In vivo recordings of cone photoreceptor outputs in a tetrachromate reveal efficient ancestral strategy for color processing.
Abstract
For color vision, retinal circuits separate information about intensity and wavelength. In vertebrates that use the full complement of four “ancestral” cone types, the nature and implementation of this computation remain poorly understood. Here, we establish the complete circuit architecture of outer retinal circuits underlying color processing in larval zebrafish. We find that the synaptic outputs of red and green cones efficiently rotate the encoding of natural daylight in a principal components analysis–like manner to yield primary achromatic and spectrally opponent axes, respectively. Blue cones are tuned to capture most remaining variance when opposed to green cones, while UV cone present a UV achromatic axis for prey capture. We note that fruitflies use essentially the same strategy. Therefore, rotating color space into primary achromatic and chromatic axes at the eye’s first synapse may thus be a fundamental principle of color vision when using more than two spectrally well-separated photoreceptor types.
INTRODUCTION
In visual scenes, information about wavelength is fundamentally entwined with information about intensity because the spectrum of natural light is highly correlated (1–3). Accordingly, wavelength information must be extracted by comparing the signals from at least two spectrally distinct photoreceptors, in a process generally referred to as “color opponency” (4). To this end, most animal eyes use up to five spectral types of photoreceptors for daylight vision, with around four being the norm for vertebrates [reviewed in (4, 5)]. However, our knowledge of how the signals from four or more spectral types of photoreceptors are harnessed at a circuit level to extract this specific chromatic information remains limited.
Increasing the diversity of available spectral photoreceptors exponentially expands the diversity of theoretically detectable spectral contrasts. However, there is a law of diminishing returns: In natural scenes, some spectral contrasts are much more abundant than others. For efficient coding (6–8), animal visual systems should therefore prioritize the specific contrasts that are particularly prevalent in their natural visual world.
Here, we explored how zebrafish extract wavelength and intensity information from their natural visual world. Similar to many surface-dwelling fish, already their larvae use the “full” ancient tetrachromatic cone photoreceptor complement comprising red, green, blue, and ultraviolet (UV) cones (9). Their retinal circuits can be noninvasively monitored and manipulated in the live animal (10) to provide insights into the computation of color in the intact circuit. We asked three questions: (i) What is the in vivo spectral tuning of zebrafish cone outputs at the synapse, (ii) what is the circuit implementation, and (iii) how does this specific tuning support efficient sampling and decomposition of natural light?
Unexpectedly, we found that two of the four cone types (green and blue) are strongly opponent, while the remaining two (red and UV) are essentially non-opponent, despite feeding into the same horizontal cell (HC) network. We go on to show how this spectral tuning is anatomically and functionally implemented at the circuit level using HCs. Furthermore, comparison of the spectral tuning of the four cone types to the spectral statistics of natural light showed that this specific cone tuning arrangement allows zebrafish to effectively “solve” a major fraction of the basic wavelength discrimination problem already at the first synapse of their visual system: Red cones encode “color-invariant” achromatic information. Green cones encode “brightness-invariant” spectral information. Blue cones provide a second chromatic axis that can be further optimized by possible opposition to green cones downstream, while UV cones by themselves provide a secondary “UV-achromatic” signal, presumably for prey capture (11). These findings also strongly imply that ancestral vertebrate circuits for color vision are built upon the opponent signals from green and blue cones, which are lost in mammals including humans (4).
Last, zebrafish are not alone in using such an efficient strategy. By linking the spectral tuning of Drosophila melanogaster photoreceptors (12) with hyperspectral natural imaging data (13), we note that fruitflies use essentially the same strategy. However, their spectral tunings are systematically blue-shifted compared to those of zebrafish, presumably to acknowledge the relatively blue-shifted statistics of natural light in air (14). Together, our findings highlight a potentially general circuit-level mechanism of vision whereby incoming light is decomposed into “color” and “grayscale” components at the earliest possible site.
RESULTS
Spectral tuning of zebrafish cones in vivo
To determine spectral tuning functions of the larval zebrafish’s four cone types (red, green, blue, and UV) (9), we custom-built a hyperspectral full-field stimulator based on an earlier design (fig. S1, A and B) (15). A diffraction grating was used to reflect the light from 14 light-emitting diodes (LEDs) (peaks, 360 to 655 nm) into a collimated fiber optic that was pointed at the live zebrafish’s eye mounted under a two-photon (2P) microscope. To avoid spectral cross-talk with the 2P imaging system, we line-synchronized each LED’s activity with the scanner retrace (16, 17). Together, this arrangement permitted spectrally oversampling the much broader cone opsins (fig. S1, B and C) during in vivo 2P imaging in the eye. All stimuli were presented as wide-field flashes from dark.
Green and blue cones, but not red and UV cones, display strong spectral opponency
We generated four cone type–specific SyGCaMP6f lines (Fig. 1, A and B), where the calcium biosensor GCaMP6f was fused to synaptophysin (18) such that it localized in the presynaptic terminals of each of the four cone photoreceptors. This allowed us to measure the spectral tuning of cones at the level of their presynaptic terminals (pedicles), i.e., their output (Fig. 1, C and D). Here, cones connect with other cones via gap junctions (19), with HCs that provide both feedback and feedforward inhibition (20), as well as with bipolar cells that carry the photoreceptor signal to the feature extracting circuits of the inner retina (21). We did not study rods, as these are functionally immature in zebrafish larvae (22, 23).
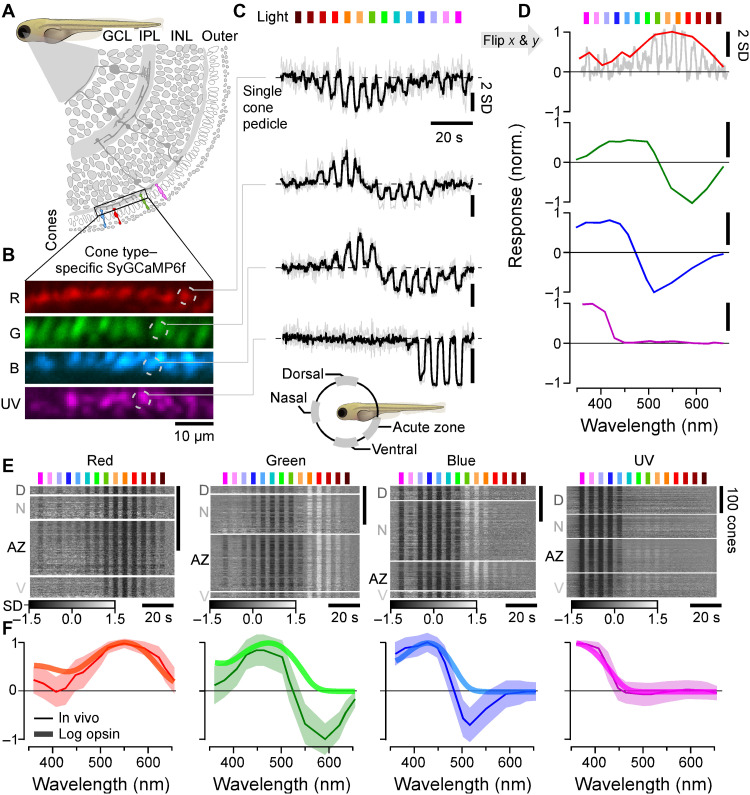
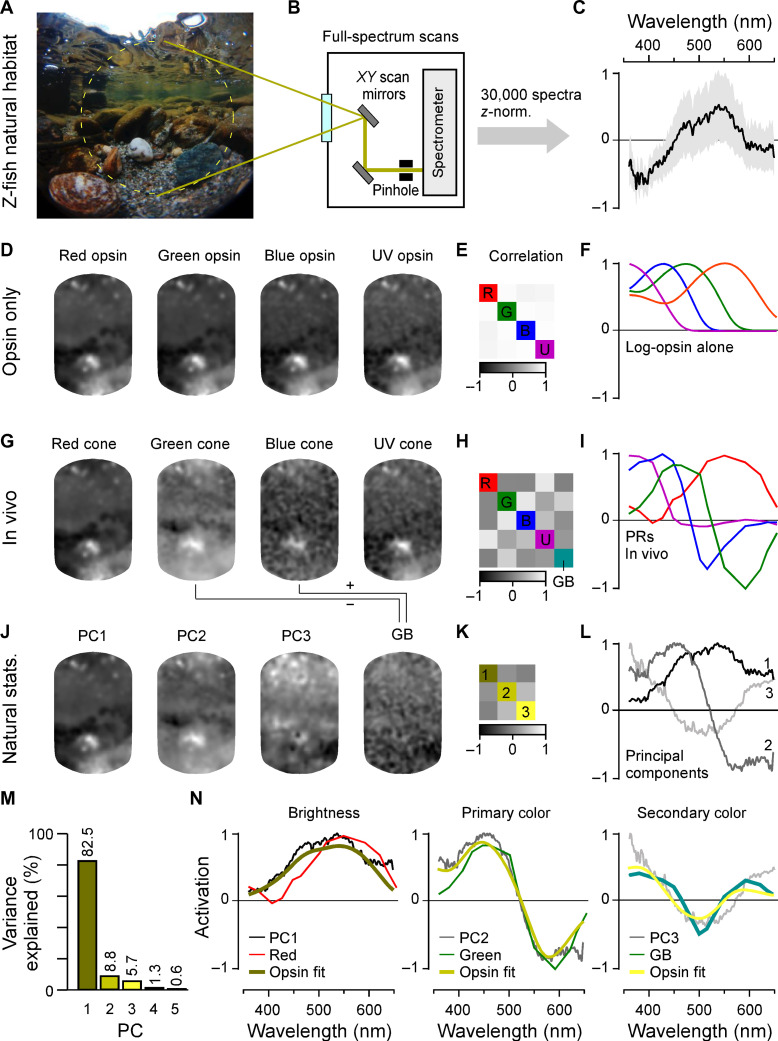
Fig. 1. In vivo spectral tuning of larval zebrafish cones and HC block.
(A) Schematic of larva zebrafish retina, with position of cone pedicles highlighted [adapted from (82)]. (B and C) Example scans of the four spectral cones (B) (Methods) with single pedicle response examples for each (C) to 3-s flashes of light from each of the 14 LEDs [see fig. S1 (A to C)]. The means superimposed on individual repeats are shown. (D) Example spectral responses summarized from (C). Note that in this representation, both the x and y axes are flipped relative to the raw responses. (E and F) Population responses of each cone type recorded in different parts of the eye (D, dorsal; N, nasal; AZ, acute zone; V, ventral; see schematic inset above for anatomical reference; vertical scale bars indicate n = 100 cones; see also fig. S1G) (E) and population mean ± 95% confidence intervals with log-transformed respective opsin template superimposed (F) (Methods). Heatmaps (E) are time-inverted to facilitate comparison to summary plots (F); grayscale bars are in z scores. Darker shades indicate a drop in calcium relative to baseline, indicative of a cone’s “intrinsic” light response, while lighter shades indicate a rise in calcium, indicative of sign-inverted inputs from the outer retinal network (see Fig. 2).
From fluorescence traces, we extracted tuning functions (Methods), inverting both the x and y axes (Fig. 1D, inset). The inversions were done to display tuning functions from short to long wavelengths as is conventional and to compensate for the fact that vertebrate photoreceptors hyperpolarize in response to light (24). We adhered to the time inversion henceforth to facilitate comparison between raw data and summary plots (e.g., Fig. 1E). We systematically measured tuning functions for n = 409, 394, 425, and 431 individual red, green, blue, and UV cones, respectively (n = 9, 11, 12, and 7 fish). A total of n = 172, 288, 312, and 410 recordings, respectively, passed a quality criterion (Methods and fig. S1, D to G) and were kept for further analysis.
Because the larval zebrafish eye is both structurally and functionally asymmetrical (11, 14, 25–28), we always sampled from four different regions of the eye’s sagittal plane: dorsal (D), nasal (N), ventral (V), and the area temporalis [acute zone, also known as “strike zone” (14)]. With exceptions noted below (see also Discussion), we found that the spectral tuning of cones was approximately eye position invariant (fig. S1G). For further analysis, we therefore averaged across cones irrespective of their position in the eye (Fig. 1, E and F).
On average, red and UV cones had approximately monophasic (non-opponent) output tuning functions that were largely in line with the tuning function of their respective log-transformed opsins (Methods). Such a log transform is expected from the nature of signal transfer between outer segment phototransduction to synaptic calcium in the pedicle (12, 29, 30). Red cones were broadly tuned and never exhibited opponency (Fig. 1F, left). Some individual red cones hyperpolarized in response to all tested wavelengths (Fig. 1E, left; cf. fig. S1G). Nevertheless, on average, red cone sensitivity was weakly suppressed in the UV range compared to the log-transformed opsin template (Discussion). In contrast, all UV cones were narrowly tuned up to the short-wavelength cutoff imposed by the eye optics (~350 nm). Their tuning curve near perfectly matched the respective opsin template (Fig. 2F, right). UV cones in the acute zone and ventral retina exhibited weak but significant opponency to mid-wavelengths (fig. S1G and Discussion).
