High-dimensional single-cell landscape of immune alterations during HTLV-1 infection and leukemogenesis identifies hallmarks of premalignant and malignant T-cell states and the accompanying shift of systemic immune state toward myeloid and immunosuppressive.
Abstract
Premalignant clonal expansion of human T-cell leukemia virus type-1 (HTLV-1)–infected cells occurs before viral carcinogenesis. Here we characterize premalignant cells and the multicellular ecosystem in HTLV-1 infection with and without adult T-cell leukemia/lymphoma (ATL) by genome sequencing and single-cell simultaneous transcriptome and T/B-cell receptor sequencing with surface protein analysis. We distinguish malignant phenotypes caused by HTLV-1 infection and leukemogenesis and dissect clonal evolution of malignant cells with different clinical behavior. Within HTLV-1–infected cells, a regulatory T-cell phenotype associates with premalignant clonal expansion. We also delineate differences between virus- and tumor-related changes in the nonmalignant hematopoietic pool, including tumor-specific myeloid propagation. In a newly generated conditional knockout mouse model recapitulating T-cell–restricted CD274 (encoding PD-L1) gene lesions found in ATL, we demonstrate that PD-L1 overexpressed by T cells is transferred to surrounding cells, leading to their PD-L1 upregulation. Our findings provide insights into clonal evolution and immune landscape of multistep virus carcinogenesis.
Significance:
Our multimodal single-cell analyses comprehensively dissect the cellular and molecular alterations of the peripheral blood in HTLV-1 infection, with and without progression to leukemia. This study not only sheds light on premalignant clonal expansion in viral carcinogenesis, but also helps to devise novel diagnostic and therapeutic strategies for HTLV-1–related disorders.
Introduction
Oncogenic viral infections, such as human T-cell leukemia virus type-1 (HTLV-1), are responsible for 10% to 15% of human cancers worldwide (1). These viruses can induce carcinogenesis by various mechanisms, such as chronic inflammation, cellular transformation, and immune suppression (2). Accumulating evidence suggests that clonal expansion of infected cells precedes malignancy and contributes to tumorigenesis (3, 4). Besides viral carcinogenesis, recent studies described somatic mutations in normal tissues, such as the hematopoietic and gastrointestinal systems, pointing to a critical role of premalignant clonal expansion in diverse oncogenic processes (5, 6). However, the biological properties and molecular basis of premalignant clonal expansion are not well characterized.
HTLV-1 is a retrovirus infecting approximately 5 to 20 million people worldwide, particularly in endemic regions (7). This virus preferentially targets CD4+ T cells and is associated with a variety of diseases, ranging from inflammatory disorders to adult T-cell leukemia/lymphoma (ATL). ATL is an aggressive T-cell neoplasm evolving after a long latency of >30 to 50 years. Even before ATL onset, premalignant HTLV-1–infected clones bearing driver mutations can be detected (4), offering a unique opportunity to functionally dissect premalignant clonal expansion in multistep viral carcinogenesis. However, as only a small proportion (usually <1%–5%) of peripheral blood mononuclear cells (PBMC) of asymptomatic carriers (AC) contain integrated proviral DNA (8), the phenotypical and functional evaluation of HTLV-1–infected cells is difficult.
Efficient cytotoxic T-lymphocyte (CTL) response to HTLV-1 has been reported to limit the proviral load and the risk of associated diseases (9). By contrast, ATL patients are severely immunocompromised, frequently contracting opportunistic infections (10). These observations emphasize the relevance of immune dynamics in HTLV-1–related diseases. This view is supported by genetic studies showing frequent alterations in immune molecules, such as structural variations (SV) truncating the CD274 3′-untranslated regions (UTR) in ATL (11, 12). However, although many efforts have been devoted to CTL characterization, the entire composition and functional heterogeneity of the immune microenvironment in HTLV-1 infection and ATL remain elusive.
Recent advances in single-cell analysis provide an avenue to elucidate phenotypic features at a cellular resolution, allowing profound understanding of tumor heterogeneity and microenvironment in cancer (13). Particularly, cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) is a recently developed tool for exploring transcriptomic and surface proteomic states in the same single cell (14). Also, single-cell T/B-cell receptor (TCR/BCR) sequencing (scTCR/BCR-seq) enables detection of T/B-cell clonal expansion patterns through identical TCR/BCR sequence detection (15), which can be utilized to track clonal evolution in lymphoid malignancy.
Here, to investigate the cellular and molecular architecture in HTLV-1 infection and ATL, we performed multimodal single-cell analysis, evaluating transcriptome and many surface markers. Combined with scTCR-seq, our approach deciphers functional heterogeneity of HTLV-1–infected premalignant cells and the hierarchy and clonal evolution of malignant cells. Simultaneous mRNA and protein assessment delineates discordant regulation of their expression levels, particularly in myeloid lineage. In addition, we reveal variegated alterations in the immune microenvironment, including a novel mode of PD-L1 upregulation in surrounding cells by CD274 SV-harboring tumors.
Results
Landscape of the Multicellular Ecosystem in HTLV-1 Infection and ATL
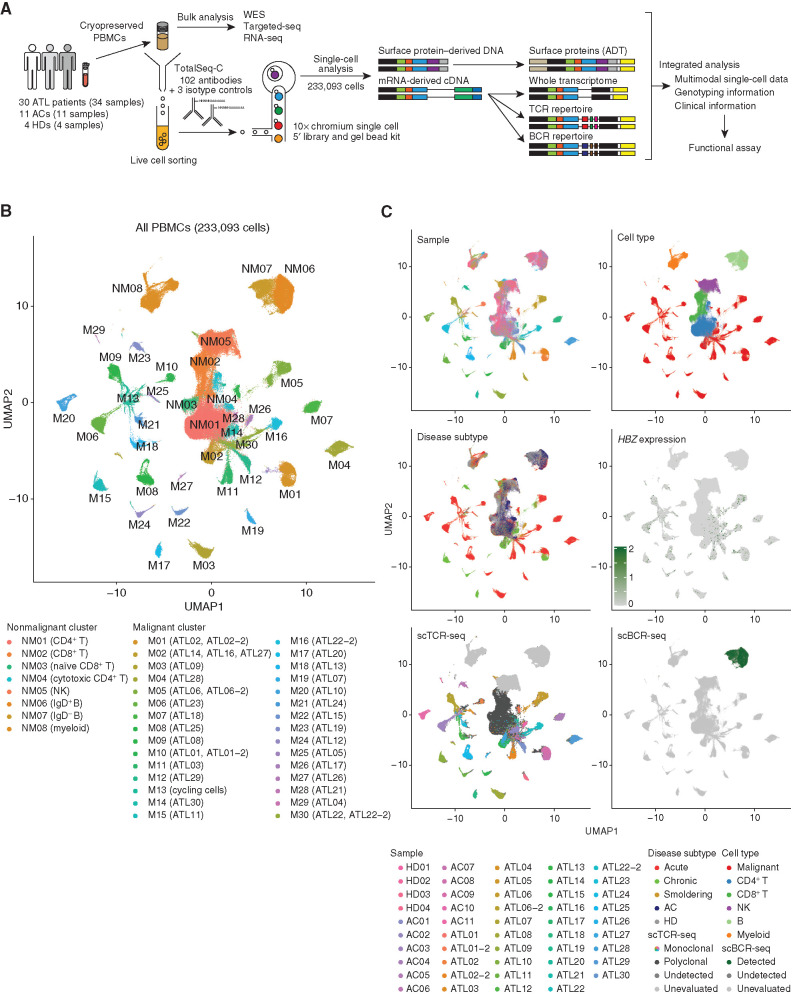
To delineate the multicellular ecosystem of HTLV-1 carrier state and ATL, we constructed transcriptomic, surface phenotypic, and immune repertoire maps using droplet-based CITE-seq combined with TCR/BCR-seq at the single-cell level. For CITE-seq analysis, we simultaneously generated 10x Genomics 5′ single-cell RNA-sequencing (scRNA-seq) and antibody-derived tag sequencing (scADT-seq) libraries from 233,093 PBMCs obtained from 30 ATL patients (34 samples, including 4 sequential ones), 11 HTLV-1 ACs, and 4 healthy donors (HD; Fig. 1A; Supplementary Table S1). Using targeted sequencing (targeted-seq), whole-exome sequencing (WES), and single-nucleotide polymorphism (SNP) array of bulk samples, somatic mutations and/or copy-number alterations (CNA) were examined for all ATL samples, most (n = 32) of which also underwent bulk RNA-seq (Supplementary Table S1). Among the analyzed 102 ADTs, we removed 32 (31%) because of insufficient separation of known positive and negative lineages and/or poor sensitivity (Supplementary Table S2). On average, 1,422 informative genes [4,477 unique molecular indexes (UMI)] and 64 antibodies (2,745 UMIs) were detected per cell (Supplementary Table S2). TCR and BCR sequences were detected in 85% and 96% of T and B cells, respectively. These quality control parameters were comparable among patient groups (Supplementary Fig. S1A).
Figure 1.
Entire landscape of single-cell CITE-seq and TCR/BCR-seq. A, Scheme of the overall study design. Targeted-seq, targeted sequencing; WES, whole-exome sequencing. B, UMAP plot of 233,093 cells from PBMCs of 4 HD, 11 AC, and 34 ATL samples; 30 malignant (M) and 8 nonmalignant (NM) clusters determined by TCR clonality are shown in different colors. C, UMAP plots (same as A) colored by sample, cell type, disease subtype, HBZ mRNA level, scTCR-seq, and scBCR-seq. See also Supplementary Fig. S1.
