Abstract
Many antitumor and antibacterial drugs inhibit DNA topoisomerases by trapping covalent enzyme-DNA cleavage complexes. Formation of cleavage complexes is important for cytotoxicity, but evidence suggests that cleavage complexes themselves are not sufficient to cause cell death. Rather, active cellular processes such as transcription and/or replication are probably necessary to transform cleavage complexes into cytotoxic lesions. Using defined plasmid substrates and two-dimensional agarose gel analysis, we examined the collision of an active replication fork with an antitumor drug-trapped cleavage complex. Discrete DNA molecules accumulated on the simple Y arc, with branch points very close to the topoisomerase cleavage site. Accumulation of the Y-form DNA required the presence of a topoisomerase cleavage site, the antitumor drug, the type II topoisomerase, and a T4 replication origin on the plasmid. Furthermore, all three arms of the Y-form DNA were replicated, arguing strongly that these are trapped replication intermediates. The Y-form DNA appeared even in the absence of two important phage recombination proteins, implying that Y-form DNA is the result of replication rather than recombination. This is the first direct evidence that a drug-induced topoisomerase cleavage complex blocks the replication fork in vivo. Surprisingly, these blocked replication forks do not contain DNA breaks at the topoisomerase cleavage site, implying that the replication complex was inactivated (at least temporarily) and that topoisomerase resealed the drug-induced DNA breaks. The replication fork may behave similarly at other types of DNA lesions, and thus cleavage complexes could represent a useful (site-specific) model for chemical- and radiation-induced DNA damage.
Type II DNA topoisomerases are involved in diverse cellular processes such as replication, transcription, recombination, chromosome condensation, and the maintenance of genome stability. These enzymes transform the topological state of DNA by a strand passage reaction (for reviews, see references 6, 65, and 66). After binding to one segment of duplex DNA, a type II topoisomerase cleaves the duplex with a four-base stagger while covalently attaching to both 5′ ends via phosphotyrosine linkages. This reaction intermediate, with the enzyme covalently linked to broken DNA, is referred to as the cleavage complex. After passage of a second duplex segment through the break, the topoisomerase reverses the phosphotyrosine linkages and rejoins the cleaved DNA ends.
Mammalian type II topoisomerases are targets of many important antitumor drug classes, including aminoacridines, anthracenediones, anthracyclines, ellipticines, and epipodophyllotoxins (for reviews, see references 10, 13, and 55). In addition, bacterial type II topoisomerases are targets of clinically important antibacterial agents (quinolones and flouroquinolones) (17, 28). Remarkably, all these antitumor and antibacterial agents inhibit the enzyme by trapping the cleavage complex. The simplest model is that the drug, localized precisely at the site of DNA cleavage, prevents topoisomerase from rejoining DNA breaks.
Results from different systems suggest that the cytotoxicity of topoisomerase inhibitors depends on trapping of the cleavage complex rather than loss of enzyme activity (for reviews, see references 13, 17, and 44). First, Escherichia coli cells that contain both nalidixic acid-sensitive and -resistant DNA gyrase are sensitive to nalidixic acid (24, 33). This result implies that in the presence of drug, the drug-sensitive gyrase causes cytotoxicity even though the resistant gyrase is enzymatically active. Second, bacteriophage T7 growth is inhibited by nalidixic acid even though T7 does not require DNA gyrase for growth, and this inhibition is alleviated by heat inactivation of a thermosensitive gyrase subunit A (36). Third, mutational inactivation of bacteriophage T4 topoisomerase, which is not totally essential for growth, causes drug resistance (50). Fourth, overproduction of type II topoisomerase in Saccharomyces cerevisiae causes hypersensitivity to antitumor drugs (52). Fifth, in various systems, recombinational repair reduces sensitivity to topoisomerase inhibitors, consistent with the cleavage complex (or some derivative) causing DNA damage (18, 32, 33, 50, 52).
The drug-induced cleavage complex itself is apparently not a cytotoxic lesion; cellular processes are probably required to cause cytotoxicity (15). For example, inhibition of protein synthesis protects cells from cytotoxicity but does not prevent the formation of cleavage complexes (59). In addition, mammalian cells treated with a topoisomerase poison survive much better when they are simultaneously treated with dinitrophenol, an uncoupler of oxidative phosphorylation that reduces the intracellular ATP pool (39). Since dinitrophenol has little effect on cleavage complex formation, an ATP-requiring process (e.g., transcription or DNA replication) subsequent to cleavage complex formation is apparently involved in the cytotoxicity of topoisomerase poisons.
Transcription appears to play a role in the killing of certain mammalian cells by 4′-(9-acridinylamino)methanesulfon-m-anisidide (m-AMSA) because the transcription inhibitor cordycepin protects G1-phase cells (15). The involvement of transcription, however, may not be universal. Yeast cells arrested by α factor are transcription competent and yet resistant to m-AMSA and the epipodophyllotoxin etoposide (53). This result suggests that DNA replication might be important for cytotoxicity. Indeed, S-phase yeast cells are more readily killed by m-AMSA than are G1-phase yeast cells (53). Furthermore, the cytotoxicity of m-AMSA can be reduced or abolished if mammalian S-phase cells are cotreated with the DNA polymerase inhibitor aphidicolin (15, 27, 67). These results argue that some interaction between the drug-induced cleavage complex and the replication apparatus can be very important in cell death mediated by topoisomerase inhibitors.
We analyzed the collision of a replication fork with a drug-stabilized cleavage complex in vivo by using the bacteriophage T4 model system. Phage T4 encodes a type II DNA topoisomerase that is inhibited by many of the same antitumor drugs that block the mammalian enzyme. The T4 topoisomerase was demonstrated to be the physiological target for m-AMSA (31), and inhibition of phage growth is probably dependent on cleavage complex formation (see above) (50). Recombinational repair of damage derived from the drug-induced cleavage complex is important in both the bacteriophage T4 and mammalian systems, which also argues that T4 is a useful model system for these studies. Bacteriophage T4 is obviously not a suitable model system for all aspects of topoisomerase inhibitor action, since it lacks the cell cycle checkpoints and programmed cell death responses seen in the more complex mammalian systems. Nevertheless, T4 provides many experimental advantages for this study, including the availability of mutants in all important replication and recombination genes, well-defined replication origins, a unique cytosine modification that marks DNA at the time of replication, and a type II topoisomerase that has been carefully analyzed for DNA sequence recognition (19, 38). Here, we use a strong topoisomerase cleavage site and a cloned T4 replication origin to examine the in vivo consequence of a collision between a replication fork and an m-AMSA-induced type II topoisomerase cleavage complex.
MATERIALS AND METHODS
Materials.
m-AMSA was provided by the Drug Synthesis and Chemistry Branch, National Cancer Institute. Oligonucleotides were purchased from National Biosciences. Nylon blotting membrane was obtained from Schleicher & Schuell, the random-primed labeling kit was obtained from Boehringer Mannheim, and Sequenase version 2.0 was obtained from United States Biochemicals. Restriction enzymes and T4 ligase were purchased from New England Biolabs.
Bacterial and phage strains.
The host for all infections was an acrA::Tn10-kan derivative of E. coli NapIV (hsdMK+ hsdRK− hsdSK+ rglB1 nonsuppressing) (51). The acrA mutation eliminates a multidrug efflux pump so that the infected cells are more sensitive to m-AMSA (J. George and K. Kreuzer, unpublished data). Phage strains used in this study were T4 K10 (amB262 [gene 38] amS29 [gene 51] nd28 [denA] rIIPT8 [denB-rII deletion]) (61), K10-46/uvsX (as K10, with amB14 [gene 46] and am11 [gene uvsX]) (37), K10-39 (as K10, with amN116 [gene 39]) (49) and T4 dC (amC87 [gene 42], amE51 [gene 56], NB5060 [denB-rII deletion], W7 [gene alc], and probably an uncharacterized denA mutation) (62).
