HIV-1 proviruses in the first introns of STAT3 and LCK can contribute to the development of T cell lymphomas.
Abstract
Retroviruses cause cancers in animals by integrating in or near oncogenes. Although HIV-1 infection increases the risk of cancer, most of the risk is associated with immunodeficiency and coinfection by oncogenic virus (Epstein-Barr virus, Kaposi sarcoma herpesvirus, and human papillomavirus). HIV-1 proviruses integrated in some oncogenes cause clonal expansion of infected T cells in vivo; however, the infected cells are not transformed, and it is generally believed that HIV-1 does not cause cancer directly. We show that HIV-1 proviruses integrated in the first introns of signal transducer and activator of transcription 3 (STAT3) and lymphocyte-specific protein tyrosine kinase (LCK) can play an important role in the development of T cell lymphomas. The development of these cancers appears to be a multistep process involving additional nonviral mutations, which could help explain why T cell lymphomas are rare in persons with HIV-1 infection.
INTRODUCTION
Insertional mutagenesis by retroviruses is a common cause of oncogenesis in animals (1). Integration of a provirus in or near an oncogene can enhance the expression of the oncogene, affect the nature of the expressed oncogene protein, or both. Some retroviruses, including HIV-1, are not known to cause cancers by insertional mutagenesis. Chronic HIV-1 infection increases the risk of certain type of cancers, including Hodgkin’s lymphoma, non-Hodgkin’s lymphoma, Kaposi’s sarcoma, and human papillomavirus–associated cancers (2). Much of this increase in cancer incidence in persons with HIV-1 is a result of HIV-1–induced immunodeficiency, which allows greater replication of viruses that cause human cancers, including Epstein-Barr virus, Kaposi sarcoma herpesvirus, and human papillomavirus. As would be expected if these viruses are the primary cause of many of the cancers that arise in persons with HIV-1, much of the increased cancer incidence involves cell types that are not readily infected by HIV-1.
We and others showed that integration of an HIV-1 provirus in oncogenes can cause clonal expansion of infected cells; however, the infected cells were not transformed (3, 4). We recently did a much larger analysis and identified a total of seven oncogenes in which an HIV provirus can cause clonal expansion of the infected cells (5). If HIV-1 integration can lead directly to cancers, then the most likely progenitor would be a CD4+ T cell. T cell lymphomas are rare, both in HIV-1–negative and HIV-1–positive individuals, although HIV-1 infection does increase the risk of T cell lymphomas (6). We show here that HIV-1 proviruses integrated in the signal transducer and activator of transcription 3 (STAT3) and lymphocyte-specific protein tyrosine kinase (LCK) genes can play a direct role in the development of T cell lymphomas.
There have been a small number of reports that HIV-1 proviruses were integrated in oncogenes in human tumors; however, the mechanisms of oncogenic transformation were not defined (7–10). Most of the proviruses described in those reports were not integrated in either the STAT3 or the LCK gene. There is one report that a B cell lymphoma had a defective HIV-1 provirus integrated in the first intron of the STAT3 gene. The provirus was in the same orientation as the gene, and the cells in the B cell lymphoma expressed STAT3 (8), although no RNA analysis was reported for the B cell lymphoma. Even if, in that particular B cell lymphoma, an HIV-1 provirus integrated into the STAT3 gene contributed to the formation of the lymphoma, B cells are not normally infected by HIV-1, and it is not clear to what extent HIV-1 proviruses contribute to the formation of B cell lymphomas in persons with HIV-1. It was recently reported that, in T cells infected with HIV-1 in culture, integration of an HIV provirus in the STAT3 gene can cause clonal expansion of the infected cell (11). However, there are two important differences between what was reported for the in vitro experiments and the in vivo data we report here. First, despite the fact that proviruses integrated in the STAT3 gene can cause clonal expansion of T cells in culture, there is strong evidence that the integration of a provirus either in the STAT3 or LCK genes is not able to cause the clonal expansion of T cells in vivo (5). Proviruses integrated in seven oncogenes have been shown to cause the expansion and/or increase the survival of infected T cells. However, neither STAT3 nor LCK are among the seven oncogenes. Having found the proviruses in STAT3 and LCK in the lymphomas, we reexamined the dataset in (5) and confirmed that there was no evidence for a positive selection for cells with proviruses integrated in either STAT3 or LCK. In addition, in almost all of the cases in which a provirus in the STAT3 gene caused clonal expansion in vitro, the provirus was integrated in the second intron, which would, in the simplest model, lead to the production of a truncated form of the STAT3 protein (11). In contrast, in the tumors we describe here, the provirus is integrated in the first intron of the STAT3 gene, which would lead to the expression of a full-length version of the STAT3 protein.
RESULTS
Some of the T cell lymphoma samples have high levels of HIV-1 DNA
We obtained, from the AIDS and Cancer Specimen Resource (ACSR), samples of lymphomas or lymphoproliferative disorders from 13 HIV-1–positive individuals and two control samples from HIV-1–negative individuals (see Table 1). Genomic DNA from the samples was tested for the presence of high levels of HIV-1 DNA (Table 1). The DNA extracted from three of the samples was too degraded, and these samples were excluded from further analysis. As expected, the B cell and natural killer cell lymphomas and the control tissues had little or no HIV-1 DNA. Although some of the T cell lymphomas had very low levels of HIV-1 DNA, T cell lymphomas from three donors had high levels of HIV-1 DNA, including two samples from donor 1 (specimens 1A and 1B), one sample from donor 11 (specimen 11), and two samples from donor 12 (specimens 12A and 12B). These five lymphoma samples were subjected to HIV-1 integration site analysis (3, 12).
Table 1. Lymphoma samples and ratio of HIV LTR DNA : human β-globin copy number.
| Donor ID | Summary of specimen type and diagnostic pathology | Ratio (HIV LTR DNA:β-globin) |
| 1A* | Skin, high-grade T cell lymphoma (frozen tissue) | 2.88 |
| 1B* | Skin second site, high-grade T cell lymphoma (frozen tissue) |
Not done |
| 2 | Lymph node, high-grade T cell lymphoma (frozen tissue) |
0.0026 |
| 3 | Lymph node, angioimmunoblastic, lymphadenopathy (frozen tissue) |
0.009 |
| 4 | Lymph node, Burkitt lymphoma (frozen tissue) | 0.00004 |
| 5 | Lymph node, Burkitt lymphoma (frozen tissue) | 0.002 |
| 6 | Lymph node, angioimmunoblastic T cell lymphoma (pathology slide) |
0.089 |
| 7 | Lymph node, large cell lymphoma T cell type (pathology slide) |
Degraded DNA |
| 8 | Right tonsil biopsy-anaplastic large lymphoma, ALK negative (pathology slide) |
Degraded DNA |
| 9 | Skin, anaplastic large lymphoma, ALK positive (pathology slide) |
0.00 |
| 10 | Skin, T cell lymphoma (pathology slide) | 0.004 |
| 11 | Lymph node, cutaneous anaplastic large cell lymphoma (FFPE tissue) |
6.62 |
| 12A* | Skin, anaplastic large cell lymphoma (FFPE tissue) |
10.36 |
| 12B* | Skin second site, anaplastic large cell lymphoma (FFPE tissue) |
3.27 |
| 13 | Ethmoid sinus, blastic natural killer cell lymphoma (FFPE tissue) |
Degraded DNA |
| 14 | Control human heart (FPPE tissue) | 0.00 |
| 15 | Control human heart (FFPE frozen) | 0.00 |
*Two donors (1 and 12) from which two tumor samples were taken (A and B).
Integration site analysis of the HIV-1 proviruses in the five lymphoma samples with high levels of HIV-1 DNA
The two specimens from donor 1 (samples 1A and 1B) were cryopreserved with optimal cutting temperature compound. The three samples from donors 11 and 12 were formaldehyde-fixed and paraffin-embedded (FFPE) tissues. We recovered HIV-1 integration sites from all five samples; however, the numbers of integration sites recovered from the FFPE samples were much lower than that from the cryopreserved samples. Table 2 describes the integration sites that were recovered (a complete list is given in table S1). Each of the five samples had an HIV-1 provirus integrated in the first intron of the gene for STAT3. In all five samples, the lymphoma cells that carried the proviruses integrated in the STAT3 gene had undergone clonal expansion, as evidenced by repeated recovery of an identical integration site from the specimen. In addition, samples 11, 12A, and 12B each had a provirus integrated in the first intron of the gene for LCK, and the lymphoma cells that carried the LCK proviruses had clonally expanded (Table 2).
Table 2. Integration sites in the T cell lymphomas.
The positions of the integration sites in the human genome refer to positions in the hg19 version of the human genome. Breakpoints were used to determine how often the same integration site was obtained in the analysis (see Materials and Methods). If there is more than one breakpoint for a particular integration site, then that site came from a cell that had clonally expanded.
| Sample |
Integration site, provirus
orientation in the host genome |
Provirus orientation on the
host gene |
Integration sites isolated |
Integration sites/
breakpoints identified |
| Tumor 1A | chr17, −40,500,566 | +STAT3 | 3LTR/5LTR | ~1000 |
| Tumor 1A | All others | ~8000 | ||
| Tumor 1B | chr17, −40,500,566 | +STAT3 | 3LTR/5LTR | ~500 |
| Tumor 1B | All others | ~750 | ||
| Tumor 12A | chr17, −40,502,259 | +STAT3 | 3LTR/5LTR | 16 |
| Tumor 12A | chr1, +32,724,529 | +LCK | 3LTR/5LTR | 10 |
| Tumor 12A | All others | 15 | ||
| Tumor 12B | chr17, −40,502,259 | +STAT3 | 5LTR | 13 |
| Tumor 12B | chr1, +32,729,544 | +LCK | 5LTR | 38 |
| Tumor 12B | All others | 2 | ||
| Tumor 11 | chr17, −40,506,110 | +STAT3 | 3LTR | 2 |
| Tumor 11 | chr1, +32,738,734 | +LCK | 5LTR | 2 |
| Tumor 11 | chr3, −45,742,822 | +SACM1L | 3LTR/5LTR | 7 |
| Tumor 11 | All others | 12 |
Characterization of the lymphoma samples from donor 1
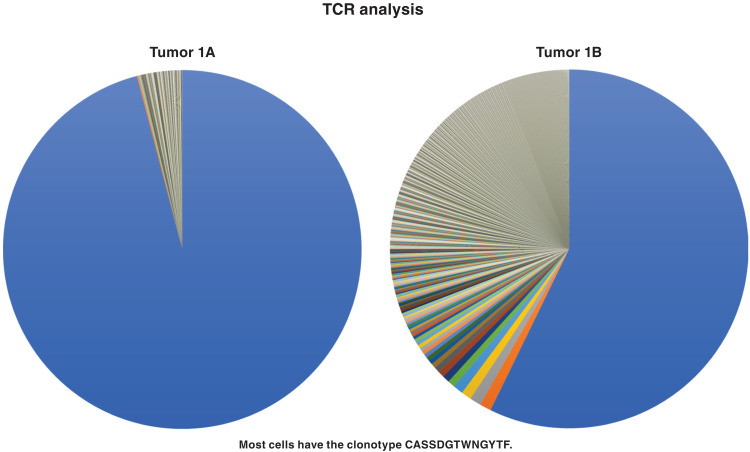
Samples 1A and 1B were cutaneous nodules of anaplastic large cell T cell lymphoma (ALCL) collected from a 48-year-old HIV-1–positive male. To determine the clonal composition of T cells in the tumor, we performed T cell receptor (TCR) sequencing. A dominant T cell clone was found in sample 1A: 96% of the T cells had a rearranged TCR beta (TCRB) with the clonotype CASSDGTWNGYTF in the CDR3 region (Fig. 1). We developed a droplet digital polymerase chain reaction (ddPCR) assay that was specific for the sequence of this recombinant TCRB receptor (Materials and Methods) and quantified this specific TCRB relative to a human reference gene. In tumor 1A, 86% of all the cells in the sample were from this T cell clone. This agrees with the histopathology of the lymphoma nodule, which showed that sample 1A was largely composed of malignant cells (Fig. 2). The identical rearranged TCRB was present in sample 1B, although both the histopathology and the TCRB analysis showed that the fraction of the sample that was malignant was considerably smaller (Figs. 1 to 3) than in sample 1A (discussed below).