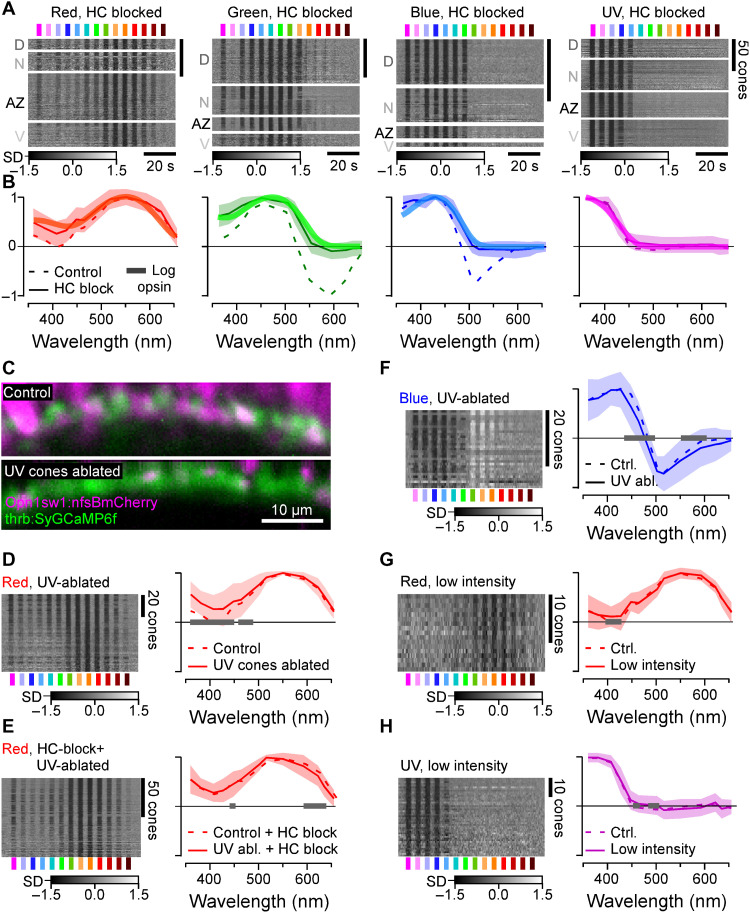
Fig. 2. Opsin-like cone responses in the absence of HCs.
(A and B) Population responses of each cone type during pharmacological blockage of HCs (A) (Methods) and population mean ± 95% confidence intervals with log-transformed respective opsin template superimposed (B) (Methods). (C) Pharmacogenetic UV cone ablation in the background of red cone GCaMP labeling before (top) and 24 hours after 2-hour treatment of metronidazole (10 mM) application (bottom) (Methods). (D and E) Red cone tunings after UV cone ablation (n = 77) (D) and after additional pharmacological HC blockage (n = 103) (E). Heatmaps (left) and means ± SD (solid lines and shadings) and analogous data in the presence of UV cones (dotted, from Figs. 1F and 2B) are shown. Note that the 361-nm LED was omitted in this experiment. (F) As (D), but here, recording from blue cones (n = 30). (G and H) Red (n = 17) (G) and UV cone tunings (n = 43) (H) at about ninefold reduced overall stimulus-light intensities (solid lines and shadings; Methods), compared to tunings at “standard” light intensities (from Fig. 1F). Gray bars on the x axis in (D) to (H) indicate significant differences based on the 99% confidence intervals of the fitted generalized additive models (GAMs) (Methods). Note that heatmaps (A and D to H) are time-inverted to facilitate comparison to summary plots (B and D to H). Grayscale bars are in z scores.
Unlike red and UV cones, the in vivo output tuning functions of green and blue cones did not match their log-transformed opsin templates. Instead, these cones consistently exhibited strong spectral opponency to mid- and/or long-wavelength light (Fig. 1, E and F, middle). Here, blue cones had a highly consistent zero crossing at 483 ± 1 nm, while most green cones inverted at 523 ± 1 nm (mean, 95% confidence intervals; Methods). Green cones in the acute zone were slightly long wavelength shifted with a zero crossing at 533 ± 1 nm (fig. S1G and Discussion). To our knowledge, these are the first direct in vivo measurements of cone pedicles’ spectral tuning functions in a vertebrate.
Spectral tuning of zebrafish cones is fully accounted for by expressed opsins and HC feedback
The nature of phototransduction in cone photoreceptors dictates that the absorption of photons leads to a drop in synaptic calcium. Accordingly, light-driven increases in synaptic calcium (Fig. 1F) must come from a sign-inverting connection from other cones, most likely via HCs (31, 32). We therefore decoupled HCs by pharmacologically blocking the glutamate output from cones using 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) (Methods and Fig. 2, A and B). This completely abolished all spectral opponency and increased the UV response amplitude of red cones. As a result, now, all four cone tuning functions were fully accounted for by the respective log-transformed opsins (Fig. 2, A and B, and fig. S2A). Our results further implied that heterotypical cone-cone gap junctions, if present, do not strongly contribute to spectral cone tuning. In support, cone tunings were essentially invariant to additional genetic ablation of UV cones in the absence of HCs (Fig. 2, C to F). Moreover, reducing overall stimulus brightness to probe for possible response saturation had no major effects on tuning functions (Fig. 2, G and H). Together, our results strongly suggest that in vivo, the spectral tuning of all zebrafish cones is driven by the expressed opsin variant and shaped only by specific connections with HCs relaying feedforward signals from other cones. What are these HC connections?
A connectome of the larval zebrafish outer retina
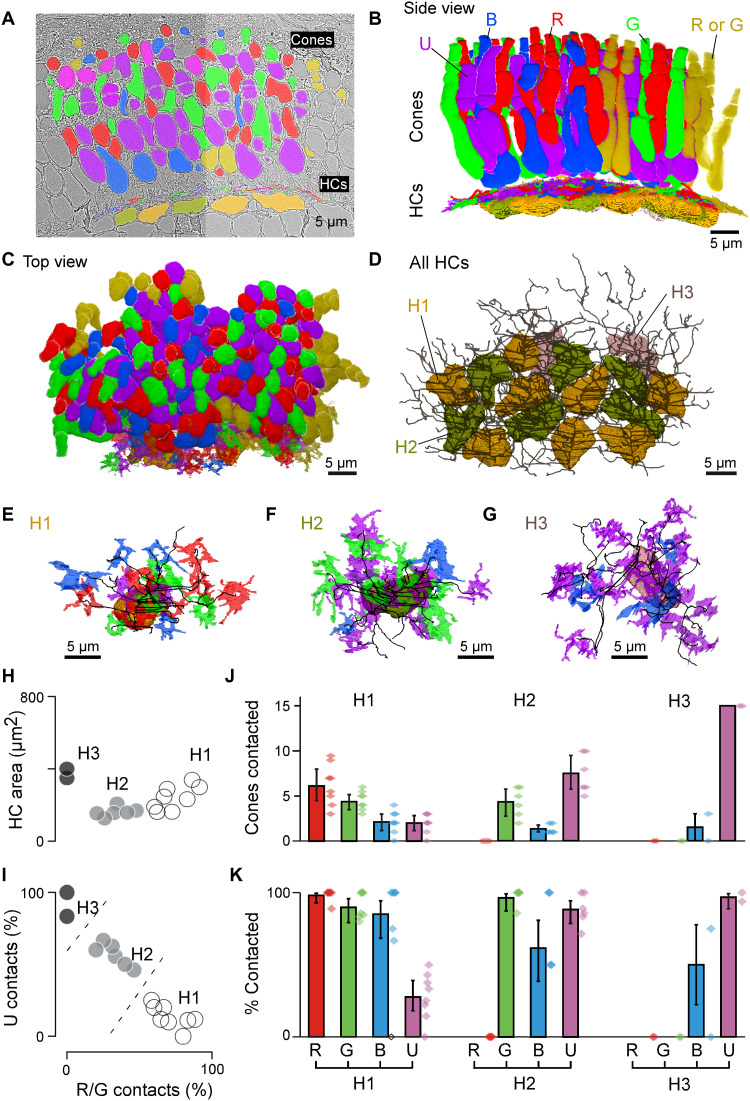
Light microscopy studies in adult zebrafish have described at least three types of cone HCs (H1 to H3), which contact R/G/B/(U), G/B/U, and B/U cones, respectively (31, 33). However, for larval zebrafish, HC types and their connections to cones are not known except for H3 (34). To complete this gap in knowledge, we used a connectomics approach based on a combination of serial section electron microscopy (EM) (Fig. 3) and confocal imaging (fig. S3 and Methods). In total, we reconstructed a 70 μm by–35 μm by–35 μm patch of larval outer retina in the acute zone, which comprised n = 140 cones and n = 16 HCs (Fig. 3, A to D). UV and blue cones were identified directly in the EM volume on the basis of their characteristic outer plexiform layer (OPL)–proximal mitochondrial pockets (UV; fig. S3A) and somata (blue, fig. S3B), respectively. This allowed initially sorting cones into three groups: UV, blue, and red/green. Next, we traced each HC’s dendritic tree and identified their connections to cones belonging to each of these cone groups (Fig. 3, D to K, and fig. S3, C to H). Relating each HC’s relative connectivity to UV cones to their connections to red/green cones allowed separating HCs into three groups (Fig. 3I and fig. S3G), which were verified by clustering the HCs on all extracted features (Methods). These were dubbed H1, H2, and H3, on the basis of their similarity to known adult HC types (33, 35, 36). The same classification was then further confirmed by confocal microscopy (fig. S3, D to H). Of these, some HCs reliably contacted all red/green cones within their dendritic field and were presumed to be H1. Other HCs systematically avoided approximately half of these cones. These were presumed to be H2s given that that this type of HC contacts green but not red cones. In line with confocal data (fig. S3), this allowed disambiguating red cones (contacted only by H1) from green cones (contacted by both H1 and H2). With the exception of n = 14 of 66 red-green cones that could not be unequivocally allocated because of their location at the edge of the volume [yellow, counted as 0.5 red and 0.5 green in Fig. 3 (B and D)], this completed cone type identifications.
Fig. 3. Connectomic reconstruction of outer retinal circuitry.
(A) Example vertical EM section through the outer retina, with cones and HCs painted. Cones are color coded by their spectral identity, with “yellow cones” indicating red or green cones at the section edge that could not be unequivocally attributed (Methods); HCs: H1, yellow/brown; H2, dark green; and H3, light pink. (B to D) Full volumetric reconstruction of all cones and skeletonized HCs in this patch of retina, shown from the side (B), top (C), and HCs only (D). (E to G) Example individual HCs classified as H1 (E), H2 (F), and H3 (G) with connecting cone pedicles. (H to K) Quantification of HC dendritic area (H) (cf. fig. S3G) and cone contacts (J and K) shown as absolute numbers with bootstrapped 95% confidence intervals (J) and percentage of cones in dendritic territory with binomial confidence intervals (I and K).
From here, we quantified each HC groups’ connections to the four cone types. This revealed that H1 contacted essentially all red, green, and blue cones within their dendritic fields but imperfectly avoided UV cones (Fig. 3, J and K). In contrast, H2, by definition, never contacted red cones but contacted all other cones including UV cones. Last, H3 was strongly dominated by UV cone contacts, with a small contribution from blue cones. H3 never contacted red or green cones. Together, this confirmed that essential features of adult HC connectivity are already present in larvae and, moreover, contributed cone-weighting information for the three HC types. We next asked how this specific HC connectivity matrix underpins cone spectral tunings.
