Abstract
Older adults in nursing homes (NHs) have increased frailty, medication, and antimicrobial exposures, all factors that are known to affect the composition of gut microbiota. Our objective was to define which factors have the greatest association with the NH resident gut microbiota, explore patterns of dysbiosis and compositional changes in gut microbiota over time in this environment. We collected serial stool samples from NH residents. Residents were assessed using the Mini Nutritional Assessment tool and Clinical Frailty Scale. Bacterial composition of resident stool samples was determined by metagenomic sequencing. We used mixed-effect random forest modeling to identify clinical covariates that associate with microbiota. We enrolled and followed 166 residents from 5 NHs collecting 512 stool samples and following 15 residents for > 1 year. Medications, particularly psychoactive and antihypertensive medications, had the greatest effect on the microbiota. Age and frailty also contributed, and were associated with increased and decreased diversity, respectively. The microbiota of residents who had lived in the NH for > 1 year were enriched in inflammatory and pathogenic species and reduced in anti-inflammatory and symbiotic species. We observed intraindividual stability of the microbiome among older adults who had lived in the NH already for >1 year followed with sample collections 1 year apart. Older adult NH gut microbiome is heavily influenced by medications, age, and frailty. This microbiome is influenced by the length of NH residency with dysbiosis becoming evident at 12 months, however, after this point there is demonstrated relative stability over time.
Keywords: Antibiotics, Frailty, Gut microbiome, Medications, Nursing home, Residents
As we understand how important the microbiome is toward maintaining health, it is important to understand the factors influencing the microbiome of older adults aged 65 years and older in the nursing home (NH). Factors that disrupt the microbiome are common in NHs, including high use of antimicrobials and more frequent hospitalizations (1). In NHs, multidrug-resistant infections are more common and NHs may serve as a reservoir for introducing drug-resistant pathogens into other health care settings (2). Antibiotic exposure is highly prevalent in the NH (3) and it rapidly and profoundly affects the gut microbiota, reducing overall diversity and shifting composition away from beneficial species (4). Antibiotics are not the only medication that influences microbiome composition as we now know many prescription drugs have a notable impact on the overall architecture of the intestinal microbiome (5). Additionally, more than 75% of NH residents have at least 2 of the 10 most common chronic medical conditions (6). These medical conditions have strong associations with the gut microbiome composition, such as high blood pressure (7), Alzheimer’s disease (8), and diabetes (9).
NH residents tend to be frailer than community-dwelling older adults and are exposed to many more medications. Polypharmacy, a condition in which a person takes 5 or more daily medications (10), is widespread within U.S. NHs (11). Polypharmacy is associated with a number of adverse health outcomes experienced by NH older adults such as falls, adverse drug reactions, increased length of hospital stays, and mortality (12). We now know that nonantibiotic, polypharmacy itself, independent of specific drug classes, has been shown to represent a determinant of gut microbiota composition that has detrimental clinical consequences and is associated with a reduction in microbiome diversity and gut dysbiosis (13).
Frailty is a major public health problem for an aging society and its prevalence and associations with gut dysbiosis (14) make it another critical factor among NH residents. Frailty is a quantifiable syndrome that encompasses loss of physical, psychological, and social abilities, and leaves one less able to recover from a stressful event, such as injury or illness. This multidimensional syndrome has losses in reserves of energy, physical ability, cognition, and health that leads to vulnerability (15). About half of all NH older adults are frail and upwards of 40% can be defined as pre-frail (16).
Given that dysbiosis of the gut microbiome intersects several critical issues facing NH older adults, we sought to gain a better understanding of how clinical factors contribute to the composition of the NH older adult microbiome. Accordingly, we followed a cohort of older adults from multiple NH facilities to investigate the influence of the following: (a) medical and demographic factors; (b) frailty and age-related factors on microbiome composition; and (c) the temporal stability of the older adult microbiome during NH residency. Our goal was to contribute to the understanding of the NH microbiome and highlight how the microbiome can serve as a potential therapeutic target to reduce disease burden among NH residents.
Method
Study Setting and Population
This prospective cohort study was approved by the institutional review board at the University of Massachusetts Medical School. This cohort consists of NH residents ≥65 years of age who resided in 1 of the 5 NH facilities located in central Massachusetts. We enrolled residents who had been residing at a NH facility and did not have any diarrheal illness or antimicrobial exposure within the preceding 4 weeks. It is important to note that residents within each NH facility were provided the same daily foods to eat. No residents in this study suffered from dysphagia or had a feeding tube. Any residents with antimicrobial exposure or a diarrheal illness before the first sample collected through the final sample collected were excluded from this analysis. A subset of willing participants were enrolled a second time 1 year after their initial stool sample was collected. These participants met the same criteria described above. This study was approved by the University of Massachusetts Medical School’s Institutional Review Board (IRB docket H00010892). All participants, or the participant’s legally authorized representative, gave informed consent to participate in this study.