Fig. 1. TCRB analysis of the high-grade cutaneous T cell lymphoma from donor 1 (samples 1A and 1B were from different skin nodules).
DNA extracted from the samples was analyzed for the TCRB rearrangements that are associated with the maturation of T cells (see Material and Methods). The same predominant rearranged TCRB was present in both samples. The predominant gene made up a greater fraction of the rearranged TCRB genes in sample 1A than 1B. The predominant TCRB clonotype is given in the figure.
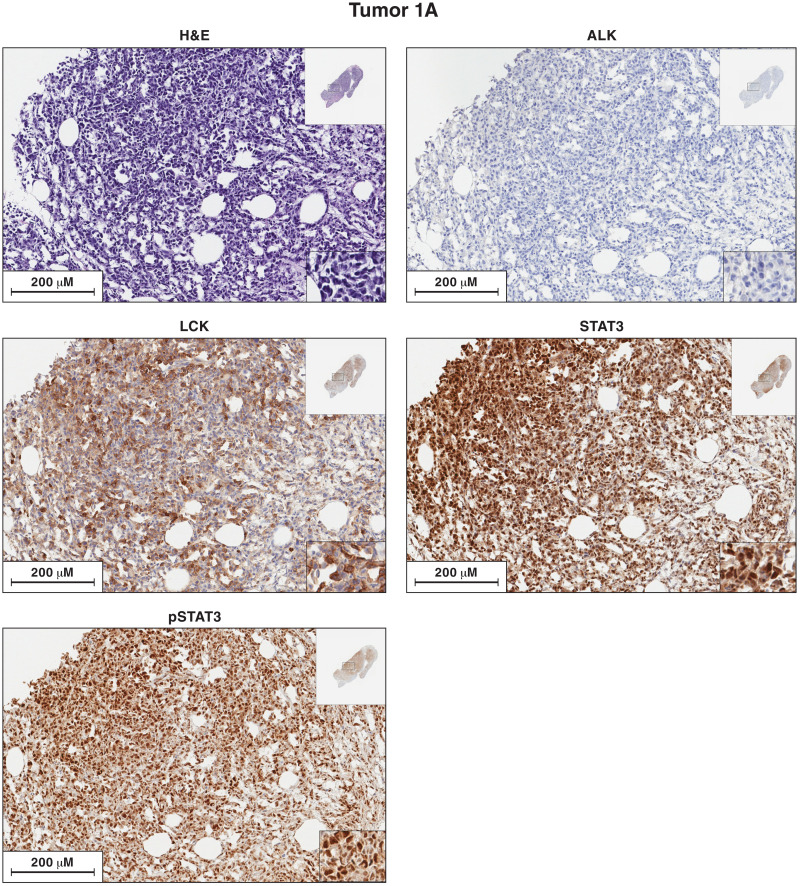
Fig. 2. Antibody staining of sections of sample 1A for the expression of ALK, LCK, STAT3, and pSTAT3.
Each image is labeled to show which lymphoma sample it came from and what the section was stained for. The procedures used to antibody stain the sections of the lymphoma samples are given in Materials and Methods. In the single-stained slides, the bound antibodies were detected using horseradish peroxidase (HRP). Sections from each of the lymphoma samples were hematoxylin and eosin (H&E) stained. Additional sections from each lymphoma were reacted separately with antibodies to ALK, LCK, STAT3, and phosphorylated STAT3 (pSTAT3). The images in the figures show a portion of the section. For each stained section from a particular lymphoma, the same section was chosen for the figure. The inset at the top right shows the whole section, and the inset at the bottom right shows a small part of the image at higher magnification. The images can be expanded to show more detail.
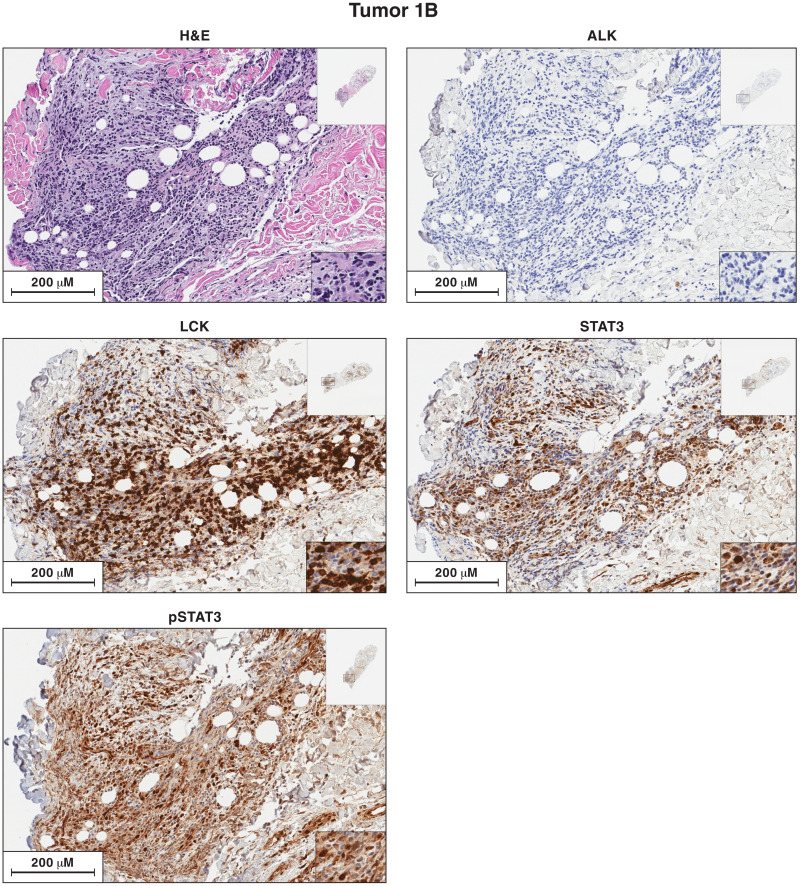
Fig. 3. Antibody staining of sections of sample 1B for the expression of ALK, LCK, STAT3, and pSTAT3.
Each image is labeled to show what the section was stained for. The procedures used to antibody stain the sections of the lymphoma samples are given in Materials and Methods. Sections from each of the lymphoma samples were H&E stained. Additional sections from each lymphoma were reacted separately with antibodies to ALK, LCK, STAT3, and pSTAT3. The images in the figures show a portion of the section. For each stained section from a particular lymphoma, the same section was chosen for the figure. The inset at the top right shows the whole section, and the inset at the bottom right shows a small part of the image at higher magnification. The images can be expanded to show more detail.
The lymphoma cells in sample 1A have an HIV-1 provirus integrated in the first intron of STAT3 gene
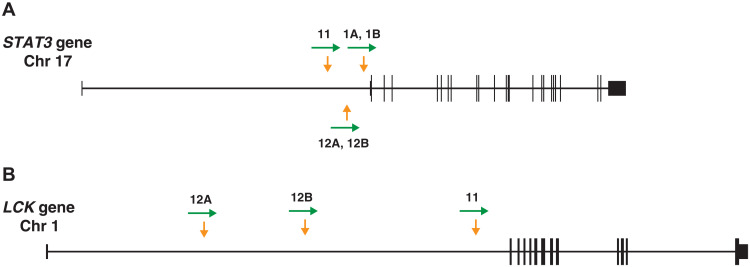
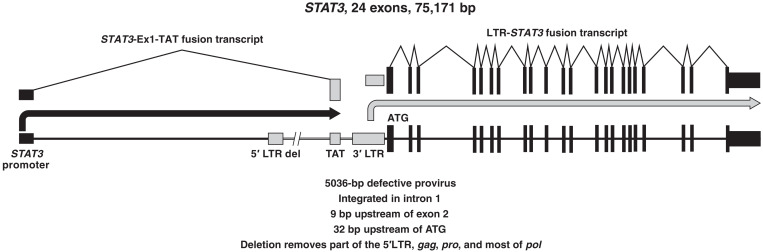
Of the ~9000 integration sites recovered from lymphoma sample 1A (Table 2), ~10% were from a provirus integrated at position chr17:40,500,566, in the first intron of the STAT3 gene, 9 base pairs (bp) upstream of exon 2 (hg19). The integration site is just 32 bp upstream of the translation start site of the STAT3 gene in exon 2 (Table 2 and Fig. 4A).
Fig. 4. Integration sites in the T cell lymphomas from donors 1 (samples 1A and 1B), 11, and 12 (samples 12A and 12B).
(A) Map of the STAT3 gene, derived from the hg19 version of the human genome using the UCSC browser. The vertical arrows (yellow) show the sites of HIV integration in the genomic DNA extracted from the lymphomas. The horizontal arrows (green) show the orientation of the HIV-1 provirus. The exons are shown as boxes; the coding regions are shown as taller boxes than the noncoding regions. (B) Similar to (A), showing the integration sites in the LCK gene.