Plasmid construction.
For these studies, we used a strong m-AMSA-inducible topoisomerase cleavage site (topo site) that had been designed based on an analysis of T4 topoisomerase sequence recognition (19, 20). Plasmid pGH2, containing this strong site, was constructed by ligating the following duplex oligonucleotide to the large BamHI-SalI fragment of pBR322: 5′GATCCAAGCTAAAGTTATATAACTTTATTCAAGG3′ 3′GTTCGATTTCAATATATTGAAATAAGTTCCAGCT5′
Plasmid pGH2-01 was constructed by ligating a purified PstI-ClaI fragment [containing the T4 origin, ori(34) (ori)] from plasmid pKK061-6 (46) to the large purified PstI-ClaI fragment of pGH2 (which contains the topo site). A control plasmid, pGH4, lacking both the topo site and the T4 origin, was constructed by cleaving pGH2 with BamHI and SalI, filling in the ends with Klenow polymerase, purifying the large fragment from low-melting-temperature agarose, and then ligating the fragment into a circle. Another control plasmid, pGH4-01, which has the ori but not the topo site, was constructed as follows. Plasmid pGH2-01 was cleaved with PstI and ClaI, and the small fragment containing the T4 origin was purified. Similarly, plasmid pGH4 was cleaved with the same two enzymes and the large fragment containing the BamHI-SalI deletion was purified. The two fragments were ligated and transformed.
T4 infections and DNA preparations.
NapIV acrA cells containing the indicated plasmid were grown with vigorous shaking at 37°C to a cell density of 4 × 108 per ml and then infected with the indicated bacteriophage T4 strain at a multiplicity of 3 PFU per cell. After 4 min at 37°C, without shaking, for adsorption, the cells were incubated for 2 min with shaking and then m-AMSA was added at 10 μg/ml (unless otherwise indicated). The infected cells were then incubated with vigorous shaking at 37°C for 18 min. Infected cells from 1 ml of the culture were collected by centrifugation, and the pellet was frozen in a dry ice-ethanol bath. The frozen pellet was thawed in 300 μl of cleavage lysis buffer (50 mM Tris-HCl [pH 7.8], 10 mM disodium EDTA, 100 mM NaCl, 0.2% sodium dodecyl sulfate [SDS]). Proteinase K was added to 0.5 mg/ml, and the suspension was incubated at 65°C for 2 h. The total nucleic acid was extracted sequentially with phenol and chloroform-isoamyl alcohol (24:1) and then dialyzed overnight at 4°C against TE buffer (10 mM Tris-HCl [pH 7.8], 1 mM disodium EDTA).
The experiment in Fig. 8 compared two different extraction conditions. One aliquot (1 ml) of infected cells was added to 1 ml of 2× reversal lysis buffer (final concentrations: 50 mM Tris-HCl [pH 7.8], 10 mM disodium EDTA, 100 mM NaCl, 1% Triton X-100, 1.8 mg of lysozyme per ml) and placed at 65°C for 20 min. After this incubation for reversal of cleavage complexes, SDS (0.2%) and proteinase K (0.5 mg/ml) were added and the sample was incubated at 65°C for an additional 100 min. A second aliquot (1 ml) of infected cells was added to 200 μl of 6× cleavage lysis buffer (final concentrations: 50 mM Tris-HCl [pH 7.8], 10 mM disodium EDTA, 100 mM NaCl, 0.2% SDS) and incubated at 37°C for 20 min. The sample was then incubated at 65°C for an additional 100 min. Nucleic acids from both samples were purified as described above.
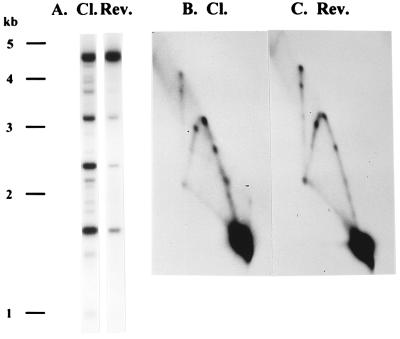
FIG. 8.
Reversal of topoisomerase cleavage does not affect the production of branched DNA. E. coli cells harboring plasmids containing both the ori and the topo site were infected with K10, treated with m-AMSA, and then lysed with either reversal lysis buffer (Rev.) or cleavage lysis buffer (Cl.) as described in Materials and Methods. The AseI-digested samples were analyzed on a one-dimensional gel (A) and on a two-dimensional gel (B and C). Note that most of the topoisomerase-mediated cleavage was reversed by the reversal conditions (A), yet the amount of unique branched DNA did not increase (B and C).
Agarose gel electrophoresis and Southern hybridization.
Nucleic acid samples were treated with the indicated restriction enzymes before being subjected to gel electrophoresis. The one-dimension gels contained 1% agarose and were run in 0.5× TBE buffer (1× TBE contains 89 mM Tris-HCl, 89 mM borate, and 2.0 mM disodium EDTA) at 2.8 V/cm for 15 h. The two-dimension gel protocol was from Friedman and Brewer (21). Briefly, the first-dimension gel was a 0.4% agarose gel run in 1× TBE buffer for 30 h at 1 V/cm at room temperature (21°C). The desired gel lane was sliced from the first-dimension gel and cast across the top of a second-dimension 1% agarose gel, which was run in 1× TBE containing ethidium bromide (0.3 μg/ml) for 15 h at 6 V/cm in the cold (4°C). For Southern hybridization, agarose gels were transferred to a nylon blotting membrane by the downward sponge method (adapted from reference 48). The probe for the Southern blots consisted of pBR322 DNA labeled with [α-32P]dATP by using the random-primed DNA-labeling kit.
RESULTS
Inhibition of DNA replication by m-AMSA.
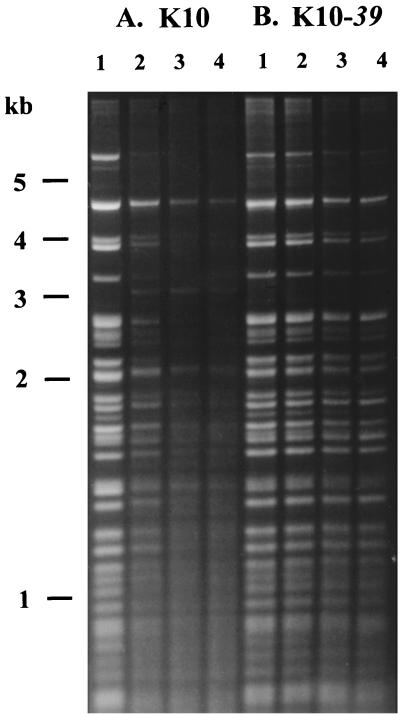
The antitumor drug m-AMSA blocks T4 growth by targeting the phage-encoded type II topoisomerase (31). Nevertheless, this enzyme is not essential for T4 DNA replication, presumably because the host DNA gyrase can substitute for it (45). We therefore began by testing whether m-AMSA inhibits T4 DNA replication and, if so, whether the inhibition is caused by reduction of the topoisomerase catalytic activity or stabilization of the cleavage complex. Bacterial cells were infected with either topoisomerase-proficient bacteriophage T4 (strain K10) or an isogenic topoisomerase-deficient mutant (K10-39am) and treated with various levels of m-AMSA. DNA was collected after 24 min of infection and analyzed by restriction enzyme digestion followed by gel electrophoresis. A large majority of the ethidium bromide-stained DNA in this gel consists of phage DNA that has been replicated during this infection, since T4 replication predominates during the infection and E. coli DNA replication is shut off. Even at the lowest level of m-AMSA, T4 DNA replication was greatly inhibited in the K10 infection (Fig. 1A). However, m-AMSA had much less effect on DNA replication in the K10-39am infection (compare Fig. 1A and B). These results argue that m-AMSA does not inhibit T4 DNA replication by reducing the topoisomerase catalytic activity. If this had been true, the extent of T4 DNA replication should have been equal in the K10-39am infection with or without drug and in the K10 infection with m-AMSA. Instead, the absence of topoisomerase actually protected phage DNA replication from m-AMSA. These results argue strongly that formation of the cleavage complex inhibits T4 DNA replication.