Dimensionality reduction analysis applied to scRNA-seq identified a total of 38 clusters, consisting of 30 malignant and 8 nonmalignant ones as determined by TCR clonality (Fig. 1B; Supplementary Fig. S1B; Supplementary Table S3). Nonmalignant clusters consisted of two CD4+ T-cell clusters, two CD8+ T-cell clusters, two B-cell clusters, one natural killer (NK)–cell cluster, and one myeloid cell cluster based on mRNA and ADT levels of canonical lineage markers (Supplementary Fig. S1C). Within the malignant clusters, different ATL samples were segregated into different clusters, suggestive of interpatient heterogeneity, although sequential samples were grouped into the same clusters (Fig. 1B and C). By contrast, nonmalignant cells, including those from ACs and HDs, were classified into the same clusters according to their lineage, suggesting their transcriptomic similarity among patients. Almost all malignant clusters expressed HTLV-1 HBZ (Fig. 1C). Although almost no cells were classified into malignant clusters in ACs or HDs, malignant cells accounted for 15% to 99% (median 82%) of the total cells in ATL, which were lower in smoldering than in other subtypes (Supplementary Fig. S1D and S1E). Single-cell mutation and CNA estimation revealed somatic alterations almost exclusively in malignant clusters, consistent with the bulk sequencing and SNP array results (Supplementary Fig. S1F and S1G), confirming successful separation of malignant from nonmalignant cells.
Lineage-Specific Regulation of mRNA and ADT Levels
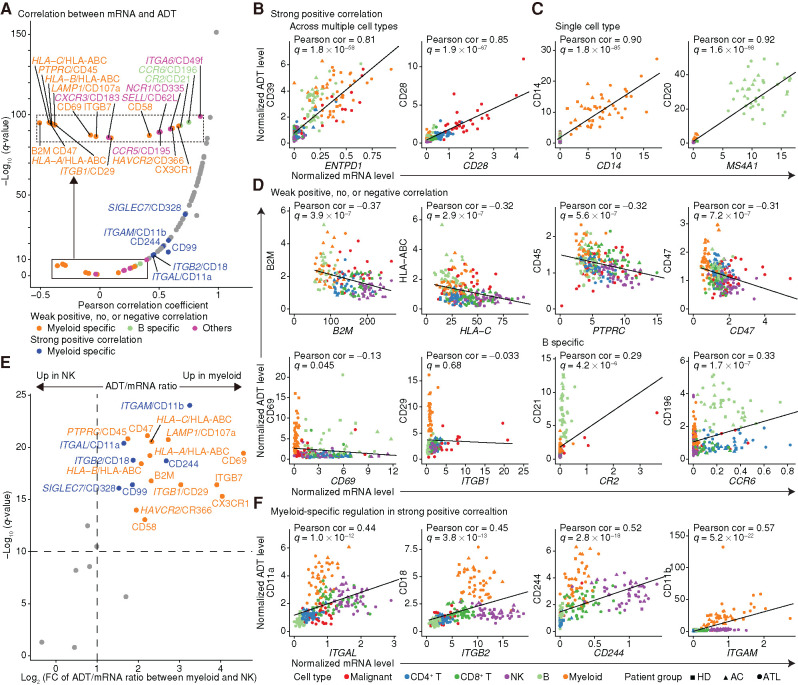
Protein abundance is a more direct determinant of cellular functions but cannot necessarily be inferred from mRNA level due to posttranscriptional processes (16). CITE-seq analysis in healthy, virally infected, and leukemic states enables a detailed comparison of mRNA and ADT level changes in various cell types. At the single-cell level, ADT was less prone to dropout than mRNA, conferring higher sensitivity for many surface markers (Supplementary Fig. S2A; Supplementary Table S2). As expected, more than two thirds of genes showed strong positive correlations between mRNA and ADT levels when averaging expression within clusters (Fig. 2A), suggesting that overall expression of many surface markers is transcriptionally regulated. These included surface markers showing a moderate to high expression in multiple cell types, including ENTPD1/CD39 and CD28, for which mRNA and ADT levels were highly correlated within and across lineages (Fig. 2B; Supplementary Fig. S2B). Lineage-specific markers, such as MS4A1/CD20 and CD14, were also observed, showing elevated expression of both mRNA and ADT levels in a single cell type (Fig. 2C; Supplementary Fig. S2B).
Figure 2.
Comparison of mRNA and ADT levels across cell types. A, Correlation coefficients and their significance for normalized mRNA and ADT levels. Genes with weak positive, no, or negative correlations (q ≥ 1 × 10−10) were depicted separately. Dots are colored if genes are under lineage-specific posttranscriptional regulation; otherwise, they are colored gray. The inset (dotted box) represents an enlarged view of the solid box. B–D, mRNA and ADT levels for representative genes showing a strong positive correlation across multiple cell types (B) and in a single cell type (C), and a weak positive, no, or negative correlation (D), with shapes and colors indicating patient group and cell type. E, Volcano plot comparing the ratio of ADT to mRNA levels between myeloid (NM08) and NK (NM05) cells. Fold change (FC) was calculated as the ADT/mRNA ratio in myeloid cells divided by that in NK cells when the average mRNA level was ≥0.2 in both cell types. ADTs with q < 10−10 and log2(FC) ≥ 1 were considered significant and colored by correlation significance. Mann–Whitney U test. F, mRNA and ADT levels of representative genes with dissociated ADT upregulation in myeloid cells. B–D and F, Each dot represents an average expression value for each cell type (≥30 cells) in each sample. Pearson correlation (cor) test with Benjamini–Hochberg correction. Fitted linear regression line is shown. See also Supplementary Fig. S2.
However, there were important exceptions: Several surface markers, such as major histocompatibility complex (MHC) class I molecules and PTPRC/CD45, exhibited weak positive, no, or even negative correlations between mRNA and ADT levels (Fig. 2A and D). For these markers, some cell types displayed substantial upregulation of ADT levels despite low to moderate mRNA levels, whereas other cell types showed low ADT abundance irrespective of mRNA level. Among these markers, myeloid cell–specific dissociated ADT elevation was observed for 11 markers, including CD47, CD69, and ITGB1/CD29, whereas CCR6/CD196 and CR2/CD21 showed dissociated ADT upregulation only in B cells (Fig. 2D; Supplementary Fig. S2B), suggesting lineage-specific posttranscriptional regulation.
As such dissociated ADT upregulation was frequently observed between myeloid and NK cells, we compared the ratio of ADT to mRNA level between these cell types for surface markers including those showing strong positive correlations (Fig. 2E). This analysis identified an additional six markers, including ITGAL/CD11a and ITGB2/CD18, showing higher ADT levels in myeloid cells than NK cells with equal or higher mRNA levels (Fig. 2F; Supplementary Fig. S2B), which included a lineage marker or functional protein in myeloid cells (17). The dissociated ADT upregulation in myeloid cells was validated using other public CITE-seq data sets (Supplementary Fig. S2C–S2E). Our approach systematically delineated lineage-specific regulation of surface marker expression, demonstrating the involvement of posttranscriptional regulatory mechanisms, operating particularly in myeloid cells.
Phenotypic and Transcriptomic Characterization of Malignant and Nonmalignant CD4+ T Cells
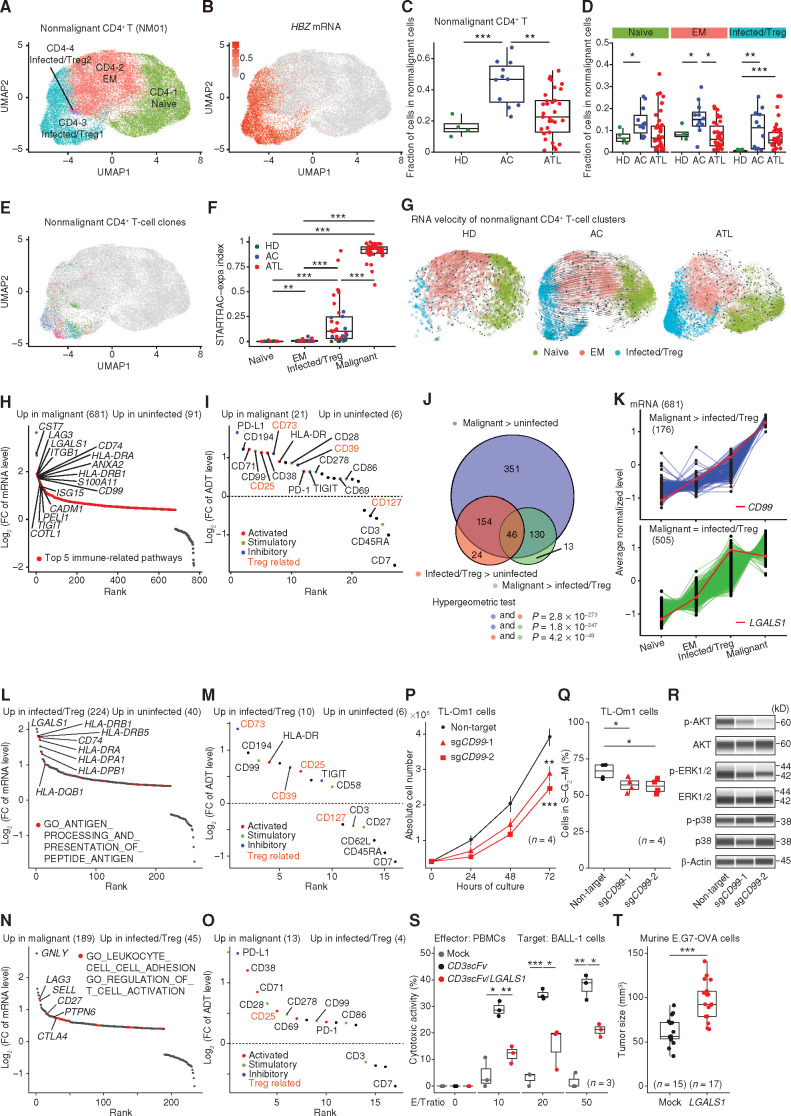
Then, we comprehensively investigated phenotypic and transcriptomic features of malignant and nonmalignant CD4+ T-cell clusters, as distinguished by TCR clonality. Within the nonmalignant CD4+ T-cell cluster (NM01, 35,668 cells), we identified four subclusters. Subclusters CD4-3 and -4 were characterized by HBZ, FOXP3, and CADM1 mRNA levels and the CD45RO+CD25+CD7−CD194+ immunophenotype, suggestive of the regulatory T-cell (Treg)–like features (Fig. 3A and B; Supplementary Fig. S3A–S3D; Supplementary Table S4). We therefore referred to these subclusters as HTLV-1–infected/Tregs, as previously reported (18). Subcluster CD4-1 and CD4-2 showed typical naïve CD45RA+CD7+CD62L+ and effector/memory (EM) CD45RO+CD127+ immunophenotypes, respectively, together with corresponding mRNA levels (Supplementary Fig. S3B and S3C; Supplementary Table S4). In total, about one third of nonmalignant CD4+ T cells (NM01) were classified into HTLV-1–infected/Treg clusters in ATL and ACs, but only 4% in HDs, which were found to be HBZ negative (Fig. 3C and D; Supplementary Fig. S3A and S3E). These results suggest that the HTLV-1–infected/Treg clusters consisted mainly of HTLV-1–infected cells, although a small number of normal Tregs were also present. The fraction of HTLV-1–infected/Tregs was highly correlated with HTLV-1 proviral load in ACs (Supplementary Fig. S3F), confirming the reliability of our estimation.