In sample 1A, there were, on average, ~4.5 proviruses per cell (Table 1), suggesting that the lymphoma cells had been heavily superinfected by HIV-1. We amplified HIV-1 DNA from the sample and sequenced the proviruses. The sequences of the HIV-1 proviruses were diverse (average pairwise difference is 1.5%), which suggests that there were multiple superinfecting viruses. Of the 14 proviruses that were sequenced, 6 were judged to be intact, based on analysis by ProSeq-IT (13). A tree representing the phylogeny of the proviruses is shown in fig. S1. The tree also shows the provirus integrated in STAT3 and sequences of viral genomes obtained from RNA sequencing (RNAseq) data from samples 1A and 1B. The presence of high levels of 2 Long Terminal Repeat (2LTR) circle junctions and free ends of unintegrated linear viral DNA in the samples shows that there was ongoing HIV-1 replication involving the lymphoma at the time it was taken from the donor (table S2). Although we suspect that the donors were not on antiretroviral therapy when the lymphoma samples were taken, we were not able to determine whether or not they were at the time of sampling.
To determine the frequency of the lymphoma cells that carried the STAT3 provirus, we designed a ddPCR assay that was specific for the host-virus junction of this provirus and compared it to a human reference gene. Approximately 82% of the cells in the lymphoma sample carried the STAT3 provirus, which is similar to the 86% of the cells that had the CASSDGTWNGYTF TCRB clonotype.
Because there was considerable overlap in the breakpoints used to enumerate the STAT3 provirus in the integration site analysis (12), the 1:8 ratio of the STAT3 integration sites relative to the non-STAT3 sites is likely an underestimate of the frequency of the STAT3 integration sites in the sample. Although we recovered more than 8000 additional integration sites from sample 1A, only 4 of the additional integration sites were recovered 3 or more times, and none were recovered more than 13 times (see table S1). The observations that the secondary integration sites are diverse and that most were recovered only once show that the lymphoma was superinfected by HIV-1 relatively late in the course of its growth.
The provirus in STAT3 in lymphoma 1A is defective
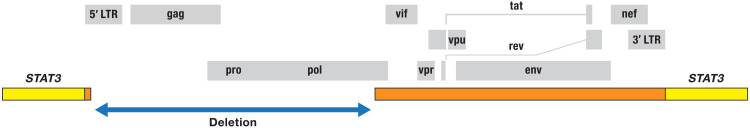
The STAT3 provirus was selectively amplified, and the sequence of the entire provirus was determined (data file S1). The provirus is missing a large portion of the 5′ LTR, including the normal start site for transcription, the major splice donor, all of gag, pro, and almost all of pol (Fig. 5). The open reading frames encoding the Rev, Vpu, Tat, and Env proteins are intact.
Fig. 5. Structure of the provirus integrated in STAT3 in the lymphomas from donor 1 (samples 1A and 1B).
The diagram at the top shows the organization of an intact HIV-1 provirus; the open reading frames for the viral genes are shown as gray boxes. The blue arrow indicates the extent of the deletion that removes most of the 5′ LTR, all of gag, pro, and most of pol. The extent of the viral sequences remaining in the provirus integrated in STAT3 is shown by orange bars in the middle of the figure. The small orange bar is the remnant of the 5′ LTR.
In lymphoma sample 1A, the provirus in STAT3 drives the expression of an LTR-STAT3 fusion transcript
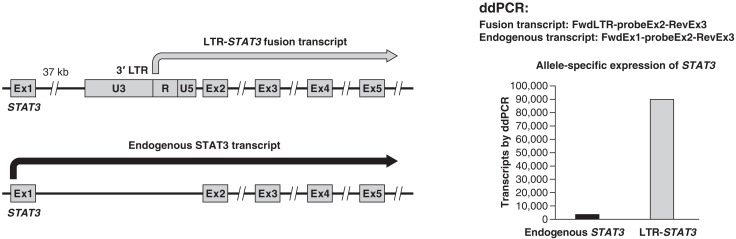
STAT3 is an oncogene that plays an important role in T cell regulation and is frequently mutated and/or activated in T cell malignancies (14). The HIV-1 provirus integrated in STAT3 in sample 1A is inserted 9 nucleotides (nt) upstream of the exon 2 splice acceptor. Given the structure of the provirus, the 3′ LTR could drive the expression of STAT3. Normally, the promoter activity of the 3′ LTR is low, presumably because of interference from transcription that originates in the 5′ LTR. However, the provirus that is integrated in STAT3 in sample 1A has a defective 5′ LTR (Fig. 5). Complementary DNA (cDNA) was made from RNA isolated from sample 1A. Primers that matched sequences in the 3′ LTR and downstream primers in STAT3 were used to look for the fusion transcripts. As shown in Figs. 6 and 7, an LTR-STAT3 fusion transcript was readily detected in the sample. To ensure that the recovered sequences were derived from a fusion RNA, independent reactions were run with forward primers that matched sequences in the LTR and reverse primers that matched exons 3 and 4 of STAT3 (fig. S2). The LTR-STAT3 fusion transcript was also confirmed by RNAseq. The RNAseq data were used to map the 5′ end of the fusion transcript to the R region of the 3′ LTR (fig. S3). The provirus was integrated close enough to exon 2 of STAT3 to disrupt important elements of the splice acceptor. The LTR-driven STAT3 fusion transcript retains 9 bp of intron 1 and continues directly into exon 2 (fig. S3). The STAT3 ATG is only 23 nt downstream from the splice acceptor in exon 2. Of the two STAT3 alleles, only one is disrupted by the provirus, and the second allele should not be affected. We compared the levels of the normal STAT3 and the LTR-fusion STAT3 transcripts; the level of the LTR-driven STAT3 fusion transcript is about 30-fold higher than the normal transcript (Fig. 7).
Fig. 6. Expression of two fusion mRNAs: STAT3-Tat and 3′LTR-STAT3.
The diagram at the bottom shows the provirus whose structure is shown in Fig. 4 integrated into the STAT3 gene. The black and gray arrows in the middle show the transcription of the fusion RNA message for tat (black) and STAT3 (gray). The diagram at the top shows how the transcripts are spliced to produce the mature messages.
Fig. 7. The 3′ LTR of the provirus drives the overexpression of STAT3.
There is a normal allele of STAT3 in tumor 1A. A diagram showing the two STAT3 alleles, and their expression, is on the left. A ddPCR assay (see main text and Materials and Methods) was developed that distinguishes the transcripts from each other and from genomic DNA (which is unspliced). As shown on the right, the LTR-driven STAT3 is expressed at about a 30-fold higher level than the normal transcript.
The STAT3 promoter drives the expression of an HIV-tat fusion transcript
Efficient elongation of HIV-1 transcripts requires the Tat protein (15). After the lymphoma was superinfected by HIV-1, Tat could be supplied in trans by the superinfecting proviruses. However, as previously discussed, there is good evidence that superinfection happened late in growth of the lymphoma. How then was tat expressed before the lymphoma was superinfected? There is an intact tat open reading frame in the defective provirus. We asked whether the STAT3 promoter could be used to express a STAT3-tat fusion RNA. Reverse transcription PCR (RT-PCR) showed that the lymphoma cells in sample 1A produced a fusion RNA in which exon 1 of STAT3 is spliced to the A3 splice acceptor of HIV-1 (Fig. 6 and fig. S4). A3 is normally used in the production of the spliced tat messages (16). Because STAT3 exon 1 is noncoding, the STAT3-Ex1-tat fusion transcript should be able to express Tat. It is therefore likely that the STAT3-Ex1-tat transcript was able to produce enough Tat protein to allow the LTR-STAT3 fusion transcript to be expressed before the lymphoma cells were superinfected by HIV-1.
There is an insertion mutation in the activated allele of STAT3 in lymphoma sample 1A
Exome sequencing data from lymphoma 1A showed that there was, in one of the STAT3 alleles, an insertion of 9 nt that would cause three amino acids (VIK) to be inserted into the STAT3 DNA binding domain between what were initially amino acids 365 and 366. The inserted DNA is a short duplication of an AT-rich sequence (fig. S5). Although matched normal tissue or blood samples were not available from donor 1, analysis of sample 1B (which had a larger proportion of normal tissue) showed that the 9-nt insertion in STAT3 mutation was a somatic mutation rather than a germline mutation. The STAT3 allele containing the VIK insertion was expressed at a much higher level than the wild-type STAT3 allele, indicating that the mutant STAT3 allele was driven by the HIV-1 provirus. Although most oncogenic mutations in STAT3 in T cell lymphomas are in the Src homology 2 domain (SH2 domain) (14), there are rare activating mutations in the DNA binding domain of STAT3 (17).
Lymphoma sample 1B is related to sample 1A
Lymphoma sample 1B was taken from a different cutaneous site from donor 1. In the 1B sample, the major T cell clone made up a smaller fraction of the total number of cells (33% in 1B versus 82% in 1A, estimated by ddPCR), which was confirmed by immunohistochemical analysis of sections of the two specimens (see Fig. 3). Sample 1B lymphoma was similar to sample 1A in that it contained a large clone of cells with the same TCR (TCRB clonotype CASSDGTWNGYTF), the same predominant integration site in STAT3, and the 9-nt insertion mutation in STAT3. Thus, both the 1A and 1B ALCL nodules were derived from the same HIV-1–infected cell. Similar to sample 1A, the cells in sample 1B were superinfected by HIV-1; however, the fraction of the STAT3 integration sites compared to all the other integration sites was higher in sample 1B (approximately 400 of almost 1300 integration sites). Although integration site analysis identified a few small clones of infected cells in lymphoma 1B, none of the non-STAT3 integration sites that were recovered from sample 1B matched any of the integration sites found in lymphoma 1A, which strengthens the argument that both lymphomas were superinfected late in the course of their growth.
The STAT3 activation in lymphoma samples 1A and 1B is associated with phosphorylated pSTAT3 protein in the nuclei of the malignant cells
We prepared sections of lymphomas 1A and 1B and stained them with hematoxylin and antibodies to ALK (anaplastic lymphoma receptor tyrosine kinase), STAT3, phosphorylated STAT3 (pSTAT3), and LCK (Figs. 2 and 3). As expected, on the basis of the molecular analyses, the section of the lymphoma 1A sample was composed primarily of lymphocytes with a malignant appearance, and the lymphoma 1B sample had a much larger fraction of nonmalignant cells. The lymphoma cells in both samples (1A and 1B) expressed high levels of STAT3 protein. An antibody that was specific for the activated form of STAT3 (phosphorylated on Y705, pSTAT3) also strongly stained the nuclei of the lymphoma cells (discussed below). There is a small insert in the lower right of the images that is shown at higher magnification. In addition, the images can be expanded to show more detail.
ALK translocations are frequently seen in ALCL, and activated ALK is thought to phosphorylate STAT3 (14, 18–20). To determine whether activated ALK was present, we analyzed RNAseq and exome sequencing data from tumors 1A and 1B. We found no ALK mutations or translocations, ALK RNA was almost undetectable, and both lymphoma samples 1A and 1B were ALK negative by antibody staining. These results suggest that another tyrosine kinase (or kinases) plays a role in the activation of STAT3 in lymphomas 1A and 1B.