FIG. 1.
Inhibition of T4 replication by m-AMSA. Nonsuppressing E. coli cells were infected with either bacteriophage K10 (encoding wild-type topoisomerase) (A) or K10-39am (topoisomerase-deficient mutant) (B). Phage attachment was allowed for 4 min, and m-AMSA was added 2 min later (at 0, 5, 15, and 50 μg/ml in lanes 1 to 4, respectively). After an additional 18 min of infection, DNA was harvested. The DNA was purified, digested with AseI and HaeIII, and subjected to electrophoresis through a 1% agarose gel, and the resulting fragments were visualized by ethidium bromide staining. These bacterial cells also harbored a plasmid containing a T4 origin; plasmid replication is analyzed in detail below (Fig. 2).
Plasmid replication and m-AMSA-induced cleavage during T4 infection.
To examine the encounter of replication forks and m-AMSA-induced cleavage complexes, a plasmid model system was used because proper controls can easily be constructed and because larger amounts of DNA can be visualized. We constructed a series of closely related plasmids that differ only in whether they contain a cloned strong m-AMSA-inducibe topoisomerase cleavage site (topo site) and/or the T4 replication origin ori(34) (ori) (20, 46). Drug-induced cleavage of the plasmids was analyzed after plasmid-bearing cells were infected with T4 strain K10, which produces modified DNA during replication (see below). Although these plasmids contain the ColE1 origin, they do not replicate during a T4 infection unless a T4 origin is present on the plasmid (5) (see below).
DNA from the drug-free control infections produced the expected linear AseI fragments from all four plasmids (Fig. 2A, lanes 1 to 4). For the two ori-containing plasmids, this band consists of both T4-replicated plasmid and residual plasmid that was not replicated during the infection. To visualize only the plasmid DNA that had been replicated by T4, we took advantage of the cytosine modifications that are introduced during T4-directed DNA replication (by direct incorporation of 5-hydroxymethyl dCMP [Hm-dCMP]). AseI, which was used to linearize the plasmid DNA, cleaves DNA regardless of whether the cytosine residues are modified. However, HaeIII cannot cleave Hm-dCMP-containing DNA, and the plasmids contain numerous HaeIII recognition sites. Therefore, by including HaeIII in the digest, all unreplicated plasmid DNA is cleaved into small fragments that run off the gel but all T4-replicated plasmid DNA is unaffected. In this case, Southern hybridization with the plasmid probe revealed only T4-replicated DNA. As expected, T4-replicated plasmid DNA was generated only with the ori-containing plasmids (Fig. 2A, lanes 5 to 8).
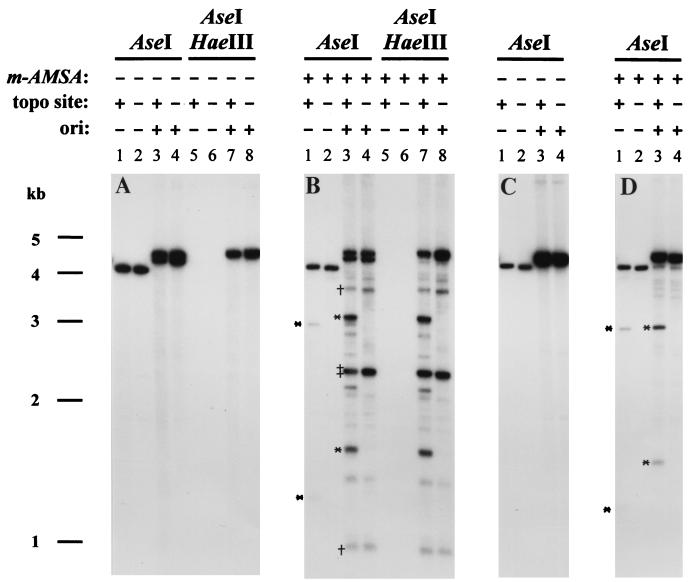
FIG. 2.
Replication and topoisomerase cleavage of plasmid substrates. DNA was purified after T4 infection either in the presence (B and D) or absence (A and C) of m-AMSA (10 μg/ml for K10 and 2.5 μg/ml for T4 dC infections [same conditions as described in Fig. 1 and Materials and Methods]). The infecting phage was T4 K10, which produces modified DNA (A and B), or T4 dC, which produces unmodified DNA (C and D). The presence or absence of the topo site and the ori on each plasmid is indicated. Each sample was digested with the indicated restriction enzymes and subjected to agarose gel electrophoresis and Southern hybridization with pBR322 as the probe. The cleavage products from the cloned m-AMSA-induced cleavage site are indicated by asterisks. One relatively strong m-AMSA-induced cleavage site in the vector generates two partner fragments, indicated by daggers, whereas another generates two comigrating partner fragments, indicated by a double dagger. In comparing the cleavage products generated from the cloned topo site in the ori versus non-ori plasmids (highlighted with asterisks in lanes 1 and 3 of panels B and D), the smaller cleavage products differed in size due to the presence or absence of the cloned ori. In the K10 infections (panel B), the larger cleavage product from the ori-containing plasmid migrated more slowly than that from the non-ori plasmid because the former DNA is modified.
When m-AMSA was added to the infections, topoisomerase cleavage products smaller than the linear plasmid DNA were evident (Fig. 2B). Consider first the non-ori plasmids. When the non-ori plasmid contained the cloned topo site, most of the m-AMSA-induced cleavage was at this site, generating the two expected partner fragments after AseI digestion; these two fragments disappeared when the topo site was absent (Fig. 2B, lanes 1 and 2). The patterns with the ori-containing plasmids are more complex. In the AseI digest of the ori- and topo site-containing plasmid, the same two partner fragments were detected, along with several additional sublinear fragments (Fig. 2B, lane 3). These include fragments generated by at least two other relatively strong topo sites in the vector (Fig. 2B, lanes 3 and 4). The additional cleavage products must have been generated from the replicated plasmid DNA, because they were resistant to HaeIII (lanes 7 and 8). Furthermore, these additional cleavage fragments were not detected when the DNA was treated with only PstI, which linearizes unreplicated DNA but cannot cleave T4-modified DNA (data not shown).