H1 HCs likely underlie most spectral tuning
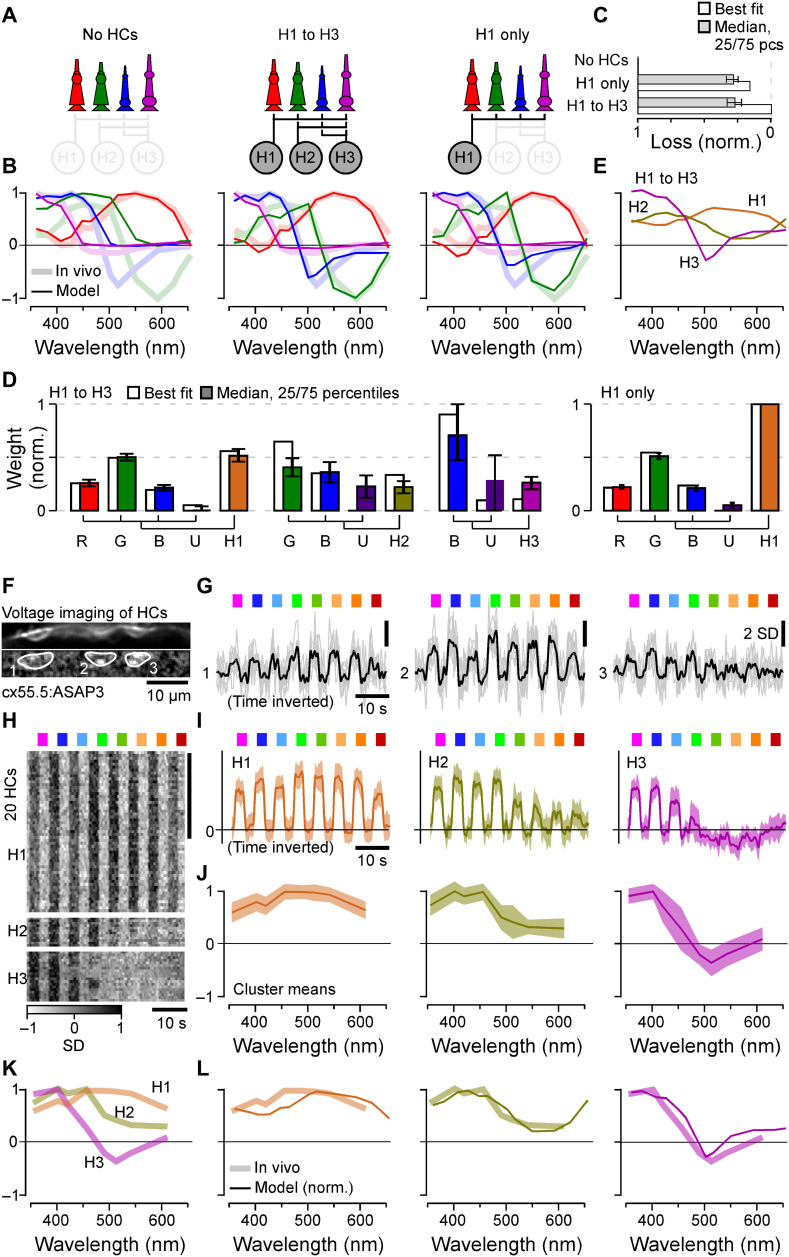
To explore how the three HC types contribute to spectral cone tunings, we first set up a series of functional circuit models for all possible combinations of HCs (Methods). These linear models included the established connectivity structure (Fig. 3K) and were driven by the cone tunings in the absence of HCs (Fig. 2A), with the goal of explaining cone tunings in the presence of HCs (Fig. 1F). We computed posteriors for the model parameters using likelihood-free inference (37) based on the cone tunings, and we assumed sign-preserving connections from cones to HCs but sign-inverting connections from HCs to cones. The model recapitulated well the in vivo tuning functions of all cones when simultaneously drawing on all three HC types. However, almost the same fit quality was achieved when using H1 alone (Fig. 4, A to D; cf. fig. S4, A to C), while H2 mainly fine-tuned the blue and UV cones and H3 had negligible impact on any cone tunings (fig. S4A). Any model that included H1 outperformed any model that excluded H1 (fig. S4, A to C). H1, where present, also consistently provided the strongest feedback among HCs (Fig. 4D and fig. S4C). Together, modeling therefore suggests that H1-like HCs are the main circuit element underlying the in vivo spectral tuning of zebrafish cones. Moreover, the inferred relative cone type weighting for H1 approximated their anatomical connectivity established by EM (Fig. 4J), with the exception of green cones that had stronger-than-expected weights (Fig. 4D), possibly uncovering an increased synaptic gain at this site.
Fig. 4. Spectral tuning of cones by HCs.
(A to E) Linear model of spectral tuning in an outer retinal network composed of four cone types and three HC types, with maximum connectivity matrix defined as in Fig. 3K (Methods). Cone tunings are initiated on the basis of in vivo data during HC block (Fig. 2B). Different HC combinations include (A) (from left) the following: no HCs, all HCs, and H1 only. In each case, the model computes resultant cone tunings (solid lines) superimposed on in vivo data in the absence of HC block (shadings, from Fig. 1F) (B) and reconstruction quality (C) as loss relative to the peak performance for the full H1 to H3 model (loss = 0) and in the absence of HCs (loss = 1) and normalized weights such that cones contributing to a given HC, and HCs contributing to the full model, each add up to 1 (D). In addition, resultant HC tunings are shown for the full H1 to H3 model (E). (F to J) In vivo voltage imaging of HC somata’s spectral tuning (Methods). (F and G) Example scan (F) [average image (top) and local response correlation (83) and regions of interest (ROIs; bottom)] and responses (G) (mean superimposed on individual repeats shown for the three HC somata in this scan, of which ROIs 1 and 2 responded broadly across all tested wavelength, while ROI 3 exhibited a clear short-wavelength preference). (H to J) Results of clustering of mean responses from n = 86 ROIs (H) (n = 15 fish) with cluster means (I) and extracted tuning functions (J) (means ± SD). (K and L) Mean tunings of in vivo HC clusters [(K), from (J)] and superposition of each modeled [solid lines, from (E)] and measured [shading, from (K)] HCs. Note that raw (G) and averaged (I) HC responses and the summary heatmap (H) are time inverted to facilitate comparison with summary plots (J to L). The grayscale bar in (H) is in z scores.
Next, we sought to verify the model by experimentally measuring the spectral tunings of HCs and comparing these to the predicted HC tunings from the full model (Fig. 4E). For this, we used in vivo 2P voltage imaging of HC somata using the genetically encoded voltage biosensor ASAP3 (Fig. 4, F to L) (38). The choice of voltage over calcium imaging was motivated by a lack of detectable calcium responses in the somata of larval HCs (Methods). In total, recordings from n = 86 HCs that passed a quality criterion (Methods) were sorted into three clusters (Methods). The largest cluster exhibited a spectrally broad, monophasic response that closely matched the model’s prediction for H1 [Fig. 4L; see also (31, 35)]. Next, short-wavelength biased clusters 2 and 3 closely matched the model’s prediction for H2 and H3, respectively (31, 35).
Efficient encoding of achromatic and chromatic contrasts in natural light
To explore how the specific in vivo cone tuning functions may support zebrafish vision in nature, we next computed the distribution of achromatic and chromatic content of light in their natural habitat. For this, we used a total of n = 30 underwater hyperspectral images (1000 pixels each: 30,000 spectra) previously recorded in a zebrafish natural habitat in Northern India (Fig. 5, A to C) (13, 14). Using one example scan for illustration (Fig. 5A), we first computed each cone’s view of the world in the absence of outer retinal feedback by taking the dot product of each log-transformed opsin spectrum with each pixel spectrum (Fig. 5, D to F). In this configuration, the intensity-normalized representations of the scene by each of the four cones were extremely similar as expected from high spectral correlations in natural light (Fig. 5D). In contrast, when the same scene was computed for the intact outer retinal network by taking the in vivo cone tuning functions (from Fig. 1F), the different cones instead delivered much more distinct images (Fig. 5, G to I).
Fig. 5. In vivo cone tunings efficiently represent statistics of natural light.
(A to C) Hyperspectral data acquisition from zebrafish natural visual world. A 60° window around the visual horizon of an example scene recorded in the zebrafish natural habitat (A) was sampled at 1000 equispaced points with a custom-built spectrometer-based scanner (13) (B) to yield 1000 individual spectral readings from that scene. (C) Summary of the pooled and z-normalized data from n = 30 scenes (30,000 spectra) with mean ± SD [data from (14)]. Photo credit (A): Tom Baden, University of Sussex. (D to L) Reconstructions and analysis of the example scene as seen through different spectral filters: (D to F) log-opsin spectra, (G to I) cone in vivo tunings, and (J to L) based on the first three PCs that emerge from the hyperspectral data shown in (C). From left to right: (D, G, and J) example scene [from (A)] reconstructed on the basis of opsin/in vivo/PC tunings as indicated, (E, H, and K) correlation matrices between these respective reconstructions, and (F, I, and L) the actual tunings/PCs. A fifth element GB (for “green/blue”) is computed for in vivo tunings as contrast between green and blue cone tunings (cf. fig. S5). (M) Percent variance explained by the first five PCs (l). (N) Superposition of cone in vivo tunings (colored lines), PCs, and a linear R/G/B/U log-opsin fit to the respective PC (yellow; Methods). The latter fit can be seen as the biologically plausible optimum match to a given PC that can be achieved in a linear regime.
Next, to determine the spectral axes that optimally captured the variance of natural light in the zebrafish’s natural underwater world (Discussion), we used principal components analysis (PCA) across the spectra of all n = 30,000 pixels in the dataset (Fig. 5, C and J to L). Because of the strong spectral correlations in natural light, the first component (PC1) captured the achromatic (“black and white”) image content, while subsequent components (PC2, PC3, etc.) captured the major chromatic (color) axes in decreasing order of importance (1, 6). Together, PC1 to PC3 accounted for 97% of the natural spectral variance (Fig. 5M). We computed what the example scene would look like if sampled by detectors that were directly based on the first three PCs. We found that scenes processed by PC1 and PC2 (Fig. 5J) were highly reminiscent of the scenes sampled by in vivo red and green cones, respectively (Fig. 5G). Next, PC3 was not obviously captured by either of the remaining blue or UV cones in isolation; however, it did approximately resemble the scene when reconstructed by a green/blue cone opponent axis (“GB,” turquoise; Discussion). PC3 could be approximated by a variety of cone combinations; however, all best matches (ρ = 0.97; Methods) required opposing green and blue cones (fig. S5).
Direct superposition of these cone output spectra with the respective PCs further illustrated their notable match (Fig. 5N). These cone spectra were also well matched by a direct fit to the PCs when using the four cones’ opsin templates as inputs (Fig. 5N, yellow; Methods). Here, our rationale was that these opsin fits present a biologically plausible optimum for mimicking the PCs.
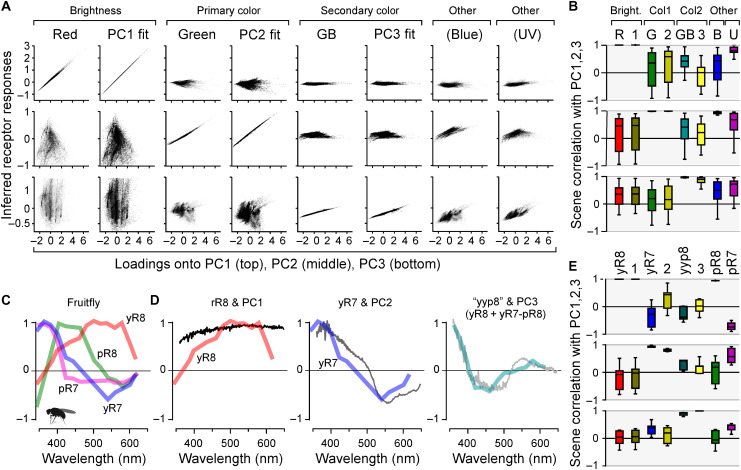
To quantitatively explore this match and its consequences for the encoding of natural light, we next computed how each of the 30,000 individual collected spectra would activate red and green cones and the GB axis. We then plotted these activations against the respective loadings of PC1 to PC3 for these spectra (Fig. 6A). In each case, we also computed the same metric for the best log-opsin fits to the PCs. This confirmed the excellent performance of the system for separating achromatic from chromatic information under natural light. Red cone activation correlated almost perfectly (mean ρ > 0.99; 2.5/97.5 percentiles 0.99/>0.99) with spectral loadings against PC1 (Fig. 6A, top left; cf. Fig. 6B, top left) but was uncorrelated with either PC2 (ρ = −0.16, −0.89/0.88) or PC3 (ρ = 0.29, −0.34/0.91) (Fig. 6, A and B, middle and bottom left). Moreover, red cone performance was near indistinguishable from that of the opsin fit against PC1 (ρ > 0.99, >0.99/>0.99), which was used as a biologically plausible benchmark of optimality (Fig. 6, A and B, second column). Accordingly and despite the minor differences in short-wavelength activation of the red cone action spectrum compared to PC1 and its opsin fit (Fig. 5N, left; Discussion), red cones encoded natural achromatic contrast (i.e., “brightness,” PC1) with negligible contamination of chromatic information (i.e., PC2 and PC3). In contrast, activation of green cones was highly correlated with PC2 (ρ = 0.99, 0.98/>0.99) but uncorrelated with either PC1 (ρ = −0.15, −0.88/0.88) or PC3 (ρ = 0.14, −0.66/0.81; Fig. 6, A and B, columns 3). Again, their performance was near indistinguishable from that of the respective opsin fit (Fig. 6, A and B, columns 4). Accordingly, green cone activation carried no information about brightness but instead encoded an efficient primary chromatic signal.
Fig. 6. Encoding of natural achromatic and chromatic contrast.
