeficiency virus type-1 infected patients treated with zidovudine. J Med Virol. 1992;37:241–246. doi: 10.1002/jmv.1890370402. [DOI] [PubMed] [Google Scholar]
- 8.Kemp S D, Kohli A, Larder B A. Genotypic characterization of an HIV-1 mutant co-resistant to AZT and 3TC. J Acquired Immune Defic Syndr. 1995;10:S25. . (Abstract.) [Google Scholar]
- 9.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphism observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 10.Kuritzkes D R, Quinn J B, Shugarts D L, Griffin A, Bakhtiari M, Poticha D, Eron J J, Fallon M A, Rubin M. Drug resistance and virologic response in NUCA 3001, a randomized trial of lamivudine (3TC) versus zidovudine (ZDV) versus ZDV plus 3TC in previously untreated patients. AIDS. 1996;10:975–981. doi: 10.1097/00002030-199610090-00007. [DOI] [PubMed] [Google Scholar]
- 11.Leigh Brown A J, Richman D D. HIV-1: gambling on the evolution of drug resistance? Nat Med. 1997;3:268–271. doi: 10.1038/nm0397-268. [DOI] [PubMed] [Google Scholar]
- 12.Nijhuis M, Boucher C A B, Schipper P, Leitner T, Schuurman R, Albert J. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease inhibitor therapy. Proc Natl Acad Sci USA. 1998;95:14441–14446. doi: 10.1073/pnas.95.24.14441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nijhuis M, Schuurman R, Boucher C A B. Homologous recombination for rapid phenotyping of HIV. Curr Opin Infect Dis. 1997;10:475–479. [Google Scholar]
- 14.Perrin L, Telenti A. HIV treatment failure: testing for HIV resistance in clinical practice. Science. 1998;280:1871–1873. doi: 10.1126/science.280.5371.1871. [DOI] [PubMed] [Google Scholar]
- 15.Rosenblum B B, Lee L G, Spurgeon S L, Khan S H, Menchen S M, Heiner C R, Chen S M. New dye-labeled terminators for improved DNA sequencing patterns. Nucleic Acids Res. 1997;25:4500–4504. doi: 10.1093/nar/25.22.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antiviral News. 1997;5:129. [Google Scholar]
- 17.Schuurman R. State of the art of genotypic HIV-1 drug resistance. Curr Opin Infect Dis. 1997;10:480–484. [Google Scholar]
- 18.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, Kwok S, Sninsky J, Boucher C A B. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 19.Syvanen A C, Aalto-Setala K, Harju L, Kontula K, Soderlund H. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics. 1990;8:684–692. doi: 10.1016/0888-7543(90)90255-s. [DOI] [PubMed] [Google Scholar]