Activated STAT3 enhances the expression of known target genes in lymphoma sample 1A
The high level of the pSTAT3 protein suggested that the STAT3 pathway was activated in tumors 1A and 1B. RNAseq analysis of sample 1A showed that there were increases in the RNA levels for a number of known STAT3 target genes. For example, the level of the RNA for the SOCS3 gene (suppressor of cytokine signaling 3) was up-regulated approximately 70-fold in the lymphoma cells (table S3). RNAseq also showed major up-regulation in expression of the TNFRSF8 gene (tumor necrosis factor receptor superfamily member 8) (18), which encodes CD30, the best-known histologic marker of ALCL (table S3). The genes for the cytotoxic proteins perforin and granzyme B were also notably up-regulated (see Discussion).
Lymphoma samples from individuals 11 and 12
To determine whether HIV-1 proviral insertions in STAT3 are a common mechanism of gene activation in T cell lymphomas, we analyzed three other lymphoma samples from two additional donors (11 and 12). A cutaneous ALCL (sample 11) was obtained from a 3-year-old female child. Two samples (12A and 12B) were obtained from different cutaneous ALCL nodules from a 34-year-old white male; the biopsies were done approximately 5 months apart. Samples 11, 12A, and 12B were all FFPE prepared tissue. The DNA that was recovered from these samples was small (ca. 200 bp), and although we identified HIV-1 integration sites from all three samples, we were neither able to characterize the structure of the corresponding proviruses nor analyze the RNA.
An HIV-1 provirus is integrated in intron 1 of STAT3 in the lymphoma from individual 12
In both lymphoma samples 12A and 12B, there was a clone of cells with an HIV-1 provirus integrated in the first intron of STAT3 at the same site (chr17:40,502,259). The provirus is in the same orientation as the gene (Fig. 4) ~1700 nt upstream of exon 2 (Table 2 and Fig. 4). We isolated both the 5′ LTR and the 3′ LTR junctions of the STAT3 provirus multiple times from both samples, showing that the lymphoma is composed of a clone of HIV-1–infected cells.
There are HIV-1 proviral insertions in LCK in the lymphoma samples from donor 12
We obtained additional integration sites from lymphoma samples 12A and 12B, most of which were found only once, suggesting that these lymphomas, similar to lymphomas 1A and 1B, were superinfected late in their development. However, it appears, in both lymphoma samples, that there was a superinfection event that happened relatively early in the development of the lymphoma. In sample 12A, there was a clone of cells with an HIV-1 provirus integrated in the first intron of the LCK gene. LCK is a Src family tyrosine kinase that was originally identified as a target of murine leukemia virus (MLV) insertional activation in mouse T cell lymphomas (21, 22). LCK plays an important role in the TCR signaling (23) and can phosphorylate STAT3 (24).
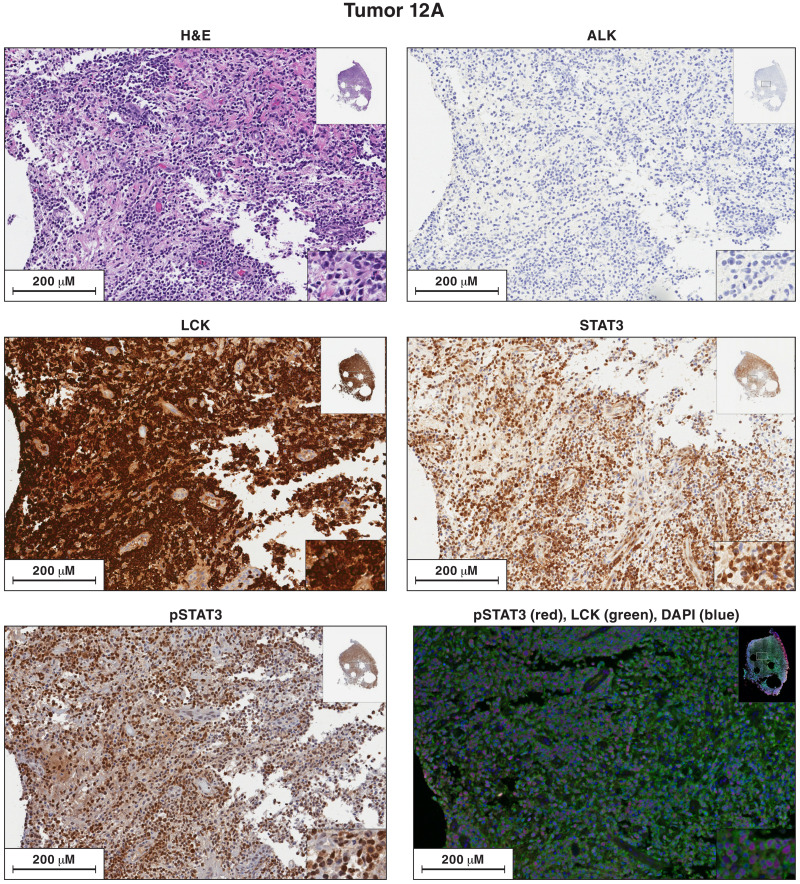
Similar to STAT3, it is the second exon of LCK that is the first coding exon, raising the possibility that HIV-1 proviruses in exon 1 activated LCK and STAT3 in a similar way. Because the DNA was degraded, we were not able to determine the structure of the provirus integrated in the LCK gene in sample 12A. We isolated the integration site for the provirus in LCK in lymphoma 12A multiple times, showing that the LCK-infected cells had clonally expanded. We cannot be sure, based on the integration site data alone, whether the same cells have both the STAT3 and LCK proviruses. However, antibody staining (Fig. 8) shows that most of the cells in lymphoma 12A have high levels of both LCK and pSTAT3, suggesting that most of the cells in the lymphoma carry both the STAT3 and LCK integrated proviruses (see Discussion).
Fig. 8. Antibody staining of sections of sample 12A for the expression of ALK, LCK, STAT3, and pSTAT3.
Each image is labeled to show what the section was stained for. The procedures used to singly and doubly antibody stain the sections of the lymphoma samples are given in Materials and Methods. In the single-stained slides, the bound antibodies were detected using HRP. In the dual-stained slides, the bound antibodies were detected by immunofluorescence. Sections from each of the lymphoma samples were H&E stained. Additional sections from each lymphoma were reacted separately with antibodies to ALK, LCK, STAT3, and pSTAT3. Sections from tumor 12A were costained with antibodies to STAT3 and LCK. The images in the figures show a portion of the section. For each stained section from a particular lymphoma, the same section was chosen for the figure. The inset at the top right shows the whole section, and the inset at the bottom right shows a small part of the image at higher magnification. The images can be expanded to show more detail.
Lymphoma sample 12B has a provirus in exactly the same site in the STAT3 gene as lymphoma sample 12A. This suggests that both ALCL nodules arose from the same progenitor cell. Lymphoma sample 12B also has a provirus integrated in the LCK gene; however, the provirus is in a different site in LCK than in sample 12A. Although the integration site in LCK in lymphoma 12B is also in the first intron and the provirus is in the same orientation as the gene, the integrations sites in LCK in samples 12A and 12B are about 5000 bp apart. These results suggest that the insertional activation of STAT3 preceded the insertional activation of LCK. In contrast to lymphoma sample 12A, for which the integration site analysis showed clear evidence of a late secondary HIV-1 infection, in lymphoma sample 12B, only 2 of 53 integration sites that we recovered were not in STAT3 or LCK (see Discussion). We did not have a sample of 12B that could be used to look for STAT3 and LCK expression by antibody staining.
The lymphoma sample from donor 11 also has proviruses integrated in STAT3 and LCK
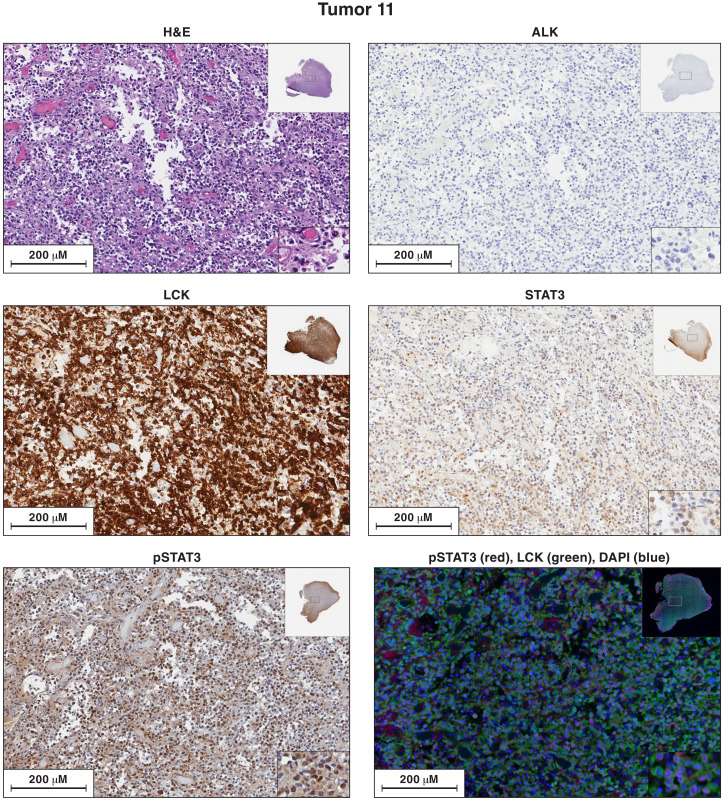
Analysis of lymphoma sample 11 supports the hypothesis that insertional activation of LCK by an HIV-1 provirus can potentiate the effects of insertional activation of STAT3. In sample 11, there is a clone of cells with a provirus integrated in the first intron of the STAT3 gene, in the same orientation as the gene, and there is a clone of cells with a provirus in the first intron of LCK gene, in the same orientation as the gene. Because we recovered relatively few integration sites from sample 11, we cannot be certain that both the LCK provirus and the STAT3 provirus are present in the same cells. However, that interpretation is supported by having three lymphomas in which there are proviruses integrated in both genes. In addition, antibody staining showed that most of the cells in lymphoma sample 11 express high levels of LCK and pSTAT3 (Fig. 9).
Fig. 9. Antibody staining of sections of sample 11 for the expression of ALK, LCK, STAT3, and pSTAT3.
