Abstract
The unconventional T cell compartment encompasses a variety of cell subsets that straddle the line between innate and adaptive immunity, often reside at mucosal surfaces and can recognize a wide range of non-polymorphic ligands. Recent advances have highlighted the role of unconventional T cells in tissue homeostasis and disease. In this Review, we recast unconventional T cell subsets according to the class of ligand that they recognize; their expression of semi-invariant or diverse T cell receptors; the structural features that underlie ligand recognition; their acquisition of effector functions in the thymus or periphery; and their distinct functional properties. Unconventional T cells follow specific selection rules and are poised to recognize self or evolutionarily conserved microbial antigens. We discuss these features from an evolutionary perspective to provide insights into the development and function of unconventional T cells. Finally, we elaborate on the functional redundancy of unconventional T cells and their relationship to subsets of innate and adaptive lymphoid cells, and propose that the unconventional T cell compartment has a critical role in our survival by expanding and complementing the role of the conventional T cell compartment in protective immunity, tissue healing and barrier function.
The selection through evolution of a system as complex as T cell immunity and its requirement for survival leaves no doubt about its role and importance in vertebrates. Conventional T cells, which are the central actors of adaptive immunity, target and clear infectious non-self. Whereas conventional T cells primarily recognize specific peptides that are presented by the classical polymorphic major histocompatibility complex (MHC) class I and class II molecules, the unconventional T cell compartment encompasses subsets of T cells that cover the sensing of a diverse range of self and non-self molecules. These include lipids sensed by natural killer T (NKT) cells, CD1a-, CD1b- or CD1c-restricted T cells and T cell receptor (TCR)γδ T cells; metabolites sensed by mucosal-associated invariant T (MAIT) cells and TCRγδ T cells; peptides sensed by H2-M3-restricted T cells, Qa-1-restricted T cells, HLA-E-restricted T cells and TCRαβ CD8αα intraepithelial T lymphocytes (IELs); and self-surface proteins sensed by various subsets of TCRγδ T cells. Numerous previous reviews have focused on TCRαβ1–11 and TCRγδ unconventional T cells12,13. Because of its highly redundant nature and the difficulty of demonstrating the requirement for any specific unconventional T cell subset, it is essential to discuss the unconventional T cell compartment as a whole with its unique features and functions. Why vertebrates have also evolved an unconventional T cell compartment, and what the importance of this compartment is in health and disease, remain highly debated questions in biology.
In this Review, we will attempt to provide order to a complex unconventional T cell compartment, while drawing comparisons with the more prominent conventional T cell compartment. We will discuss unconventional T cells with regard to their abundance, their niches and their stability, and will position them in terms of where they fit in an immune response, with an emphasis on human immunity. For example, for TCRγδ T cells we will focus on the butyrophilin (BTN)- and butyrophilin-like (BTNL)-reactive subsets. Finally, we will discuss the evolution and timescale of unconventional T cells relative to adaptive immunity, emphasize the value of redundancy as revealed by their level of genetic conservation across individuals, but also highlight aspects of these T cells that are indispensable to survival.
Classifying unconventional T cells
Class of ligand
The central requirement for a functional immune response is the ability to respond to foreign agents that breach the barriers of the host. Similar to conventional T cells, some unconventional T cell subsets also engage in non-self recognition. For instance, unconventional HLA-E-restricted T cells in humans14 recognize foreign peptides15. However, non-self recognition by unconventional T cells extends beyond peptides, and includes the recognition of canonical non-self molecules that are present across bacterial species, including foreign lipid moieties16–18, bacteria-derived vitamin B metabolites19,20, bacteria-derived formylated peptides21,22 and phosphoantigens23. Therefore, one can think of the antigens driving these unconventional cells as a form of microbial extended self.
The same set of machinery that facilitates the recognition of non-self is also used for the recognition of self. CD1 molecules can load self-lipids and CD1a autoreactive T cells have been characterized in human skin24,25. Fetal Vγ9Vδ2 T cells take on an effector profile26, suggesting that the recognition of self phosphoantigens is a key part of their biology. Although HLA-E can present foreign peptides in the context of certain viral infections14,27, under homeostatic conditions it actually serves to present peptides derived from MHC class I leader sequences28 to primarily regulate T cells29 and natural killer (NK) cells30 through the engagement of NKG2–CD94 receptors. However, the potential role of T cells with specificity for HLA-E in immunity remains poorly understood31. The recognition of self is of course not reserved to unconventional T cells, as regulatory T cells are known to recognize self-peptides32.
Moreover, the unconventional T cell compartment can recognize non-polymorphic ligands that have no capacity to present antigens. This is particularly true for the TCRγδ T cell compartment—for example, the biology of a subset of mouse skin-resident TCRγδ T cells depends on recognition of SKINT-1 (ref.33), and the biology of a subset of intraepithelial intestine-resident cells depends on the molecules BTNL1 and BTNL6 (hereafter, BTNL1/6) in mice34 or the human equivalents BTNL3 and BTNL8 (hereafter, BTNL3/8) in the colon34 and small intestine35. TCRγδ T cells can recognize other non-antigen-presenting molecules such as MICA36, ULBP437, T1038 and T2239, but the biological implications of these interactions are less clear than those of SKINT-1 and the BTNLs.