Figure 3.
Cellular and molecular features of malignant and nonmalignant CD4+ T cells. A, Subclustering of nonmalignant CD4+ T cells (NM01). B, Normalized HBZ mRNA level (after ALRA imputation) on UMAP plots in A. C and D, The fraction of total nonmalignant CD4+ T cells (NM01; C) and naïve (CD4-1), EM (CD4-2), and infected/Treg (CD4-3 and -4) CD4+ T cells (D) in nonmalignant cells (NM01–08) for each sample. E, T-cell clones (present in ≥5 cells) on UMAP plots in A. Each color represents a distinct TCR clonotype. F, Clonal expansion levels of each CD4+ T cluster (≥30 cells) for each sample. G, Steady-state RNA velocity of nonmalignant CD4+ T clusters from HD, AC, and ATL samples. H and I, Differentially expressed mRNAs [q < 0.01 and log2(FC) > 0.4; H] and ADTs [q < 0.01 and log2(FC) > 0.3; I] between uninfected (CD4-1 and -2) and malignant (M01–30) clusters. FC, fold change. J, Overlap of upregulated mRNAs among comparisons between uninfected (CD4-1 and -2), infected/Treg (CD4-3 and -4), and malignant (M01–30) clusters. Hypergeometric test. K, Gene-expression patterns among CD4+ T clusters with higher (top) or comparable (bottom) mRNA levels in malignant (M01–30) compared with infected/Treg (CD4-3 and -4) clusters among those upregulated in malignant (M01–30) compared with uninfected (CD4-1 and -2) clusters. CD99 and LGALS1 are shown in red. L–O, Differentially expressed mRNAs (L and N) and ADTs (M and O) between uninfected (CD4-1 and -2) and infected/Treg (CD4-3 and -4; L and M) clusters and between infected/Treg (CD4-3 and -4) and malignant (M01–30; N and O) clusters. H, L, and N, Red color represents upregulated mRNAs related to indicated pathways. I, M, and O, Different colors represent different sets of T-cell–related ADTs. Treg-related ADTs are shown in orange. H, I, and K–O, Numbers of significant mRNAs or ADTs are shown in parentheses. P and Q, Growth curve (mean ± SD; P) and the frequency of cells in S–G2–M phase (Q) for TL-Om1 cells transduced with indicated single-guide RNAs (sgRNA; n = 4). R, Phospho (p)-AKT, AKT, p-ERK1/2, ERK1/2, p-p38, p38, and β-Actin (loading control) expressions for the same TL-Om1 cells. S, Cytotoxic activity of PBMCs (effector: E) against BALL-1 cells transduced with indicated plasmids (target: T) at various E/T ratios (n = 3). CD3scFV, anti-CD3 single-chain variable fragment. T, Tumor volume of mock- or LGALS1-transduced E.G7-OVA tumors at 6 days after transplantation (n = 15–17). C, D, F, P, Q, S, and T, Welch t test. ***, P < 0.0005; **, P < 0.005; *, P < 0.05. See also Supplementary Figs. S3 and S4.
Among 23,457 uninfected CD4+ T cells (CD4-1 and -2) expressing at least one TCR (either α or β chain), 63% of cells expressed paired productive TCR αβ chains, whereas only one or more than two TCRs were detected in 28% and 8% of cells, respectively (Supplementary Fig. S3G; Supplementary Table S5), consistent with previous studies (19, 20). Clonal analysis by the single T-cell analysis by RNA-seq and TCR tracking-expansion (STARTRAC-expa) index (15) revealed almost exclusive clonal expansions in HTLV-1–infected/Treg clusters (Fig. 3E and F; Supplementary Table S5), indicating a clear biological disparity between HTLV-1–infected and uninfected cells. The STARTRAC-expa index was proportional to the fraction of HTLV-1–infected/Tregs in ACs (Supplementary Fig. S3F), suggesting that HTLV-1–infected cell expansion can be attributed to clonal proliferation. Overall, the proportion of CD4+ T cells among nonmalignant cells was elevated in ACs (Fig. 3C). Interestingly, uninfected CD4+ T cells, including naïve and EM cells, were increased in ACs compared with HDs (Fig. 3D), suggesting that HTLV-1 infection affects even uninfected cells. RNA velocity and pseudotime trajectory analyses (21) showed rapid differentiation from naïve to EM only in ACs (Fig. 3G; Supplementary Fig. S3H). These findings suggest that HTLV-1 infection not only intrinsically but also extrinsically regulates T-cell proliferation.
Next, we investigated the similarities and differences of mRNA and ADT levels among uninfected, infected/Treg, and malignant clusters. First, by comparing with uninfected cells (CD4-1 and -2), we identified 681 upregulated and 91 downregulated genes in malignant cells (M01–30; Fig. 3H; Supplementary Table S6). These malignant signature gene expressions were more strongly altered in aggressive than in indolent subtypes (Supplementary Fig. S4A and S4B). Functional enrichment analysis showed upregulation of various immune-related pathways. Comparison of ADT levels revealed co-upregulation of a wide range of immune-related molecules, including inhibitory molecules (PD-L1, CD73, and CD39) and activation markers (CD71, CD25, and CD38) in malignant cells (Fig. 3I; Supplementary Table S6). Besides these reported molecules, malignant cells also exhibited increased expression of stimulatory molecules (CD99, CD28, and CD278). These molecules were frequently coexpressed in malignant cells (Supplementary Fig. S4C), suggesting their synergistic function. By contrast, naïve T-cell markers, such as CD45RA and CD7, were downregulated in malignant cells as reported (18). Interestingly, among CD4+ T-cell clusters, despite their mRNA upregulation, malignant cells exhibited decreased CD3 and CD62L ADT abundance (Supplementary Fig. S4D and S4E), suggesting their tumor-specific posttranscriptional regulatory mechanism.
Interestingly, many differentially expressed mRNAs showed sequential expression changes from uninfected to malignant cells (Fig. 3J and K; Supplementary Fig. S4F and S4G). Among the upregulated genes, 176 genes showed higher expression in malignant (M01–30) than infected/Treg clusters (CD4-3 and -4), whereas the remaining 505 gene levels were comparable (Fig. 3J and K). A substantial proportion of these genes were upregulated in infected/Treg clusters (CD4-3 and -4) and remained high or even increased (154 and 46 genes) in malignant cells (M01–30), suggesting that ATL cells acquire additional malignant phenotype during leukemogenesis while maintaining the transcriptomic features of HTLV-1–infected cells. Comparison between nonmalignant clusters identified 224 upregulated and 40 downregulated genes in infected/Tregs (CD4-3 and -4) relative to uninfected cells (CD4-1 and -2), most of which remained high and low in malignant cells, respectively (Fig. 3L; Supplementary Table S6). Interestingly, MHC class II pathways were enriched in the upregulated genes, suggesting, together with higher HLA-DR ADT level in infected/Tregs (Fig. 3M), that MHC class II upregulation may contribute to HTLV-1–infected cell proliferation by stimulating T–T interaction and intracellular signaling (22). Importantly, some differentially expressed mRNAs and ADTs, including CD73 and CD99, showed a better discriminatory power between uninfected and infected/Tregs than known molecules (Fig. 3M; Supplementary Table S6), pointing to their potential as an HTLV-1 infection marker.
By comparison with nonmalignant infected/Tregs (CD4-3 and -4), 189 upregulated and 45 downregulated genes were identified in malignant cells (M01–30; Fig. 3N; Supplementary Table S6). Pathways regulating T-cell adhesion and activation, such as LAG3 and CTLA4, were significantly enriched in the upregulated genes. Moreover, comparison of ADT levels depicted elevated expression of costimulatory and coinhibitory molecules, particularly those related to somatic alterations (PD-L1 and CD28; Fig. 3O; Supplementary Fig. S4H; Supplementary Table S6), suggesting further deregulation of immune-modulatory property in malignant cells. These cells also showed coordinated upregulation of several activation markers (CD38, CD71, and CD25), suggesting their highly activated state. Therefore, although both HTLV-1 infection and malignant transformation alter the activity of diverse immune pathways, they affect a different set of molecules that are supposed to collaborate in multistep carcinogenesis by HTLV-1.
CD99 is a transmembrane protein involved in T-cell activation and adhesion, but its function is poorly understood (23). CD99 expression was higher in infected/Tregs (CD4-3 and -4) compared with uninfected cells (CD4-1 and -2) and further elevated in malignant cells (M01–30; Fig. 3K). Bulk RNA-seq data showed a stronger CD99 expression in aggressive rather than indolent subtypes (Supplementary Fig. S4I). Notably, CRISPR-mediated CD99 knockout inhibited cell proliferation in vitro and caused a decrease in cycling cells in the TL-Om1 ATL cell line (Fig. 3P and Q; Supplementary Fig. S4J). Among proliferation-related signaling pathways, CD99 disruption suppressed AKT activation (Fig. 3R). LGALS1 encodes Galectin-1, a secreted protein exerting pleiotropic immune functions (24). LGALS1 was upregulated in infected/Tregs (CD4-3 and -4) and remained high in malignant cells (M01–30; Fig. 3K). Bulk RNA-seq data showed equivalent LGALS1 expression between aggressive and indolent subtypes (Supplementary Fig. S4K). Lentiviral overexpression of LGALS1 on BALL-1 cells significantly attenuated the cytotoxic activity of anti-CD3/CD28–stimulated PBMCs (Fig. 3S; Supplementary Fig. S4L). In addition, LGALS1 overexpression promoted tumor growth in transplantation of murine E.G7-OVA T-cell lymphoma (Fig. 3T; Supplementary Fig. S4M). In summary, these data corroborated the single-cell data that highly upregulated immune proteins play a pivotal role in ATL pathogenesis.