One model to explain the additional sites of m-AMSA-induced cleavage in the replicated (modified) DNA is that the T4 topoisomerase recognizes additional sites when DNA contains modified cytosine residues (35, 56). If this is true, these additional sites should not be recognized when the replicated DNA is generated during infections by T4 dC, a multiple-mutant phage strain which replicates with unmodified cytosine residues (40). The ori-containing plasmids replicated extensively in T4 dC infections, as judged by the ori-dependent increase in the amount of linearized plasmid DNA in the AseI digests (Fig. 2C, lanes 1 to 4). Also as expected, the replicated plasmid DNA was sensitive to HaeIII (data not shown). The m-AMSA-induced cleavage patterns from the T4 dC infections were very simple, with only the cloned topo site generating the two expected partner fragments regardless of the presence of the ori (Fig. 2D, lanes 1 and 3). Therefore, the T4 cytosine modifications led to recognition of the additional cleavage sites by T4 topoisomerase in the K10 infections above. This conclusion was confirmed by in vitro cleavage assays with purified T4 topoisomerase (data not shown). For the experiments described below, the most important conclusion is that the cloned topo site is the only strong site in the plasmid during T4 dC infections and is one of about three strong sites during infections by phages that make modified DNA (e.g., strain K10).
Unique Y-form DNAs generated at the topo site in the presence of m-AMSA.
Since the m-AMSA-induced cleavage complexes formed at the cloned topo site can be visualized easily, we next analyzed the collision between a replication fork and the topoisomerase cleavage complex by two-dimensional (neutral-neutral) gel electrophoresis (8, 21). In this method, DNA fragments are separated by mass in the first dimension (left to right) and by both mass and shape in the second dimension (top to bottom). Because of their branched structures, a series of replicative intermediates generates unique arc shapes with reduced migration in the second dimension.
The branched DNA structures generated from T4 ori-containing plasmids during T4 infection are primarily intermediates of rolling-circle replication. Indirect evidence suggests that such plasmids begin replication in the theta forms, which are then converted into rolling circles that replicate in either direction (2–4) (Fig. 3A). The two different directions of rolling-circle replication generate two families of Y-form intermediates after the plasmid is linearized by restriction digests in vitro (Fig. 3E), but both families fall on the simple Y arc in the two-dimensional gel. This arc emanates from the linear monomer spot, reaches a peak that contains intermediates with branches near the middle of the restriction fragment, and then returns to the diagonal of linear DNA at twice the size of the plasmid (almost fully replicated intermediates).
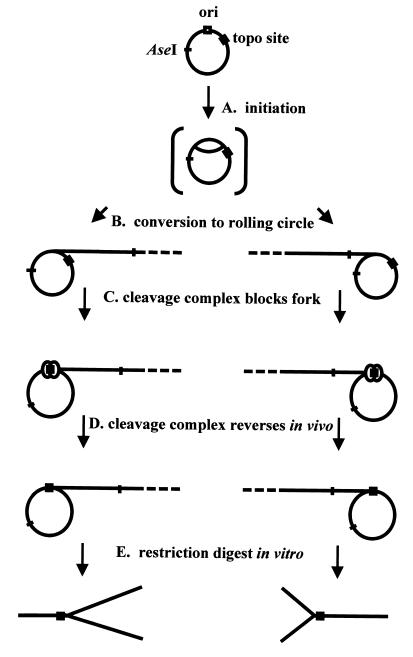
FIG. 3.
Model for generation of Y-form DNAs. The possible steps involved in generating Y-form DNA from an ori-containing plasmid are diagrammed. (A) The plasmid initiates replication in a presumed theta form. (B) The theta form then converts into a rolling circle, possibly by means of a DNA break generated within the bubble region of the theta. (C) Rolling-circle replication proceeds until it is blocked by an m-AMSA-induced topoisomerase cleavage complex (depicted as two circles at the topo site). (D) The m-AMSA-induced topoisomerase cleavage complex reverses in vivo, but the replication fork does not immediately restart. (E) After a restriction digest to linearize the DNA, two unique Y-form DNAs are generated from the two directions of rolling-circle replication.
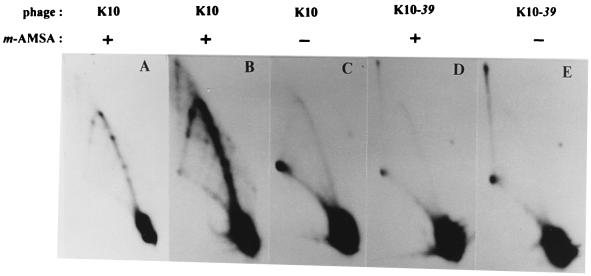
When DNA was prepared from K10-infected cells harboring plasmids that contained both the ori and the topo site, the simple Y arc was greatly enhanced by the presence of m-AMSA (compare Fig. 4A and B). Furthermore, a series of unique Y-form DNAs appeared as discrete spots along the simple Y arc in the infections with m-AMSA. Importantly, two of these spots depended on the presence of the cloned topo site (compare Fig. 4B and C). The migration of these two Y-form DNAs in the first-dimension gel (relative to size markers) indicated that each contains a branch close to the cloned topo site. The two spots represent the two different orientations of the branch, presumably resulting from rolling circles that replicate in each of two directions. The additional spots most probably reflect Y-form DNA with the branches at the additional strong topoisomerase cleavage sites in the plasmid vector (see the cleavage site analysis [above]). Since the unique Y-form DNAs depend on the presence of m-AMSA and accumulate at the topo site, we propose that they consist of blocked replication forks. The proposed pathway for the formation of blocked replication forks is diagrammed in Fig. 3, and various aspects of this pathway are discussed and defended throughout this paper. Ignoring the strong spots, the entire length of the Y arc is much stronger in the presence of m-AMSA than in its absence (compare Fig. 4A and B). We believe that this increased Y-form DNA is also caused by blocked replication forks. The plasmid vector contains numerous weak topoisomerase cleavage sites, and cleavage complexes located at these sites could produce a relatively smooth Y arc by blocking replication forks. (In addition, the analysis of a topoisomerase-deficient mutant [below] argues that the increase in the Y arc is not caused by a loss of topoisomerase activity [see Fig. 6].) Overexposure of a blot from a K10 infection without m-AMSA showed a simple Y arc without spots, presumably representing the normal intermediates of rolling circle replication (Fig. 4D) (4).
FIG. 4.
Two-dimensional agarose gel electrophoresis of plasmid DNA from a bacteriophage K10 infection. E. coli cells harboring plasmids with or without the cloned topo site were infected with bacteriophage K10. For this and all subsequent experiments, attachment was for 4 min and m-AMSA (10 μg/ml) was added 2 min later where indicated. After an additional 18 min of infection, the DNA was harvested, purified, digested with AseI (which linearizes the plasmid), and subjected to two-dimensional gel electrophoresis. The first-dimension gel was run from left to right, and the second-dimension gel was run from top to bottom. Plasmid DNA forms were visualized by Southern hybridization with a plasmid probe. The arrows depict the two unique Y-form DNAs that result from the cloned topo site. Panel D shows an overexposure of a blot with DNA from a K10 infection of E. coli harboring the topo site-containing plasmid in the absence of m-AMSA.
FIG. 6.
Requirement for T4 topoisomerase. E. coli cells harboring the plasmid with both the ori and the topo site were infected with either K10 (wild type) or K10-39am (topoisomerase-deficient mutant) in the presence or absence of m-AMSA. DNA was purified, digested with AseI, and subjected to two-dimensional gel analysis. Panel A is a lighter exposure of panel B, while panels B to E are matched exposures.
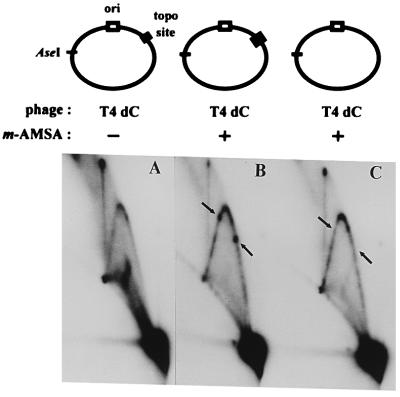
As demonstrated above, only a single strong topoisomerase cleavage site is recognized on the plasmid during infections by T4 dC. When T4 dC infections were analyzed by two-dimensional gel electrophoresis, the unique Y-form DNAs again depended on the presence of m-AMSA, and, as predicted, the only strong spots occurred at the two positions corresponding to the cloned topo site (Fig. 5).