Thus, the unconventional T cell compartment as a whole is capable of surveillance through the TCR at all levels of cellular immunity—via the recognition of non-self- and self-derived antigens as well as non-antigen-presenting self-ligands—akin to how innate immunity operates.
T cell receptors
Unconventional T cells can be classified into three pools on the basis of their TCR usage and diversity. The first pool is characterized by semi-invariant TCR usage and includes the most-characterized unconventional T cell subset—the type I NKT cell—which expresses an invariant Vα14-Jα18 (TRAV11, TRAJ18) and Vα24-Jα18 (TRAV10, TRAJ18) TCR α-chain in mice and in humans, respectively40, to recognize CD1d loaded with the prototypical lipid antigen α-galactosylceramide4,41. Similarly, MAIT cells express an invariant Vα19-Jα33 (TRAV1–2, TRAJ33)42 and Vα7.2-Jα33–20-12 (TRAV1–2, TRAJ33, TRAJ12, TRAJ20)43 TCR α-chain in mice and humans, respectively, to recognize the vitamin B precursor 5-OP-RU loaded on MHC class I-related protein (MR1)19,44. For both of these unconventional T cell subsets, whereas the use of the TCR α-chain is largely fixed, that of the TCR β-chain is variable but constrained. Similarly, germline-encoded mycolyl-reactive (GEM) T cells express an invariant TRAV1–2 and TRAJ9 TCR α-chain to sense mycobacterial mycolates loaded on human CD1b45,46. Notably, TCRγδ T cell subsets also tend to be characterized by their limited TRGV and TRDV gene-segment use, and this is reflected in their ligand specificities; for example, Vγ5 TCRs are restricted to SKINT-133, and TCRs that use the Vγ chains Vγ7 in mice and Vγ4 in humans are restricted to BTNL1/6 and BTNL3/8, respectively34,35. However, these TCRγδ T cell subsets exhibit a high CDR3 diversity35,47. Given this conserved restriction on TCR–ligand pairs, it is likely that TCRs and their ligands coevolved over time48.
The second pool of unconventional T cells is characterized by diverse TCR usage with regard to both TCR gene-segment use and CDR3 sequence, and includes subsets such as H2-M3-restricted T cells49, HLA-E-restricted T cells50 and TCRαβ CD8αα IELs that recognize class I and II molecules51,52 and non-classical MHC class I molecules53. Of note, this second pool is tailored—similarly to conventional T cells—to the recognition of peptides.
Finally, the third pool of unconventional T cells is characterized by T cells with diverse TCRs that can bind CD1 and MR1. This pool includes type II NKT cells1, MR1-restricted T cells54 and TCRγδ T cells55–57. The presence of such cells is probably the consequence of the diversity present within the naive TCR repertoire, as well as the diversity of antigens that such ligands can present, which allows for naive T cell clones being selected in the periphery by these monomorphic MHC class I-like molecules.
Modes of ligand recognition
Since the pioneering studies detailing the first αβTCR–peptide–MHC-Ia structures 25 years ago58,59, we have learned a lot about the molecular basis that underpins αβTCR engagement. Numerous structural studies have shown that αβTCRs can recognize peptide–MHC-Ia complexes in various docking modes, mostly with a canonical docking polarity60 (Fig. 1a)—albeit with two notable exceptions61,62. Nevertheless, αβTCRs have been universally shown to simultaneously co-recognize the peptide and MHC63, which represents a central tenet of the MHC-restricted T cell response.
Fig. 1 |. Comparison of TCR docking modes.