Functional Heterogeneity of HTLV-1–Infected Nonmalignant and Malignant CD4+ T Cells
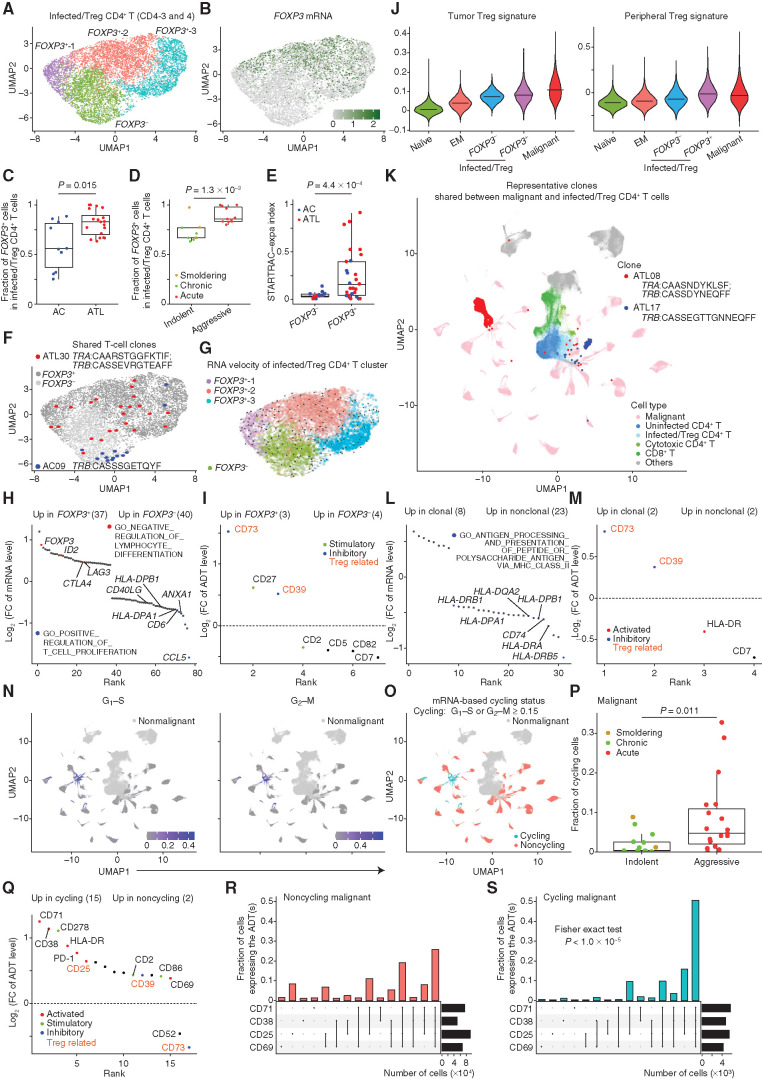
To evaluate the functional heterogeneity of HTLV-1–infected T cells, we performed subclustering of these cells, identifying three FOXP3+ clusters and one FOXP3− cluster (Fig. 4A and B; Supplementary Fig. S5A; Supplementary Table S4). Although all samples contained a varying degree of both FOXP3+ and FOXP3− HTLV-1–infected cells, the proportion of FOXP3+ cells was elevated in ATL patients compared with ACs and was higher in aggressive versus indolent subtypes (Fig. 4C and D). FOXP3+ cells displayed a higher degree of clonal propagation than FOXP3− cells by STARTRAC-expa index (Fig. 4E; Supplementary Fig. S5B; Supplementary Table S5), suggesting that the FOXP3+ phenotype is a premalignant state likely resulting in clonal evolution. Importantly, FOXP3+ and FOXP3− cells shared TCRs in several ATL patients and ACs, but no unidirectional flow was demonstrated between these cells by RNA velocity and pseudotime trajectory analyses (Fig. 4F and G; Supplementary Fig. S5C and S5D), evidently demonstrating the phenotypic plasticity between FOXP3+ and FOXP3− states. Compared with FOXP3− cells, FOXP3+ cells exhibited the upregulation of transcriptional programs negatively regulating lymphocyte activation, such as CTLA4 and LAG3, and CD73 and CD39 immunosuppressive proteins (Fig. 4H and I; Supplementary Fig. S5E and S5F; Supplementary Table S6), suggesting that FOXP3+ cells resemble Tregs with highly suppressive activity. This phenotype was validated by elevation of established Treg gene signature scores (ref. 25; Fig. 4J). These findings suggest that FOXP3+ Treg–like cluster expansion precedes ATL development, favoring premalignant clonal proliferation.
Figure 4.
Functional heterogeneity of HTLV-1–infected nonmalignant and malignant CD4+ T cells. A, Subclustering of infected/Treg CD4+ T clusters (CD4-3 and -4). B, Normalized FOXP3 mRNA level on UMAP plot in A. C and D, The fraction of FOXP3+ cells in infected/Treg clusters (CD4-3 and -4) for each sample grouped by disease state (C) and by disease subtype (D). E, Clonal expansion levels of FOXP3− and FOXP3+ cells for each AC and ATL sample. F, T-cell clones shared between FOXP3− (light gray) and FOXP3+ (dark gray) cells. G, Steady-state RNA velocity of infected/Treg clusters (CD4-3 and -4). H and I, Differentially expressed mRNAs [q < 0.01 and log2(FC) > 0.4; H] and ADTs [q < 0.01 and log2(FC) > 0.3; I] between FOXP3+ and FOXP3− cells. FC, fold change. J, Tumor Treg (left) and peripheral Treg (right) signature scores for each CD4+ T-cell cluster. Lines indicate medians. K, Representative clones shared between malignant (M01–30) and infected/Treg (CD4-3 and -4) clusters on UMAP plot in Fig. 1B, colored by cell type. L and M, Differentially expressed mRNAs (L) and ADTs (M) between clonal and nonclonal cells of infected/Treg clusters (CD4-3 and -4). H and L, Red and blue colors represent upregulated and downregulated mRNAs related to indicated pathways, respectively. N and O, G1–S (left) and G2–M (right) signature scores (N) and their based cycling status (G1–S or G2–M score ≥ 0.15; O) of malignant cells on UMAP plots in Fig. 1B. P, The fraction of cycling cells in malignant cells for each ATL sample, grouped and colored by disease subtype. Q, Differentially expressed ADTs between cycling and noncycling cells. I, M, and Q, Different colors represent different sets of T-cell–related ADTs. Treg-related ADTs are shown in orange. R and S, UpSet plot showing overlap of T-cell activation ADTs expressed (normalized expression ≥ 0.5) in noncycling (R) and cycling (S) cells. C–E and P, Welch t test. H–M and Q, Numbers of significant mRNAs or ADTs are shown in parentheses. See also Supplementary Fig. S5.
Next, we phenotypically characterized clonally expanded cells within HTLV-1–infected/Treg clusters (CD4-3 and -4). Indeed, many clones harbored the identical TCRs with malignant clonal cells in many ATL patients (17 of 34 samples; Fig. 4K), confirming that ATL frequently develops from expanded nonmalignant clones. More than one third of ACs harbored somatic mutations related to ATL and/or clonal hematopoiesis of indeterminate potential (refs. 5, 11; Supplementary Table S7), suggesting that these mutations may cooperate with HTLV-1 infection to drive premalignant clonal expansions. Interestingly, multiple MHC class II pathway genes were modestly reduced in clonal cells relative to nonclonal cells (Fig. 4L; Supplementary Fig. S5G; Supplementary Table S6). This reduction was confirmed by a slight decrease of HLA-DR ADT level (Fig. 4M), suggesting that HTLV-1–infected T cells may downregulate excessive MHC class II molecules for clonal expansion, thereby avoiding MHC class II–mediated immunity or anergy (22).
Although intertumor heterogeneity is evident in ATL, intratumor differences were also demonstrated by a principal component analysis (PCA) of mRNA levels, which showed the significant association of PC1 and PC2 with cell-cycle genes (Supplementary Fig. S5H and S5I). For more precise characterization, we used G1–S and G2–M signature scores (26) and found a small proportion of cycling cells in each tumor (0%–33%; mainly in M13 cluster), which were higher in aggressive than indolent subtypes (Fig. 4N–P; Supplementary Fig. S5J). As expected, cycling cells showed stronger expression of activation markers, such as CD71 and CD38, than noncycling cells, most of which were simultaneously positive (Fig. 4Q–S; Supplementary Fig. S5K). Interestingly, CD52 and CD73 were downregulated in cycling cells (Supplementary Fig. S5K), suggesting their negative impact on cycling malignant cells. Thus, these results suggest that transcriptomic and immunophenotypic heterogeneity is related to cell cycling and activation in malignant cells.