(A) Computed responses of in vivo cones, the GB axis, and each respective log-opsin PC fit [all from Fig. 5 (I and N)] to each of the n = 30,000 individual natural spectra, plotted against each spectrum’s loadings onto PC1 (top row), PC2 (middle row), and PC3 (bottom row), as indicated. Responses were plotted on the y axis, and PC loadings were plotted on the x axis. In general, a column that shows a near-perfect correlation in one row, but no correlation in both other rows (e.g., column 1), can be seen as a tuning function that efficiently captures the respective PC (e.g., column 1 shows that red cones efficiently represent PC1 but not PC2 or PC3). (B) Corresponding summary statistics from (A), based on scene-wise Spearman correlations. (C) Spectral tuning functions of Drosophila R7/8 photoreceptors as measured in vivo at their synaptic output [data from (12)]. (D) Comparison of Drosophila tuning functions with the first three PCs that emerge from terrestrial natural scenes [data from (13)]. Here, PC3 is matched with a yyp8 axis as indicated [cf. fig. S6 (D to F)]. (E) Summary stats of Drosophila photoreceptor responses to each of the n = 4000 individual terrestrial natural spectra plotted against their respective PC loadings.
Next, both activation of the GB-opponent axis and of the corresponding opsin fit correlated strongly with PC3 (ρ = 0.95, 0.80/0.99 and ρ = 0.79, 0.18/0.99, respectively) but not with PC1 (ρ = 0.38, −0.25/0.92 and ρ = −0.08, −0.76/0.52) or PC2 (ρ = 0.31, −0.62/0.91 and ρ = −0.15, −0.80/0.57; Fig. 6, A and B, columns 5 and 6). Accordingly, contrasting the signals of blue and green cones offers the theoretical possibility to build an efficient secondary chromatic signal in downstream circuits (Discussion). Notably, blue cones in isolation correlated mainly with PC2 (ρ = 0.94, 0.85/0.99) rather than PC1 (ρ = 0.18, −0.74/0.91) or PC3 (ρ = 0.43, −0.33/0.90) (Fig. 6, A and B, columns 7), suggesting that they could potentially serve to provide an alternative route to encoding primary chromatic information.
Last, UV cones mainly correlated with PC1 (ρ = 0.80, 0.51/0.99), suggesting that this ultrashort-wavelength channel may serve to provide a secondary achromatic signal (Fig. 6, A and B, columns 8). However, its performance in doing so was substantially inferior to that of red cones, suggesting that its primary function is not the encoding of achromatic brightness per se but rather to specifically detect short-wavelength signals. Here, their weak but significant opponency to spectrally intermediate signals may serve to accentuate contrast against an otherwise “gray” background (Discussion).
Together, it appears that larval zebrafish effectively “rotate” color space already at their visual system’s first synapse signal along an achromatic axis (red cones) and a primary chromatic axis (green cones), with the added possibility to build an efficient secondary chromatic axis by opposing green and blue cones downstream. Together, this system captures at least 91.3% of spectral variance in natural scenes when using red and green cones alone and, potentially, up to 97% if including green-blue opponency. Elegantly, it also leaves UV cones to serve independent visual functions, such as prey capture of UV-bright microorganisms (Discussion) (11).
A comparison to spectral processing in fruitflies
A conceptually similar decomposition of natural light may also be used in D. melanogaster (Fig. 6, C to E, and fig. S6), the only other tetrachromatic species where in vivo spectral tuning functions of photoreceptor outputs are available (12). In these flies, R1-6 photoreceptors express a mid-wavelength–sensitive opsin and are generally considered an achromatic channel, while R7/8-type photoreceptors are associated with color vision (39). We therefore compared spectral tuning curves of the four varieties (yR8, yR7, pR8, and pR7) of Drosophila R7/8-type photoreceptors [Fig. 6C; taken from (12)] with the PCs that emerged from natural spectra of n = 4 daytime field and forest scenes (13), each comprising 1000 individual spectra as before (Fig. 6, D and E; fig. S6, A to G; and Discussion).
Like for zebrafish, this showed that their spectral tuning curves were well approximated by the first three terrestrial PCs: PC1 and yR8 (ρ > 0.99, 0.99/>0.99), PC2 and yR7 (ρ = 0.93, 0.91/0.98), and lastly PC3 by jointly opposing both yR8 and yR7 against pR8 (for simplicity, “yyp8,” ρ = 0.72, 0.60/0.84; Fig. 6, D and E; cf. fig. S6, D to G). Compared to zebrafish, the spectral matches between photoreceptor action spectra and natural PCs were however slightly worse, which may, in part, be linked to the use of a smaller natural imagery dataset and to the comparatively lower spectral resolution information currently available in flies. Nevertheless, this general match was made possible by the fact that, in line with the relatively increased predominance of short-wavelength light above the water (fig. S6A), all terrestrial PCs (fig. S6B) and corresponding action spectra (fig. S6G) were blue-shifted relative to those of aquatic environments and of zebrafish, respectively.
Together, this suggests that “rotating” color space into primary achromatic and chromatic axes (i.e., PC1 to PC2) as early as possible, while leaving the ultrashort-wavelength system largely isolated, may be a fundamental principle of color vision when using more than two spectrally well-separated photoreceptor types, in a notable example of convergent evolution (Discussion).
DISCUSSION
Our physiological recordings from cones (Figs. 1 and 2) and HCs (Fig. 4, F to L), linked to synaptic level EM reconstructions (Fig. 3) and computational modeling (Fig. 4, A to E), provide a comprehensive in vivo account of spectral processing for an efficient decomposition of natural light (Figs. 5 and 6) at the visual system’s first synapse in a tetrachromatic vertebrate.
Linking retinal color opponency to the PCs of natural light spectra
Using PCA of light spectra for understanding the encoding of natural scenes by animal visual systems has a long tradition, for example, in information-theoretic considerations by Buchsbaum and Gottschalk in 1983 (1). This seminal work described how the three primaries of the human eye (long-, mid-, and short-wavelength sensitive: L/“red,” M/“green,” and S/“blue,” respectively) can be efficiently combined to derive one achromatic and two chromatic axes with none, one, and two zero crossings, respectively. These theoretically optimal channels corresponded well to psychophysically determined opponent mechanisms in human vision and were later shown to capture much of the spectral variance in natural light (3). However, in contrast to zebrafish, the circuit mechanisms that enable this notable link between the human primaries and perception that involve multiple levels of computation across both the retina and the brain remain incompletely understood: First, many retinal ganglion cells (RGCs) and their central targets, including in visual cortices, are mid/long-wavelength biased and non-opponent and encode achromatic contrasts (40). Second, inherited from probably nonselective retinal wiring, midget circuits carry “red-yellow” or “green-yellow” spectral information that is thought to be decoded into a primary “red-green” color-opponent axis in the central brain by mechanisms that remain largely unsolved. Third, at least three types of “blue-yellow” RGCs contrast the signals from blue cones against the sum of red and green cones. This RGC opponency is mainly achieved at the level of RGC dendrites, by contrasting the signals of approximately non-opponent inner retinal neurons (41).
In addition, primate blue cones themselves are yellow-blue opponent due to feedforward inputs of red/green cone inputs via HCs (42), reminiscent of the strategies used by zebrafish cones. However, primate blue and red/green cones are homologous to zebrafish UV and red cones, respectively (43, 44), and the underlying HC is H2 (41). Accordingly, spectral opponency in primate blue cones is presumably linked to the weak but significant mid-wavelength opponency of zebrafish UV cones rather than the much stronger opponency of zebrafish green or and blue cones (Fig. 1F).
Beyond primates, comparative circuit knowledge of vertebrate retinas for spectral processing is sparse and mainly restricted to dichromatic mammals (4). Among tetrachromats that retain ancestral green and blue cones, measurements of spectral responses in adult HCs of diverse species of fish (5, 31, 35, 45) are in good agreement with our in vivo HC data in larval zebrafish. Moreover, zebrafish inner retinal neurons (9, 14, 25, 46) display both non-opponent and a wide diversity of opponent responses that generally prioritize simple short-versus-long wavelength computations over more complex combinations, broadly in agreement with predictions from theory (1). However, in the absence of systematic and spectrally resolved sensitivity measurements of zebrafish inner retinal neurons, it has not been possible to explicitly link their properties to the variance in natural visual light. In addition, direct in vivo spectral measurements of zebrafish cone photoreceptor outputs have remained outstanding.
Among invertebrates, D. melanogaster stands out as the only tetrachromatic species where spectrally resolved photoreceptor output tuning functions are available (12). As discussed, these reveal a conceptual match to those of zebrafish, even down to circuit implementation involving a single HC-like feedback neuron, all despite their eyes having evolved independently since long before the emergence of image-forming vision in any animal. Here, the authors draw on Buchsbaum and Gottschalk’s ideas on efficient encoding (1) to suggest that like for zebrafish bipolar cells (14), the Drosophila R7/8 single and double zero crossings can be conceptually matched with opsin-based primary and secondary color axes, respectively. However, how this link would look like in practice for the encoding of spectral variance in natural light remained unclear. Here, we extend these theoretical links to directly show how like in zebrafish, Drosophila PC1 and PC2 are each well captured by a single receptor, while capturing PC3 requires possible opposing of multiple receptors downstream.
Achromatic signaling
Natural scenes are generally dominated by achromatic over chromatic contrasts (6) and biased to mid or long wavelengths. Accordingly, an efficient achromatic encoder should approximate the resultant mid/long-wavelength biased mean spectrum of light in a non-opponent manner, as is the case for both zebrafish red cones (Fig. 2N) and for Drosophila yR8 photoreceptors (Fig. 6D). Here, the quality of the spectral match primarily affects the maximal achievable signal to noise of the encoder rather than its ability to encode brightness per se (47, 48). Accordingly, despite their minor respective mismatches compared to the mean of available light (see below), both zebrafish (Fig. 6, A and B) and Drosophila implementations (Fig. 6, D and E) capture PC1 well. For the same reason, also other non-opponent photoreceptors, such as Drosophila R1-6 (49) as well as vertebrate rods or “true” double cones in many nonmammalian vertebrates, are generally thought to capture achromatic signals (4). However, in all these cases, the presumed non-opponent nature at the level of their synaptic output in vivo remains to be confirmed.
In both zebrafish red cones and in Drosophila yR8, the largest mismatch to their natural environment’s PC1 was in the UV range (Figs. 2N and 6D). Here, it is tempting to speculate that their low short-wavelength sensitivity is linked to a need to isolate behaviorally critical “general” achromatic signals from those that incur specifically in the UV range. In the case of zebrafish, UV-specific signals carry key visuo-ecological relevance, in that they can report the presence of prey (11), a rare feature that is unlikely to be captured in our scene-wide data of natural spectra (see also discussion on UV signaling below).
Ultimately, the signals from red cones must be read out by downstream circuits, in a manner that approximately preserves their spectral tuning. This could principally occur via a private channel, as potentially provided by mixed-bipolar cells, which, in adults, receive direct inputs only from red cones and from rods (50). However, most zebrafish bipolar cells receive direct inputs from more than one cone type, presumably mixing their spectral signals. Nevertheless, a PC1-like signal does filter all the way to the brain where it forms the dominant off-response (48).
Primary chromatic signaling
In natural scenes, all spectral variance that is not captured by PC1 is chromatic, with any subsequent components capturing progressively smaller fractions of the remaining variance in a mutually orthogonal manner. Accordingly, PC2 and PC3 are maximally informative about primary and secondary spectral contrasts, respectively, while at the same time being uninformative both about brightness (i.e., PC1) and about each other. Here, we found that zebrafish green cones (Fig. 2N) and Drosophila yR7 photoreceptors both provide a good match to their respective environment’s PC2 (Fig. 6D). In the case of zebrafish, this match was close to perfect: When challenged with natural spectra, green cones were highly informative about PC2 but uninformative about PC1 or PC3. Accordingly, like for red cones (discussed above), the visual system would be well served to read out the signal from green cones in a private line at least once so as to preserve this already efficient chromatic signal. Green cones are anatomically the only cones in the zebrafish retina known to have such an arrangement: Two of more than 20 zebrafish bipolar cell “morpho types,” both stratifying in the traditional “off-stratum” of the inner plexiform layer (IPL), make exclusive contacts to green cones (50). Potentially in agreement, we previously identified a small but well-defined population of singly color-opponent bipolar cell responses in this part of the IPL (14).
Further chromatic signaling
Beyond PC1 and PC2, most of the remaining spectral variance was captured by PC3, which presents a triphasic spectral response with two zero crossings. However, neither of the remaining blue and UV cones exhibited such a tuning. Of these, blue but not UV cones were strongly opponent, nevertheless suggesting their important role in spectral processing. Accordingly, we explored why blue cones did not directly capture PC3. For this, we returned to our HC model, this time, immediately optimizing red, green, and blue cones to match PC1, PC2, and PC3, respectively. To complete the model, UV optimization was left unchanged to again target its own in vivo tuning function. Using this strategy, it was possible to produce only weakly distorted red, green, and UV cone spectra. However, the model failed to directly capture PC3 using blue cones, and the mild relative distortion of green cone spectral tuning was sufficient to noticeably degrade their ability to capture PC2 (fig. S6, H to K). This tentatively suggests that the specific connectivity of the outer retina, constrained by the four principal zebrafish cone opsins, is poorly suited to additionally produce a PC3-like spectral response.