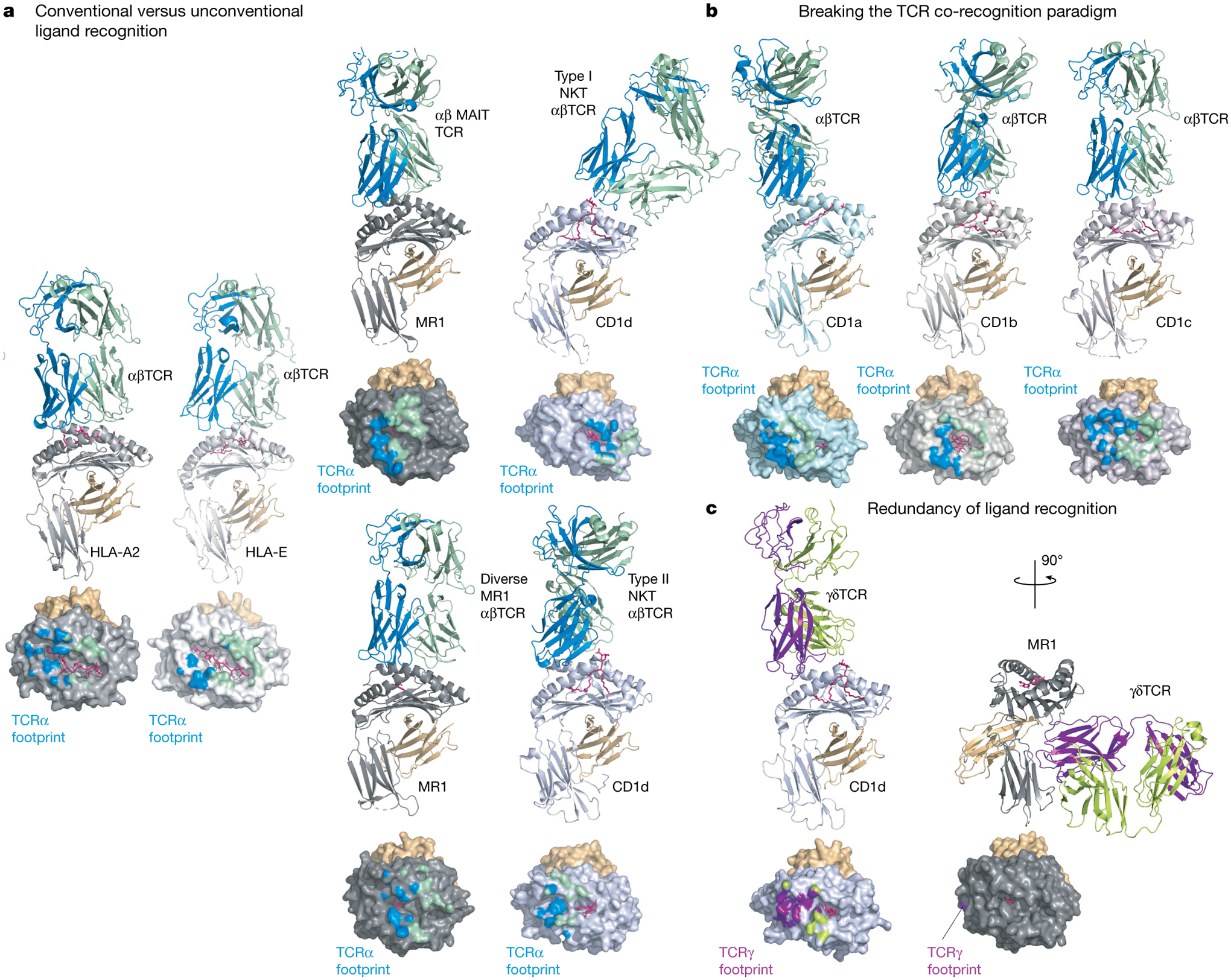
Experimentally determined TCR-binding modes are shown as cartoon representations for a selection of MHC class I or class I-like antigen-presenting molecules. In each panel the MHC-I subunit or equivalent is coloured as follows: light grey (HLA-A2), white (HLA-E), dark grey (MR1), light blue (CD1a), steel (CD1b), light pink (CD1c) or blue-white (CD1d). The respective antigens are coloured pink and associated β2-microglobulin orange; the interacting TCR subunits are coloured either blue (α-subunit) and green (β-subunit) or purple (δ-subunit) and lemon (γ-subunit). Below each structure is a surface representation of the antigen-presenting MHC-I molecules coloured according to their TCR subunit-recognition surfaces. a, Conventional versus unconventional ligand recognition. From left to right: a tumour-associated MART-peptide antigen in complex with HLA-A2 or HLA-E in complex with an αβTCR67,138; MR1-presenting vitamin metabolites recognized by a MAIT TCR70, a diverse αβTCR74 and CD1d in complex with type I139 and type II72 NKT TCRs. b, Breaking the TCR co-recognition paradigm. From left to right: an autoreactive αβTCR in complex with a self-lipid presented by CD1a25; CD1b in complex with a mycobacterial lipid recognized by a GEM TCR46; and an autoreactive TCR recognizing CD1c75. c, Redundancy of ligand recognition by alternative unconventional T cell subsets. The diversity of γδTCR recognition is shown with a CD1d-reactive γδTCR using a relatively standard docking mode55 (left) or the more radical recognition of the underside of MR1 (right)57.
With regard to unconventional TCRs, whether they would adopt the general principles of TCR–peptide–MHC recognition was unknown. The MHC fold has shown remarkable plasticity, having adapted to present lipid- and metabolite-based antigens (by the CD1 family and by MR1, respectively)64. Moreover, whereas MHC-Ia is highly polymorphic, MHC-Ib, CD1 and MR1 show extremely limited polymorphism, yet represent targets for αβTCR and γδTCR recognition65. Although MHC-Ib molecules are considered primarily a ligand for NK cells66, they can also be recognized by TCRαβ T cells, in which the αβTCR can bind peptide–MHC-Ib in a similar manner to TCR–peptide–MHC-Ia binding67 (Fig. 1a). The first insight into how unconventional TCR recognition differs from conventional TCRs came from the structure of the type I NKT TCR–CD1d–lipid complex68,69. In this structure, the type I NKT TCR was perched towards the extreme end of the CD1d antigen-binding cleft, adopting a parallel docking mode. The TCR–MR1–metabolite recognition of MAIT cells was more analogous to TCR–peptide–MHC-I recognition. The invariant TCR α-chain bias observed in type I NKT cells and MAIT cells was attributable to specificity contacts with CD1d–lipid and MR1–metabolite, respectively68,70,71 (Fig. 1a), indicating that germline-encoded recognition is a common feature of type I NKT and MAIT cells—an observation echoed by GEM TCR recognition of a mycobacterial antigen presented by CD1b46 (Fig. 1b).