ATL Clonal Evolution Revealed by scTCR-seq
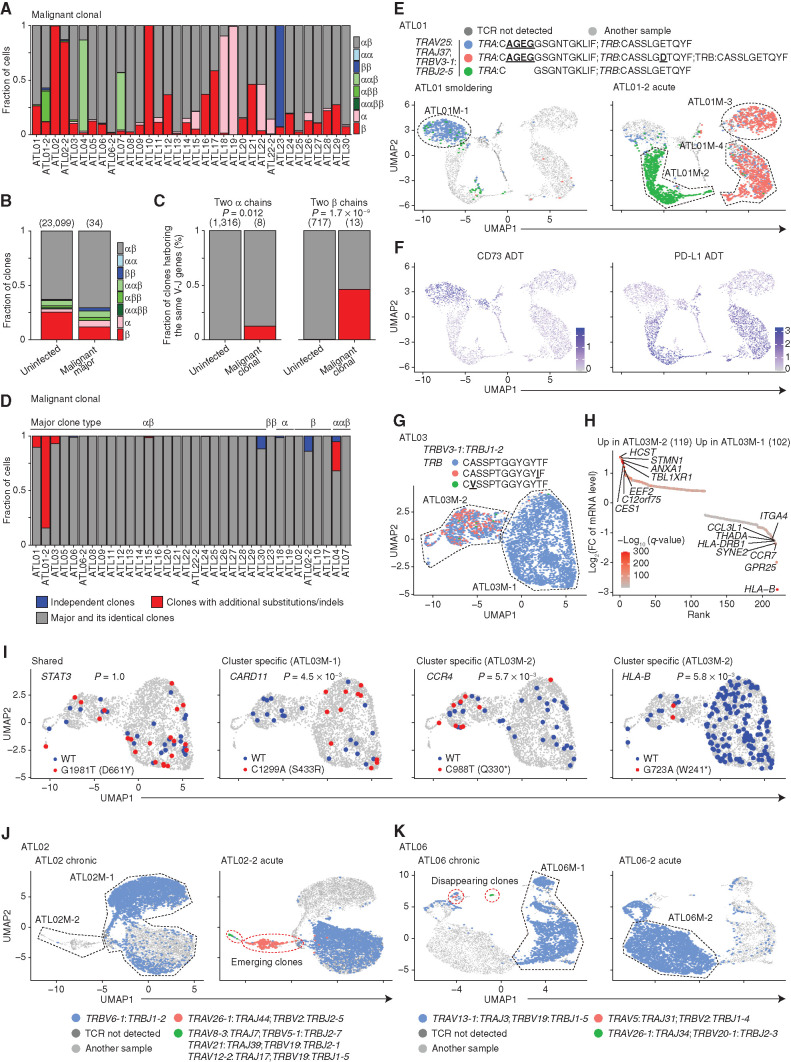
To further delineate intratumor heterogeneity, we analyzed scTCR-seq data in malignant clusters (M01–30), where most expanded (= major) clones expressed two TCRs in 25 samples (24 αβ and 1 ββ), and only one or three TCRs were mainly detected in 6 (2 α and 4 β) and 3 samples (2 ααβ and 1 αββ; Fig. 5A; Supplementary Fig. S6A; Supplementary Table S5). These numbers were proportional to the TCR distribution of uninfected CD4+ T cells (CD4-1 and -2; Fig. 5B), suggesting that although malignant clones present diverse TCR expression, they mostly reflect the physiologic TCR variation. There were, however, notable exceptions: In uninfected CD4+ T cells (CD4-1 and -2), different TCR V-J pairs were almost uniformly found in clones expressing two α or β chain (ααβ or αββ; Fig. 5C), likely resulting from allelic inclusion (27). By contrast, 36% of malignant cells (M01–30) with two α or β chain expression harbored the same pairs (Fig. 5C; Supplementary Fig. S6B), suggesting TCR duplication. Indeed, TRB amplification was detected in a sample (ATL01-2) predominantly expressing the same TRB V-J pair (Supplementary Fig. S6C). In this case, two slightly different TRB complementarity-determining region 3 (CDR3) sequences were detected in the dominant clone (mainly in ATL01M-3 and -4; Fig. 5D and E; Supplementary Table S5). Considering that a dominant clone harbored the former CDR3 sequence in the smoldering phase (ATL01), these results indicate that a CDR3 substitution occurred after TRB duplication. TRA CDR3 deletion was also identified in another major clone from the acute phase (ATL01-2). Different transcriptomic and immunophenotypic patterns, such as reciprocal expression of CD73 and PD-L1, were shown among major clones (Fig. 5F; Supplementary Fig. S6D–S6G). Interestingly, sequential comparison depicted that the same clones were present as a subclone in the smoldering phase and expanded after chemotherapy, suggesting different therapeutic sensitivity among ATL clones (Fig. 5E).
Figure 5.
ATL clonal evolution revealed by scTCR-seq. A, The fraction of malignant clonal cells (≥5 cells) expressing different numbers of productive TCR α and β chains in each ATL sample. B, The fraction of clones expressing different numbers of productive TCR α and β chains in uninfected CD4+ T-cell and most expanded (= major) malignant clones. C, The fraction of clones harboring the same V-J gene pairs in uninfected CD4+ T and malignant cell clones expressing two productive α (left) or β (right) chains. B and C, Numbers of TCR clonotypes are shown in parentheses. D, The fraction of malignant clonal cells from major and its identical clones (gray), clones with additional substitutions/indels (red), or independent clones with different V-J pairs (blue) for each ATL sample. E and F, Distribution of expanded TCR clonotypes in ATL01 (left) and ATL01-2 (right; E) and normalized CD73 (left) and PD-L1 (right) ADT levels (F) on UMAP plots of malignant cells from both samples. G, Distribution of expanded TCR clonotypes on UMAP plot of ATL03 malignant cells. E and G, Underlined, bold text indicates CDR3 mutations. H, Differentially expressed mRNAs [q < 0.01 and log2(FC) > 0.4] between ATL03M-1 and ATL03M-2 malignant clusters. Red color gradient indicates q-value. FC, fold change. I, Somatic STAT3, CARD11, CCR4, and HLA-B mutations and their references on UMAP plots in G, detected by scRNA-seq data. Asterisk (*) indicates stop codon. WT, wild-type. C and I, Fisher exact test. J and K, Distribution of expanded TCR clonotypes in ATL02 (left) and ATL02-2 (right; J) and in ATL06 (left) and ATL06-2 (right; K) on UMAP plots of malignant cells from both samples of each patient. Red dotted lines surround clones emerging (J) or disappearing (K) after progression. E, G, J, and K, Black dotted lines surround subclusters. See also Supplementary Fig. S6.
Another example was ATL03, harboring several minor clones with additional TRB CDR3 substitutions with the same V-J pair (Fig. 5G). HLA-B expression loss was found in a cluster encompassing these minor clones (ATL03M-2), where an HLA-B nonsense mutation, together with other cluster-specific mutations, was identified by single-cell mutation analysis (Fig. 5H and I; Supplementary Fig. S6H). Similar CDR3 mutations were found in subclones from five samples (Fig. 5D), most of which were validated by bulk WES or RNA-seq (Supplementary Table S5), suggesting that subclonal acquisition of TCR mutations is common in ATL evolution.
Besides such TCR subclones, subclones with different V-J pairs were found in seven samples (21%; Fig. 5D; Supplementary Table S5), suggesting that independent subclones frequently arise in the same patient. In a sample harboring the largest subclone (ATL30), transcriptomic and immunophenotypic differences between independent clones were confirmed (Supplementary Fig. S6I–S6K). Moreover, sequential analysis of ATL02 revealed the emergence of an independent clone after acute transformation, which showed the enhanced malignant signature, together with other samples after progression (Fig. 5J; Supplementary Fig. S6L–S6N). By contrast, in ATL06, whereas a dominant clone remained, several independent subclones disappeared after acute transformation (Fig. 5K), suggesting their clonal selection. Thus, independent subclones with different biological features and clinical behavior frequently develop in ATL.
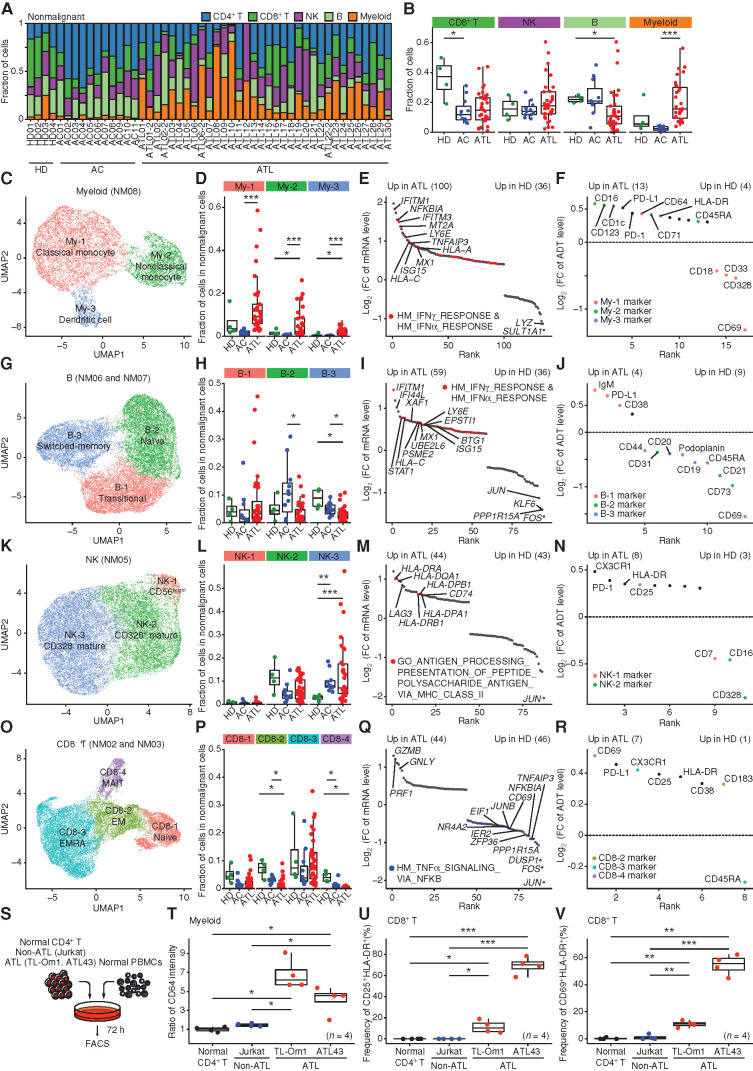
Dynamic Changes in the Nonmalignant Hematopoietic Pool of HTLV-1 Infection and ATL
HTLV-1 infection may induce various microenvironmental changes, whereas additional modulation can occur through ATL leukemogenesis. To uncover the complexity of the HTLV-1–related immune ecosystem, we evaluated nonmalignant populations. Among their changes, the most prominent was a marked increase of myeloid cells in ATL, particularly in aggressive subtypes, despite a decreasing trend in ACs (Fig. 6A and B; Supplementary Fig. S7A), pointing to the relevance of the myeloid lineage in the nonmalignant hematopoietic pool of ATL. Subclustering of the myeloid cluster (NM08, 11,698 cells) revealed three subclusters: CD14+ classic monocytes (My-1), CD16+ nonclassic monocytes (My-2), and CD1c+ dendritic cells (My-3; Fig. 6C; Supplementary Fig. S7B–S7D; Supplementary Table S4). All these subcluster fractions, particularly of nonclassic monocytes and dendritic cells, were increased in ATL (Fig. 6D). Functional enrichment analysis in total myeloid cells showed the upregulation of interferon (IFN) γ and α pathways in ATL patients compared with HDs (Fig. 6E; Supplementary Table S6). Although ADT levels largely reflected the subcluster fraction shift, upregulation of activation markers (including CD64) and immune checkpoints (such as PD-1) was observed in ATL (Fig. 6F; Supplementary Table S6), suggesting the invigoration of increased myeloid lineage. Of note, CD69, an early activation marker, was downregulated in both ACs and ATL patients, suggesting differential regulation of CD69 from other activation markers.