Each image is labeled to show what the section was stained for. The procedures used to singly and doubly antibody stain the sections of the lymphoma samples are given in Materials and Methods. In the single-stained slides, the bound antibodies were detected using HRP. In the dual-stained slides, the bound antibodies were detected by immunofluorescence. Sections from each of the lymphoma samples were H&E stained. Additional sections from each lymphoma were reacted separately with antibodies to ALK, LCK, STAT3, and pSTAT3. Sections from tumor 11 were costained with antibodies to STAT3 and LCK. The images in the figures show a portion of the section. For each stained section from a particular lymphoma, the same section was chosen for the figure. The inset at the top right shows the whole section, and the inset at the bottom right shows a small part of the image at higher magnification. The images can be expanded to show more detail.
Having found that LCK was expressed in samples 11, 12A, and 12B, we asked whether LCK was expressed in samples 1A and 1B. LCK immunostaining was evident in both samples (Figs. 2 and 3). The levels of LCK expression, based on immunostaining, appeared to be higher in lymphoma 1B, but the levels of LCK RNA were not higher in the lymphoma samples than in normal T cells. The RNAseq data also showed that there was a high level of mRNA for another Src family kinase, HCK (hematopoietic cell kinase), in both samples 1A and 1B. HCK and/or LCK could have been responsible for phosphorylating the STAT3 protein on Y705 in the lymphoma from donor 1 (see Discussion).
In the sample from donor 11, there was another provirus in cells that had clonally expanded (Table 2 and table S1). The provirus was integrated in the first intron of the SACM1L gene (suppressor of actin mutations 1-like), in the same orientation of the gene. The SACM1L gene differs from LCK and STAT3 in that it is not a known oncogene, and it is not clear what role, if any, the provirus integrated in SACM1L played in the development of the lymphoma. In addition to the proviruses that were acquired early in the development of the lymphoma (the STAT3, LCK, and SACM1L proviruses), the lymphoma sample also contained additional proviruses for which there was no evidence that they were in clonally expanded cells, suggesting that, similar to lymphoma samples 1A, 1B, and 12A, lymphoma sample 11 was superinfected late in its growth.
Four of the five T cell tumors have a high viral load
One of the unexpected findings was that, based on the ratio of HIV sequences to a host gene (Table 1), all five of the T cell lymphomas had very high levels of HIV-1 proviral DNA (on average, several proviruses per cell). The integration site data show that four of the five lymphomas were superinfected late in their development. In the case of lymphoma 1A, it is clear that the superinfecting virus was diverse, and there was evidence of active ongoing replication involving both lymphoma samples 1A and 1B. High levels of HIV-1 infection tend to kill cells, both because some HIV-1 proteins are toxic and because the host’s immune system can kill cells that express viral proteins. There are two possible mechanisms that might have helped the lymphoma cells escape surveillance by the immune system. The malignant cells in both samples 1A and 1B express a high level of cytotoxic genes such as granzyme B and perforin, which could kill other immune cells. The lymphoma cells also express a high level of PDL1 (CD274), which could inhibit normal T cell responses.
DISCUSSION
The fact that T cell lymphomas are rare can be explained by the need for multiple mutations to accumulate before cells are able to proliferate and become fully malignant (14, 18). In people without HIV-1 infection, all of the mutations needed to form a T cell malignancy accumulate in the genome without the activation of an oncogene by HIV-1 proviral insertion. In ALK-negative ALCL tumors, a combination of convergent mutations and activated tyrosine kinases can lead to oncogenic activation of STAT3 (19). We show here that, in some people with HIV-1 infection, proviral activation of STAT3 and, in some cases, both STAT3 and LCK is able to substitute for one or two of the steps required to generate T cell lymphomas. The activation of STAT3 seems to be a common step in both viral and nonviral T cell malignancies. Activation involves the efficient expression of STAT3 and, in many cases, an activating mutation in STAT3. In addition, the STAT3 protein needs to be phosphorylated on tyrosine. In T cell malignancies that do not involve HIV-1, ALK is often an activating tyrosine kinase for this step (20). In ALK-negative T cell malignancies, other kinases, including LCK, can play that role (24, 25). It is possible that the activating tyrosine kinases have other important roles in T cell malignancies in addition to phosphorylating STAT3. In the T cell lymphomas we describe here, the activating tyrosine kinase is apparently either LCK (11, 12A, and 12B) or HCK and/or LCK (1A and 1B). In the lymphoma samples that did not have a provirally activated LCK (1A and 1B), the overexpressed STAT3 had acquired a somatic mutation (a 9-nt/three–amino acid insertion; see fig. S6) (discussed below). Viewed this way, the HIV insertional activation of STAT3 and LCK contributes to the group of mutations that are required to generate a T cell malignancy. The number of HIV-1 proviral insertions that directly contribute to the formation of the T cell lymphomas can be none (in lymphomas in which there are no proviruses) or, in the lymphomas we describe here, either one (1A and 1B) or two (11, 12A, and 12B).
We identified two of the events that were involved in the generation of the T cell lymphomas in two of the three individuals (11 and 12). In these lymphomas, there were two HIV-1 proviral insertions, one in STAT3 and the second in LCK. In addition, in lymphoma samples 12A and 12B, although the STAT3 provirus was in exactly the same site in both lymphomas, the LCK proviruses were in different sites, which shows that STAT3 was activated before LCK in the development of the lymphoma in donor 12.
In lymphoma samples 1A and 1B, only one of the steps in the generation of the tumor involved an HIV-1 provirus. Insertion of an HIV-1 provirus caused STAT3 to be overexpressed at both the protein and the RNA level, and there was abundant STAT3 protein phosphorylated on Y705 in the nuclei of cells in both of the lymphoma samples. The overexpressed STAT3 was functional: Genes that are known targets for STAT3 were overexpressed. In both the 1A and 1B lymphoma samples, there was a three–amino acid insertion in the DNA binding domain of the provirally activated STAT3 allele. Most STAT3-activating mutations are in the SH2 domain (14); however, mutations in the DNA binding domain of STAT3 can be tumorigenic (17). Because transformed cells make up less than half of sample 1B, we know that the 9-nt/three–amino acid insertion happened during the development of the lymphoma. The three–amino acid insertion in the DNA binding domain of STAT3 could have an effect that is similar to the rare activating mutation H410R, which was found in leukemia (17). Both the H410R mutation and the insertion of VIK add a charged residue in the STAT3 DNA binding domain, which could increase the affinity of the STAT3 protein for DNA. Thus, it is likely that the VIK insertion in STAT3 was one of the nonviral somatic mutations that contributed to the development of the lymphoma in donor 1.
As mentioned earlier, STAT3 activation requires that the protein be both expressed and phosphorylated. ALK is frequently coexpressed with STAT3 in ALCL (20); however, none of the lymphoma samples we analyzed showed any evidence of ALK expression, either at the RNA or the protein level. It appears instead that a Src family kinase (LCK and/or HCK) could be involved, either directly or indirectly. LCK was first reported to be activated by MLV in mouse T cell lymphoma (21, 22). LCK can also be activated by the insertion of a provirus in rat lymphomas (26), and overexpression of LCK causes T cell lymphomas in transgenic mice (27). In most of the provirally induced rat and mouse lymphomas, a provirus is integrated in the first intron of LCK, upstream of the first coding exon. In three of the lymphoma samples described here (tumors 11, 12A, and 12B), there is an HIV-1 provirus integrated in the first intron of LCK, and the protein is expressed at a high level. Although no HIV-1 proviruses were found in either the LCK or HCK genes in lymphoma samples 1A or 1B, LCK was expressed at both the RNA and the protein level, and HCK RNA was overexpressed.
In retrovirally induced malignancies in animals, if the retrovirus replicates efficiently, then insertion of a provirus in or near an oncogene usually leads to the efficient formation of cancer in most or all of the animals in less than a year (1). However, despite the fact that HIV-1 replicates efficiently, infects large numbers of cells in people, and integrates into a number of oncogenes, HIV-1–associated T cell malignancies are rare. At least part of the explanation, which has already been presented, lies in the fact that, in the human T cell lymphomas, additional nonviral mutations are required. There is also an additional factor that might make it more difficult for HIV-1 to induce T cell malignancies: HIV-1 Tat, which is required for efficient HIV-1 proviral expression. Tat binds to Tar, an RNA structural element in the U5 region of the HIV-1 LTR. When Tat is bound to Tar, it recruits host factors that permit the efficient expression of viral RNA (15). If STAT3 and LCK are activated by HIV-1 promoter insertion, then the Tat/Tar system needs to be either functional or circumvented.
The HIV-1 provirus that activated STAT3 in lymphomas 1A and 1B is missing most of the 5′ LTR; however, the Tat reading frame is open, and an RNA that could be translated to produce Tat is expressed from the STAT3 promoter. There are, in both lymphoma samples 1A and 1B, high levels of STAT3 RNA expressed from the promoter in the 3′ LTR. The lymphoma samples we have from donor 1 were heavily superinfected, so at the time lymphoma 1A and 1B samples were taken from the patient, there were high levels of Tat RNA in the lymphoma cells from the superinfecting virus. However, our results show that the lymphoma cells were superinfected late in the development of the tumor. It seems likely that, before the lymphoma cells were superinfected, the STAT3-tat fusion RNA led to the expression of sufficient Tat to allow the efficient expression of the HIV-1–STAT3 fusion RNA. Thus, the provirus that led to the overexpression of STAT3 in lymphoma samples 1A and 1B needed to have special characteristics, both in terms of its location within the intron and the deletion it carries, to allow Tat to be expressed, which would, in turn, allow the 3′ LTR to drive a high-level expression of STAT3.
By contrast, in the lymphomas that were obtained from individuals 11 and 12, it seems unlikely that an unspliced readthrough RNA could be initiated from the 3′ LTR that could cause the overexpression of either STAT3 or LCK. In these lymphomas, the proviruses are located far enough from the first coding exon (exon 2 in both genes) that it would be difficult to express a functional LCK or STAT3 mRNA by a direct readthrough mechanism. Although it is formally possible, in the lymphomas from individuals 11 and 12, that the proviruses could cause the overexpression of STAT3 and LCK genes by enhancer insertion, the location of all of the activating proviruses in the first intron, just upstream of the first coding exon, and the fact that all the proviruses are oriented in the same direction as the gene make that less likely than a promoter insertion mechanism. If the mechanism of activation is promoter insertion, then the HIV-STAT3 and HIV-LCK fusion RNAs could be spliced messages that originate in the 5′ LTR. High-level expression of spliced fusion mRNA would require efficient readthrough of the 3′ LTR. It is possible that the 3′ LTRs of the proviruses that integrated at a distance from the second exon of STAT3 and LCK were defective; it is also possible that, in these lymphomas, there are some other mechanisms that allow the RNA polymerase to read through the 3′ LTRs with reasonable efficiency. However, efficient expression of a fusion RNA from either LTR would also require that Tat be expressed. It is possible to imagine defective proviruses whose structures would fulfill those requirements; however, additional data from unfixed samples will be required to determine the mechanisms by which HIV-1 proviruses can activate STAT3 and LCK when the proviruses are integrated farther upstream in the first intron.