Data Collection
We conducted baseline and end-of-study medical record abstraction for factors associated with key study outcomes. These factors included the following: age, nutritional status, comorbidities, medications, and frailty (17). Specifically, prior history of hospitalizations within 1 year prior to enrollment, and antibiotic exposures 6 months prior; daily and “as needed” (PRN) medications; polypharmacy defined as 5 or more daily medications (10); and age, sex, race, and length of NH stay were recorded. Resident age collapsed into 4 groups—65–74, 75–84, 85–94, and ≥95 years. Comorbidity and frailty were assessed during baseline interviews with corroborating input from family or facility staff by trained research staff. We used the Charlson Comorbidity Index, a widely used instrument designed to measure the burden of medical diseases and predict mortality (18). Frailty was categorized according to the validated and widely utilized Canadian Study of Health and Aging’s 7-point Clinical Frailty Scale. This instrument has been previously applied to define signatures of frailty in the gut microbiota (19). We assessed nutritional status using the Mini Nutritional Assessment (MNA) tool, which is used routinely in NH residents (20). Residents were categorized as normal, at risk, or malnourished based on the MNA survey administered to the residents by trained research staff or the nurse caring for the resident if mentally impaired. Assessments were also made only at baseline and at the end of the study.
Sample Collection and Processing
We collected monthly stool samples from each resident with a goal of 4 consecutive months. DNA was extracted from stool samples using the PowerMag Soil DNA Isolation Kit on an epMotion 5075 TMX liquid handling workstation according to manufacture protocols (MO BIO Laboratories, #27100-4-EP). Sequencing libraries were constructed using the Nextera XT DNA Library Prep Kit (Illumina, Inc., #FC-131–1096) and sequenced on a NextSeq 500 Sequencing System as 2 × 150 nucleotide paired-end reads.
Sequence Processing and Analysis
Shotgun metagenomic reads were first trimmed and quality filtered to remove sequencing adapters and host contamination using Trimmomatic and Bowtie2, respectively, as part of the KneadData pipeline version 0.7.2 (https://huttenhower.sph.harvard.edu/kneaddata/). Reads were then profiled for microbial taxonomic abundances using Metaphlan2 (21) (as in our previous work) (8,19).
Statistical and Computational Analysis
We performed traditional unsupervised correspondence analysis (nonmetric multidimensional scaling and unsupervised hierarchical clustering) to first determine sample similarity with respect to the clinical covariates of interest. Permutation Multivariate Analysis of Variance (PERMANOVA) was performed to evaluate inter- and intraindividual variation in microbial abundance. To determine the contribution of each covariate to changes in microbiome composition, we performed mixed-effect random forest regression modeling in R. Specifically, for every microbe, we predicted its relative abundance as a function of the available clinical covariates and using patient ID as random effect. We ran permutated importance analysis to determine the significance of associations between clinical covariates and microbial abundances. We deemed as significant microbiome–covariate associations with false discovery rate (FDR) adjusted p-value less than .05. To determine the effect of repeated enrollment, we used mixed-effect random forest classification to predict enrollment window (first or second) as a function of species abundance. Again, permuted importance analysis was used to determine species whose abundance is significantly associated (FDR < 0.05) with classifying a sample according to enrollment. The R package ggplot2 was used for visualization of the relationship between clinical covariates and modeling-selected microbiome features. We want to note that our data set consists of repeated samples (eg, multiple samples belonging to the same individuals), so application of any methods (including Spearman’s or Linear discriminant analysis Effect Size) would be a statistical mistake. In order to fully use the entire data sets and not having to average across patient samples, we need to use a mixed-effect modeling type of approach. We could not use linear mixed-effect models because of the proportional nature of the microbiome data (relative abundances). Furthermore, we need a method that can include data of different modalities. The advantage of using mixed-effect random forest is that it is basically the only method that allows for longitudinal of repeated samples, does not care about the underlying distribution of the data (it is nonparametric), and allows for including covariates that are on different scales or are of different nature.
Results
NH Facilities and Prevalence of Polypharmacy
Over a 3-year time period, we collected 512 stool samples from 166 older adults across 5 NH facilities with an average of 3.1 stool samples per resident. The average age was 86.2 years (SD 9.1) with 18.1% men. There were prevalent exposures within the preceding 6 months to both antibiotics (18.7%) and hospital admissions (23.5%). Significant differences were observed in gender, polypharmacy, and clinical scores for medical comorbidities, frailty, and malnutrition across the 5 NH sites (Table 1).
Table 1.
Demographics and Clinical Scores by Nursing Home Site
| Resident Characteristic* | Site 1 n = 35 |
Site 2 n = 20 |
Site 3 n = 39 |
Site 4 n = 51 |
Site 5 n = 21 |
p Value |
|---|---|---|---|---|---|---|
| Age (mean [SD]) (years) | 86.8 (8.3) | 82 (9.3) | 86.1 (8.4) | 86.4 (10.8) | 88.6 (5.7) | .21 |
| Male | 0 (0.0) | 4 (20.0) | 15 (38.5) | 8 (15.7) | 3 (14.3) | .001 |
| Polypharmacy | 19 (54.3) | 14 (70.0) | 31 (79.5) | 32 (62.7) | 21 (100) | .003 |
| Antibiotic exposure | 6 (17.1) | 6 (30.0) | 3 (7.7) | 9 (17.6) | 7 (33.3) | .10 |
| Hosp Exp | 7 (20.0) | 3 (15.0) | 7 (17.9) | 11 (21.6) | 4 (19.0) | .98 |
| Clinical Scores | ||||||
| CCI (mean [SD]) | 0 (0.0) | 0.1 (0.4) | 1.6 (2.6) | 0.5 (1.0) | 2.7 (2.7) | <.001 |
| CFS (mean [SD]) | 5.6 (1.0) | 6.6 (1.1) | 6.5 (0.9) | 7.0 (0.6) | 6.8 (1.0) | <.001 |
| MNA (mean [SD]) | 1.6 (0.7) | 2.2 (0.8) | 2.2 (0.7) | 2.3 (0.6) | 2 (0.6) | <.001 |
Notes: χ 2 test was used to compare categoric variables and analysis of variance for continuous variables. CCI = Charlson Comorbidity Index; CFS = Clinical Frailty Scale; Hosp Exp = hospital exposure past year; MNA = The Mini Nutritional Assessment; SD = standard deviation.