Nevertheless, blue cones did exhibit a single zero crossing that differed from that of green cones, meaning that two zero crossings could be readily achieved in a linear model that opposed green and blue cone signals (fig. S5). We showed that such an arrangement would, at least in theory, allow building a spectral filter that closely captures PC3 while producing only poorly correlated responses to PC1 and PC2. Such a PC3-like filter is observed at the level of the brain, which mainly opposes UV and red “on” signals with spectrally intermediate blue/green “off” signals (48). However, how this brain response is set up at the level of the retina remains unclear. Last, a PC3-like signal could also be achieved in Drosophila by opposing their two mid-wavelength–sensitive yR7 and pR8 photoreceptors; however, in this case, the best match was achieved when in addition recruiting the more broadly tuned yR8 alongside yR7 (fig. S6, D and F).
A private channel for detecting UV signals
Unlike red, green, or blue cones, the final output of zebrafish UV cones appeared to not be central to support dominant achromatic nor chromatic processing. UV cones also use a nearly UV-exclusive HC (H3,Figs. 3 and 4) (31, 34), likely for temporal tuning (11), while barely contributing to the signals of H1 and H2 (Fig. 4D). Accordingly, outer retinal UV circuits appear to approximately signal in isolation from those of the remaining cones. Similarly, direct contributions from the UV-sensitive pR7 photoreceptors were also not required to approximate the first three PCs that emerge from the natural spectral world of Drosophila (Fig. 6, D and E). In both cases, these photoreceptors contrasted their strong, short-wavelength exclusive response with weaker opposition at most other wavelengths. From here, it is tempting to speculate that these UV systems may serve to detect, rather than necessarily to spectrally contrast, the presence of strongly UV-biased objects against a “naturally gray” background. Such a detector would be invaluable for reporting the presence of the UV-bright single-celled microorganisms when illuminated by the sun, which larval zebrafish feed on (11). To our knowledge, a similarly specific visuo-ecological purpose of UV vision in Drosophila remains unknown. More generally, UV light can be highly informative about edges in space, as it tends to accentuate objects’ silhouettes against bright backgrounds (51–53).
In zebrafish, previous work has highlighted a key role of UV vision across the retina and brain leading to behavior (11, 14, 25, 48, 54, 55). Most notably, the retina’s acute zone (26) is dominated by UV-sensitive circuits (14). Here, most bipolar cell terminals respond primarily to UV stimulation, and only some, in addition, respond to other wavelengths (14), a general pattern that is recapitulated also at the level of the RGCs (25) to drive a strong UV response in the brain (48, 56, 57), which filters all the way to spinal circuits (57, 58). Nevertheless, despite this profound functional dominance, no anatomical study has reported the presence of UV cone–dedicated bipolar cells, for example, in the case of green cones (see above) (50). While it remains unknown whether such connectivity specifically exists in the acute zone, it seems clear that more broadly across the retina, the signals from UV cones are mixed with those of other cones. How this connectivity serves to support the diverse visuo-ecological needs of zebrafish UV vision will be important to address in the future.
Regional differences in cone spectral tuning
Unlike many other aspects of larval zebrafish retinal structure (14, 25–27, 59) and function (11, 14, 25), the overall spectral tuning of zebrafish cones was eye region invariant (fig. S1G). Nevertheless, small but significant regional variations were observed in all cone types. Of these, the most notable differences occurred in red and green and, to a smaller extent, also in UV cones. Red cones and, to a weaker extent, also other cones exhibited relatively narrowed tuning ventrally and broadened tunings dorsally. These differences might help in keeping cones within operational range despite the large difference in absolute amount light driving them: bright direct skylight versus dimmer reflected light from below, respectively. Next, among green cones, the acute zone exhibited the strongest short-wavelength response, resulting in a long-wavelength shift in their zero crossing. This finding is conceptually in line with an increase in absolute light sensitivity among UV cones in this part of the eye (11); however, a possible visuo-ecological purpose of this shift remains to be established. Last, mid-wavelength opponency among UV cones was strongest in the acute zone and ventrally, which may be linked to the behavioral need to contrast UV-bright prey against a spectrally intermediate but bright background in the upper-frontal parts of visual space (11, 60). In contrast, larval zebrafish rarely pursue prey below or behind them (60, 61), as surveyed by dorsal and nasal UV cones, respectively.
How might these small but significant regional differences in spectral tuning be brought about? One possibility may relate to expressed opsin variants: Unlike blue and UV cones, which each express a single opsin variant, larval zebrafish red and green cones can express up to two (red) or four (green) variants with slightly shifted absorption spectra (62). Among red cones, the dominant LWS-2 variant (λmax = 548 nm) is supplemented by LWS-1 expression (λmax = 558 nm) at the retina’s ventral pole (63), which may partly explain the ventral long-wavelength shift observed at the level of function (fig. S1G); however, it cannot explain the general “narrowing” of the red cone response in this part of the eye. Among green cones, RH2-1 (λmax = 467 nm) is dominant across the retina but supplemented nasally by the relatively long-wavelength shifted RH2-3 (λmax = 488 nm) and RH2-4 (λmax = 505 nm) (62, 63); however, no corresponding long-wavelength shift of the nasal green cone response was noted in our physiological recordings (fig. S1G). Notably, the regional bias across opsin variants becomes more pronounced with age (63), and it is possible that any corresponding functional effects were too subtle to reliably detect in the larval stage. Instead, we wondered how eye region differences in cone tunings might be achieved by leveraging outer retinal circuits. To explore this, we again returned to our HC model, this time, fitting it individually to only the subsets of recordings from each of the four regions. This revealed that the same anatomically established maximal connectivity matrix (Figs. 3 and 4) served well to produce any of these regional differences by minimally shifting their relative weights (table S1). Accordingly, it seems likely that the same principal HC network produces these regional variations in tuning based on minor rebalancing of its relative input strengths.
METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, T.B. (t.baden@sussex.ac.uk).
Experimental model and subject details
Animals
All procedures were performed in accordance with the U.K. Animals (Scientific Procedures) Act 1986 and approved by the animal welfare committee of the University of Sussex. For all experiments, we used 7- to 8-day post fertilization (dpf) zebrafish (Danio rerio) larvae. The following previously published transgenic lines were used: Tg(opn1sw1:nfsBmCherry) (34), Tg(opn1sw1:GFP) (64), Tg(opn1sw2:mCherry) (65), and Tg(thrb:Tomato) (66). In addition, Tg(opn1sw2:SyGCaMP6f), Tg(LCRhsp70l:SyGCaMP6f), and Tg(thrb:SyGCaMP6f) lines were generated by injecting pBH-opn1sw2-SyGCaMP6f-pA, pBH-LCRhsp70l-SyGCaMP6f-pA, or pBH-thrb-SyGCaMP6f-pA. Tg(cx55.5:nlsTrpR,tUAS:ASAP3) line was generated by coinjecting pBH-cx55.5-nlsTrpR-pA and pBH-tUAS-ASAP3-pA plasmids into single-cell stage eggs. Injected fish were out-crossed with wild-type fish to screen for founders. Positive progenies were raised to establish transgenic lines.
All plasmids were made using the Gateway system (Thermo Fisher Scientific, 12538120) with combinations of entry and destination plasmids as follows: pBH-opn1sw2-SyGCaMP6f-pA [pBH (34) and p5E-opn1sw2 (34), pME-SyGCaMP6f (11), and p3E-pA (67)], pBH-LCRhsp70l-SyGCaMP6f-pA [pBH and p5E-LCRhsp70l, pME-SyGCaMP6f, and p3E-pA; pBH-thrb-SyGCaMP6f-pA: pBH and p5E-1.8thrb (66), pME-SyGCaMP6f, and p3E-3.2thrb (66)], and pBH-tUAS-ASAP3-pA [pBH and p5E-tUAS (68), pME-ASAP3, and p3E-pA]. Plasmid p5E-LCRhsp70l was generated by inserting a polymerase chain reaction (PCR)–amplified locus control region (LCR) for green opsins (RH2-1 to RH2-4) (69) into pME plasmid and subsequently inserting a PCR-amplified zebrafish 0.6-kb hsp70l gene promoter region (70) downstream of LCR. pME-ASAP3 was made by inserting a PCR-amplified ASAP2s fragment (71) and subsequently introducing L146G, S147T, N149R, S150G, and H151D mutations (38) in pME plasmid.
Animals were housed under a standard 14:10 day/night rhythm and fed three times a day. Animals were grown in 0.1 mM 1-phenyl-2-thiourea (PTU; Sigma-Aldrich, P7629) from 1 dpf to prevent melanogenesis. PTU has been linked with autophagy (72) and decreases thyroxine (73, 74), which is implicated in the development of cone function (75). To minimize these possible adverse effects, care was taken to use the lowest concentration of PTU sufficient to prevent skin pigmentation (74). For 2P in vivo imaging, zebrafish larvae were immobilized in 2% low–melting point agarose (Fisher Scientific, BP1360-100), placed on a glass coverslip, and submerged in fish water. Eye movements were prevented by injection of α-bungarotoxin (1 nl of 2 mg/ml; Tocris, catalog no. 2133) into the ocular muscles behind the eye. For some experiments, CNQX (~0.5 pl, 2 mM; Tocris, catalog no. 1045) or meclofenamic acid sodium salt (~0.5 pl, 5 mM; Sigma-Aldrich, catalog no. M4531) in artificial cerebrospinal fluid was injected into the eye.
Method details
Light stimulation
With fish mounted on their side with one eye facing upward toward the objective, light stimulation was delivered as full-field flashes from a spectrally broad liquid waveguide with a low numerical aperture (0.59; Newport, 77555), positioned next to the objective at ~45°. To image different regions in the eye, the fish was rotated each time to best illuminate the relevant patch of photoreceptors given this stimulator geometry. The other end of the waveguide was positioned behind a collimator-focusing lens complex (Thorlabs, ACL25416U-A and LD4103), which collected the light from a diffraction grating that was illuminated by 14 spectrally distinct LEDs (details on LEDs below). Following the earlier design (15), the specific wavelength and relative angle of each LED to the diffraction grating defined the spectrum of light collected by the collimator to be ultimately delivered to the fish’s eye, according to
where α is the angle of light incident to the diffraction grating, λ is the wavelength (in nm), β is the first-order diffraction exit angle, and G is the diffraction grating’s groove density. Moreover, each LED was individually collimated (Signal Construct SML 1089, LT-0454) and attached to a rail (Thorlabs, XE25L450/M and XE25L225/M) by a three-dimensional printed holder (available at https://github.com/BadenLab/HyperspectralStimulator).
An Arduino Due (Arduino) and LED driver (Adafruit TCL5947) were used to control and drive the LEDs, respectively. Each LED could be individually controlled, with brightness defined via 12-bit depth pulse width modulation. To time-separate scanning and stimulating epochs, a global “blanking” signal was used to switch off all LEDs during 2P scanning but enable them during the retrace, at line-rate of 500 Hz [see also (16, 17); the stimulator code is available at https://github.com/BadenLab/HyperspectralStimulator].
LEDs used were as follows: Multicomp Pro, MCL053RHC; Newark, C503B-RAN-CZ0C0AA1; Roithner, B5-435-30S; Broadcom, HLMP-EL1G-130DD; Roithner, LED-545-01; TT Electronics, OVLGC0C6B9; Roithner, LED-490-06; Newark, SSL-LX5093USBC; and Roithner, LED450-03, VL430-5-1, LED405-03V, VL380-5-15, and XSL-360-5E. Effective LED peak spectra as measured at the sample plane were 655, 635, 622, 592, 550, 516, 501, 464, 448, 427, 407, 381, 361, and 360 nm, respectively. Their maximal power outputs were, respectively (in μW), as follows: 1.31, 1.06, 0.96, 0.62, 1.26, 3.43, 1.47, 0.44, 3.67, 0.91, 0.24, 0.23, 0.04, and 0.20. From here, the first 10 LEDs (655 to 427 nm) were adjusted to 0.44 μW, while the four UV-range LEDs were set to a reduced power of 0.2 μW (407, 381, and 360 nm) or 0.04 μW (361 nm). This relative power reduction in the UV range was used as a compromise between presenting similar power stimulation across all LEDs while, at the same time, ameliorating response saturation in the UV range as a result of the UV cones’ disproportionately high light sensitivity (11, 25). In this regard, we took advantage of the strong spectral overlap between the two shortest-wavelength LEDs (360 and 361 nm) to probe this wavelength range at two intensities (0.2 and 0.04 μW, respectively).
From here, all spectral tuning functions were based on the responses to the 13 spectrally distinct LEDs, excluding the response to low-power 361-nm LED. This strategy yielded biologically highly plausible spectral sensitivity functions in all cones that closely resembled their underlying opsin’s tuning when pharmacologically isolated from HCs (Fig. 2B). Nevertheless, UV cones weakly but consistently undershot their opsin template at the shortest tested wavelength (360 nm), hinting that they may have approached their saturation point at this wavelength and power. In agreement, the 0.04-μW, 361-nm LED elicited only mildly lower response amplitudes in UV cones compared to the 0.2-μW, 360-nm LED (Rlow = 0.88 ± 0.14; Rhigh = 0.96 ± 0.06; errors in SD; difference P << 0.001, Wilcoxon signed-rank test). In contrast, all other cones responded much more weakly to the low-power UV LED: blue cone (Rlow = 0.35 ± 0.16; Rhigh = 0.67 ± 0.21), green cone (Rlow = −0.12 ± 0.24; Rhigh = 0.09 ± 0.32), and red cone (Rlow = −0.02 ± 0.27; Rhigh = 0.21 ± 0.27; all low-high pairs, P << 0.001), suggesting that these cones were not near their UV saturation points.