Although there are complexities involved in the requirement that an HIV provirus would need to express Tat to allow it to cause the overexpression of STAT3 and LCK, there are data that show that it is not really difficult for HIV-1 proviruses to induce the expression of several other oncogenes in vivo. Many individuals on antiretroviral therapy (ART) have expanded clones of nonmalignant T cells in which there is an HIV-1 provirus integrated in BACH2, MKL2, MKL1, IL2RB, MYB, POU2F1, or STAT5B (5). It appears likely that, in most or all of these cases, the expression of the oncogenes was driven by HIV-1 promoter insertion. The fact that clones in which there is nonmalignant HIV-1 proviral activation of one of these seven oncogenes are frequently seen in those on ART is due, in part, to the large numbers of infected cells in these individuals. Integration site data obtained from peripheral blood mononuclear cells (PBMCs) that were infected with HIV-1 in vitro (5) show that STAT3 is a reasonably good target for HIV-1 integration, and LCK is a less-favored target. Thus, it is likely that many persons with HIV-1 carry cells that have an HIV-1 provirus in the first intron of STAT3 and integrations in the first intron of LCK. There is no reason to think that it is more difficult for an HIV-1 provirus to activate the expression of STAT3, LCK, or the seven oncogenes. Thus, the data showing that the seven oncogenes are activated in many persons with HIV-1 suggest that expression of STAT3 and LCK has probably been activated by the insertion of an HIV-1 provirus in many people who have not developed T cell lymphomas. This reinforces the idea that additional nonviral mutations are required for the formation of T cell malignancy.
If CD4+ T cells that have a provirally activated STAT3 oncogene and, in some cases, a provirally activated LCK oncogene subsequently acquire other relevant mutations, then the frequency of T cell lymphomas in persons with HIV-1 could increase over time. Most cancers are diseases of the elderly, and persons with HIV-1 are now living much longer, as a result of highly effective ART. It may take many years for a T cell to accumulate the full set of mutations that are required to convert it into a malignant cell. It now appears that proviral activation of STAT3 and LCK can be two components in a larger collection of mutations that can ultimately give rise to T cell lymphomas. Whether ART can prevent the activation of STAT3 and LCK by proviral insertions, particularly if ART is initiated early in the course of infection, is a critical question that can be answered by continued surveillance for T cell lymphomas in persons with HIV-1 and by additional studies of the molecular mechanisms that allow HIV-1 proviruses to contribute to the generation of T cell malignancies.
MATERIALS AND METHODS
Clinical specimens
All lymphoma tissue biospecimens were provided by the ACSR without donor identifiers. The work described here was determined to be exempt from human research oversight by the Institutional Review Boards at the participating sites and laboratories.
Isolation of total nucleic acid from fresh-frozen tissues
Total nucleic acid was isolated from fresh-frozen tissues using a genomic DNA extraction protocol that has been previously described (28). Briefly, the optimal cutting temperature compound that had been added to the tissues was washed off five times with 5 mM tris-HCl (pH 7.6). Guanidinium hydrochloride supplemented with proteinase K [3 M guanidinium hydrochloride (Sigma-Aldrich, USA), 50 mM tris-HCl (pH 7.6), 1 mM calcium chloride, and proteinase K (1 mg/ml) (Thermo Fisher Scientific, USA)] was added, and the tissues were sonicated for approximately 10 s to disperse the sample, followed by incubation at 42°C for 1 hour. Guanidinium thiocyanate supplemented with glycogen [6 M guanidinium thiocyanate (Sigma-Aldrich, USA), 50 mM tris-HCl (pH 7.6), 1 mM EDTA, and glycogen (600 μg/ml) (Roche, Switzerland)] was added followed by incubation at 42°C for 10 min. The nucleic acids were precipitated by addition of 100% isopropanol, washed with 70% ethanol, and resuspended in 5 mM tris-HCl (pH 7.6).
Isolation of total nucleic acid from FFPE tissues
Total nucleic acid from the FFPE tumor samples was isolated with the QIAamp DNA FFPE Tissue kit following the manufacturer’s protocol.
Integration site analysis
HIV proviral integration site analysis was carried out as described (3, 12), with DNA isolated from either fresh-frozen tissue or FFPE tissue. The integration site recovery from the FFPE tissue was much less efficient than it was from the frozen tissue. The sequences of the primers used for each tumor are given in table S4.
TCRB analysis
TCRB sequencing was performed on DNA isolated from tumors 1A and 1B using an ImmunoSEQ hsTCRB kit and ImmunoSEQ Analyzer from Adaptive Biotechnology (Seattle, WA), following the manufacturer’s suggested protocol. We were not able to recover TCRB sequences using DNA from the FFPE samples.
Droplet Digital PCR
ddPCR was performed using the Bio-Rad QX200-ddPCR system following the manufacturer’s suggested protocols. The STAT3 integration site in the clonally expanded cells in tumors 1A and 1B was confirmed by targeted PCR using primers that matched host sequences and primers that matched sequences at the integration site junction (see table S5). To quantify the fraction of cells in the sample that carried the provirus at this integration site, ddPCR was performed to determine the fraction of the cells with the STAT3 integration site and the predominant TCRB locus. For TCRB quantification, T1-STAT3-TCR forward (TGCCAGGCCCTCACATA), T1-STAT3-TCR reverse (GAACCGAAGGTGTAGCCATT), and T1-STAT3-TCR PRB (/56-FAM/CAGTACCTC/ZEN/TGTGCCAGCAGTGAC/3IABkFQ/) were used. For STAT3_IS_chr17_40,500,566, STAT3-5LTRjunction forward (CTGGGACTTGTGGTGAACAT), STAT3-5LTRjunction-PRB (/56-FAM/TCCCTGATT/ZEN/GGCAGAATTACACACCA/3IABkFQ/), and STAT3-5LTRjunction reverse (TCTCTGCTGTCCCTGTAATAAAC) were used.
The host gene MKL2 was used to normalize the DNA input in the ddPCR assays (MKL2 forward, 5′-AGATCAGAAGGGTGAGAAGAATG-3′; MKL2 reverse, 5′-GGATGGTCTGGTAGTTGTAGTG-3′; MKL2 probe, 5′-/56-HEX/TGTTCCTGC/ZEN/AACTGCAGATCCTGA/3IABkFQ/-3′). Because cells were diploid, the cell number was calculated by using half of the MKL2 counts.
RNA sequencing
RNAseq was carried out using total nucleic acid from tumors 1A and 1B and nucleic acid from CD8-depleted PBMCs from an uninfected donor using a NEBNext rRNA depletion kit and a NEBNext Ultra II Directional RNA library preparation kit for Illumina following the manufacturer’s protocol (New England Biolabs, Ipswich, MA). Illumina 2 × 150-bp paired-end sequencing was carried out on a NovaSeq. Analysis of the sequences of the fusion transcripts of HIV and host genes was performed using CLC Genomics Workbench (Qiagen, Germantown, MD).
Exome sequencing
Whole-exome sequencing was performed on DNA from tumors 1A and 1B using the Agilent SureSelect Human All Exon V7 for preparation of the library, which was sequenced on an Illumina NovaSeq with 2 × 150-bp paired-end reads.
CNV analysis
DNA from tumor 1B was used for chromosome copy number variation analysis using the Affymetrix OncoScan CNV array. No abnormal amplifications or deletions were found.
Analysis of the HIV-STAT3 fusion transcripts
The HIV-LTR–driven STAT3 fusion transcripts and the STAT3 promoter–driven Exon1-HIV-tat fusion transcripts were identified by RNAseq, and their structures were confirmed by targeted RT-PCR. For targeted RT-PCR, 200 ng of total nucleic acid was subjected to reverse transcription using an ABI High-Capacity RNA-to-cDNA kit that includes random octamers and an oligo(dT)-16 primer. We supplemented the kit with a STAT3 exon 6–specific primer (CTTGCATGTCTCCTTGACTCTT). The resulting cDNA was used for PCR amplification. To avoid amplification of the genomic DNA that was present with the cDNA, PCR primers were designed to anneal to STAT3 exons (exons 3, 4, and 5) that are far away from the HIV integration site in genomic DNA. A combination of forward primers from the R region of the HIV-LTR (CCCACTGCTTAAGCCTCAATAA) and the U5 region of the HIV-LTR (ACTAGAGATCCCTCAGACCATT) with reverse primers from STAT3-Exon3 (TCTTCGTAGATTGTGCTGATAGAG), STAT3-Exon4 (CAGTCTGTAGAAGGCGTGATT), and STAT3-Exon5 (GGACATCCTGAAGGTGCTG) were used and produced bands of the expected sizes. The STAT3-Exon1-HIV-tat fusion transcripts were amplified with the STAT3-Exon1 primer (AACCGGATCCTGGACAGGCA) and the HIV-tat primer (CTTGATGAGCCTGACAGTCT). ddPCR was performed using the Bio-Rad QX200 ddPCR system to quantify the endogenous STAT3 transcripts and the HIV-LTR–driven STAT3 fusion transcripts. cDNA was generated using the ABI High-Capacity RNA-to-cDNA kit. Again, the primers and probes were designed to be on different exons to avoid amplifying sequences from contaminating genomic DNA. The expression of endogenous STAT3 was measured using a STAT3-Exon1 forward primer (CCT CTG CCG GAG AAA CA), a STAT3-Exon2 probe (/56-FAM/CTT GAC ACA /ZEN/CGG TAC CTG GAG CAG /3IABkFQ/), and a STAT3-Exon3 reverse primer (CAC CAA AGT GGC ATG TGA TTC). The HIV-LTR-STAT3 fusion transcript was measured using an HIV-LTR-R forward primer (CCC ACT GCT TAA GCC TCA ATA A), STAT3-Exon2 probe (/56-FAM/CTT GAC ACA /ZEN/CGG TAC CTG GAG CAG /3IABkFQ/), and a STAT3-Exon3 reverse primer (CAC CAA AGT GGC ATG TGA TTC).
Immunohistochemistry
Tumor samples were stained using antibodies to ALK, STAT3, pSTAT3, and LCK. Paraffin serial sections (5 μm) were separately stained for ALK, LCK, and STAT3 on a BOND RX (Leica Biosystems) autostainer using the BOND Polymer Refine DAB Detection Kit. Endogenous peroxidase was quenched by placing slides in 3% H2O2 for 10 min. Following heat-induced epitope retrieval (HIER) with BOND ER1, sections were incubated for 30 min with antibodies to ALK (diluted 1:200; Novocastra, #NCL-L-ALK) or STAT3 (diluted 1:50; Abcam, #ab31370). Following HIER with BOND ER2, sections were incubated for 30 min with antibody to LCK (diluted 1:100; Abcam, #ab227976). Immunohistochemical staining for pSTAT3 was performed manually. Antigen retrieval with EDTA (15 min at 95°C) was performed, and then 5% normal goat serum was applied to slides for 1 hour. Following overnight incubation at 4°C with antibody to pSTAT3 (diluted 1:100; Cell Signaling Technology, #9145), staining was completed with biotinylated goat anti-rabbit immunoglobulin G (IgG), ABC horseradish peroxidase (ABC-HRP) (Vector Labs), and 3,3′-diaminobenzidine (DAB). After hematoxylin counterstain, slides were dehydrated, and coverslips were added.