FIG. 5.
Two-dimensional agarose gel analysis of plasmid DNA from bacteriophage T4 dC infection. E. coli cells harboring plasmids with or without the cloned topo site were infected with bacteriophage T4 dC as described in the legend to Fig. 4, except that the concentration of m-AMSA was 2.5 μg/ml. The arrows depict the two unique Y-form DNAs that result from the cloned topo site. We do not understand the large amount of branched DNA in the T4 dC infection without m-AMSA; note that the unique spots are only present with m-AMSA.
If these unique Y-form DNAs are generated from topoisomerase cleavage complexes, they should depend on the presence of the T4 topoisomerase. We therefore compared DNA from infections by wild-type and topoisomerase-deficient (gene 39am) phages in the presence and absence of m-AMSA. As above, an intense Y arc with unique spots was detected from the wild-type infection in the presence of m-AMSA (Fig. 6A and B; panel A is underexposed compared to the other panels) but not from the topoisomerase-deficient infection (Fig. 6C). The faint arc without spots in Fig. 6C through E presumably represents simple replication intermediates (4). We conclude that the unique Y-form DNAs depend on m-AMSA, the topo site, and the presence of a functional topoisomerase.
Replication is needed for generation of Y-form DNAs.
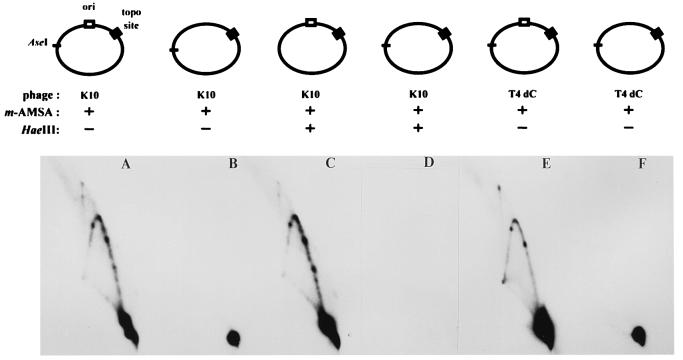
We next asked whether the cloned ori is also important in the generation of the unique Y-form DNAs during infection by either T4 K10 or T4 dC. In both infections, only the ori-containing plasmids produced the intense Y-arc with strong spots (Fig. 7A, C, and E), consistent with the model that these represent blocked replication forks.
FIG. 7.
Requirement for T4 origin and DNA replication. E. coli cells harboring plasmids with the topo site, either with or without the ori, were infected with either K10 (A through D) or T4 dC (E and F). DNA samples were digested with AseI, in either the presence or absence of HaeIII, and subjected to two-dimensional gel electrophoresis.
We also asked whether the DNA within the spots on the Y arc had been replicated by treating DNA from the K10 infections with HaeIII, which cleaves unreplicated DNA into small pieces (see above). The pattern was unchanged (compare Fig. 7A and C), and therefore the branched Y-form molecules must have been replicated throughout the length of all three arms. The origin dependence and the replicated status of the Y-form DNA strongly support the conclusion that the Y-form DNAs arise from blocked replication forks.
Why are Y-form DNAs not cleaved at the topo site?
If the unique Y-form DNAs arise from replication forks that were blocked at a cleavage complex, why is the Y form intact? The infected cells were lysed in the presence of SDS, which should disrupt any topoisomerase cleavage complex and reveal the staggered double-stranded break. One explanation is that the topoisomerase resealed the cleaved DNA within the cleavage complex before cell lysis and yet the replication fork did not restart (Fig. 3D) (see Discussion). A different explanation is that the topoisomerase resealed the cleaved DNA during cell lysis in some fraction of the Y molecules, in spite of the presence of SDS. In this case, perhaps a large proportion of Y molecules are lost during extraction due to SDS-induced DNA cleavage, and a large increase in the amount of Y-form DNA would be obtained if we could prevent this SDS-induced cleavage. To explore these two possibilities, we set up cell lysis conditions which would disfavor SDS-induced cleavage but favor resealing of the DNA within the cleavage complex.
Purified topoisomerase can reseal broken DNA within the cleavage complex in vitro upon dilution of drug, brief heating to 65°C, or treatment with EDTA and/or high salt (a phenomenon known as reversal) (29, 54, 57, 63). In addition, at least upon brief heat treatment at 65°C, cleavage complexes can reverse on chromosomal DNA within mammalian cells (30).
We tested reversal conditions with purified T4 topoisomerase in vitro and found that a large fraction of cleavage complexes could be reversed to intact plasmid DNA by treatment with Triton X-100 detergent (1%) at 65°C (data not shown). We next compared both DNA cleavage and production of Y-form DNA when m-AMSA-treated, infected cells were lysed either under these reversal conditions or under the standard conditions that should not favor resealing. About 80% of the total cleavage complexes underwent reversal, demonstrating that the reversal conditions worked well for in vivo samples (Fig. 8A). However, the unique Y-form DNAs appeared in similar amounts in the two samples (compare Fig. 8B and C). These results argue that intact Y-form DNA is not created by resealing during the cell lysis procedure. Rather, we believe that topoisomerase resealing happens in vivo prior to cell lysis (see Discussion).
Y-form DNAs are not dependent on recombinational repair.
One concern about our interpretation of these results is that, in theory, Y-form DNAs could also arise from recombination. The m-AMSA-induced topoisomerase cleavage complex or some derivative thereof is repaired by a recombinational mechanism (see the introduction), which could perhaps lead to Y-form intermediates with the branches near topoisomerase cleavage sites.
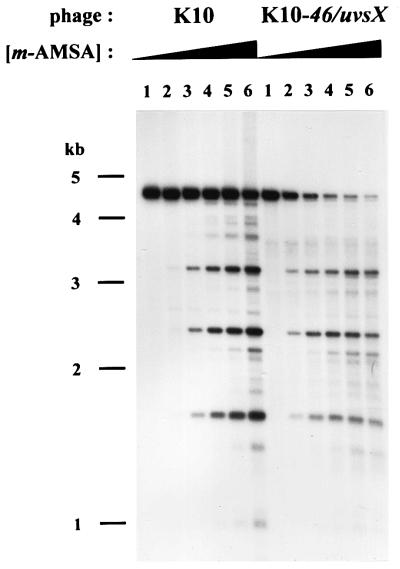
To ask whether the Y-form DNAs are recombination intermediates, we analyzed a T4 K10-46/uvsX double mutant, which is strongly deficient in recombination and recombinational repair. Since the majority of T4 DNA replication occurs through a recombination-dependent mode and therefore relies on recombination proteins gp46 and UvsX, we began by comparing the extent of plasmid replication as a function of the m-AMSA dose in the wild type (K10) and the recombination mutant (K10-46/uvsX). Particularly at the higher levels of drug, the recombination mutant produced much less replicated plasmid DNA (Fig. 9) (see Discussion). The ratio of intact linear DNA to topoisomerase-cleaved DNA was also lower for the recombination mutant than for the wild-type as the m-AMSA dose increased.
FIG. 9.
Titration of m-AMSA in wild-type and recombination-deficient mutant infection. E. coli cells harboring the plasmid with both the ori and topo site were infected with either K10 or K10-46/uvsX in the presence of increasing concentrations of m-AMSA (0, 1, 2, 3, 5, and 10 μg/ml in lanes 1 through 6, respectively). DNA was purified from the infected cells, digested with AseI and HaeIII, and subjected to agarose gel electrophoresis. The resulting fragments were visualized by Southern hybridization with pBR322 as the probe.