Together, it therefore remains possible that measured cone tuning functions relatively underestimate UV components; however, this effect is likely to be very small in non-UV cones that dominate “traditional” color vision in zebrafish (Discussion). The exact slope of the cones’ UV response also had negligible impact on their relative matches with PCs or their contributions to the HC network, in line with an only weak interdependence of the outer retina’s UV versus red/green/blue cone systems (see Discussion).
2P calcium and voltage imaging
All 2P imaging was performed on a Movable Objective Microscope (MOM)–type 2P microscope [designed by W. Denk, Max Planck Institute (MPI), Martinsried; purchased through Sutter Instruments/Science Products] equipped with a mode-locked Ti:sapphire laser (Chameleon Vision-S, Coherent) tuned to 960 nm for SyGCaMP6f and ASAP3 imaging. To measure HC tuning functions, we first expressed GCaMP6f in HCs. However, while we observed strong light-driven calcium responses at their dendritic tips, adjacent to cone terminals and thus indicative of local processing, we did not observe robust calcium responses in the HC soma (as a proxy of global processing). This lack of somatic calcium responses could be due to a putative lack of voltage-gated calcium channels in larval HC somata [unlike, e.g., in adult mouse (76)]. Instead, we therefore measured voltage responses using the genetically encoded voltage sensor, ASAP3 (38), which presumably also gave a more direct readout of HC global function. We used two fluorescence detection channels for SyGCaMP6f/ASAP3 (F48x573, AHF/Chroma) and mCherry (F39x628, AHF/Chroma) and a water immersion objective (W Plan-Apochromat 20x/1.0 DIC M27, Zeiss). For image acquisition, we used custom-written software [ScanM, by M. Mueller (MPI, Martinsried) and T. Euler (Center for Integrative Neuroscience (CIN), Tuebingen)] running under IGOR Pro 6.3 for Windows (WaveMetrics). Recording configurations were as follows: UV cone SyGCaMP6f, 128 × 128 pixels (2 ms per line, 3.9 Hz) or 256 × 256 pixels (2 ms per line, 1.95 Hz); all other cones SyGCaMP6f and HC ASAP3, 256 × 256 pixels (2 ms per line, 1.95 Hz).
Preprocessing and extraction of response amplitudes of 2P data
Regions of interest (ROIs), corresponding to individual presynaptic cone terminals, were defined automatically on the basis of local thresholding of the recording stack’s SD (typically >25) projection over time, followed by filtering for size and shape using custom-written scripts running under IGOR Pro 6.3 (WaveMetrics), as used previously (11). Specifically, only ellipsoidal ROIs (<150% elongation) of sizes 2 to 5 μm2 were further analyzed. For ASAP3 recordings, ROIs were manually placed to follow the shape of individual HC somata. Calcium or voltage traces for each ROI were extracted and z-normalized on the basis of the time interval of 1 to 6 s at the beginning of recordings before presentation of systematic light stimulation. A stimulus time marker embedded in the recording data served to align the traces relative to the visual stimulus with a temporal precision of 2 ms.
Following the approach used in (77), a quality criterium (QC) of how well a cell responded to a stimulus was computed as
where C is the T by R response matrix (time samples by stimulus repetitions) and 〈〉x and Var[]x denote the mean and variance across the indicated dimension, respectively. If all trials are identical such that the mean response is a perfect representative of the response, then QC is equal to 1. If all trials are random with fixed variance, then QC is equal to 1/R. For further analysis, we used only cells that responded well to the stimulus (QC > 0.4 for SyGCaMP6f or QC > 0.32 for ASAP3) (see also fig. S2B).
After filtering out poorly responsive cells using QC, outliers were removed using PCA. Because, in all cone types, PC1 explained >80% variance of the data, we computed the loading values of the PC1 of cone tuning function within each cone type and defined outliners as the cones with PC1 loading below 1.25 times the length of the 97th percentile departure from the mean.
To extract response amplitudes to each stimulus wavelength, an exponential curve was fit to the entire rising (or falling, for hyperpolarizing responses) phase during each stimulus presentation, with the maximum value of the fitted curve taken as the response amplitude. Because cones are intrinsically “off-cells” (i.e., hyperpolarize to light), we then sign-inverted extracted amplitude values such that off-responses would yield positive amplitude readings and vice versa for on-responses. However, for voltage imaging, because ASAP3 fluorescence intensity increases as cells hyperpolarize, we preserved the polarity of the response amplitudes.
Immunostaining and confocal imaging
Larval zebrafish (7 to 8 dpf) were euthanized by tricane overdose and then fixed in 4% paraformaldehyde (Agar Scientific, AGR1026) in phosphate-buffered saline (PBS) for 30 min at room temperature (RT). After three washes in PBS, whole eyes were enucleated and the cornea was removed by hand using the tip of a 30-gauge needle. Dissected and fixed samples were treated with PBS containing 0.5% Triton X-100 (Sigma-Aldrich, X100) for at least 10 min and up to 1 day, followed by the addition of primary antibodies. After 3 to 5 days of incubation at 4°C, samples were washed three times with PBS and 0.5% Triton X-100 solution and treated with secondary antibodies. After 1 day of incubation, samples were mounted in 1% agar in PBS on a coverslip, and subsequently, PBS was replaced with mounting media (VECTASHIELD, H-1000) for imaging. For HC imaging (fig. S6, C to F), the retina was flat-mounted with the photoreceptors facing the coverslip. For cone side-view imaging (fig. S6A), the lens was kept attached to the retina to maintain the spherical shape of the retina, with the whole “retina ball” mounted with the lens side facing the coverslip. All presented data were imaged in the acute zone.
Primary antibodies were zpr-1 antibody [mouse, 1:100; Zebrafish International Research Center (ZIRC)]. Secondary antibodies were DyLight647 anti-mouse (donkey, 1:500; Jackson ImmunoResearch Laboratories). Confocal image stacks were taken on a TCS SP8 (Leica) with a 63× oil immersion objective (HC PL APO CS2, Leica). Typical voxel size was 90 nm and 0.5 μm in xy and z, respectively. Contrast, brightness, and pseudo-color were adjusted for display in Fiji [National Institutes of Health (NIH)]. To sparsely label HCs, plasmids pCx55.5:Gal4 and pUAS:MYFP were coinjected into one-cell stage eggs (78).
UV cone ablation
Larval zebrafish were immersed in fish water containing 10 mM metronidazole (Met) for 2 hours to ablate nfsB-expressing UV cones. Following Met treatment, zebrafish were transferred into fish water without Met and fed regularly until used for 2P imaging.
EM data acquisition, reconstruction, and annotation
A larval zebrafish (8 dpf) was euthanized by tricane overdose, and then, a small incision on a cornea was made using 30-gauge needle in a fixative solution containing 4% glutaraldehyde (Agar Scientific, AGR1312) in 0.12 M cacodylate buffer (pH 7.4). The tissue was immediately transferred into a 1.5-ml tube with the fixative, centrifuged at 3000 rpm for 3 min, and further fixed in the fixative overnight on a shaker at RT. Subsequently, the tissue was washed three times in 0.12 M cacodylate buffer (pH7.4) and incubated in a solution containing 1.5% potassium ferrocyanide and 2% osmium tetroxide (OsO4) in 0.1 M cacodylate buffer [0.66% lead in 0.03 M aspartic acid (pH 5.5)] for 1 hour. After washing, the tissue was placed in a freshly made thiocarbohydrazide (TCH) solution (0.1 g of TCH in 10 ml of double-distilled H2O heated to 600°C for 1 hour) for 20 min at RT. After another rinse, at RT, the tissue was incubated in 2% OsO4 for 30 min at RT. The samples were rinsed again and stained en bloc in 1% uranyl acetate overnight at 40°C, washed, and stained with Walton’s lead aspartate for 30 min. After a final wash, the retinal pieces were dehydrated in a graded ice-cold alcohol series and placed in propylene oxide at RT for 10 min. Last, the sample was embedded in Durcupan resin. Semithin sections (0.5 to 1 μm thick) were cut and stained with toluidine blue, until the fiducial marks (box) in the ganglion cell layer (GCL) appeared. The block was then trimmed and mounted in a serial block-face scanning electron microscope (GATAN/Zeiss, 3View). Serial sections were cut at a thickness of 50 nm and imaged at an xy resolution of 5 nm. Two tiles, each about 40 μm by 40 μm with an overlap of about 10%, covering the entire photoreceptor, and HC layers in a side view at the acute zone were obtained. The image stacks were concatenated and aligned using TracEM (NIH). The HCs and cones were traced or painted using the tracing and painting tools in TracEM2 (79).
Clustering of HCs in EM and confocal data
To validate the ad hoc group assignment based on UV contacts (HC area) and R/G contacts for the EM (Fig. 3, H and I) and confocal data (fig. S3G), we used mixture of Gaussian (MoG) clustering on all extracted features. These features (area size and number of contacts to R/G, B, and U for EM and area size, tip density, and number of contacts to R, G, and B/U for CM) were z-normalized and clustered in the same framework as the HC recordings (see below). The MoG clusters did coincide with the ad hoc group assignment.
Opsin templates and log transforms
For the log-transformed opsin templates (Figs. 1F and 2B), we assumed a baseline activation (represented by b in Eq. 2) and fit a linear transformation to take the arbitrary scaling of the recordings into account. We then optimized the function fa,b,c to minimize the mean squared error (MSE) between fa,b,c(opsin) and the data of the HC block condition for each cone type
| (1) |
where y is the mean of the HC block condition and f is the function
| (2) |
For the optimization, we used the Python package scipy.optimize.minimize (version 1.4.1). The inverse of this procedure is shown in fig. S4A, where the mean of HC block condition is fitted in the same way to the opsin curves of each cone with the function
| (3) |
The data distribution (25th, 50th, and 75th percentiles) is then calculated by passing each individual HC block recording through the optimized function f′.
Model of cone and HC interaction
We modeled cone-HC interactions as a linear model and included the established (Fig. 3K) connectivity pattern for the three types of HC as a (3 × 4) connectivity matrix W, where wij indicates connection strength from cone type j to HC type i. Furthermore, we assumed the feedback strength per connection of each HC type to be constant for all cones and defined it as a diagonal matrix A. To compute the effective feedback, this matrix is then weighted by the relative connection strength per cone and HC, represented in a (4 × 3) matrix F, with . This represents the strength from HC type j to cone of type i. Here, we assume a symmetric connectivity pattern that is justified by the symmetrical cone mosaic in zebrafish. With these definitions, we can formulate the model recurrently as follows: The inputs to the HCs is defined as
where κ(λ) represents the raw activity in the synapse, which still has to be shifted according to the baseline. The summed outputs of the HCs are computed as
Last, the raw activity in the synapses is computed as
where o(λ) represents the wavelength-dependent opsin activation.
The same formulas hold for computing the baseline of the cones, for which o(λ) was set to 1, which accords to the applied normalization on the recorded data. The final output of the model is the tuning curves κ shifted to the cone-specific baselines and normalized.
The same normalization procedure was applied to the shown HC spectra, which are the normalized spectra Hin(λ). In the reduced models, in which we only included specific types of HCs, we set the corresponding entries in the weight matrix W to zero but did not change the model otherwise.
Model input
To extract the cone tuning curves from the experimental data for the model, we computed the mean amplitude of each bright and dark 3-s interval but excluded in each interval the first second as adaption time. We then took, for every individual trace, the difference of each bright interval to its preceding dark interval based on these means. Last, we averaged over these values for each cone type and experimental condition and, by assuming smooth tuning functions, interpolated (using the SciPy function scipy.interpolate.interp1d) the data to an equidistant resolution of 1 nm.
As input to our model, we took the normalized traces of the blocked HC condition. This normalization can be interpreted as a maximal dark current of 1 and a minimal current of 0 during activation. The input acted as “opsin sensitivity” curves o(λ) of the cones. We decided to use these curves instead of the theoretical available opsin tuning curves since we have a pure linear model and, as shown in Fig. 2B, these traces are a good proxy for the log-transformed opsin templates, which is the effective activation for this linear model. All spectral tuning curves of the cones were normalized to have a maximal absolute value of one.
Fitting procedure
We used the Sequential Neural Posterior Estimation method (also called SNPE-B) described in (37) (code available at https://github.com/mackelab/delfi, version 0.5.1) with small modifications that were already applied in (80) to fit our model.