Figure 6.
Dynamic changes in the nonmalignant hematopoietic pool of HTLV-1 infection and ATL. A and B, The fraction of each cell type in nonmalignant cells is shown for each sample (A) and compared among patient groups (B). C–R, Evaluation of myeloid (NM08; C–F), B (NM06 and NM07; G–J), NK (NM05; K–N), and CD8+ T (NM02 and NM03; O–R) cells. C, G, K, and O, Subclustering of myeloid (C), B (G), NK (K), and CD8+ T (O) cells. D, H, L, and P, The fraction of cells in each myeloid (D), B (H), NK (L), and CD8+ T (P) subcluster in nonmalignant cells (NM01–08) among patient groups. E, I, M, and Q, Differentially expressed mRNAs [q < 0.01 and log2(FC) > 0.4] between HD- and ATL-derived myeloid (E), B (I), NK (M), and CD8+ T (Q) cells. Red and blue colors represent upregulated and downregulated mRNAs related to indicated pathways respectively. FC, fold change; HM, hallmark. F, J, N, and R, Differentially expressed ADTs [q < 0.01 and log2(FC) > 0.3] between HD- and ATL-derived myeloid (F), B (J), NK (N), and CD8+ T (R) cells. Each color represents each related subcluster. E, F, I, J, M, N, Q, and R, Numbers of significant mRNAs or ADTs are shown in parentheses. S, Experimental design for coculture assay. T–V, Relative CD64 expression in myeloid cells (T) and the fraction of CD25+HLA-DR+ (U) and CD69+HLA-DR+ (V) cells in CD8+ T cells cocultured with normal CD4+ T, Jurkat (non-ATL), TL-Om1, or ATL43 cells (ATL). B, D, H, L, P, and T–V, Welch t test. ***, P < 0.0005; **, P < 0.005; *, P < 0.05. See also Supplementary Figs. S7 and S8.
B cells were decreased in ATL compared with HDs and ACs (Fig. 6B), suggesting impaired humoral immunity. Within the B-cell clusters (NM06 and NM07, 24,907 cells), three distinct subclusters emerged: CD38+IgM+ transitional (B-1), CD27−IgD+ naïve (B-2), and CD27+IgD− switched-memory (B-3) B cells (Fig. 6G; Supplementary Fig. S7E–S7G; Supplementary Table S4). Among these, switched-memory B cells were most significantly decreased in ATL (Fig. 6H), suggesting attenuated germinal center reaction. Like myeloid cells, ATL patients showed stronger expression levels of IFN pathway genes and a lower CD69 level, together with ADT levels corresponding to the subcluster changes (Fig. 6I and J; Supplementary Table S6). IFN pathway activation was also observed in NK and CD8+ T cells (Supplementary Fig. S7H), suggesting that this is considered a pervasive phenomenon in immune cells in ATL.
Subclustering of the NK cell cluster (NM05, 20,662 cells) identified three subclusters: CD56bright (NK-1), CD328 (SIGLEC7)+ mature (NK-2), and CD328− mature (NK-3) NK cells (Fig. 6K; Supplementary Fig. S7I–S7K; Supplementary Table S4). Although the proportion of total NK cells was comparable, ACs and ATL patients showed a larger fraction of CD328− NK cells than HDs (Fig. 6B and L). These results were confirmed by decreased CD328 ADT level in total NK cells in ATL. ATL-derived NK cells also showed MHC class II pathway activation and increased CX3CR1 ADT level (Fig. 6M and N; Supplementary Table S6). Therefore, NK cell dysfunction seems to occur in ACs and be further enhanced in ATL patients (28, 29).
Similarly, within CD8+ T cells, four typical subclusters (NM02 and NM03, 21,595 cells) were present: naïve (CD8-1), EM (CD8-2), EM reexpressing CD45RA (EMRA; CD8-3), and mucosal-associated invariant T (MAIT; CD8-4) cells (Fig. 6O; Supplementary Fig. S8A–S8C; Supplementary Table S4). ACs and ATL patients had or tended to have a smaller fraction of total CD8+ T cells than HDs (Fig. 6B). Although the frequency of EMRA cells was maintained, EM and MAIT cells were decreased, particularly in ATL (Fig. 6P). Despite the lack of clear velocity trajectories in HDs and ACs, ATL patients showed a strong directional flow from naïve to EM and eventually EMRA, which were considered to be less proliferative (Supplementary Fig. S8D and S8E). There were a few clonal CD8+ T cells in HDs, whereas ATL exhibited a more significant clonal expansion (Supplementary Fig. S8F and S8G; Supplementary Table S5). Although no remarkable pathway was upregulated in ATL, TNFα signaling was significantly downregulated (Fig. 6Q; Supplementary Table S6). Furthermore, ATL-derived CD8+ T cells showed stronger expression of activation markers, such as CD69, CD25, and HLA-DR (Fig. 6R; Supplementary Table S6). Thus, ACs have decreased EM cells, which is further aggravated by ATL development despite induction of activation markers.
To elucidate whether ATL cells themselves can induce phenotypic changes in nonmalignant cells, we performed coculture assay with normal PBMCs (Fig. 6S). Unlike control cells, ATL cell lines significantly enhanced CD64 expression in myeloid cells and increased the frequency of activated (CD25+HLA-DR+ and CD69+HLA-DR+) CD8+ T cells (Fig. 6T–V). These results highlight the substantial difference between virus- and tumor-induced immune responses, including selective increase in myeloid cells and decrease in B cells and aggravation of HTLV-1–induced dysfunction of NK and CTL responses in ATL.
Transfer of Genetically Overexpressed PD-L1 Proteins into Nonmalignant Cells
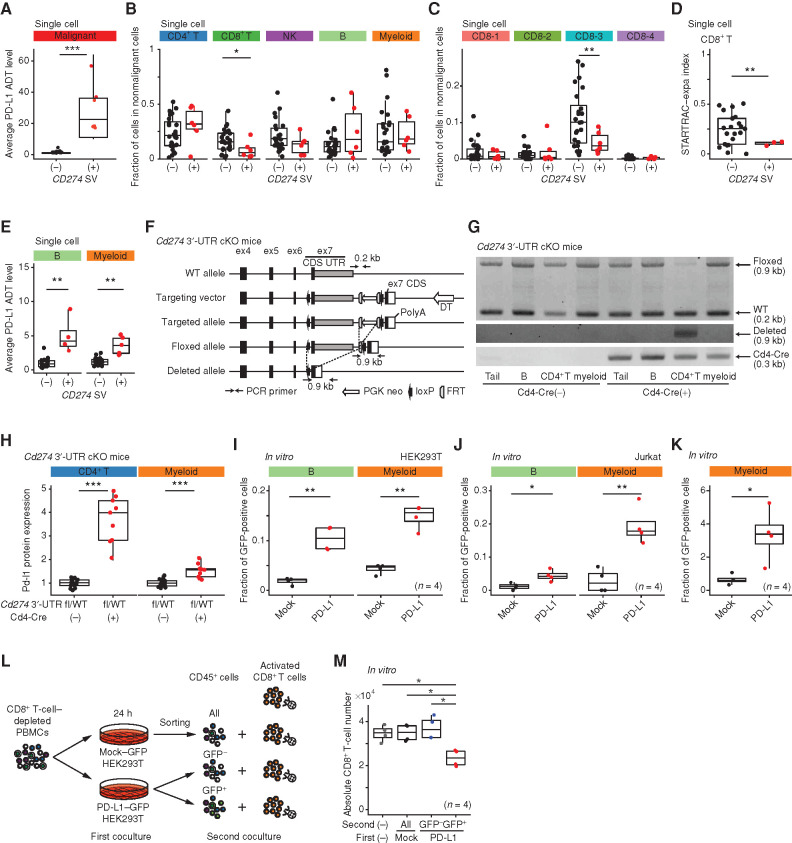
To disclose the effect of genetic alterations on the nonmalignant hematopoietic cells, we focused on CD274 SVs leading to PD-L1 overexpression (12). In this cohort, there were eight samples harboring CD274 SVs, detected by targeted-seq and/or RNA-seq (Fig. 7A; Supplementary Table S1). These showed decreased proportion of CD8+ T cells, particularly EMRA cells (Fig. 7B and C), confirming the previous result in solid cancers (12). In addition, CD274 SVs restrained clonal expansion of CD8+ T cells (Fig. 7D). Conspicuously, in patients with CD274 SVs, PD-L1 ADT level was increased in nonmalignant cells, particularly in B and myeloid cells, despite almost no mRNA level change (Fig. 7E; Supplementary Fig. S8H), suggesting that PD-L1 overexpression in tumor cells can upregulate PD-L1 level in surrounding cells. To further elucidate this phenomenon, we analyzed a newly generated mouse model recapitulating CD274 3′-UTR truncation [Cd274 3′-UTR conditional knockout (cKO); Fig. 7F; Methods]. We crossed these mice with CD4-cre transgenic mice and validated recombination of the Cd274 allele and resultant Pd-l1 overexpression in CD4+ T cells (Fig. 7G). These mice exhibited almost no phenotype compared with wild-type (WT) mice (Supplementary Fig. S8I and S8J). Importantly, Cd274 3′-UTR truncation in CD4+ T cells caused a moderate elevation of Pd-l1 expression in myeloid cells independent of the mRNA level (Fig. 7H; Supplementary Fig. S8K), validating the human single-cell data results. To investigate whether overexpressed PD-L1 proteins in tumor cells are released into surrounding cells, we performed PBMC coculture (Supplementary Fig. S8L). Compared with control protein, PD-L1–GFP fusion protein was more efficiently transferred from HEK293T and Jurkat T cells into B and myeloid cells and expressed on the cell surface (Fig. 7I and J; Supplementary Fig. S8M). In addition, the conditioned medium obtained from PD-L1–GFP fusion–overexpressing HEK293T cells induced GFP expression in the treated cells, unlike only GFP overexpression (Fig. 7K), suggesting that the biological property of PD-L1 promotes the intercellular protein transfer at least partly through indirect mechanisms. Importantly, the transferred PD-L1–GFP fusion inhibited the proliferation of activated CD8+ T cells (Fig. 7L and M). Together, genetically overexpressed PD-L1 in tumor cells can serve as a major contributor of PD-L1 upregulation in nonmalignant hematopoietic pool.