*Data are presented as the number (%), unless otherwise specified.
Medication Exposure
Polypharmacy was prevalent with 70.5% of residents taking 5 or more daily medications. Approximately half of the cohort (43.4%) took 8 or more daily medications and 87.3% reported at least 2 daily medications; only 6.6% reported no daily medications. The 5 most frequently encountered medication classes were antidepressants, beta blockers, acid-reducing medications, statins, and calcium channel blockers (Supplementary Table 1). We did note significant differences in medications taken by residents dependent on site. Daily probiotic use (14%), however, was similar across all sites with Lactobacillus acidophilus being most prevalent. Hypertension (67.1%), hypercholesteremia (40.8%), COPD (11.0%), and mild cerebrovascular disease (10.4%) were the most prevalent medical conditions.
Microbiome Composition Correlates with Clinical Factors Including Age and Frailty
We sought to explain how microbiome composition varies with older adult covariates demographic, clinical, and medication data. Microbiome alpha diversity (determine as Shannon and Inverse Simpson diversity metrics) calculated from the metagenomic profiles of longitudinally collected samples were modeled using linear mixed-effects modeling, with clinical covariates as fixed effects and patient ID as random effect. Microbiome alpha diversity was negatively associated with frailty, and positively associated with age (after adjusting for sex, nutrition, location, time in the facility, polypharmacy, and 6-month history of antibiotic exposure; Table 2).
Table 2.
Mixed-effects Modeling Using Shannon Index and Inverse Simpson Index with Clinical Covariates
| Shannon Index | Inverse Simpson | |||
|---|---|---|---|---|
| Covariate | B Coefficient | p Value | B Coefficient | p Value |
| Age* | .0105449 | .009 | .086833 | .022 |
| Sex | .1032911 | .27 | .7653592 | .39 |
| Frailty* | −.0876286 | .039 | −.9267414 | .020 |
| Malnutrition | −.0176967 | .76 | .0768242 | .89 |
| Polypharmacy | −.0730231 | .34 | −.2927881 | .68 |
| Antibiotic exposure | −.0413258 | .63 | −1.087216 | .18 |
| Time at facility | −.0000411 | .98 | −.0033293 | .79 |
| Facility | −.0659972 | .50 | −.3106582 | .73 |
| Floor | .0292822 | .42 | .1522867 | .64 |
Note: *Denote variables with statistical significance (p < .05).
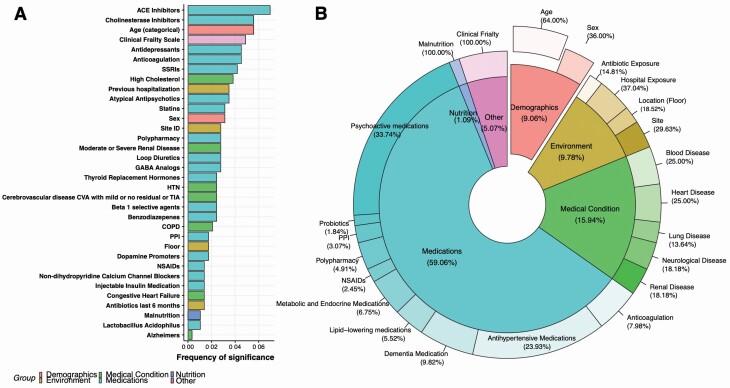
To determine associations between the relative abundance of each microbial species detected from the metagenomic sequencing and the clinical covariates, we performed mixed-effect random forest modeling (see Method). This method is agnostic to the distribution of the data and allows for assessment of predictive accuracy and identification of statistically significant associations through permutation analysis. Our results revealed a mix of medications and demographic, environmental, and medical conditions to be strong predictors of microbiome composition. Medications were the most prevalent predictors with 12 specific medications being significant (FDR adjusted p < .05). These medications, in order of strength of predicting variable, included angiotensin-converting enzyme (ACE) inhibitors, cholinesterase inhibitors, antidepressants, anticoagulation, serotonin reuptake inhibitors, atypical antipsychotics, statins, loop diuretics, gamma-aminobutyric acid analogs, thyroid replacement medications, beta 1 selective agents, and benzodiazepines. Also significant were hypertension, kidney disease, and cerebrovascular disease (medical conditions); age and gender (demographic); hospital exposure (environmental); and frailty and malnutrition (Figure 1A). The 2 strongest predictors are ACE inhibitors used for the reduction of blood pressure, and cholinesterase inhibitors for the treatment of Alzheimer’s and other neurological diseases. Only 10% of residents were taking either ACE or cholinesterase inhibitors but, notably, both of these medications predicted higher abundances of pathobionts such as Pseudomonas aeruginosa (22) associating with ACE inhibitors, and Bacteroides thetaiotaomicron (23) associating with cholinesterase inhibitors and also predicted higher abundances of bacterial species that produce butyrate or are considered anti-inflammatory (ie, Faecalibacterium prausnitzii (24) with ACE inhibitors and Ruminococcus torques (25) with cholinesterase inhibitors; Supplementary Figure 1).