Fluorescent double staining of pSTAT3 and LCK was performed manually by incorporating the OPAL 7-Color Manual kit (PerkinElmer). Slides were placed in EDTA (pH 9.0) (Agilent, #S2367) and heated in a microwave processor for 10 min to 100°C and then held for 15 min at 20% power. Slides were rinsed, and 5% normal goat serum was applied for 1 hour. Following overnight incubation at 4°C with pSTAT3 (diluted 1:50; Cell Signaling Technology, #9145), staining was continued with biotinylated goat anti-rabbit IgG, ABC-HRP (Vector Labs), and OPAL 650 Fluorophore (PerkinElmer). Slides were rinsed and then placed again in EDTA (pH 9.0) (Agilent, #S2367) and heated in a microwave processor for 10 min to 100°C and then held for 15 min at 20% power. After cooling, normal horse serum was applied for 20 min. Following a 30-min incubation with an LCK antibody (diluted 1:50; Abcam, #ab227976), staining was continued with ImmPRESS rabbit HRP reagent (Vector Labs) and OPAL 520 Fluorophore (PerkinElmer). Slides were rinsed, 4′,6-diamidino-2-phenylindole (DAPI) stained, and coverslips were added with ProLong Gold Antifade Reagent (Invitrogen).
HIV LTR assay
Briefly, ddPCR was used to amplify sequences from the HIV LTR and human β-globin. Nucleic acids extracted from the 8E5 cell line were used as a standard to quantify LTR copies, and human genomic DNA (Sigma-Aldrich) was used as a standard to quantify the β-globin copies. The LTR and β-globin copy numbers were determined using the Bio-Rad QuantaSoft ddPCR platform. Lymphoma samples with a high ratio of LTR copies to β-globin copies were chosen for further analysis.
Amplification and sequencing of the full-length provirus and adjacent host sequences
The full-length provirus that was integrated in STAT3 in samples 1A was 1B was selectively amplified using a nested PCR approach that generated two amplicons that overlapped by 79 bases in the Nef and envelope regions of HIV. Having determined the integration site and orientation of the provirus in STAT3, nested primers were prepared that matched the host sequences adjacent to the provirus. The primers were designed using sequence data in the UCSC Human Genome Browser (human genome assembly reference hg19) (29). Host-specific primers upstream of the 5′ end of the provirus were paired with HIV envelope-specific reverse primers, while downstream host primers were paired with HIV Nef-specific forward primers. The near full-length (NFL) provirus was also amplified by a single round of PCR using a single primer set that spanned the two host-provirus junctions. The amplification reactions were performed using 2× RANGER DNA Polymerase Mix (Bioline) according to the manufacturer’s instructions with the following modifications for the nested PCR: 35 cycles for the first PCR round, followed by 30 cycles for the nested PCR. The PCR1 product was diluted 1:9, and 2 μl was transferred to the PCR2 master mix. The same conditions were used to amplify the NFL provirus using the junction PCR primers for 48 cycles. After the PCR reactions were complete, products were monitored using an ultraviolet transilluminator and 1× GelRed nucleic acid stain (Biotium). Samples were taken from positive wells and fractionated on 0.7 to 1% agarose gel in sodium borate buffer to determine the size of the amplified products. DNA from positive wells was purified using 0.6 to 1× KAPA Pure Beads (KAPA Biosystems) according to the manufacturer’s instructions. Products from multiple independent PCR reactions were combined from the Host-5′HIV-Env and the junction PCR products. Multiple reaction products from the Nef-3′HIV-Host region were also combined and sequenced separately for comparison. Purified PCR products were sequenced using either Sanger (nested PCR products) or an Illumina platform. The three amplicons were aligned to give the sequence of the full-length STAT3 provirus. There were no sequence differences in the overlapping sequences.
Primers used are as follows: 5′ STAT3 outer, AGACCTGACACCTGTGTTG; 5′ STAT3 inner, GCAATGGCTACTTCTAGATTGTTTACC; 3′ STAT3 outer, AACTGCCGCAGCTCCATTG; 3′ STAT3 inner, TGTAGCTGATTCCATTGGGC; nef F1, GCCACAGCCATAGCAGTAGCTGAGGGG; nef F2, CCTAGAAGAATAAGACAGGGCTTGGAAAG; env R1, ACATGGAGCAATCACAAGTAGCAA; env R2, GGGTGGGAGCAGTATCTCGAGAC; 5′ LTR junction, CATATGCACACTTTGGTTTTGGAA; 3′ LTR junction, GGTCCCAACTGTAAACCTGCTA.
Multiple displacement amplification
Genomic DNA was diluted to the proviral endpoint in a final volume of 210 μl in water, and 2 μl was seeded across all the wells of a 96-well plate. Briefly, genomic DNA was denatured with 2 μl of freshly prepared 0.2 M KOH/18.75 mM EDTA for 3 min at room temperature and then neutralized with 2 μl of 0.3 M tris-HCl (pH 7.5)/0.2 M HCl on an ice for 1 min. After neutralization, 19 μl of ice-cold multiple displacement amplification (MDA) mastermix [final concentrations: 10 mM tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM (NH4)2SO4, 6 mM dithiothreitol, bovine serum albumin (100 ng/μl), 3 mM deoxynucleotide triphosphates, 0.6 M trehalose, 20 μM phosphorothioate-terminated random nanomers, 40 μM hg19-specific random decamers, and phi29 polymerase (0.3 U/μl)] was added to the plate on ice and mixed thoroughly, and the plate was incubated on a thermal cycler at 40°C for 20 hours, followed by 65°C for 10 min, and then held at 4°C (30). After MDA, the amplified DNA was purified by adding 20 μl of suspended paramagnetic KAPA Pure Beads (KAPA Biosystems) to the plate, mixing, and incubating for 7 min. The supernatant was removed and discarded, and the beads were washed twice with 100 μl of 80% ethanol for >30 s for each wash. After washing, the ethanol was removed, and beads were dried for 30 s, followed by resuspension in 40 μl of 5 mM tris-HCl (pH 8.0) and incubation at 37°C for 5 min to elute the DNA. The supernatants containing the recovered DNA were transferred to a clean plate.
Near full-length proviral amplification and sequencing of proviruses recovered from the MDA samples
The DNA recovered from the MDA wells was screened for proviral sequences using NFL proviral amplification and sequencing (NFL-PAS). Nested PCR was performed by diluting 2 μl of the recovered MDA DNA with 8 μl of 5 mM tris-HCl (pH 8.0). The proviruses in the diluted MDA DNA (2 μl) were selectively amplified using the 2× RANGER Polymerase PCR kit (Bioline) in 10-μl reactions with 400 nM HIV NFL-PAS primers [PCR1, 5′-AGTCAGTGTGGAAAATCTCT*A*G-3′ (forward) and 5′-GAGGGATCTCTAGTTACCAG*A*G-3′ (reverse); PCR2, 5′-GTGGAAAATCTCTAGCAGT*G*G-3′ (forward) and 5′-TTACCAGAGTCACACAACAG*A*C-3′ (reverse)] (* represents phosphorothioate bonds), using the following PCR program for both PCR reactions: (i) 95°C for 3 min, (ii) 98°C for 10 s, (iii) 57°C for 10 min, (iv) steps (ii) and (iii) repeated 29×, (v) 57°C for 10 min, and (vi) 10°C hold. After the first PCR, 80 μl of 5 mM tris-HCl (pH 8.0) was added to each well, and 2 μl of the diluted reaction was used in the nested PCR reaction. After nested PCR, 40 μl of 5 mM tris-HCl (pH 8.0) was added to each well, and 5 μl was taken from each well and added to a plate in which each well contained 15 μl of 1× GelRed nucleic acid stain. The plate was read at 302 nm on a transilluminator, and samples from the positive wells were fractionated on 0.8% sodium borate agarose gel by electrophoresis for 20 min at 250 V. DNA from the wells that had well-defined single amplification products was purified by adding 36 μl of KAPA Pure Beads, mixing, and incubating for 5 min. The supernatant was removed and discarded, and the beads were washed twice with 200 μl of 80% ethanol for >30 s each. After washing, the ethanol was removed, and the beads were dried for 2 min. The beads were resuspended in 40 μl of 5 mM tris-HCl (pH 8.0) and incubated at room temperature for 5 min to elute the DNA. The supernatants that contained the eluted DNA were then transferred to a clean plate. NFL-PAS amplicons were sequenced on Illumina MiSeq using an in-house TruSeq workflow using dual indexes and paired-end 250-bp reads (31).
Sequence assembly, statistics, and phylogenetic analyses
MiSeq reads from NFL amplicons were assembled into consensus sequences by CLC Genomics Workbench v12 using the de novo assembler (QIAGEN, Redwood City, CA, USA). RNAseq reads from tumors 1A and 1B were mapped to the HIV-1 genome, and full-length consensus sequences were built by CLC Genomics Workbench v12. HIV-1 NFL sequences with mixtures of sequences were excluded, and the remaining consensus sequences were analyzed for drug resistance mutations in the HIV-1 protease, reverse transcriptase, and integrase genes using the Stanford University HIV Drug Resistance Database online software (https://hivdb.stanford.edu/hivdb/by-sequences/). Inferred intactness was determined by the ProSeq-IT online software from the National Cancer Institute Proviral Sequencing Database. Hypermutated sequences were identified using the Hypermut online software from the Los Alamos HIV Database (www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html). NFL consensus sequences, the sequence of the STAT3 provirus, and the RNAseq sequences from tumors 1A and 1B were aligned by Sequencher v5.4.6 (Gene Codes, Ann Arbor, MI, USA) to the STAT3 proviral sequence, which included INDELs. Sequences were trimmed to an equal length (the size of the STAT3 proviral sequence) and exported as concatenated FASTA files containing all the trimmed NFL sequences, the STAT3 proviral sequence, and the RNAseq sequences from tumors 1A and 1B (Gene Codes, Ann Arbor, MI, USA). Phylogenetic analyses were performed using MEGA v6.0.6 (32). Neighbor-joining p-distance phylogenetic trees were rooted to subtype B with bootstrapping at 1000 replicates per tree using MEGA 6.0.6. MEGA v6.0.6 was also used to calculate the average pairwise distance. Only the HIV-1 DNA sequences that were not hypermutated were used in the analyses.