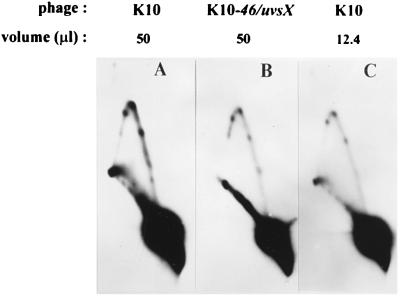
For the analysis of Y-form DNA above, our standard level of m-AMSA was 10 μg/ml (equivalent to lane 6 in Fig. 9). However, for comparisons of K10 and K10-46/uvsX infections, this level is problematic for the following reasons. First, there was very little replicated plasmid DNA at the higher drug level in the recombination mutant infection. Second, the mutant infection was probably more aberrant than the wild-type infection at high levels of drug, because phage DNA replication was more strongly inhibited (data not shown) (the stronger inhibition of phage DNA replication would presumably lead to lower levels of replication and recombination proteins). Third, a higher fraction of the replicated DNA was cleaved by topoisomerase at the high concentration of m-AMSA in the mutant infection, and thus a larger proportion of the Y-form DNA would presumably be destroyed due to topoisomerase cleavage complexes in one of the three arms (unrelated to the cleavage complex that led to branch formation). For these reasons, we analyzed Y-form DNA by two-dimensional gel electrophoresis of DNA from infections with a relatively low drug level, 2 μg/ml (equivalent to lane 3 in Fig. 9). When equal volumes of DNA were compared, the recombination mutant infections were found to produce somewhat less Y-form DNA than the wild-type infections did (Fig. 10A and B). However, Y-form DNA cannot exist without unit-length plasmid DNA, and the recombination mutant infection has less replicated unit-length plasmid DNA even at this low drug level (see Fig. 9). It seems more fair to compare the ratios between the amount of Y-form DNA and replicated unit-length linear DNA. We therefore quantitated the replicated unit-length plasmid DNA and equalized this amount between the pair of infections by loading decreased amounts of the wild-type DNA sample. When this equalization was applied, the amount of Y-form DNA was unaffected by the recombination mutations (Fig. 10B and C). These results argue that the unique Y-form DNAs seen on the simple Y arc are not recombination intermediates.
FIG. 10.
Branched DNA in the recombination-deficient mutant infection. E. coli cells harboring the plasmid with both the ori and the topo site were infected with either K10 or K10-46/uvsX in the presence of m-AMSA at 2 μg/ml. Equal volumes of DNA (50 μl) were compared in panels A and B. To allow a comparison of wild-type and mutant infections at similar levels of intact replicated monomer plasmid, samples from each infection were equalized by changing the volume of the K10 infection DNA that was digested with AseI. Equalization was calculated after quantitating the unit-length linear fragments in Fig. 9 (lane 3) by using an Ambis direct radioisotope imaging system. Note that the unit-length linear spots are approximately the same intensity in panels B and C.
DISCUSSION
Type II topoisomerases such as bacterial DNA gyrase act in front of DNA replication forks to relieve the overwinding produced from unwinding of the parental helix (42). Here we showed that m-AMSA-induced cleavage complexes can arrest the progression of T4 replication forks in vivo, consistent with the placement of the type II topoisomerase in front of the replication fork. Since DNA molecules containing the arrested replication forks were found to be intact, we infer that the replication apparatus must have been inactivated upon encountering the cleavage complex and that the topoisomerase subsequently resealed the DNA breaks prior to cell lysis (Fig. 3). Since the cytotoxicity of topoisomerase inhibitors is known to involve DNA replication, these arrested replication forks may become cytotoxic lesions if they are not restarted or repaired.
Previous reports have shown that replication forks can be blocked by bound proteins, such as terminator protein binding at ter sites or centromere protein-DNA complexes (1, 23, 26). The replication fork barrier in yeast or human rDNA is also probably a protein-mediated blockage (9, 34, 43). In addition, two previous reports suggest that a drug-induced type II topoisomerase cleavage complex can block a replication fork. Using mammalian cells, Catapano et al. (11) measured a reduction in the extent of replication downstream from a cleavage site but did not directly analyze the state of DNA at the cleavage site. Using an in vitro E. coli replication system, Hiasa et al. (25) presented evidence that replication forks are blocked by quinolone-induced topoisomerase IV cleavage complexes; they did not attempt to map the position of the blocked fork with respect to particular topoisomerase IV cleavage sites.
In this study, we detected an accumulation of unique Y-form DNA, appearing as spots along the simple Y arc, when m-AMSA, topoisomerase, and a topoisomerase cleavage site were all present. Several results argue strongly that the unique Y-form DNA originates from replication forks blocked at topoisomerase cleavage complexes. First, a T4 origin was required on the plasmid to generate the unique Y-form DNA molecules. Second, the unique Y-form DNA spots persisted after HaeIII digestion, demonstrating that all three arms of the branched DNA were replicated during the T4 infection. Third, deletion of the strong topo site led to the disappearance of two spots (one for each direction of replication). Fourth, the spots were present in the analysis of DNA from a recombination-deficient infection, arguing against the possibility that recombination generates the unique Y-form DNA.
A somewhat surprising aspect of our results is that the Y-form DNA molecules are not actually cleaved at the topo site. Instead, the results indicate that the topoisomerase-mediated DNA cleavages are resealed after fork blockage but before cell lysis. There was no noticeable increase in the amounts of unique Y-form DNAs when the cells were lysed under conditions that favored resealing of the enzyme-mediated breaks. In vivo resealing is a reasonable possibility because drug-induced cleavage complex formation is an equilibrium process rather than an absolute trapping of the cleaved intermediate. If the resealing event happens in vivo, why do the Y-form DNAs not disappear due to the restart of the replication fork? Perhaps the collision between the replication fork and the topoisomerase cleavage complex leads to disassembly of one or more components of the replication complex such that restart is delayed or prevented.
The above model assumes that the fork-blocking lesion at the topo site is a cleavage complex complete with topoisomerase-bound DNA breaks. A different interpretation is that m-AMSA stabilizes another type of topoisomerase-DNA complex that does not contain any DNA breaks. To our knowledge, no one has detected the induction of stable complexes without DNA breaks by this general class of topoisomerase inhibitors. Furthermore, in their in vitro study, Hiasa et al. (25) found that a mutant topoisomerase IV, which binds DNA but cannot cleave, does not block replication in the presence of inhibitor.
The topoisomerase cleavage complex, or some derivative thereof, can be repaired by a recombinational mechanism (see the introduction). During phage T4 infection, this repair depends on the products of the 46 and uvsX genes (50). Since recombination could, in principle, produce Y-form DNA intermediates, we tested the involvement of gp46 and UvsX in the production of the Y-form DNA. When we equalized the replicated monomer fragments between the wild-type and 46/uvsX infections, we found a comparable intensity of the Y-form DNAs. This result argues strongly that the Y-form DNAs do not arise from recombination and is consistent with the stalled-fork model presented above. Nevertheless, there was a fairly dramatic difference in the level of plasmid replication between the recombination mutant and wild-type infections, particularly as the concentration of m-AMSA increased (Fig. 9). This result suggests that the continued replication of the ori-containing plasmid in the presence of m-AMSA depends on the recombinational repair system that is blocked in the 46/uvsX infection. In other words, we propose that these recombination proteins assist the restart of replication after the blockage event. One plausible scenario is that the Y-form DNA is converted into an overt DNA break and that the DNA break (after a strand invasion reaction) serves to initiate a new round of recombination-dependent plasmid replication. Previous studies demonstrated that recombinational repair of endonuclease-generated breaks involves extensive DNA replication triggered from the break (22, 49).