Figure 1.
Clinical and demographic variables that predict microbiome composition. Results from mixed-effect random forest modeling using all demographic and clinical variables (including each medication taken and clinical scoring systems for frailty, nutrition, and comorbidities) to predict species abundance in the gastrointestinal tract. (A) Ranking of forest predictors based on frequency of significance (# of times a predictor is found to be significantly associated with a microbe over the total number of modeled species). Significance was determined by running permutated importance analysis calculations and using an FDR adjusted p-value of .05. (B) Pie chart to illustrate the distribution of the clinical factors found by the modeling to be significantly associated with the NH microbiome. Drugs were grouped by treatment class. NH = nursing home.
We noted similar patterns with the other medications, where for each medication, the model predicts a mixture of bacterial species with known beneficial and detrimental benefits to human health to be associated. Regarding demographics, both frailty and age were significant predictors of microbiome composition. Increasing frailty and age both associated with higher abundances of bacteria associated with disease and lower abundances of species often referred to as symbiotic (see Supplementary Figure 2A for age and Supplementary Figure 2B for frailty). Increasing frailty had higher abundances of pathogens and inflammatory-associated microbiota such as Bacteroides dorei (26), and Flavonifractor plautii (27), and lower abundances of butyrate producing organisms such as Bacteroides vulgatus (28), Anaerostipes hadrus (29), and F prausnitzii (24). Among older residents’ higher abundances of pathogens (Shigella species) (30) and inflammatory-associated microbiota (Bacteroides eggerthii and B dorei) (26) and lower abundances of symbiotic microbiota (Bifidobacterium adolescentis and Eubacterium hallii) (31,32) was seen, however, conversely, higher abundances of the key butyrate producer F prausnitzii (24) were also seen in older residents.
After grouping medications according to indications for treatment, we sought to determine the extent to which the relative influence on microbiome composition could be explained by treatment of medical conditions (Figure 1B). The relative abundance of species in the NH microbiome is over 50% (representing the number of species in where a factor belonging to a certain group is predicted by the modeling to have significant association over the total number of modeled species) and is influenced by medications followed next by medical conditions (15.9%), demographics (9.1%; including age and gender), environment (9.8%), frailty (5.1%), and finally malnutrition (1.1%). Environmental factors included recent hospital admissions, and antibiotic use along with location either by site (NH facility) or floor/wing. Interestingly, the floor/wing location within a NH influenced just over 2% (or 19% of the environmental variables) of the microbiome after adjusting for the other covariates, and which also included NH site. The site variable would correlate with the diet provided as all residents within a site eat the same meals. Among medication the psychoactive, antihypertensive, metabolic, and dementia medications are predicted to have the greatest influence on microbiome composition.
Microbiome Composition Changes Occur Over Length of Stay Within the NH
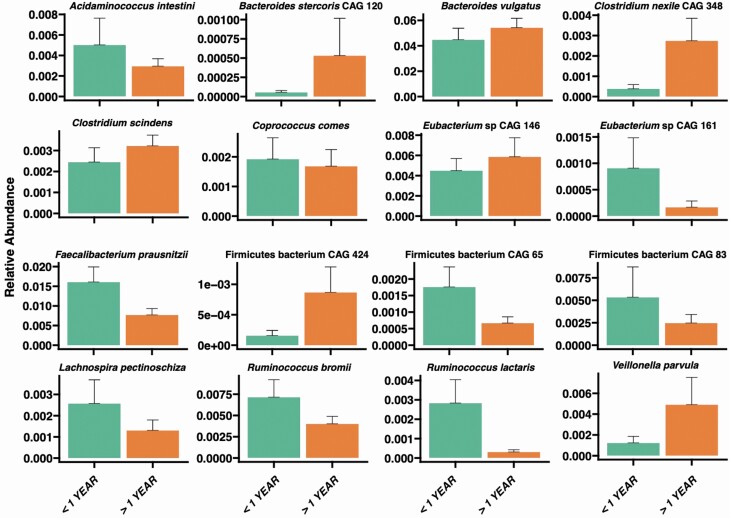
We explored the effect of the length of NH residency on microbiome composition. It has been previously shown that the NH microbiome differs from the microbiome of community-dwelling older adults; it takes more than 1 year before new NH residents adopt a NH microbiome (17). Using repeated-samples random forest classification (8), we analyzed older adults with less than or greater than 1 year of NH residence. For older adults with residence >1 year, species associated with inflammation were increased in abundance (ie, B vulgatus and Veillonella parvula) (26,33) and species with anti-inflammatory and symbiotic relationships were decreased (ie, F prausnitzii and Eubacterium species) (24,34) (Figure 2). We also noted lower abundances of anti-inflammatory microbiota such as Ruminococcus bromii and Ruminococcus lactaris (35).
Figure 2.
Species significantly associated with length of nursing home (NH) residence: Species level composition differs between older adults with less than, or greater than, 1 year NH residence. Data are presented as the mean among each residents’ average abundance with whiskers corresponding to the 95th percentiles. Mixed-effect random forest modeling was used to identify significant associations (FDR adjusted p < .05) using participants ID as a random effect to account for repeated sampling.