In comparison to MAIT cells and type I NKT cells, a more diverse unconventional TCR repertoire directed against MR1 and CD1 has been observed, which has manifested in more-varied docking strategies atop their respective antigen-presenting molecules, with many features analogous to that of conventional TCR–peptide–MHC recognition72–74 (Fig. 1a). Some CD1 family members and MR1 also represent ligands for γδTCRs. γδTCR recognition of CD1d–lipid demonstrated how the γδTCR bound the CD1d molecule and co-recognized the lipid55,56 (Fig. 1c), whereas a subset of γδTCRs was shown to bind ‘down under’ the antigen-binding platform of MR1, and thereby not interact with the metabolite-based antigen57 (Fig. 1c). This break of the TCR co-recognition paradigm also appears as a feature of autoreactive αβTCRs towards CD1a and CD1c, whereby the αβTCR sat atop the antigen-binding platforms but nevertheless, via distinct mechanisms, did not contact the lipid25,75 (Fig. 1b). Thus, for some unconventional TCRs, a distinguishing feature is the lack of requirement for the co-recognition of antigen and antigen-presenting molecule, which raises questions relating to thymic selection, specificity of response and whether such features could be exploited for therapeutic purposes.
Development and gain of effector programs
Conventional T cell development takes place in the thymus, in which cells are positively selected on self-peptide–MHC complexes, whereas the acquisition of effector programs occurs in the periphery on foreign-peptide–MHC complexes. The unconventional T cell compartment does not fit into this paradigm. By focusing on where and how a given T cell subset acquires its effector program, we can divide unconventional T cells into three groups (Fig. 2).
Fig. 2 |. Classification of non-classical T cells on the basis of central or peripheral development.
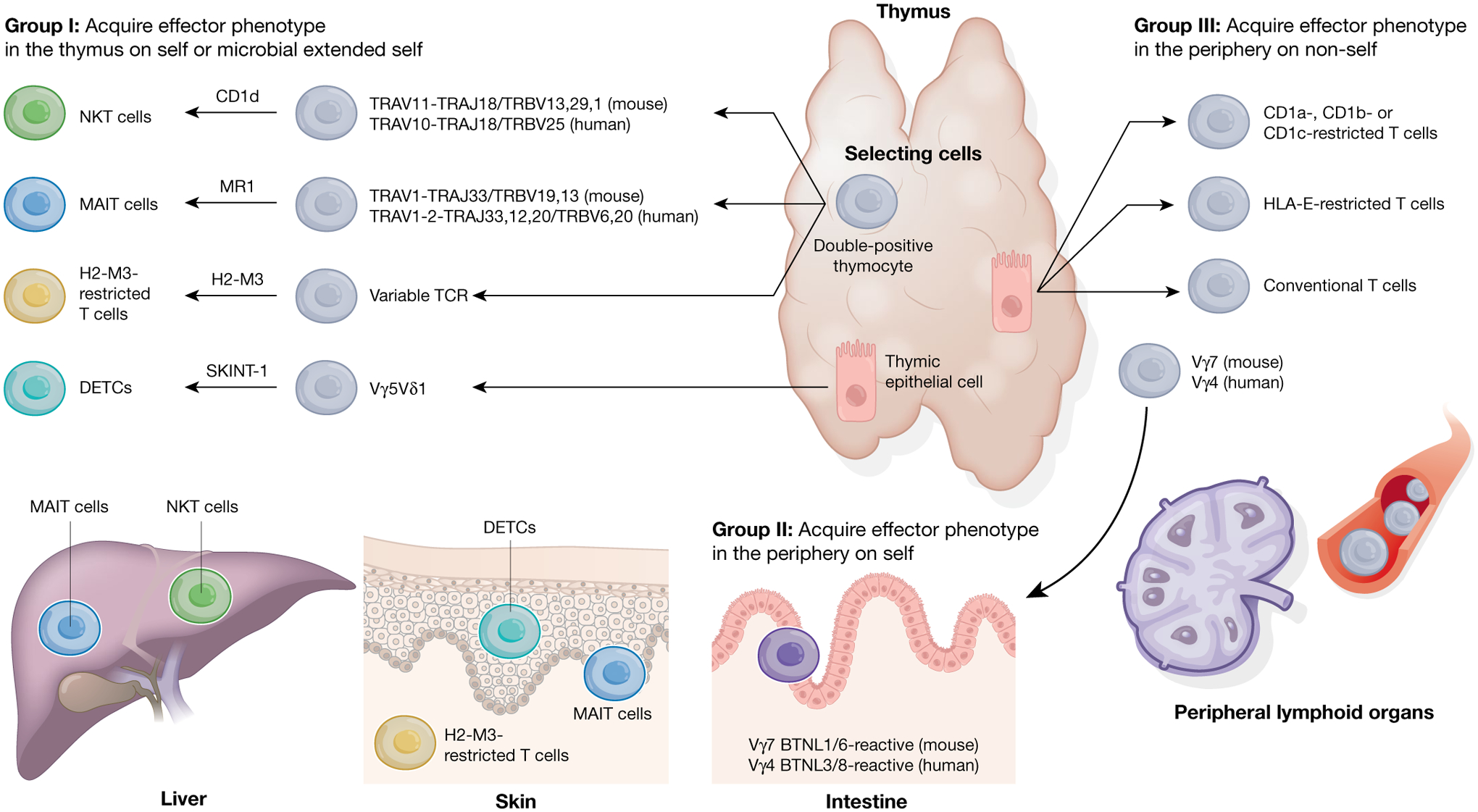
Unconventional T cells can be broadly separated into three groups largely based on their selection and differentiation patterns and how that affects their acquisition of effector programs. Group I unconventional T cells, which are classified by their acquisition of effector functions in the thymus, include NKT, MAIT and H2-M3-restricted T cells, and DETCs. Uniquely for NKT, MAIT and H2-M3-restricted T cells, this process takes place on double-positive thymocytes and requires the SAP pathway. These cells ultimately seed tissues such as the skin and liver, in which they exert their effector functions. Group II unconventional T cells include BTNL-reactive TCRγδ T cells, which leave the thymus naive and acquire effector functions in the periphery on tissue-specific self-ligands. Finally, group III unconventional T cells follow the conventional T cell path by leaving the thymus naive and only acquire effector functions once they encounter their cognate foreign antigen in the periphery. Grey cells represent naive T cells; different coloured cells represent effector T cells.