Briefly, SNPE-B draws parameters {θi}i ∈ I over several rounds r = 1, …, R from a (proposal) prior and evaluates the model for these parameters. For the evaluations ei(θi),the discrepancy function xi(ei) = D(ei) is computed and a mixture density network (MDN) qϕ(θ, x) is trained on the data pairs {(θi, xi)}i ∈ I. The posterior pr(θ∣x0) is then calculated as qϕ(θ∣x = x0) and used as a new proposal prior in the next sampling round: . We took the MSE between model output and the data as discrepancy function. This implies x0 = 0, but as our data are noisy, our model cannot get to a MSE of zero. This would mean that the MDN has to extrapolate to unreached discrepancy values, which could lead to an unstable behavior. As a consequence, we took as x0 the 0.01 percentile of {xi}i ∈ I in each round. This evaluation of qϕ(θ∣x = x0) can be understood as the posterior over the parameters for the “best possible” model evaluations. Testing for different percentiles in a reasonable range did not change the results. We took the same approach for setting an adaptive bandwidth for the kernel [see also (80)]. As, for a few models, the posteriors became slightly worse after some rounds, we compared post hoc the posterior distributions of each round and took the one with the smallest 1st percentile of its samples.
We ran SNPE-B over five rounds, with 200,000 samples per round. The prior was a multivariate normal distribution with mean of 1n and covariance of 0.25 · Idn, where n is the number of model parameters, ranging from 11 (all HCs) to 3 (only H2). We chose three Gaussian components for the MoG and an MDN with two hidden layers with 100 nodes each. In each round, the network was trained for 600 epochs with a minimum batch size of 500, and continuous learning started in round two. To let the MDN focus on regions of low discrepancy, we used a combined uniform half-Gaussian kernel that was constant 1 up to x0 and decayed then as a half-Gaussian. The scale of the half-Gaussian part was, in each round, chosen as the 20th percentiles of the discrepancy values. For the presented tuning curves, 100,000 samples were drawn from the final posterior and the model was evaluated.
HC clustering based on spectral tuning
To identify functional clusters, we used a MoG model (sklearn.mixture. GaussianMixture, version 0.21.2) with three components and diagonal covariance matrices on the preprocessed tuning curves (n = 86), which were additionally normalized to have maximal value of one. Aiming for a stable clustering, we ran the algorithm 1000 times with different random seeds and chose the ones with the smallest Bayesian Information Criterion (BIC) and, under these, chose the partition that appeared most often. The different runs did not change the general shape of the cluster means, but the specific assignment was variable for some traces. With this procedure, we got a partition with n = 12, 19, and 55 elements, which were allocated to the known functional tunings for HCs of adult zebrafish (31, 35).
Natural imaging data analysis
The hyperspectral data were element-wise multiplied with a deuterium light source–derived correction curve [S.x]. The data were restricted to the domain of 360 to 650 nm and z-normalized within a given scan. Here, the long-wavelength end of the domain was decided on the basis of the long-wavelength opsin absorption curve; the short-wavelength end was dictated by the sensitivity of the spectrometer. The hyperspectral PCs were obtained using the scikit-learn 0.22.1 implementation of the PCA algorithm. Only the first three components are displayed.
Hyperspectral measurement points were spatially aligned within the scan according to the scan raster [see (13, 14) for details]. Pixel brightness is the projection of a given PC or mean of the convolution with the opsin absorption or the observed cone response curves, respectively. Presented images were smoothed using a Gaussian filter (σ = 2px). Sum of squares difference was taken between pairs of z-normalized images and their negatives. The lowest sum of squares (equal to the highest correlation, either with the original or the negative) is displayed. Smoothing did not significantly affect this measure.
To statistically compare scene reconstructions by different sets of tuning functions (fig. S6, A to C), we used two parallel strategies. First, we computed the correlation coefficient between reconstructions by the different channels (e.g., in vivo red cone versus green cone) as indicated for each of n = 30 scenes, thus yielding 30 correlation coefficients for each combination of channels in each condition. Among each comparison, we then computed the mean and SD, as shown.
Second, to capture the multivariate dependence directly, we computed the mutual information under Gaussian assumption, MI = ∑ih(xi) − h(x)~ log det [2 π e C], where C is the correlation matrix of the scene representations in the different channels (e.g., 4 × 4 in vivo: red, green, blue, and UV cone). As the diagonal of C is constant and equal to 1, the mutual information is proportional to the latter quantity. We normalized this quantity by the mutual information of the opsin set of tuning functions.
Linking opsin and photoreceptor spectra to PCs
Measured in vivo spectra of cones and their underlying log-transformed opsin templates (Fig. 1F) were linearly combined to provide least squares fits to the respective underwater spectral PCs (Figs. 5N and 6D and figs. S5 and S6). The same procedure was also used to match Drosophila R7/8 spectra [Fig. 6C, from (12)] to the PCs that emerged from natural distribution of light above the water. Next, to compare the expected responses of in vivo photoreceptors, their linear combinations (in case of PC3, see below), and their respective log-opsin constructs to natural light, individual natural light pixel spectra (n = 30,000) were multiplied with the respective sensitivity curves. In each case, pixel spectra were first z-normalized within the scene, and products were summed over all wavelengths. This procedure produced “responses” (Fig. 6A), which were plotted against the respective loadings of each spectrum onto PC1, PC2, and PC3 (in rows 1, 2, and 3, respectively). From here, scene-wise summary statistics were computed on the basis of Spearman correlation coefficients (Fig. 6, B and E).
To arrive at in vivo photoreceptor combinations that best approximated PC3s [zebrafish, fig. S5 (A to C); Drosophila, fig. S6 (E and F)], we assessed the spectral matches to them by several plausible linear combinations of in vivo photoreceptor tunings based on least squares. In both cases, the best fits required opposing the two spectrally intermediate receptors. For zebrafish, this “GB fit” performed as well as any combination of more complex fits that, in addition, used red or UV cones, so we used this simplest GB fit for further analysis. In case of Drosophila, best performance required also adding the long-wavelength sensitive receptor to yield an yR8 + yR7-pR8 axis (short, yyp8). In each case, performance as shown in figs. S5C and S6F (top) was evaluated on the basis of the mean scene-wise Spearman correlation coefficient between the resultant spectral axis, as described above. The weights needed to build these PC3-like tunings based on photoreceptor types are plotted below as abs(max)-normalized for better comparison.
Quantification and statistical analysis
Statistics
No statistical methods were used to predetermine sample size. Owing to the exploratory nature of our study, we did not use randomization or blinding.
We used generalized additive models (GAMs) to analyze the relationships between wavelength and cone activity under different experimental conditions (Fig. 2, D to H, and fig. S2). GAMs can be understood as an extension to the generalized linear model by allowing linear predictors, which depend on smooth functions of the underlying variables (81). We used the mgcv-package (version 1.8-31) in R on an Ubuntu 16.04.6 LTS workstation with default parameters. We modeled the dependence of the variable of interest as a smooth term with 13 degrees of freedom. The models explained ~59 to 82% of the deviance. Statistical significance for differences between the dependence of activation in the different experimental conditions was obtained using the plot_diff function of the itsadug-package for R (version 2.3). Significance of opponency (fig. S1G) and zero crossings of the tuning curves (Fig. 1G and fig. S1G) were also calculated on the basis of GAMs with “zone” as an additional predictive variable and grouping where applicable.
Acknowledgments
We thank S. Knecht and R. Wong for EM volume acquisition. We thank T. Euler for critical feedback. We would also like to acknowledge support from the FENS-Kavli Network of Excellence and the EMBO YIP.
Funding: Funding was provided by the European Research Council (ERC-StG “NeuroVisEco” 677687 to T.B.), the Wellcome Trust (Investigator Award in Science 220277/Z20/Z to T.B.), the UKRI (BBSRC, BB/R014817/1 to T.B.), the German Ministry for Education and Research (01GQ1601, 01IS18052C, and 01IS18039A to P.Be.), the German Research Foundation (BE5601/4-1, EXC 2064-390727645 to P.Be.), the Leverhulme Trust (PLP-2017-005 to T.B.), the Lister Institute for Preventive Medicine (to T.B.), and Marie Curie-Skłodowska Actions individual fellowship (“ColourFish” 748716 to T.Y.) from the European Union’s Horizon 2020 research and innovation programme. F.S.-P. was supported by the McNair Medical Foundation, startup funds from Baylor College of Medicine, the Klingenstein-Simons Fellowship Award in Neuroscience, a Welch Foundation grant (Q-2016-20190330), NIH grants (R01EB027145 and U01NS113294), and NSF grants (NeuroNex 1707359 and Ideas Lab 1935265). S. Knecht is supported, in part, by NIH grant (EY01730).
Author contributions: T.Y., P.Ba., and T.B. designed the study, with input from C.S., F.K.J., and P.Be. T.Y. generated novel lines and performed 2P data collection and preprocessing. T.Y. also performed anatomical imaging and EM tracing. T.Y. and C.S. analyzed anatomical data. P.Ba. built the light stimulator with input from F.K.J. P.Ba. and T.B. performed natural imaging data analysis, with input from P.Be. C.S. performed computational modeling of the HC cone circuit with input from P.Be. C.S. analyzed voltage recordings with input from P.Be. C.S., T.Y., P.Ba., and T.B. performed general statistical analyses, with help from P.Be. F.S.-P. provided early access to ASAP plasmids. T.B. wrote the manuscript with input from all authors.
Competing interests: F.S.-P. is an inventor on a patent related to this work filed by The Board of Trustees of the Leland Stanford Junior University, Palo Alto (United States) (no. U.S. 9,606,100 B2, filed 14 May 2015, published 28 March 2017). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Plasmids pBH-opn1sw2-SyGCaMP6f-pA, pBH-LCRhsp70l-SyGCaMP6f-pA, pBH-thrb-SyGCaMP6f-pA, pBH-cx55.5-nlsTrpR-pA, and pBH-tUAS-ASAP3-pA and transgenic lines Tg(opn1sw2:SyGCaMP6f), Tg(LCRhsp70l:SyGCaMP6f), Tg(thrb:SyGCaMP6f), and Tg(cx55.5:nlsTrpR,tUAS:ASAP3), generated in this study, are available upon request to the corresponding author. Preprocessed functional 2P imaging data, natural imaging data, EM data, HC circuit modeling data, associated summary statistics, and all code for the model and the statistical analysis of the experimental data are freely available on Data Dryad (https://doi.org/10.5061/dryad.pzgmsbcmk) and further archived via the relevant links on www.badenlab.org/resources and www.retinal-functomics.net. Code for the model and the statistical analysis of the experimental data are also available on GitHub (https://github.com/berenslab/cone_colour_tuning). Natural imaging datasets were published previously as part of (13, 14).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Table S1
REFERENCES AND NOTES
- 1.Buchsbaum G., Gottschalk A., Trichromacy, opponent colours coding and optimum colour information transmission in the retina. Proc. R. Soc. Lond. B Biol. Sci. 220, 89–113 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Chiao C.-C., Cronin T. W., Osorio D., Color signals in natural scenes: Characteristics of reflectance spectra and effects of natural illuminants. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 17, 218–224 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Lewis A., Zhaoping L., Are cone sensitivities determined by natural color statistics? J. Vis. 6, 285–302 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Baden T., Osorio D., The retinal basis of vertebrate color vision. Annu. Rev. Vis. Sci. 5, 177–200 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Baden T., Circuit-mechanisms for colour vision in zebrafish. Curr. Biol. 31, R807–R820 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Atick J. J., Li Z., Redlich A. N., Understanding retinal color coding from first principles. Neural Comput. 4, 559–572 (1992). [Google Scholar]
- 7.Simoncelli E. P., Olshausen B. A., Natural image statistics and neural representation. Annu. Rev. Neurosci. 24, 1193–1216 (2001). [DOI] [PubMed] [Google Scholar]
- 8.H. Barlow, Possible Principles underlying the transformation of sensory messages. in Sensory Communication (1961), pp 217–234.