A secondary analysis, independent of results in Figure 2, was conducted to assess changes in the microbiome within the first year of NH residence. Length of residence was binned as <1 month, 1–6 months, and >6 months. Similar patterns of increasing pathogenic species abundances were observed beginning at 1–6 months (eg, Bacteroides ovatus, Enterococcus faecalis, Haemophilus parainfluenzae, and Klebsiella pneumoniae) and decreasing anti-inflammatory species starting at 6 months (eg, F prausnitzii) (Supplementary Figure 3).
Resident’s Microbiome Is Relatively Stable Over Long Periods of Time
Out of the cohort, 15 older adults were followed over multiple years. Stool samples were first collected over a 3- to 4-month, first-enrollment period and then, after a lapse of greater than 1 year, additional samples were collected in a second enrollment. Interval changes in their health status were noted using the same methods described above. The average time between samplings was 14 months (447 days; SD 84)). Among these older adults, 5 did not have any changes in their health, 9 had medication changes, 3 became malnourished, and 1 had worsening frailty (Supplementary Table 2).
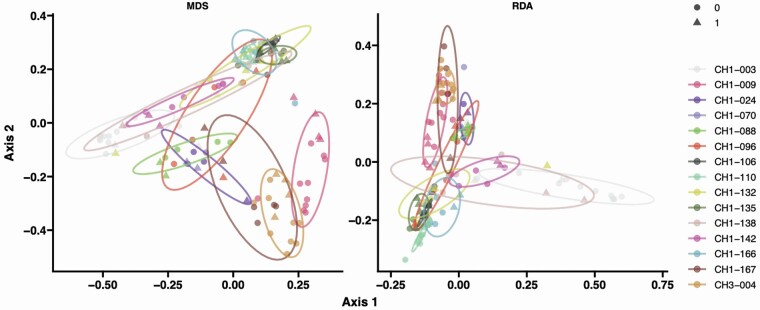
We first explored beta-diversity by ordination analysis (multidimensional scaling and redundancy analysis) using Jaccard distances for a measure of community species dissimilarity among the 15 participants from first- and second-enrollment sampling. Microbiota compositional differences were greater between individuals than within individuals over the period of enrollments demonstrating fecal microbiota stability in NH residents (PERMANOVA—Jaccard distance p < .001; Figure 3). We then tested whether alpha diversity varied between first and second enrollments according to changes in frailty, nutrition and antibiotic exposure intervals, or addition of medication classes. Antibiotic exposure was the only variable associated with decreased diversity (Shannon Index, b coef. −0.6488, p = .004; Inverse Simpson Index (b coef. −6.105988, p = .001). Interestingly, while the beta diversity analysis showed similarity between the 2-sampling periods in terms of broad community composition, a few species were found to be differential between the 2 enrollment windows by mixed-effect random forest classification modeling (Supplementary Figure 4). We could not associate these differences with changes in clinical covariates, suggesting either that the microbiome is resilient to short-term changes or there is not a specific healthy or dysbiotic pattern that emerges from these differences with the exception of antibiotic exposure. Of note, only one resident had worsening frailty limiting the analysis with this specific clinical covariate.
Figure 3.
Microbiome composition differs between nursing home older adults irrespective of time of enrollment. Samples collected from the 15 individuals (color-coded) during initial enrollment (circles) and over 1 year later (triangles). Each individual is displayed with ellipses with a 95% confidence interval. Ordination analysis using multidimensional scaling (MDS) and distance-based redundance (RDA) on the samples pairwise Jaccard distance is plotted.
Discussion
The NH contains a vulnerable group of older adults, living in confined communities, who are older, frailer, and have more comorbidities than older adults living in the community. In an attempt to better understand what factors influence the microbiome composition in NH older adults, we looked at demographic and clinical associations with the microbiome composition. We found that the high prevalence of psychoactive medications is strongly associated with microbiome composition and that other medications, such as those to treat hypertension, and other variables such as age, gender, frailty, and environmental factors all influenced microbiome composition. Interestingly, the length of NH residence correlated with increased abundances of pathogenic species and decreased abundances of anti-inflammatory or symbiotic bacterial species as early as 6 months after NH admission. Taken together, there appears to be a dysbiotic microbiome that develops over time and is strongly influenced by not only the medications that the residents are exposed to but also the resident’s frailty, age, nutrition, and length of residence.
Among Older Adults Exposed to Many Medications, Certain Classes Had a Greater Impact on Microbiome Composition
Residents in our cohort had a high prevalence of polypharmacy with 7 of 10 residents taking 5 or more daily medications. The prevalence of polypharmacy also varied significantly between NH sites, which has also been reported as typical of the NH environment (36). Medications with the strongest influence on microbiome composition were not the most commonly used. For example, the top 2 predictors of microbiome composition were medications used for the reduction of blood pressure (ACE inhibitors) and medications used for the treatment of Alzheimer’s disease (cholinesterase inhibitors) with only about 10% of the population taking either of these medication classes. In general, medications predicted 60% of the microbiome composition; specific medications, such as psychoactive medications, antihypertensives, made up a large part of this group. It is important to note that this is taking into account the resident’s medical conditions, nutrition, and demographics into the analysis which includes the categorization of polypharmacy.