The first group is characterized by cells that are selected in the thymus and acquire effector programs as a consequence of recognition of their ligand(s) in the thymus. This includes type I NKT cells and MAIT cells, which undergo positive selection on haematopoietic cells76,77, and dendritic epidermal T cells (DETCs), which are selected on thymic epithelial cells33. The acquisition of effector programs in the thymus for NKT and MAIT cells requires the transcription factor PLZF (encoded by Zbtb16 in mice)78,79, expression of the ligand on double-positive thymocytes76,77 and co-signals provided by signalling lymphocytic activation molecule (SLAM)-associated protein (SAP)80. Similarly, H2-M3-restricted T cells that acquire effector programs in the thymus are selected on haematopoietic cells81 and require SAP82. Finally, unconventional mouse TCRαβ CD8αα IELs also acquire effector programs in the thymus through selection on a diverse set of classical and non-classical MHC molecules53,83, and do not require PLZF for their development78. Notably, although expression of the ligand in the periphery is not required for the expansion and effector function of NKT cells84, it is required in mice for unconventional TCRαβ CD8αα IELs85 and probably MAIT cells86. Acquisition of effector functions in the thymus and the hardwiring for particular functional outputs has led this group of unconventional T cells to be dubbed ‘preset’ T cells that are able to colonize tissues early in life and respond rapidly to stimuli.
The second group consists of cells that are unique to the unconventional T cell compartment. These cells exit the thymus in a naive state but acquire effector programs early in life in tissues that express cognate self-ligands. This has been best described for the mouse Vγ7 subset, which acquires a unique effector profile within the first weeks of life once it engages the self-ligand BTNL1/6 in the intestinal epithelium34. In humans this is associated with the acquisition of a unique NK-cell-like program, which endows Vγ4 T cells with specific innate properties35.
The third group fits more in line with classical T cells in which effector programs are acquired in the periphery in response to engagement with cognate foreign antigen. This includes HLA-E- and Qa-1-restricted T cells87,88 and may include subsets of CD1- and MR1-reactive T cells that exhibit diverse TCR usage1,54.
Tissue-specific niches and stability
A defining feature of the majority of unconventional T cells is their intimate relationship and localization within tissues, especially mucosal sites. As was detailed above, for SKINT-1-reactive DETCs33,89 and BTNL-reactive TCRγδ T cell subsets34,35, expression of their selecting ligand determines their enrichment in the skin and in the intestine, respectively. Studies in mice have shown that NKT cells and MAIT cells exhibit enrichment in tissues such as the liver90,91, and in particular for MAIT cells in the skin86. This is presumably due to the enrichment of lipid and metabolite antigens at mucosal sites where bacteria interface with the host.
Space in tissues is limited and as a host ages, exposure to insults leads to increased occupation by adaptive tissue-resident lymphocytes92,93. Therefore, it is unsurprising that the niches that unconventional T cell subsets occupy exhibit temporal restrictions. This has been shown both for MAIT cells in the skin86 and for BTNL1/6-reactive TCRγδ T cells in the intestine34, whereby exposure to cognate ligand within the first weeks of life is required for T cell expansions and the establishment of sizeable tissue-resident niches in mice. Of note, the size of the type I NKT cell niche in the colonic lamina propria is limited by microbial signals such that germ-free mice exhibit expansions of these cells, which can only be normalized by colonization with microbiota early (and not late) in life94.