- 9.Meier A., Nelson R., Connaughton V. P., Color processing in zebrafish retina. Front. Cell. Neurosci. 12, 327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollmann J. H., The zebrafish visual system: From circuits to behavior. Annu. Rev. Vis. Sci. 5, 269–293 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Yoshimatsu T., Schröder C., Nevala N. E., Berens P., Baden T., Fovea-like photoreceptor specializations underlie single UV cone driven prey-capture behavior in zebrafish. Neuron 107, 320–337.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heath S. L., Christenson M. P., Oriol E., Saavedra-Weisenhaus M., Kohn J. R., Behnia R., Circuit mechanisms underlying chromatic encoding in Drosophila photoreceptors. Curr. Biol. 30, 264–275.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nevala N. E., Baden T., A low-cost hyperspectral scanner for natural imaging and the study of animal colour vision above and under water. Sci. Rep. 9, 10799 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann M. J. Y., Nevala N. E., Yoshimatsu T., Osorio D., Nilsson D.-E., Berens P., Baden T., Zebrafish differentially process color across visual space to match natural scenes. Curr. Biol. 28, 2018–2032.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Belušič G., Ilić M., Meglič A., Pirih P., A fast multispectral light synthesiser based on LEDs and a diffraction grating. Sci. Rep. 6, 32012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. Euler, K. Franke, T. Baden, in Neuromethods (Humana, 2019). [Google Scholar]
- 17.Zimmermann M. J. Y., M. Chagas A., Bartel P., Pop S., Prieto-Godino L. L., Baden T., LED Zappelin’: An open source LED controller for arbitrary spectrum visual stimulation and optogenetics during 2-photon imaging. HardwareX 8, e00127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreosti E., Odermatt B., Dorostkar M. M., Lagnado L., A genetically encoded reporter of synaptic activity in vivo. Nat. Methods 6, 883–889 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raviola E., Gilula N. B., Gap junctions between photoreceptor cells in the vertebrate retina. Proc. Natl. Acad. Sci. U.S.A. 70, 1677–1681 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thoreson W. B., Mangel S. C., Lateral interactions in the outer retina. Prog. Retin. Eye Res. 31, 407–441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Euler T., Haverkamp S., Schubert T., Baden T., Retinal bipolar cells: Elementary building blocks of vision. Nat. Rev. Neurosci. 15, 507–519 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Branchek T., The development of photoreceptors in the zebrafish, Brachydanio rerio. II. Function. J. Comp. Neurol. 224, 116–122 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Bilotta J., Saszik S., Sutherland S. E., Rod contributions to the electroretinogram of the dark-adapted developing zebrafish. Dev. Dyn. 222, 564–570 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Arshavsky V. Y., Lamb T. D., Pugh E. N. Jr., G proteins and phototransduction. Annu. Rev. Physiol. 64, 153–187 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Zhou M., Bear J., Roberts P. A., Janiak F. K., Semmelhack J., Yoshimatsu T., Baden T., Zebrafish retinal ganglion cells asymmetrically encode spectral and temporal information across visual space. Curr. Biol. 30, 2927–2942.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt E. A., Dowling J. E., Early retinal development in the zebrafish, Danio rerio: Light and electron microscopic analyses. J. Comp. Neurol. 404, 515–536 (1999). [PubMed] [Google Scholar]
- 27.Kölsch Y., Hahn J., Sappington A., Stemmer M., Fernandes A. M., Helmbrecht T. O., Lele S., Butrus S., Laurell E., Arnold-Ammer I., Shekhar K., Sanes J. R., Baier H., Molecular classification of zebrafish retinal ganglion cells links genes to cell types to behavior. Neuron 109, 645–662.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schröder C., Oesterle J., Berens P., Yoshimatsu T., Baden T., Distinct synaptic transfer functions in same-type photoreceptors. eLife 10, e67851 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneeweis D. M., Schnapf J. L., Photovoltage of rods and cones in the macaque retina. Science 19, 1053–1056 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Schnapf J. L., Nunn B. J., Meister M., Baylor D. A., Visual transduction in cones of the monkey Macaca fascicularis. J. Physiol. 427, 681–713 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klaassen L. J., de Graaff W., Van Asselt J. B., Klooster J., Kamermans M., Specific connectivity between photoreceptors and horizontal cells in the zebrafish retina. J. Neurophysiol. 116, 2799–2814 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapot C. A., Euler T., Schubert T., How do horizontal cells ‘talk’ to cone photoreceptors? Different levels of complexity at the cone-horizontal cell synapse. J. Physiol. 595, 5495–5506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y. N., Matsui J. I., Dowling J. E., Specificity of the horizontal cell-photoreceptor connections in the zebrafish (Danio rerio) retina. J. Comp. Neurol. 516, 442–453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimatsu T., D’Orazi F. D., Gamlin C. R., Suzuki S. C., Suli A., Kimelman D., Raible D. W., Wong R. O., Presynaptic partner selection during retinal circuit reassembly varies with timing of neuronal regeneration in vivo. Nat. Commun. 7, 10590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Connaughton V. P., Nelson R., Spectral responses in zebrafish horizontal cells include a tetraphasic response and a novel UV-dominated triphasic response. J. Neurophysiol. 104, 2407–2422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connaughton V. P., Graham D., Nelson R., Identification and morphological classification of horizontal, bipolar, and amacrine cells within the zebrafish retina. J. Comp. Neurol. 477, 371–385 (2004). [DOI] [PubMed] [Google Scholar]
- 37.J. M. Lueckmann, P. J. Goncalves, G. Bassetto, K. Öcal, M. Nonnenmacher, J. H. Macke, in Advances in Neural Information Processing Systems (NIPS, 2017). [Google Scholar]
- 38.Villette V., Chavarha M., Dimov I. K., Bradley J., Pradhan L., Mathieu B., Evans S. W., Chamberland S., Shi D., Yang R., Kim B. B., Ayon A., Jalil A., St-Pierre F., Schnitzer M. J., Bi G., Toth K., Ding J., Dieudonné S., Lin M. Z., Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnaitmann C., Pagni M., Reiff D. F., Color vision in insects: Insights from Drosophila. J. Comp. Physiol. A Neuroethol. Sensory, Neural, Behav. Physiol. 206, 183–198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Field G. D., Gauthier J. L., Sher A., Greschner M., Machado T. A., Jepson L. H., Shlens J., Gunning D. E., Mathieson K., Dabrowski W., Paninski L., Litke A. M., Chichilnisky E. J., Functional connectivity in the retina at the resolution of photoreceptors. Nature 467, 673–677 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dacey D. M., Circuitry for color coding in the primate retina. Proc.R.Soc.Lond.B. 93, 582–588 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Packer O. S., Verweij J., Li P. H., Schnapf J. L., Dacey D. M., Blue-yellow opponency in primate S cone photoreceptors. J. Neurosci. 30, 568–572 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies W. I. L., Collin S. P., Hunt D. M., Molecular ecology and adaptation of visual photopigments in craniates. Mol. Ecol. 21, 3121–3158 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Musser J. M., Arendt D., Loss and gain of cone types in vertebrate ciliary photoreceptor evolution. Dev. Biol. 431, 26–35 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Kamermans M., van Dijk B. W., Spekreijse H., Color opponency in cone-driven horizontal cells in carp retina. A specific pathways between cones and horizontal cells. J. Gen. Physiol. 97, 819–843 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong K. Y., Dowling J. E., Retinal bipolar cell input mechanisms in giant danio. III. ON-OFF bipolar cells and their color-opponent mechanisms. J. Neurophysiol. 94, 265–272 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Vorobyev M., Osorio D., Receptor noise as a determinant of colour thresholds. Proc. R. Soc. B Biol. Sci. 265, 351–358 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartel P., Janiak F. K., Osorio D., Baden T., Colourfulness as a possible measure of object proximity in the larval zebrafish brain. Curr. Biol. 31, R235–R236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharkey C. R., Blanco J., Leibowitz M. M., Pinto-Benito D., Wardill T. J., The spectral sensitivity of Drosophila photoreceptors. Sci. Rep. 10, 18242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y. N., Tsujimura T., Kawamura S., Dowling J. E., Bipolar cell-photoreceptor connectivity in the zebrafish (Danio rerio) retina. J. Comp. Neurol. 520, 3786–3802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tedore C., Nilsson D. E., Avian UV vision enhances leaf surface contrasts in forest environments. Nat. Commun. 10, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cronin T. W., Bok M. J., Photoreception and vision in the ultraviolet. J. Exp. Biol. 219, 2790–2801 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Qiu Y., Zhao Z., Klindt D., Kautzky M., Szatko K. P., Schaeffel F., Rifai K., Franke K., Busse L., Euler T., Natural environment statistics in the upper and lower visual field are reflected in mouse retinal specializations. Curr. Biol. 31, 3233–3247.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Novales Flamarique I., Diminished foraging performance of a mutant zebrafish with reduced population of ultraviolet cones. Proc. Biol. Sci. 283, 20160058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novales Flamarique I., Opsin switch reveals function of the ultraviolet cone in fish foraging. Proc. R. Soc. B Biol. Sci. 280, 20122490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guggiana Nilo D. A., Riegler C., Hübener M., Engert F., Distributed chromatic processing at the interface between retina and brain in the larval zebrafish. Curr. Biol. 31, 1945–1953.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fornetto C., Tiso N., Pavone F. S., Vanzi F., Colored visual stimuli evoke spectrally tuned neuronal responses across the central nervous system of zebrafish larvae. BMC Biol. 18, 172 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.F. K. Janiak, P. Bartel, M. R. Bale, T. Yoshimatsu, E. Komulainen, M. Zhou, K. Staras, L. L. Prieto-Godino, T. Euler, M. Maravall, T. Baden, Divergent excitation two photon microscopy for 3D random access mesoscale imaging at single cell resolution. bioRxiv 821405 [Preprint]. 29 October 2019. 10.1101/821405. [DOI]
- 59.Robles E., Laurell E., Baier H., The retinal projectome reveals brain-area-specific visual representations generated by ganglion cell diversity. Curr. Biol. 24, 2085–2096 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Mearns D. S., Donovan J. C., Fernandes A. M., Semmelhack J. L., Baier H., Deconstructing hunting behavior reveals a tightly coupled stimulus-response loop. Curr. Biol. 30, 54–69.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Bianco I. H., Kampff A. R., Engert F., Prey capture behavior evoked by simple visual stimuli in larval zebrafish. Front. Syst. Neurosci. 5, 101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chinen A., Hamaoka T., Yamada Y., Kawamura S., Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163, 663–675 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takechi M., Kawamura S., Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J. Exp. Biol. 208, 1337–1345 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Takechi M., Hamaoka T., Kawamura S., Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Lett. 553, 90–94 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Salbreux G., Barthel L. K., Raymond P. A., Lubensky D. K., Coupling mechanical deformations and planar cell polarity to create regular patterns in the zebrafish retina. PLOS Comput. Biol. 8, e1002618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki S. C., Bleckert A., Williams P. R., Takechi M., Kawamura S., Wong R. O. L., Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. U.S.A. 110, 15109–15114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C.-B., The Tol2kit: A multisite gateway-based construction kit forTol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Suli A., Guler A. D., Raible D. W., Kimelman D., A targeted gene expression system using the tryptophan repressor in zebrafish shows no silencing in subsequent generations. Development 141, 1167–1174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsujimura T., Chinen A., Kawamura S., Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 12813–12818 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halloran M. C., Sato-Maeda M., Warren J. T., Su F., Lele Z., Krone P. H., Kuwada J. Y., Shoji W., Laser-induced gene expression in specific cells of transgenic zebrafish. Development 127, 1953–1960 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Chamberland S., Yang H. H., Pan M. M., Evans S. W., Guan S., Chavarha M., Yang Y., Salesse C., Wu H., Wu J. C., Clandinin T. R., Toth K., Lin M. Z., St-Pierre F., Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. eLife 6, e25690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X. K., Kwan J. S. K., Chang R. C. C., Ma A. C. H., 1-phenyl 2-thiourea (PTU) activates autophagy in zebrafish embryos. Autophagy 17, 1222–1231 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whittaker J. R., An analysis of melanogenesis in differentiating pigment cells of ascidian embryos. Dev. Biol. 14, 1–39 (1966). [DOI] [PubMed] [Google Scholar]
- 74.Elsalini O. A., Rohr K. B., Phenylthiourea disrupts thyroid function in developing zebrafish. Dev. Genes Evol. 212, 593–598 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Deveau C., Jiao X., Suzuki S. C., Krishnakumar A., Yoshimatsu T., Hejtmancik J. F., Nelson R. F., Thyroid hormone receptor beta mutations alter photoreceptor development and function in Danio rerio (zebrafish). PLOS Genet. 16, e1008869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chapot C. A., Behrens C., Rogerson L. E., Baden T., Pop S., Berens P., Euler T., Schubert T., Local signals in mouse horizontal cell dendrites. Curr. Biol. 27, 3603–3615.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Baden T., Berens P., Franke K., Rosón M. R., Bethge M., Euler T., The functional diversity of mouse retinal ganglion cells. Nature 529, 345–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshimatsu T., Williams P. R., D’Orazi F. D., Suzuki S. C., Fadool J. M., Allison W. T., Raymond P. A., Wong R. O., Transmission from the dominant input shapes the stereotypic ratio of photoreceptor inputs onto horizontal cells. Nat. Commun. 5, 3699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cardona A., Saalfeld S., Schindelin J., Arganda-Carreras I., Preibisch S., Longair M., Tomancak P., Hartenstein V., Douglas R. J., TrakEM2 software for neural circuit reconstruction. PLOS ONE 7, e38011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oesterle J., Behrens C., Schröder C., Hermann T., Euler T., Franke K., Smith R. G., Zeck G., Berens P., Bayesian inference for biophysical neuron models enables stimulus optimization for retinal neuroprosthetics. eLife 9, e54997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.S. N. Wood, Generalized Additive Models: An Introduction with R (CRC Press, 2017). [Google Scholar]
- 82.Baden T., Euler T., Berens P., Understanding the retinal basis of vision across species. Nat. Rev. Neurosci. 21, 5–20 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Franke K., Berens P., Schubert T., Bethge M., Euler T., Baden T., Inhibition decorrelates visual feature representations in the inner retina. Nature 542, 439–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1