Our descriptive analysis demonstrates the importance of nonantibiotic medications in shaping the intestinal microbiome. Compared to daily medications, recent antibiotic exposure did not influence a significant amount of the microbiome with only about 1.5% of the composition associated with this exposure. Nonantibiotic medications have been associated with changes in microbiome composition and about 24% of marked drugs have been shown to inhibit at least one common intestinal microbiome bacterial strain in vitro (37). Many medications are well known to have specific effects on the microbiome composition. The best examples of this are antibiotic exposures, where there is a loss in diversity and shifts in taxonomy abundances (38). Other medications such as proton pump inhibitors (39), statins (40), nonsteroidal anti-inflammatory drugs (41), and atypical antipsychotics (42) have had their microbiome effects described, however generally in isolation. It is uncommon that an older adult in NH care is not taking multiple daily medications. In fact, in less than 10% of our cohort, older adults were taking only one medication each day.
Increasing Age and Frailty Associate with Microbiome Composition
After medications and medical conditions, age and frailty make up a significant proportion associating with roughly 5% of the microbiome composition. Increasing frailty was associated with decreasing diversity, a finding that has been reported in older adults before however among a cohort with younger mean age of 63 years (range 42–86) (43). We also found that increasing frailty was associated with a higher prevalence of pathogens and lower abundances of butyrate producing organisms. Frailty has been shown to associate with a loss in diversity and community-associated microbiota (17,43). Frailty is a critical aging-related state marked by diminished physiological reserve and increased risk of adverse outcomes such as disability and death (44). Recent studies are beginning to suggest that the microbiome may play a role in the pathophysiology of frailty, however, evidence of the microbiome–frailty relationship remains limited (45). Frailty and sarcopenia has also shown associations with profiles of gut microbiota that link to distinct inflammatory biomolecules (46). Here, we add more evidence of a link between frailty and a dysbiotic microbiome hallmarked by increased pathogens and a reduction in health-associated microbiota. A similar pattern is not seen when it comes to age. Among older residents’ higher abundances of pathogens and inflammatory-associated microbiota such as Bacteroides eggerthii and B dorei (26), and lower abundances of symbiotic microbiota were seen. Conversely, however, higher abundances of the key butyrate producer F prausnitzii (24) were also seen in older residents. Lower abundances of R. bromii with higher abundances of butyrate producing organisms among older NH residents have been previously noted (19). Gut microbiome diversity is known to peak around 40 years of age and then decrease with significant loss of diversity among older adults (47). We noted increasing diversity with age among NH residents. A possible explanation for this finding is that to survive to an older age a more diverse microbiome, which is more capable and resilient (48), can impart survival benefits to the older adult. This explanation also fits with F prausnitzii being more prevalent in older residents.
Among our residents, we found that the environment does play a role in the composition of the microbiome. Among health volunteers, environment has been shown to have a substantial role in shaping the microbiome (49). This is of critical importance when considering how the microbiome might influence the spread of and infection from common pathogens, such as Clostridioides difficile (50), among older adults living together in the NH. After adjusting for all other clinical and demographic variables, the local environment, in the form of the floor/wing the resident lived, predicted 30% of the environmental category or 3% overall. Recent antibiotic exposure and hospitalization also had similar predictive power and all 3 were in the top 30 predictors of microbiome composition.
Longer Time Within the NH Environment Associated with Higher Abundances of Pathogens and Lower Abundances of Symbiotic Microbiota
The length of time an older adult lived within the NH environment did correlate with decreases in a several key microbiome species. First looking at the 1-year mark as a cutoff in categorizing residents we noted increased presence of bacterial species, among older adults living in the NH >1, year that cause local inflammation such as V parvula (33) or pathogenic species such as B vulgatus and B stercoris (26). We also noted lower abundances of anti-inflammatory microbiota such as R bromii and R lactaris (35), and key commensal species including F prausnitzii (24) and Eubacterium species (34) being decreased in older adults living in the NH >1 year. We chose 1 year, a priori, to investigate microbiome-associated changes over time based off of previous investigations where it was shown that among residents living in a NH it took upwards of 1 year to have their gut microbiota profile be the furthest from community-dwelling participants (17). Given these findings, we wanted to see if there were differences noted earlier than this 1-year point. Residents were further stratified into categories of < 1 month, 1–6 months, and >6 months with findings similar to the previous time analysis where there were increasing abundances of pathogenic species starting at 1–6 months and decreases in the anti-inflammatory microbiota species starting at 6 months. This analysis suggests that there is a distinct dysbiotic microbiome pattern that develops when an individual enters the NH starting before 1 year that is hallmarked with increasing pathogens and decreasing symbiotic species.
Microbiome Stability Demonstrated Over Longer Periods of Time
Among the residents we followed for >1 year between longitudinal sampling periods, we noticed that there was not much difference in diversity or microbiome species abundances. Interval antibiotic exposure did associates with a reduction in diversity. This was not unexpected given antibiotics profound and rapid effect on the gut microbiota, with a loss of diversity (4). Older adults in the NH have been shown to have stability over short periods of time without any changes in health (19). Here, we demonstrate resilience in the microbiome over longer periods of time with changes in medications, although in a smaller cohort. We believe it takes longer exposure time for medication and health changes to impact the composition of the microbiome. We did note changes in key microbiome species (like members of the Eubacterium species) without any disenable patterns with interval medication or health changes.