Another critical question is the stability of the unconventional T cell niche with age and in conditions of inflammation, and whether it requires the ongoing expression of ligands. NKT cells, MAIT cells and BTNL3/8-reactive TCRγδ T cells can all be found in healthy adult tissues35,95,96, which highlights the stability and longevity of these compartments. However, closer inspection in the context of specific acute or chronic perturbations to these niches reveals that in the context of acute viral infection, skin-resident DETCs are locally displaced at the site of infection by conventional tissue-resident CD8 T cells generated against the virus97. Furthermore, in the context of chronic inflammation associated with coeliac disease, loss of BTNL3/8 expression in the small intestine and an expansion of TCRγδ T cells with new specificity and function is associated with a permanent displacement of Vγ4 BTNL3/8-reactive TCRγδ IELs35. Together, these examples illustrate the complexity of lymphocyte–tissue dynamics and how cross-talk and competition between different unconventional T cell subsets ultimately shapes specific niches.
Preset functional niche
The unconventional T cell compartment as a whole covers the full spectrum of T cell effector responses including chemokine-driven immune cell recruitment35,98,99, helper cytokine responses4,100, cytotoxic responses4,35,100,101 and wound healing responses35,49,86,99,102.
It is not so much the nature of the effector function mediated by unconventional T cells that sets them apart from the adaptive T cell compartment, but the nature of the ligands that drive their activation, their effector status at homeostasis and their ability to colonize tissues and respond to insults early in life (Fig. 3). The conventional T cell compartment, by generating antigen-specific ‘adaptive’ memory responses against pathogens, is best suited for sterilizing adaptive immunity, because of both its exquisite specificity and adaptability (covering all classes of microorganisms). However, this capacity is also intrinsically linked to the requirement for a T cell of a given specificity to undergo massive expansion. By contrast, the majority of unconventional T cell subsets exist as pre-expanded populations at steady state8 (Fig. 3), that recognize conserved microbial antigens19,21,22,86, constitutively expressed self-ligands such as BTNL3/834,35 or stress-induced self-ligands such as MICA36,103, which can further facilitate the establishment and maintenance of homeostasis.
Fig. 3 |. Functional niche of unconventional T cells.
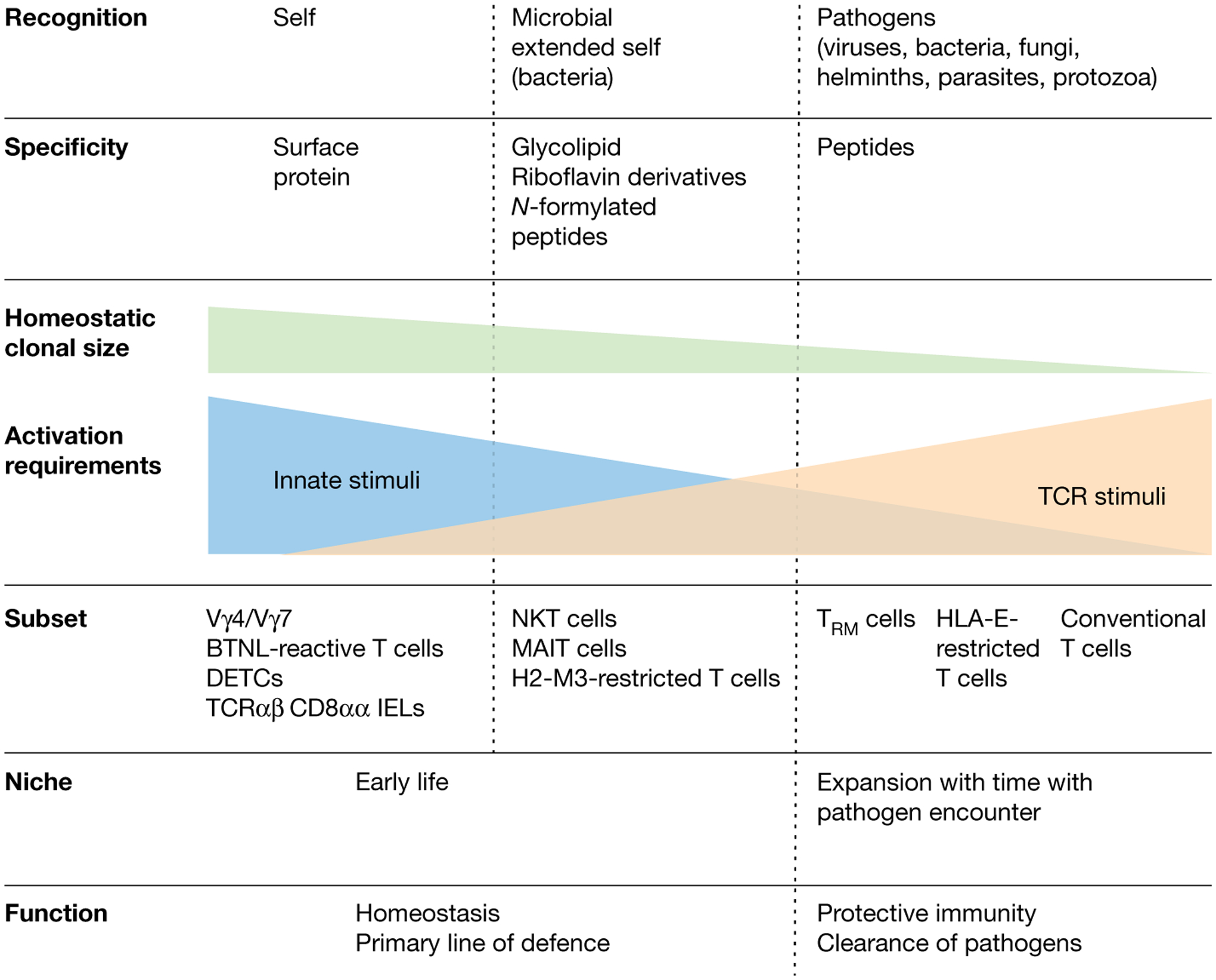
The T cell compartment is shown on a gradient from conventional T cells (right) to unconventional T cell subsets (left) according to the classifiers in bold. Classical adaptive T cells occupy a specific niche in terms of the antigenic universe they recognize (that is, MHC–peptide complexes), and they colonize tissues as tissue-resident memory T cells (TRM) only after being activated and having expanded in peripheral lymph nodes in response to an infection. By contrast, unconventional T cells recognize a broad spectrum of antigens ranging from self-molecules, to microbial extended self and non-self, to formylated peptides and to peptides. The clonal size at homeostasis for unconventional T cell subsets like NKT, MAIT and BTNL-reactive TCRγδ T cells is large, as these cells expand in tissues early in life and in the case of NKT and MAIT cells can occupy multiple tissues. The role of innate immune signals versus TCR-mediated signals varies in the activation of the different unconventional T cell subsets; innate