Acknowledgments
We would like to thank the AIDS and Cancer Specimen Resource (M. McGrath and P. Bracci) for providing tissue specimens, T. Burdette for help in preparing the manuscript, and A. Kane for help with the figures. Specimens were provided by the AIDS and Cancer Specimen Resource, funded by the National Cancer Institute. For cancer tissues obtained from the AIDS and Cancer Specimen Resource (ACSR), a request for material can be made through the ACSR at https://acsr1.com/.
Funding: This research was supported by the Intramural Research Programs of the NIH, National Cancer Institute; Leidos subcontract, 12XS547 (to J.W.M.); and contract no. HHSN261200800001E, National Cancer Institute, NIH. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services or the NIH nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Author contributions: Designed the study: J.W.M., X.W., and S.H.H. Performed integration site analysis, ddPCR, exome sequencing, and RNAseq experiment: S.G. Characterized STAT3 fusion transcripts and performed ImmunoSeq: L.S. and Z.S. Contributed to characterization of provirus: D.D. Performed immunostaining of tumor samples: D.B. and B.K. Extracted tumor DNA/RNA, screened for HIV-1 in tumors, and characterized full-length HIV-1 genomes in the tumors: A.N., L.D.B., K.W.J., E.K.H., B.S., A.H., and M.H. Analyzed the results and wrote the paper: X.W., J.W.M., and S.H.H.
Competing interests: J.W.M. is a consultant to Gilead Sciences and owns options in Co-Crystal Pharmaceuticals, Abound Bio, and Infectious Disease Connect, unrelated to the current work. The other authors declare that they have no competing interests.
Data and materials availability: The HIV tumor samples from the three donors were consumed in the analysis. No other new unique reagents were generated in the study. The pipeline used to find integration sites obtained in the analysis of the patient samples is available on the Retroviral Integration Database (RID) at https://rid.ncifcrf.gov/index.php. All of the integration site data from the tumor samples have been provided with the paper; the integration site data from the tumors are also available on the RID. The tumor HIV-1 integration data, RNAseq, and exome sequencing datasets generated in this study are available in the NCBI Sequence Read Archive (PRJNA670661, SRR12879072-SRR12879110). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Tables S2 and S5
Legends for tables S1, S3 and S4
Legend for data file S1
Other Supplementary Material for this manuscript includes the following:
Tables S1, S3 and S4
Data file S1
REFERENCES AND NOTES
- 1.N. Rosenberg, P. Jolicoeur, Retroviral pathogenesis, in Retroviruses, J. M. Coffin, S. H. Hughes, H. E. Varmus, Eds. (Cold Spring Harbor Laboratory Press, 1997), pp. 475–585. [PubMed] [Google Scholar]
- 2.Calabresi A., Ferraresi A., Festa A., Scarcella C., Donato F., Vassallo F., Limina R., Castelli F., Quiros-Roldan E.; Brescia HIV Cancer Study Group , Incidence of AIDS-defining cancers and virus-related and non-virus-related non-AIDS-defining cancers among HIV-infected patients compared with the general population in a large health district of Northern Italy, 1999-2009. HIV Med. 14, 481–490 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Maldarelli F., Wu X., Su L., Simonetti F. R., Shao W., Hill S., Spindler J., Ferris A. L., Mellors J. W., Kearney M. F., Coffin J. M., Hughes S. H., HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner T. A., McLaughlin S., Garg K., Cheung C. Y., Larsen B. B., Styrchak S., Huang H. C., Edlefsen P. T., Mullins J. I., Frenkel L. M., HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin J. M., Bale M. J., Wells D., Guo S., Luke B., Zerbato J. M., Sobolewski M. D., Sia T., Shao W., Wu X., Maldarelli F., Kearney M. F., Mellors J. W., Hughes S. H., Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy. PLOS Pathog. 17, e1009141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggar R. J., Engels E. A., Frisch M., Goedert J. J.; AIDS Cancer Match Registry Study Group , Risk of T-cell lymphomas in persons with AIDS. J. Acquir. Immune Defic. Syndr. 26, 371–376 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Herndier B. G., Shiramizu B. T., Jewett N. E., Aldape K. D., Reyes G. R., McGrath M. S., Acquired immunodeficiency syndrome-associated T-cell lymphoma: Evidence for human immunodeficiency virus type 1-associated T-cell transformation. Blood 79, 1768–1774 (1992). [PubMed] [Google Scholar]
- 8.Katano H., Sato Y., Hoshino S., Tachikawa N., Oka S., Morishita Y., Ishida T., Watanabe T., Rom W. N., Mori S., Sata T., Weiden M. D., Hoshino Y., Integration of HIV-1 caused STAT3-associated B cell lymphoma in an AIDS patient. Microbes Infect. 9, 1581–1589 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack K. D., Jin X., Yu S., Wei R., Kapp L., Green C., Herndier B., Abbey N. W., Elbaggari A., Liu Y., McGrath M. S., HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J. Acquir. Immune Defic. Syndr. 33, 308–320 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Shiramizu B., Herndier B. G., McGrath M. S., Identification of a common clonal human immunodeficiency virus integration site in human immunodeficiency virus-associated lymphomas. Cancer Res. 54, 2069–2072 (1994). [PubMed] [Google Scholar]
- 11.Yoon J. K., Holloway J. R., Wells D. W., Kaku M., Jetton D., Brown R., Coffin J. M., HIV proviral DNA integration can drive T cell growth ex vivo. Proc. Natl. Acad. Sci. U.S.A. 117, 32880–32882 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wells D. W., Guo S., Shao W., Bale M. J., Coffin J. M., Hughes S. H., Wu X., An analytical pipeline for identifying and mapping the integration sites of HIV and other retroviruses. BMC Genomics 21, 216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao W., Shan J., Hu W. S., Halvas E. K., Mellors J. W., Coffin J. M., Kearney M. F., HIV proviral sequence database: A new public database for near full-length HIV proviral sequences and their meta-analyses. AIDS Res. Hum. Retroviruses 36, 1–3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waldmann T. A., Chen J., Disorders of the JAK/STAT pathway in T cell lymphoma pathogenesis: Implications for immunotherapy. Annu. Rev. Immunol. 35, 533–550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karn J., Tackling Tat. J. Mol. Biol. 293, 235–254 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Emery A., Zhou S., Pollom E., Swanstrom R., Characterizing HIV-1 splicing by using next-generation sequencing. J. Virol. 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson E., Kuusanmaki H., Bortoluzzi S., Lagstrom S., Parsons A., Rajala H., van Adrichem A., Eldfors S., Olson T., Clemente M. J., Laasonen A., Ellonen P., Heckman C., Loughran T. P., Maciejewski J. P., Mustjoki S., Activating somatic mutations outside the SH2-domain of STAT3 in LGL leukemia. Leukemia 30, 1204–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Mel S., Hue S. S.-S., Jeyasekharan A. D., Chng W.-J., Ng S.-B., Molecular pathogenic pathways in extranodal NK/T cell lymphoma. J. Hematol. Oncol. 12, 33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crescenzo R., Abate F., Lasorsa E., Tabbo F., Gaudiano M., Chiesa N., Di Giacomo F., Spaccarotella E., Barbarossa L., Ercole E., Todaro M., Boi M., Acquaviva A., Ficarra E., Novero D., Rinaldi A., Tousseyn T., Rosenwald A., Kenner L., Cerroni L., Tzankov A., Ponzoni M., Paulli M., Weisenburger D., Chan W. C., Iqbal J., Piris M. A., Zamo A., Ciardullo C., Rossi D., Gaidano G., Pileri S., Tiacci E., Falini B., Shultz L. D., Mevellec L., Vialard J. E., Piva R., Bertoni F., Rabadan R., Inghirami G.; European T-Cell Lymphoma Study Group; T-Cell Project: Prospective Collection of Data in Patients with Peripheral T-Cell Lymphoma and the AIRC 5xMille Consortium “Genetics-Driven Targeted Management of Lymphoid Malignancies” , Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 27, 516–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing X., Feldman A. L., Anaplastic large cell lymphomas: ALK positive, ALK negative, and primary cutaneous. Adv Anat Pathol 22, 29–49 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Adler H. T., Reynolds P. J., Kelley C. M., Sefton B. M., Transcriptional activation of lck by retrovirus promoter insertion between two lymphoid-specific promoters. J. Virol. 62, 4113–4122 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voronova A. F., Sefton B. M., Expression of a new tyrosine protein kinase is stimulated by retrovirus promoter insertion. Nature 319, 682–685 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Straus D. B., Weiss A., Genetic evidence for the involvement of the lck tyrosine kinase in signal transduction through the T cell antigen receptor. Cell 70, 585–593 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Lund T. C., Coleman C., Horvath E., Sefton B. M., Jove R., Medveczky M. M., Medveczky P. G., The Src-family kinase Lck can induce STAT3 phosphorylation and DNA binding activity. Cell. Signal. 11, 789–796 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Waldmann T. A., JAK/STAT pathway directed therapy of T-cell leukemia/lymphoma: Inspired by functional and structural genomics. Mol. Cell. Endocrinol. 451, 66–70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin S., Steffen D. L., Frequent activation of the lck gene by promoter insertion and aberrant splicing in murine leukemia virus-induced rat lymphomas. Oncogene 8, 141–149 (1993). [PubMed] [Google Scholar]
- 27.Abraham K. M., Levin S. D., Marth J. D., Forbush K. A., Perlmutter R. M., Thymic tumorigenesis induced by overexpression of p56lck. Proc. Natl. Acad. Sci. U.S.A. 88, 3977–3981 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong F., Aga E., Cillo A. R., Yates A. L., Besson G., Fyne E., Koontz D. L., Jennings C., Zheng L., Mellors J. W., Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J. Clin. Microbiol. 54, 902–911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D., The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan X., Urban A. E., Palejev D., Schulz V., Grubert F., Hu Y., Snyder M., Weissman S. M., A procedure for highly specific, sensitive, and unbiased whole-genome amplification. Proc. Natl. Acad. Sci. U.S.A. 105, 15499–15504 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halvas E. K., Joseph K. W., Brandt L. D., Guo S., Sobolewski M. D., Jacobs J. L., Tumiotto C., Bui J. K., Cyktor J. C., Keele B. F., Morse G. D., Bale M. J., Shao W., Kearney M. F., Coffin J. M., Rausch J. W., Wu X., Hughes S. H., Mellors J. W., HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. J. Clin. Invest. 130, 5847–5857 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Tables S2 and S5
Legends for tables S1, S3 and S4
Legend for data file S1
Tables S1, S3 and S4
Data file S1