Several alternatives for processing topoisomerase cleavage complexes have been suggested from in vitro experiments, although none have yet been shown to be important in vivo. Howard et al. (29) showed that a helicase can disrupt a drug-induced cleavage complex, generating overt DNA breaks, and it is conceivable that a complete replication complex can do the same. Yang et al. (68) purified a yeast enzyme that is able to cleave the phosphotyrosine bond between a type I topoisomerase and DNA, generating a DNA nick. If such an enzyme is involved in processing cleavage complexes in vivo, the resulting nicks could be resealed by DNA ligase. If a similar enzyme acts on type II topoisomerase cleavage complexes, a double-strand break would probably result. Sastry and Ross (58) detected an apparently similar phosphodiesterase from human cells and also presented some evidence for a nuclease that creates double-strand breaks flanking topoisomerase cleavage complexes. Any of these processes could play important roles in the cytotoxicity and/or repair of topoisomerase-mediated DNA damage.
Based on the results presented here and prior work of other groups described above, we suggest that blocked replication forks might be the most important lesion in cytotoxicity and DNA repair of topoisomerase-mediated damage. Understanding the repair pathway could improve chemotherapy, perhaps leading to compounds that inhibit repair and thereby potentiate the topoisomerase inhibitors.
In a broader sense, the process that we are analyzing at the topoisomerase cleavage sites may occur with other forms of DNA damage. Our use of topoisomerase cleavage complexes provided a unique opportunity to analyze DNA damage in a site-specific manner in vivo. Various studies argue that replication forks are blocked by other forms of DNA damage, for example pyrimidine dimers from UV treatment (5, 64). In addition, E. coli chromosomal forks can be artificially stalled by a replicative helicase defect, perhaps mimicking DNA (or replication protein) damage (7, 41, 47, 60). Blocked forks, or some derivative thereof, may be generally cytotoxic unless repaired. Our results may also be relevant in considering the process of replication fork blockage in normal cells without overt DNA damage, which has recently become a topic of great interest. A variety of indirect results argue that many, perhaps most, E. coli replication forks initiated at oriC under normal growth conditions become blocked and need to be restarted by a recombinational process to complete replication (14). Furthermore, replication fork blockage has recently been implicated in aging: elimination of the Fob1p replication block protein prolongs the life span of yeast mother cells by preventing fork blockage (and subsequent events) in the rDNA (16). Finally, the helicases defective in the premature-aging disease Werner's syndrome and the cancer predisposition disease Bloom's syndrome have been proposed to play a role in recognizing or correcting aberrant replication structures such as blocked forks (12).
ACKNOWLEDGMENTS
We thank Michael Been, Jeffrey Dawson, Mariano Garcia-Blanco, Tao-shih Hsieh, and David Pickup for insightful discussions.
This work was supported by research grant CA60836 from National Institutes of Health/National Cancer Institute, and George Hong was supported in part by National Research Science Award 5 T32 CA09111.
REFERENCES
- 1.Bastia D, Mohanty B. Mechanisms for completing replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1996. pp. 177–215. [Google Scholar]
- 2.Belanger K G. Origin-dependent DNA replication in bacteriophage T4. Ph.D. thesis. Durham, N.C: Duke University; 1997. [Google Scholar]
- 3.Belanger K G, Kreuzer K N. Bacteriophage T4 initiates bidirectional DNA replication through a two-step process. Mol Cell. 1998;2:693–701. doi: 10.1016/s1097-2765(00)80167-7. [DOI] [PubMed] [Google Scholar]
- 4.Belanger K G, Mirzayan C, Kreuzer H E, Alberts B M, Kreuzer K N. Two-dimensional gel analysis of rolling circle replication in the presence and absence of bacteriophage T4 primase. Nucleic Acids Res. 1996;24:2166–2175. doi: 10.1093/nar/24.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger C A, Edenberg H J. Pyrimidine dimers block simian virus 40 replication forks. Mol Cell Biol. 1996;6:3443–3450. doi: 10.1128/mcb.6.10.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger J M. Type II DNA topoisomerases. Curr Opin Struct Biol. 1998;8:26–32. doi: 10.1016/s0959-440x(98)80006-7. [DOI] [PubMed] [Google Scholar]
- 7.Bierne H, Michel B. When replication forks stop. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 8.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 9.Brewer B J, Lockshon D, Fangman W L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 10.Burden D A, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 11.Catapano C V, Carbone G M R, Pisani F, Qiu J, Fernandes D J. Arrest of replication fork progression at sites of topoisomerase II-mediated DNA cleavage in human leukemia CEM cells incubated with VM-26. Biochemistry. 1997;36:5739–5748. doi: 10.1021/bi963101b. [DOI] [PubMed] [Google Scholar]
- 12.Chakraverty R K, Hickson I D. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 13.Chen A Y, Liu L F. DNA topoisomerases: essential enzymes and lethal targets. Annu Rev Pharmacol Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 14.Cox M M. Recombinational DNA repair in bacteria and the RecA protein. Nucleic Acids Res Mol Biol. 1999;63:310–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 15.D'Arpa P, Beardmore C, Liu L F. Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 1990;50:6919–6924. [PubMed] [Google Scholar]
- 16.Defossez P A, Prusty R, Kaeberlein M, Lin S J, Ferrigno P, Silver P A, Keil R L, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 17.Drlica K, Zhao X L. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng W-K, Faucette L, Johnson R K, Sternglanz R. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol Pharmacol. 1989;34:755–760. [PubMed] [Google Scholar]
- 19.Freudenreich C H, Kreuzer K N. Mutational analysis of a type II topoisomerase cleavage site: Distinct requirements for enzyme and inhibitors. EMBO J. 1993;12:2085–2097. doi: 10.1002/j.1460-2075.1993.tb05857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freudenreich C H, Kreuzer K N. Localization of an aminoacridine antitumor agent in a type II topoisomerase-DNA complex. Proc Natl Acad Sci USA. 1994;91:11007–11011. doi: 10.1073/pnas.91.23.11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman K L, Brewer B J. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 22.George J W, Kreuzer K N. Repair of double-strand breaks in bacteriophage T4 by a mechanism that involves extensive DNA replication. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenfeder S A, Newlon C S. Replication forks pause at yeast centromeres. Mol Cell Biol. 1992;12:4056–4066. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hane M W, Wood T H. Escherichia coli K-12 mutants resistant to nalidixic acid: genetic mapping and dominance studies. J Bacteriol. 1969;99:238–241. doi: 10.1128/jb.99.1.238-241.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiasa H, Yousef D O, Marians K J. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J Biol Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 26.Hill T M. Arrest of bacterial DNA replication. Annu Rev Microbiol. 1992;46:603–633. doi: 10.1146/annurev.mi.46.100192.003131. [DOI] [PubMed] [Google Scholar]