signals have a more critical role in the innate-like unconventional T cells that expand and acquire an effector phenotype either in response to self in the periphery (BTNL-reactive TCRγδ T cells) or in the thymus during their development (MR1-, CD1d- and H2-M3-restricted T cells) than in HLA-E-restricted T cells that become activated in the periphery, similarly to conventional T cells. Of note, tissue-resident memory T cells also acquire the ability to respond to innate signals after establishing residence in tissues and are distinct in that regard from circulating memory and effector memory T cells. Finally, the unconventional T cell compartment constitutes a primary line of defence and also has an important role in tissue homeostasis and healing.
The engagement at steady state by self-ligands and microbial extended-self-ligands has two major consequences. First, unconventional T cells—unlike conventional T cells that only exert effector functions at the time their TCR is engaged by a specific foreign microbial antigen—have the capacity to mediate functions at homeostasis that are important for the initiation and amplification of protective immune responses17,18,104–106, as well as for tissue healing13,49,86,102,107. A role for TCRγδ T cells in wound healing in mice107 and humans102 was proposed early on before the tissue-resident T cell field had gained traction. This has been more recently highlighted in two mouse models of wound healing that showed that wounds heal faster in mice that establish skin-resident commensal-specific MAIT cell86 and H2-M3-restricted T cell responses49 before tissue injury. Second, unconventional T cells are expanded at homeostasis, colonize tissues early in life and have the capacity to respond to innate immune signals; they can therefore have a key role in the protection of tissues against pathogens (Fig. 3). Although tissue-resident conventional CD8 T cells are also critically regulated by innate signals108, the inherent capability of innate-like T lymphocytes to respond to alarmins and stress ligands without requiring strong TCR engagement (Fig. 3) allows them not only to respond rapidly, but also to instruct the adaptive immune system as to the health status of the tissue. This role is certainly critical in the context of viral infections associated with the downregulation of MHC class I molecules109. The prominent role of innate signals alone or in combination with the TCR in the switch to protective functions and the activation of tissue-resident MAIT cells110,111, NKT cells112,113 and TCRγδ T cells114,115 has been demonstrated in mice and humans (Fig. 3). Similarly, in humans, BTNL-reactive TCRγδ T cells35 and Vδ1 TCRγδ T cells present at tumoral sites116 were shown to require IL-15 and/or the engagement of activating NK receptors to exert their full cytolytic potential. Finally, the role for innate-like T lymphocytes in the initiation of adaptive immunity and its amplification is illustrated by the adjuvant role that NKT cells can have by initiating a high-speed communication network between the innate and the adaptive immune system117. Together, the unique development, expansion and restriction of unconventional T cells endows them with the capacity to colonize tissues early in life; form a first line of defence against pathogens before adaptive tissue-resident T cells colonize tissues; and exert important homeostatic, innate, protective and healing functions at tissue sites. Their functional properties and conservation across individuals has also made them an attractive tool for immunotherapy, especially in the treatment of cancer118,119.