Figure 7.
Effects of genetically overexpressed PD-L1 on immune cells. A–E, Average PD-L1 ADT level in malignant cells (A), the fraction of each cell type (B) and each CD8+ T-cell subcluster (C) in nonmalignant cells (NM01–08), clonal expansion levels of CD8+ T cells (D), and average PD-L1 ADT level in each cell type (E) in ATL samples with or without CD274 SV. Subclusters with ≥30 cells are shown. A and E, Welch t test on log-transformed values. F, Scheme of Cd274 3′-UTR cKO mice. CDS, coding sequence; DT, diphtheria toxin. G and H, Genotyping PCR of tail and B (B220+), CD4+ T, and myeloid (Gr-1+Mac1+) cells (G) and relative Pd-l1 expression of CD4+ T and myeloid cells (H) in Cd274 3′-UTRfl/WT mice with (n = 9) or without (n = 14) Cd4-Cre. I and J, The fraction of GFP+ cells in normal B (CD19+) and myeloid (CD14+) cells cocultured with HEK293T cells (n = 4; I) and Jurkat T cells (n = 4; J) transduced with indicated plasmids. K, The fraction of GFP+ cells in myeloid cells cultured in supernatant from GFP- or PD-L1–GFP fusion–overexpressing HEK293T cells (n = 4). L, Experimental design for coculture assay. M, Absolute number of CD8+ T cells after 48 hours of coculture described in L (n = 4). B–D, H–K, and M, Welch t test. ***, P < 0.0005; **, P < 0.005; *, P < 0.05. See also Supplementary Fig. S8.
Discussion
Through multimodal single-cell analysis, we have delineated the phenotypic and functional heterogeneity of HTLV-1–infected normal and leukemic cells. HBZ expression clearly demarcates HTLV-1–infected cells from uninfected CD4+ cells, enabling their phenotypic characterization. Beyond known markers, several proteins including CD73 and CD99 can show better discriminating potential. Within HTLV-1–infected cells, there are two distinct but interchangeable subsets: FOXP3− and FOXP3+ cells. Although cell of origin of ATL remains controversial, the latter FOXP3+ Treg–like phenotype accelerates HTLV-1–infected cells into premalignant clonal expansion characterized by excessive MHC class II attenuation. By contrast, ATL cells express a diverse array of immune molecules, such as CD99 and LGALS1, which can be a potential target for immunotherapy. These observations provide unique insights into multistep viral carcinogenesis, distinguishing the malignant phenotype driven by HTLV-1 infection and ATL development.
Another notable finding is frequent TCR CDR3 mutations, enabling tracking of clonal evolution in T-cell malignancies. Moreover, the emergence of independent clones is common in ATL. scTCR-seq analysis disclosed different clinical behaviors, such as acute transformation and therapeutic resistance, between malignant clones. Simultaneous mRNA and ADT level analysis uncovered posttranscriptional regulation of surface markers, including dissociated protein upregulation for myeloid markers and ATL-specific CD3 and CD62L protein reduction. Although various posttranscriptional processes can be related, including translation rate and protein turnover control (16), our data provide the fundamental basis for understanding cell-type–specific mRNA and protein expression regulation.
Our study revealed the dynamic changes in the nonmalignant hematopoietic pool of HTLV-1 infection and ATL, demonstrating their differential effects on immune cells. Although hampered NK and CTL responses in HTLV-1 infection are further deteriorated, selective alterations occur in myeloid and B lineage in ATL, represented by myeloid expansion, particularly of nonclassic monocytes and dendritic cells. Together with recent single-cell studies (30), our findings point out an increasingly important role of various myeloid subsets and their potential as a therapeutic target. Also noteworthy is the demonstration of genetically overexpressed PD-L1 in tumor cells as a major source of PD-L1 upregulation in nonmalignant cells, providing a clue for the intricate mechanisms regulating PD-L1 expression in the tumor microenvironment. Generally, PD-L1 expression in tumor cells is induced by IFNγ, which leads to suppression of antitumor immunity (31). It is intriguing to speculate that upregulated PD-L1 in malignant cells can be transferred into the microenvironment to help shape the adaptive immune resistance. Our data combined with previous studies (32, 33) imply that somatically overexpressed proteins can spread through intercellular communication to remodel the tumor microenvironment.
Methods
Detailed materials and methods are available in the Supplementary Data.
Data and Code Availability
scRNA-seq, scADT-seq, scTCR/BCR-seq data as well as newly sequenced bulk RNA-seq, targeted-seq, and WES data have been deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001004936. All other data sets generated or analyzed during this study are included in the published article. Custom code used to analyze the single-cell data set is available at https://github.com/nccmo/ATL_scRNA_seq.
Human Samples
Primary PBMCs from 11 ACs (11 samples) and 30 ATL patients (34 samples including 4 sequential ones, which consisted of 19 acute, 12 chronic, and 3 smoldering subtypes) were collected at University of Miyazaki Hospital and Imamura General Hospital. These studies were approved by the Institutional Review Board of the National Cancer Center and other participating institutes. Written informed consent was obtained from all participants in accordance with the Declaration of Helsinki protocol. All patients were previously untreated with systemic chemotherapy at the time of peripheral blood (PB) collection, except for one patient with sequential sampling who received chemotherapy between the first (ATL01) and second (ATL01-2) PB collections. HD samples were purchased from Zen-Bio, Precision Bioservices, iQ Biosciences, and HemaCareCorporation (see also Supplementary Table S1). The available clinical characteristics and sequencing information are summarized in Supplementary Table S1.
Mice
Female C57BL/6 mice (6–10 weeks old) were obtained from CLEA Japan and maintained under pathogen-free conditions. Cd4-Cre [B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ] mice were purchased from The Jackson Laboratory. Cd274 3′-UTR cKO mice were generated by TransGenic Inc. in this study. Schematic representation of the targeting vector and the resultant allele is shown in Fig. 7F, in which 5′ homologous arm (4.7 kb), exon 7 [coding sequence (CDS) and 3′-UTR] flanked with loxP (3.3 kb), the FLP recognition target (FRT)-neomycin resistance (Neo)-FRT cassette (1.8 kb), exon 7 CDS with growth hormone poly A signal (0.3 kb), and 3′ homologous arm (3.2 kb) were arranged in order. To identify embryonic stem (ES) cells with appropriate homologous recombination, we also constructed a control vector with elongated 5′ homologous arm (5.9 kb) and 3′ homologous arm (3.8 kb) as a positive control in PCR selection. The linearized targeting vector was electroporated into RENKA (C57BL/6) ES cells. After selection with G418 antibiotic, surviving clones were expanded for PCR analysis using 5′ flanking–, 3′ flanking–, and 5′ loxP–specific primers to identify recombinant ES clones. Secondary confirmation of positive clones identified by PCR was performed by Southern blotting using hybridization technique with probes targeted against the Neo region. Positive ES clones were expanded and microinjected into ICR eight-cell stage embryos through the aggregation technique and then transplanted into C57BL/6 mice (Sankyo-Laboratory Service). Resulting chimeras with a high percentage of black coat color were crossed to a germline Flp-deleter mouse line [B6;D2-Tg(CAG-Flp)18Imeg] to eliminate the FRT-Neo-FRT cassette. Cd274 3′-UTR cKO mice were genotyped using a PCR-based assay to detect both the WT and floxed alleles. Genomic DNA was isolated from tail biopsies or PBMCs and subjected to PCR. The PCR samples were denatured at 94°C for 2 minutes, subjected to 35 cycles of amplification (98°C for 10 seconds, 62.5°C for 30 seconds, and 68°C for 3 minutes), and followed by a final extension step at 68°C for 5 minutes. PCR products were resolved by agarose gel electrophoresis. PCR primers are listed in the Key Resources Table in the Supplementary Data. Male and female 12- to 16-week-old mice were used for each animal experiment. Littermates were used as controls in all experiments. All mouse experiments were approved by the Animals Committee for Animal Experimentation of the National Cancer Center and met the Guidelines for Proper Conduct of Animal Experiments established by the Science Council of Japan.
Sample Preparation, Library Preparation, and Sequencing
PBMCs were isolated from whole blood using Ficoll-Paque PLUS (cytiva) and cryopreserved in CELLBANKER 1plus (TaKaRa Bio) at −150°C according to the manufacturer's instructions. Frozen PB samples were rapidly thawed and transferred into 10% fetal bovine serum (FBS; Biosera) containing 20 mL prewarmed RPMI-1640 (Nacalai Tesque). After centrifugation at room temperature, pellets were resuspended in 3 mL RPMI-1640 with 10% FBS, filtered using a 70-μm nylon mesh (Thermo Fisher Scientific), and stained with 0.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; DOJINDO) for live/dead staining. For cell sorting, all samples were gated based on forward and side scatter, followed by exclusion of doublets, and then gated on DAPIlow viable cells. After sorting 1 × 106 live cells, these cells were centrifuged at 300 × g at 4°C for 7 minutes, and resuspended in 50 μL Cell Staining Buffer (BioLegend) with Human TruStain FcX Blocking Solution (BioLegend) and incubated on ice for 10 minutes. The PBMCs were stained with a panel of TotalSeq-C antibodies (BioLegend), consisting of 99 antibodies (ADT Set 1) for 18 samples and 105 antibodies (ADT Set 2) for 31 samples (see Supplementary Table S2), according to the manufacturer's instructions except that each antibody was used at the amount of 0.2 μg. The scRNA-seq, scADT-seq, and scTCR/BCR-seq libraries were prepared using the Chromium Single-Cell V(D)J Reagent Kits (v1 and v1.1): Single-Cell 5′ Library and Gel Bead Kit (PN-1000006) or Chromium Next GEM Single-Cell 5′ Library and Gel Bead Kit v1.1 (PN-1000165), Chromium Single-Cell 5′ Feature Barcode Library Kit (PN-1000080), Chromium Single-Cell V(D)J Enrichment Kit, Human T-cell (PN-1000005), Chromium Single-Cell V(D)J Enrichment Kit, Human B Cell (PN-1000016), Chromium Single-Cell 5′ Library Construction Kit (PN-1000020), Chromium Single-Cell A Chip Kit (PN-120236) or Chromium Next GEM Chip G Single-Cell Kit (PN-100120), and i7 Multiplex Kit (PN-120262), Single Index Kit T Set A (PN-1000213), or Chromium i7 Multiplex Kit N, Set A (PN-1000084; 10x Genomics), according to the instructions in Chromium Next GEM Single-Cell V(D)J Reagent Kits v1.1 with Feature Barcoding Technology for Cell-Surface Protein (manual part no. CG000208 Rev A). These libraries were sequenced using the NovaSeq 6000 system with the NovaSeq 6000 S4 Reagent Kit (Illumina) at Macrogen as 150 base pair paired-end reads except for three scBCR-seq libraries (ATL12, ATL14, and ATL22) sequenced in-house using the NextSeq 500 system with the NextSeq 500/550 High Output Kit (Illumina).