- 27.Holm C, Covey J M, Kerrigan D, Pommier Y. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- 28.Hooper D C. Clinical applications of quinolones. Biochim Biophys Acta. 1998;1400:45–61. doi: 10.1016/s0167-4781(98)00127-4. [DOI] [PubMed] [Google Scholar]
- 29.Howard M T, Neece S H, Matson S W, Kreuzer K N. Disruption of a topoisomerase-DNA cleavage complex by a DNA helicase. Proc Natl Acad Sci USA. 1994;91:12031–12035. doi: 10.1073/pnas.91.25.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsiang Y-H, Liu L F. Evidence for the reversibility of cellular DNA lesion induced by mammalian topoisomerase II poisons. J Biol Chem. 1989;264:9713–9715. [PubMed] [Google Scholar]
- 31.Huff A C, Leatherwood J K, Kreuzer K N. Bacteriophage T4 DNA topoisomerase is the target of antitumor agent 4′-(9-acridinylamino) methanesulfon-m-anisidide (m-AMSA) in T4-infected Escherichia coli. Proc Natl Acad Sci USA. 1989;86:1307–1311. doi: 10.1073/pnas.86.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeggo P A, Caldecott K, Pidsley S, Banks G R. Sensitivity of Chinese hamster ovary mutants defective in DNA double strand break repair to topoisomerase II inhibitors. Cancer Res. 1989;49:7057–7063. [PubMed] [Google Scholar]
- 33.Khodursky A B, Cozzarelli N R. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J Biol Chem. 1998;273:27668–27677. doi: 10.1074/jbc.273.42.27668. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 35.Kreuzer K N, Alberts B M. Site-specific recognition of bacteriophage T4 DNA by T4 type II DNA topoisomerase and Escherichia coli DNA gyrase. J Biol Chem. 1984;259:5339–5346. [PubMed] [Google Scholar]
- 36.Kreuzer K N, Cozzarelli N R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979;140:424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreuzer K N, Engman H W, Yap W Y. Tertiary initiation of replication in bacteriophage T4. Deletion of the overlapping uvsY promoter/replication origin from the phage genome. J Biol Chem. 1988;263:11348–11357. [PubMed] [Google Scholar]
- 38.Kreuzer K N, Morrical S W. Initiation of DNA replication. In: Karam J D, editor. Molecular biology of bacteriophage T4. Washington, D.C.: ASM Press; 1994. pp. 28–42. [Google Scholar]
- 39.Kupfer G, Bodley A L, Liu L F. Involvement of intracellular ATP in cytotoxicity of topoisomerase II-targeting antitumor drugs. NCI Monogr. 1987;4:37–40. [PubMed] [Google Scholar]
- 40.Kutter E, Snyder L. Preparation of cytosine-containing T4 phage. In: Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C.: ASM Press; 1983. pp. 56–57. [Google Scholar]
- 41.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 42.Levine C, Hiasa H, Marians K J. DNA gyrase and topoisomerase IV: Biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 43.Little R D, Platt T H, Schildkraut C L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L F. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 45.McCarthy D. Gyrase-dependent initiation of bacteriophage T4 DNA replication: interactions of Escherichia coli gyrase with novobiocin, coumermycin and phage DNA-delay gene products. J Mol Biol. 1979;127:265–283. doi: 10.1016/0022-2836(79)90329-2. [DOI] [PubMed] [Google Scholar]
- 46.Menkens A E, Kreuzer K N. Deletion analysis of bacteriophage T4 tertiary origins. J Biol Chem. 1988;263:11358–11365. [PubMed] [Google Scholar]
- 47.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ming Y Z, Di X, Gomez-Sanchez E P, Gomez-Sanchez C E. Improved downward capillary transfer for blotting of DNA and RNA. BioTechniques. 1994;16:58–60. [PubMed] [Google Scholar]
- 49.Mueller J E, Clyman J, Huang Y J, Parker M M, Belfort M. Intron mobility in phage T4 occurs in the context of recombination-dependent DNA replication by way of multiple pathways. Genes Dev. 1996;10:351–364. doi: 10.1101/gad.10.3.351. [DOI] [PubMed] [Google Scholar]
- 50.Neece S H, Carles-Kinch K, Tomso D J, Kreuzer K N. Role of recombinational repair in sensitivity to an antitumor agent that inhibits bacteriophage T4 type II DNA topoisomerase. Mol Microbiol. 1996;20:1145–1154. doi: 10.1111/j.1365-2958.1996.tb02635.x. [DOI] [PubMed] [Google Scholar]
- 51.Nelson M A, Ericson M, Gold L, Pulitzer J F. The isolation and characterization of TabR bacteria: hosts that restrict bacteriophage T4 rII mutants. Mol Gen Genet. 1982;188:60–68. [Google Scholar]
- 52.Nitiss J, Wang J C. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci USA. 1988;85:7501–7505. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nitiss J L, Wang J C. Mechanisms of cell killing by drugs that trap covalent complexes between DNA topoisomerases and DNA. Mol Pharmacol. 1996;50:1095–1102. [PubMed] [Google Scholar]
- 54.Osheroff N, Zechiedrich E L. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: Trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987;26:4303–4309. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- 55.Pommier Y. DNA topoisomerase I and II in cancer chemotherapy: update and perspectives. Cancer Chemother Pharmacol. 1993;32:103–108. doi: 10.1007/BF00685611. [DOI] [PubMed] [Google Scholar]
- 56.Ripley L S, Dubins J S, deBoer J G, DeMarini D M, Bogerd A M, Kreuzer K N. Hotspot sites for acridine-induced frameshift mutations in bacteriophage T4 correspond to sites of action of the T4 type II topoisomerase. J Mol Biol. 1988;200:665–680. doi: 10.1016/0022-2836(88)90479-2. [DOI] [PubMed] [Google Scholar]
- 57.Sander M, Hsieh T-S. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 58.Sastry S, Ross B M. Mechanisms for the processing of a frozen topoisomerase-DNA conjugate by human cell-free extracts. J Biol Chem. 1998;273:9942–9950. doi: 10.1074/jbc.273.16.9942. [DOI] [PubMed] [Google Scholar]
- 59.Schneider E, Lawson P A, Ralph R K. Inhibition of protein synthesis reduces the cytotoxicity of 4′-(9-acridinylamino)methanesulfon-m-anisidide without affecting DNA breakage and DNA topoisomerase II in a murine mastocytoma cell line. Biochem Pharmacol. 1989;38:263–269. doi: 10.1016/0006-2952(89)90036-1. [DOI] [PubMed] [Google Scholar]
- 60.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 61.Selick H E, Kreuzer K N, Alberts B M. The bacteriophage T4 insertion/substitution vector system. A method for introducing site-specific mutations into the virus chromosome. J Biol Chem. 1988;263:11336–11347. [PubMed] [Google Scholar]
- 62.Snustad D P, Snyder L, Kutter E M. Effects on host genome structure and expression. In: Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C.: ASM Press; 1983. pp. 40–57. [Google Scholar]
- 63.Tewey K M, Chen G L, Nelson E M, Liu L F. Intercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984;259:9182–9187. [PubMed] [Google Scholar]
- 64.Villani G, Boiteux S, Radman M. Mechanism of ultraviolet-induced mutagenesis: Extent and fidelity of in vitro DNA synthesis on irradiated templates. Proc Natl Acad Sci USA. 1978;75:3037–3041. doi: 10.1073/pnas.75.7.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 66.Wigley D B. Structure and mechanism of DNA topoisomerases. Annu Rev Biophys Biomol Struct. 1995;24:185–208. doi: 10.1146/annurev.bb.24.060195.001153. [DOI] [PubMed] [Google Scholar]
- 67.Wilson W R, Whitmore G F. Cell-cycle-stage specificity of 4′-(9-acridinylamino)methanesulfon-m-anisidide (m-AMSA) and interaction with ionizing radiation in mammalian cell cultures. Radiat Res. 1981;87:121–136. [PubMed] [Google Scholar]
- 68.Yang S W, Burgin A B, Jr, Huizenga B N, Robertson C A, Yao K C, Nash H A. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]