Strengths and Limitations
This study had several strengths and limitations. One limitation of this study is that we had a different number of stool samples from each older adult and each older adult participated for different lengths of time. We accounted for this imbalance in our statistical approach by adjusting for each individual. A subset of individuals also participated for over 1 year. These older adults allowed us to investigate the gut microbiome composition and changes over longer periods of time in this group. This is one of the largest longitudinal cohorts of NH older adults reporting on gut microbiome composition. That being said, this study is still limited in the number of residents enrolled. A larger cohort would help us take a deeper look at the multiple levels of data and better explore other classes of medications used less frequently by NH older adults. There are potential confounding variables, specifically classes of medications the residents were taking (such as immunosuppressants) that were not evaluated in this cohort due to the small number of residents on these drugs. Men made up 18% of the study population which is lower than national averages at roughly one third of the NH population. Finally, the amount of exercise or cognitive measurements, both known to associate with microbiome composition, for each participant that was enrolled was not recorded.
Conclusions
Older adults living within a NH have a gut microbiome that is not only heavily influenced by the medications they are exposed to but also their age, frailty status, and location they are living. Their microbiome also changes over time after admittance to the facility demonstrating a likely time-dependent dysbiosis. Additionally, the NH microbiome appears stable but susceptible to antibiotic exposure. Our findings help to better describe the NH microbiome and offer a starting point from which to manipulate the gut microbiome in an attempt to improve older adult health and reduce disease burden.
Supplementary Material
Funding
J.P.H. was supported by 2 National Institutes of Health (NIH) grants from the National Institute on Aging (grants 1R03AG056356-01 and 1K23AG057790-01A1). V.B. is supported by NIH award 1R15AI112985-01A1 and by National Science Foundation (NSF) award ABI Innovation grant 1458347. B.A.M. is supported by NIH award DK056754.
Conflict of Interest
None declared.
References
- 1. Lee DC, Barlas D, Ryan JG, Ward MF, Sama AE, Farber BF. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: prevalence and predictors of colonization in patients presenting to the emergency department from nursing homes. J Am Geriatr Soc. 2002;50:1463–1465. doi: 10.1046/j.1532-5415.2002.50377.x [DOI] [PubMed] [Google Scholar]
- 2. Cassone M, Mody L. Colonization with multi-drug resistant organisms in nursing homes: scope, importance, and management. Curr Geriatr Rep. 2015;4:87–95. doi: 10.1007/s13670-015-0120-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daneman N, Bronskill SE, Gruneir A, et al. . Variability in antibiotic use across nursing homes and the risk of antibiotic-related adverse outcomes for individual residents. JAMA Intern Med. 2015;175:1331–1339. doi: 10.1001/jamainternmed.2015.2770 [DOI] [PubMed] [Google Scholar]
- 4. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Le Bastard Q, Al-Ghalith GA, Grégoire M, et al. . Systematic review: human gut dysbiosis induced by non-antibiotic prescription medications. Aliment Pharmacol Ther. 2018;47:332–345. doi: 10.1111/apt.14451 [DOI] [PubMed] [Google Scholar]
- 6. Caffrey C, Sengupta M, Park-Lee E.. Residents Living in Residential Care Facilities: United States, 2010. NCHS data brief, no 91. Hyattsville, MD: National Center for Health Statistics, 2012. [PubMed] [Google Scholar]
- 7. Jose PA, Raj D. Gut microbiota in hypertension. Curr Opin Nephrol Hypertens. 2015;24:403–409. doi: 10.1097/MNH.0000000000000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haran JP, Bhattarai SK, Foley SE, et al. . Alzheimer’s disease microbiome is associated with dysregulation of the anti-inflammatory P-Glycoprotein pathway. mBio. 2019;10:e00632–19. doi: 10.1128/mBio.00632-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singer-Englar T, Barlow G, Mathur R. Obesity, diabetes, and the gut microbiome: an updated review. Expert Rev Gastroenterol Hepatol. 2019;13:3–15. doi: 10.1080/17474124.2019.1543023 [DOI] [PubMed] [Google Scholar]
- 10. Masnoon N, Shakib S, Kalisch-Ellett L, Caughey GE. What is polypharmacy? A systematic review of definitions. BMC Geriatr. 2017;17:230. doi: 10.1186/s12877-017-0621-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dwyer LL, Han B, Woodwell DA, Rechtsteiner EA. Polypharmacy in nursing home residents in the United States: results of the 2004 National Nursing Home Survey. Am J Geriatr Pharmacother. 2010;8:63–72. doi: 10.1016/j.amjopharm.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 12. Maher RL, Hanlon J, Hajjar ER. Clinical consequences of polypharmacy in elderly. Expert Opin Drug Saf. 2014;13:57–65. doi: 10.1517/14740338.2013.827660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ticinesi A, Milani C, Lauretani F, et al. . Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep. 2017;7:11102. doi: 10.1038/s41598-017-10734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di Sabatino A, Lenti MV, Cammalleri L, Corazza GR, Pilotto A. Frailty and the gut. Dig Liver Dis. 2018;50:533–541. doi: 10.1016/j.dld.2018.03.010 [DOI] [PubMed] [Google Scholar]
- 15. Rockwood K, Song X, MacKnight C, et al. . A global clinical measure of fitness and frailty in elderly people. CMAJ. 2005;173:489–495. doi: 10.1503/cmaj.050051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kojima G. Prevalence of frailty in nursing homes: a systematic review and meta-analysis. J Am Med Dir Assoc. 2015;16:940–945. doi: 10.1016/j.jamda.2015.06.025 [DOI] [PubMed] [Google Scholar]