Statistical Analysis
Statistical analyses were performed with R3.6.1 software (The R Foundation for Statistical Computing). Comparison of categorical and continuous data was performed using the Fisher exact test and Welch t test, respectively, unless otherwise specified. All tests were two-sided, unless otherwise specified. Box plots show median (lines), interquartile range (IQR; boxes), and ± 1.5 × IQR (whiskers).
Supplementary Material
Acknowledgments
This work was supported by AMED [JP20wm0325013 (K. Kataoka), JP19ck0106261 (K. Kataoka), JP19ck0106254 (K. Shimoda), JP20ck0106542 (K. Kataoka), JP20ck0106538 (K. Shimoda), and JP20cm0106575 (K. Kataoka)], Japan Society for the Promotion of Science (JSPS) KAKENHI (19K22573 to K. Kataoka), National Cancer Center Research and Development Funds (31-A-4 to K. Kataoka), Takeda Science Foundation (K. Kataoka), Japan Leukemia Research Fund (K. Kataoka), and The Naito Foundation (K. Kataoka). The authors thank Y. Hokama, F. Ueki, and Y. Ito for technical assistance. The sequencing resources for scTCR/BCR-seq were provided by Fundamental Innovative Oncology Core at the National Cancer Center. The supercomputing resources were provided by the Human Genome Center, the Institute of Medical Science, The University of Tokyo.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
#J. Koya and Y. Saito contributed equally to this article.
##K. Kataoka is the senior author of this article.
Blood Cancer Discov 2021;2:450–67
Authors' Disclosures
N. Nakano reports personal fees from Novartis Pharma, Takeda Pharmaceutical, Kyowa Kirin, Celgene, Eisai, Nippon Shinyaku, Astellas Pharma, Bristol Myers Squibb, MSD, Otsuka Pharmaceutical, Asahi Kasei Medical, Chugai Pharmaceutical, and JIMRO outside the submitted work. A. Utsunomiya reports personal fees from Novartis Pharma, Kyowa Kirin, Daiichi Sankyo, Bristol Myers Squibb, Celgene, Pfizer, Minophagen Pharmaceutical, Janssen Pharmaceutical, HUYA Japan, JIMRO, Meiji Seika Pharma, and Otsuka Medical Devices outside the submitted work. Y. Togashi reports grants and personal fees from Ono Pharmaceutical and Bristol Myers Squibb; personal fees from Chugai Pharmaceutical and MSD; and grants from KOTAI Biotechnologies and Daiichi Sankyo outside the submitted work. S. Ogawa reports a patent for P6810396 licensed and with royalties paid from Asahi Genomics. K. Kataoka reports grants from AMED, Japan Society for the Promotion of Science, National Cancer Center Research and Development Funds, Takeda Science Foundation, Japan Leukemia Research Fund, and The Naito Foundation during the conduct of the study; grants, personal fees, and nonfinancial support from Otsuka Pharmaceutical, grants from Chordia Therapeutics, Bristol Myers Squibb, Ono Pharmaceutical, Shionogi, Teijin Pharma, Japan Blood Products Organization, Mochida Pharmaceutical, and JCR Pharmaceuticals, grants and personal fees from Chugai Pharmaceutical, Takeda Pharmaceutical, Eisai, Kyowa Kirin, and Sumitomo Dainippon Pharma, personal fees from Celgene, Astellas Pharma, Novartis, AstraZeneca, and Janssen Pharmaceutical, and other support from Asahi Genomics outside the submitted work; and a patent for genetic alterations as a biomarker in T-cell lymphomas licensed to Kyoto University and a patent for PD-L1 abnormalities as a predictive biomarker for immune-checkpoint blockade therapy licensed to Kyoto University. No disclosures were reported by the other authors.
Authors' Contributions
J. Koya: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y. Saito: Data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Kameda: Resources, writing–review and editing. Y. Kogure: Investigation, writing–review and editing. M. Yuasa: Data curation, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. J. Nagasaki: Formal analysis, validation, visualization. M.B. McClure: Investigation, writing–review and editing. S. Shingaki: Investigation, writing–review and editing. M. Tabata: Investigation, writing–review and editing. Y. Tahira: Resources. K. Akizuki: Resources. A. Kamiunten: Resources. M. Sekine: Resources. K. Shide: Resources. Y. Kubuki: Resources. T. Hidaka: Resources. A. Kitanaka: Resources. N. Nakano: Resources. A. Utsunomiya: Resources. Y. Togashi: Formal analysis, validation, visualization. S. Ogawa: Resources. K. Shimoda: Resources, funding acquisition. K. Kataoka: Conceptualization, formal analysis, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 2012;13:607–15. [DOI] [PubMed] [Google Scholar]
- 2.Butel JS.Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis 2000;21:405–26. [DOI] [PubMed] [Google Scholar]
- 3.Mason WS, Gill US, Litwin S, Zhou Y, Peri S, Pop O, et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 2016;151:986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowan AG, Dillon R, Witkover A, Melamed A, Demontis MA, Gillet NA, et al. Evolution of retrovirus-infected premalignant T-cell clones prior to adult T-cell leukemia/lymphoma diagnosis. Blood 2020;135:2023–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019;565:312–7. [DOI] [PubMed] [Google Scholar]
- 7.Ishitsuka K, Tamura K.Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol 2014;15:e517–e26. [DOI] [PubMed] [Google Scholar]
- 8.Iwanaga M, Watanabe T, Utsunomiya A, Okayama A, Uchimaru K, Koh KR, et al. Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: a nationwide prospective study in Japan. Blood 2010;116:1211–9. [DOI] [PubMed] [Google Scholar]
- 9.Bangham CR, Osame M.Cellular immune response to HTLV-1. Oncogene 2005;24:6035–46. [DOI] [PubMed] [Google Scholar]
- 10.Cook LB, Fuji S, Hermine O, Bazarbachi A, Ramos JC, Ratner L, et al. Revised adult T-cell leukemia-lymphoma international consensus meeting report. J Clin Oncol 2019;37:677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 2015;47:1304–15. [DOI] [PubMed] [Google Scholar]
- 12.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 2016;534:402–6. [DOI] [PubMed] [Google Scholar]
- 13.Baslan T, Hicks J.Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer 2017;17:557–69. [DOI] [PubMed] [Google Scholar]
- 14.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–72. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Beyer A, Aebersold R.On the dependency of cellular protein levels on mRNA abundance. Cell 2016;165:535–50. [DOI] [PubMed] [Google Scholar]
- 17.Imhof BA, Aurrand-Lions M.Adhesion mechanisms regulating the migration of monocytes. Nat Rev Immunol 2004;4:432–44. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe T.Adult T-cell leukemia: molecular basis for clonal expansion and transformation of HTLV-1–infected T cells. Blood 2017;129:1071–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stubbington MJT, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods 2016;13:329–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter JA, Preall JB, Atwal GS.Bayesian inference of allelic inclusion rates in the human T cell receptor repertoire. Cell Syst 2019;9:475–82. [DOI] [PubMed] [Google Scholar]
- 21.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature 2018;560:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holling TM, Schooten E, van Den Elsen PJ.Function and regulation of MHC class II molecules in T-lymphocytes: of mice and men. Hum Immunol 2004;65:282–90. [DOI] [PubMed] [Google Scholar]
- 23.Pasello M, Manara MC, Scotlandi K.CD99 at the crossroads of physiology and pathology. J Cell Commun Signal 2018;12:55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cedeno-Laurent F, Dimitroff CJ.Galectin-1 research in T cell immunity: past, present and future. Clin Immunol 2012;142:107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–56. [DOI] [PubMed] [Google Scholar]
- 26.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016;539:309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brady BL, Steinel NC, Bassing CH.Antigen receptor allelic exclusion: an update and reappraisal. J Immunol 2010;185:3801–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E, et al. The decreased expression of Siglec-7 represents an early marker of dysfunctional natural killer-cell subsets associated with high levels of HIV-1 viremia. Blood 2009;114:3822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sciumè G, De Angelis G, Benigni G, Ponzetta A, Morrone S, Santoni A, et al. CX3CR1 expression defines 2 KLRG1+ mouse NK-cell subsets with distinct functional properties and positioning in the bone marrow. Blood 2011;117:4467–75. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O'Brien SA, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 2020;181:442–59. [DOI] [PubMed] [Google Scholar]
- 31.Topalian SL, Taube JM, Anders RA, Pardoll DM.Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 2008;10:619–24. [DOI] [PubMed] [Google Scholar]
- 33.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq, scADT-seq, scTCR/BCR-seq data as well as newly sequenced bulk RNA-seq, targeted-seq, and WES data have been deposited in the European Genome-phenome Archive (EGA) under accession number EGAS00001004936. All other data sets generated or analyzed during this study are included in the published article. Custom code used to analyze the single-cell data set is available at https://github.com/nccmo/ATL_scRNA_seq.