- 17. Claesson MJ, Jeffery IB, Conde S, et al. . Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- 18. Austin SR, Wong YN, Uzzo RG, Beck JR, Egleston BL. Why summary comorbidity measures such as the Charlson Comorbidity Index and Elixhauser score work. Med Care. 2015;53:e65–e72. doi: 10.1097/MLR.0b013e318297429c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haran JP, Bucci V, Dutta P, Ward D, McCormick B. The nursing home elder microbiome stability and associations with age, frailty, nutrition and physical location. J Med Microbiol. 2018;67:40–51. doi: 10.1099/jmm.0.000640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guigoz Y. The Mini Nutritional Assessment (MNA) review of the literature–What does it tell us? J Nutr Health Aging. 2006;10:466–485; discussion 485. [PubMed] [Google Scholar]
- 21. Truong DT, Franzosa EA, Tickle TL, et al. . MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589 [DOI] [PubMed] [Google Scholar]
- 22. Hidron AI, Edwards JR, Patel J, et al. . NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861 [DOI] [PubMed] [Google Scholar]
- 23. Mishra S, Imlay JA. An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol Microbiol. 2013;90:1356–1371. doi: 10.1111/mmi.12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sokol H, Pigneur B, Watterlot L, et al. . Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takahashi K, Nishida A, Fujimoto T, et al. . Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion. 2016;93:59–65. doi: 10.1159/000441768 [DOI] [PubMed] [Google Scholar]
- 26. Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berger FK, Schwab N, Glanemann M, Bohle RM, Gärtner B, Groesdonk HV. Flavonifractor (Eubacterium) plautii bloodstream infection following acute cholecystitis. IDCases. 2018;14:e00461. doi: 10.1016/j.idcr.2018.e00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yoshida N, Emoto T, Yamashita T, et al. . Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. 2018;138:2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714 [DOI] [PubMed] [Google Scholar]
- 29. Kant R, Rasinkangas P, Satokari R, Pietilä TE, Palva A. Genome sequence of the butyrate-producing anaerobic bacterium Anaerostipes hadrus PEL 85. Genome Announc. 2015;3:e00224–15. doi: 10.1128/genomeA.00224-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zaidi MB, Estrada-García T. Shigella: a highly virulent and elusive pathogen. Curr Trop Med Rep. 2014;1:81–87. doi: 10.1007/s40475-014-0019-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duranti S, Ruiz L, Lugli GA, et al. . Bifidobacterium adolescentis as a key member of the human gut microbiota in the production of GABA. Sci Rep. 2020;10:14112. doi: 10.1038/s41598-020-70986-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Engels C, Ruscheweyh HJ, Beerenwinkel N, Lacroix C, Schwab C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol. 2016;7:713. doi: 10.3389/fmicb.2016.00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gouze H, Noussair L, Padovano I, et al. . Veillonella parvula spondylodiscitis. Med Mal Infect. 2019;49:54–58. doi: 10.1016/j.medmal.2018.10.001 [DOI] [PubMed] [Google Scholar]
- 34. Barcenilla A, Pryde SE, Martin JC, et al. . Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagao-Kitamoto H, Kamada N. Host-microbial cross-talk in inflammatory bowel disease. Immune Netw. 2017;17:1–12. doi: 10.4110/in.2017.17.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beloosesky Y, Nenaydenko O, Gross Nevo RF, Adunsky A, Weiss A. Rates, variability, and associated factors of polypharmacy in nursing home patients. Clin Interv Aging. 2013;8:1585–1590. doi: 10.2147/CIA.S52698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maier L, Pruteanu M, Kuhn M, et al. . Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Imhann F, Bonder MJ, Vich Vila A, et al. . Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. doi: 10.1136/gutjnl-2015-310376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J, Lee H, An J, Song Y, Lee CK, Kim K, et al. . Alterations in gut microbiota by statin therapy and possible intermediate effects on hyperglycemia and hyperlipidemia. Front Microbiol. 2019;10:1947. doi: 10.3389/fmicb.2019.01947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016;22:178.e1–178.e9. doi: 10.1016/j.cmi.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Flowers SA, Evans SJ, Ward KM, McInnis MG, Ellingrod VL. Interaction between atypical antipsychotics and the gut microbiome in a bipolar disease cohort. Pharmacotherapy. 2017;37:261–267. doi: 10.1002/phar.1890 [DOI] [PubMed] [Google Scholar]
- 43. Jackson MA, Jeffery IB, Beaumont M, et al. . Signatures of early frailty in the gut microbiota. Genome Med. 2016;8:21. doi: 10.1186/s13073-016-0275-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. Lancet. 2013;381:752–762. doi: 10.1016/S0140-6736(12)62167-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piggott DA, Tuddenham S. The gut microbiome and frailty. Transl Res. 2020;221:23–43. doi: 10.1016/j.trsl.2020.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Picca A, Ponziani FR, Calvani R, et al. . Gut microbial, inflammatory and metabolic signatures in older people with physical frailty and sarcopenia: results from the BIOSPHERE Study. Nutrients. 2019;12:1–15. doi: 10.3390/nu12010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Biagi E, Nylund L, Candela M, et al. . Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rothschild D, Weissbrod O, Barkan E, et al. . Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- 50. Buffie CG, Bucci V, Stein RR, et al. . Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.