Evolution and redundancy
Perspective on evolution
Unconventional T cell immunity is generally thought of as the primitive form of conventional ‘adaptive’ T cell immunity. A common argument presented is that limited gene diversity existed initially in the TCR locus and this limited diversity was best suited for the recognition of a limited set of non-polymorphic MHC-like molecules120. Under this premise, the unconventional T cell compartment was insufficient and thus the adaptive immune system that encompasses conventional MHC-Ia-restricted T cells evolved to support it. This is certainly a plausible scenario, but here we would like to entertain the alternative possibility whereby conventional T cell immunity evolved first and was later supplemented at different moments over the course of evolution by a variety of unconventional T cell compartments. This occurred, we suggest, because the addition of unconventional T cell compartments added both resilience—by providing the means to ensure early-life protective immunity, tissue homeostasis and barrier function—and robustness, by informing and amplifying the adaptive immune response.
The first argument can be made by studying the occurrence of conventional versus unconventional T cell immunity across species. To our knowledge, no species has been studied to date that has evidence of unconventional T cell immunity in the absence of conventional T cell immunity in terms of both ligands and TCRs, meaning that MHC and MHC-like molecules are always found together and semi-invariant TCRs are not found in the absence of diverse TCRs. There are mammalian species that lack MR1 and CD1d and which therefore lack MAIT and NKT cells120–122, and the self-ligand SKINT-1 that selects the TCRγδ T cell subset of DETCs in mice is not well-conserved123. When comparing mice and humans, subsets such as H2-M3-restricted T cells in mice are absent in humans, whose genomes do not contain the H2-M3 gene124, whereas humans have CD1a-, CD1b- and CD1c-restricted T cell subsets that are absent in mice48,65,125. Thus, it is possible that each species evolved or maintained the ligands required for the selection of particular unconventional T cells on the basis of the pressures imparted by their unique lifestyle.
In addition to the evidence provided at the species level, another approach to discussing the origins of unconventional T cell immunity is to consider the nature of the antigens that are recognized. Notably, unconventional T cell subsets such as NKT, MAIT and H2-M3-restricted T cells are all geared towards the recognition of conserved bacterial products, and have a blind spot for the direct recognition of virus-derived antigens. Although there are studies that implicate these subsets in antiviral responses106,126, the evolutionary benefit is more likely to arise from the recognition of universal bacterial products—which in some cases go as far as to guide their development, as in the case of MAIT cells127. Such a recognition could provide the host with the ability to gauge the overall bacterial load at mucosal surfaces and provide broad protective immunity early in life. However, no single unconventional T cell subset is able on its own to ensure effective protective immunity against a variety of pathogens, and adaptive immunity was shown to be required for survival in a pathogen-rich environment128. Indeed, a system favouring the broad recognition of protein-derived peptides would thus have been the more suitable first choice as it would allow the host to respond to all classes of pathogens, including bacteria, viruses, fungi, helminths, parasites and protozoans. In this framework, conventional T cell immunity would have evolved first to ensure a broad coverage of classes of pathogens, before focusing on supplementing mucosal barriers and innate immunity in an effort to maintain the optimal homeostatic relationship with the microbiota.
Framing redundancy within T cells
The majority of studies in animal models that have attempted to demonstrate a critical role for a given unconventional T cell subset in protective immunity have failed or suggested it using contrived systems. An alternative approach to gain insights into the requirement for the unconventional T cell compartment is to take a human genetics approach129,130 and query how constrained deleterious mutations are in genes that encompass the building blocks of all T cell responses (Fig. 4a). In healthy individuals, loss-of-function mutations are found in CD1d, MR1131 and BTNL3/8, but also, strikingly, in classical MHC molecules such as HLA-A and HLA-B (Fig. 4a), suggesting that for both unconventional and conventional T cells there is a substantial degree of redundancy when considering a T cell subset with a given ligand specificity. This observation probably reflects the notion of a layered immune system, whereby the same effector functions can be attained through both unconventional and conventional T cells and therefore the loss of any individual component might be tolerated (Fig. 4b). By contrast, null mutations in ZAP70, encoding a signalling molecule required for the differentiation and activation of all T cell subsets, are very rare (Fig. 4a). Other genes that are intolerant to loss-of-function mutations encode proteins that are shared not only by all T cell subsets but also by non-T cell subsets. This can be observed at the level of cell programming; for example, for transcription factors such as TBX21 that are associated with T cell differentiation, and effector molecules such as interferon-γ (IFNγ), which are relevant for both conventional and unconventional T cells as well as innate lymphoid cells. Notably, for effector molecules involved in type 2 immunity, in which IL-4, IL-5 and IL-13 may be able to compensate for one another, loss-of-function mutations are more common (Fig. 4a). The tissue-repair-associated molecule amphiregulin exhibits a strong selective constraint (Fig. 4a), which is particularly interesting when considering that unconventional T cells may be best suited for wound healing in tissues as described above.
Fig. 4 |. Conservation and redundancy within the T cell compartment.
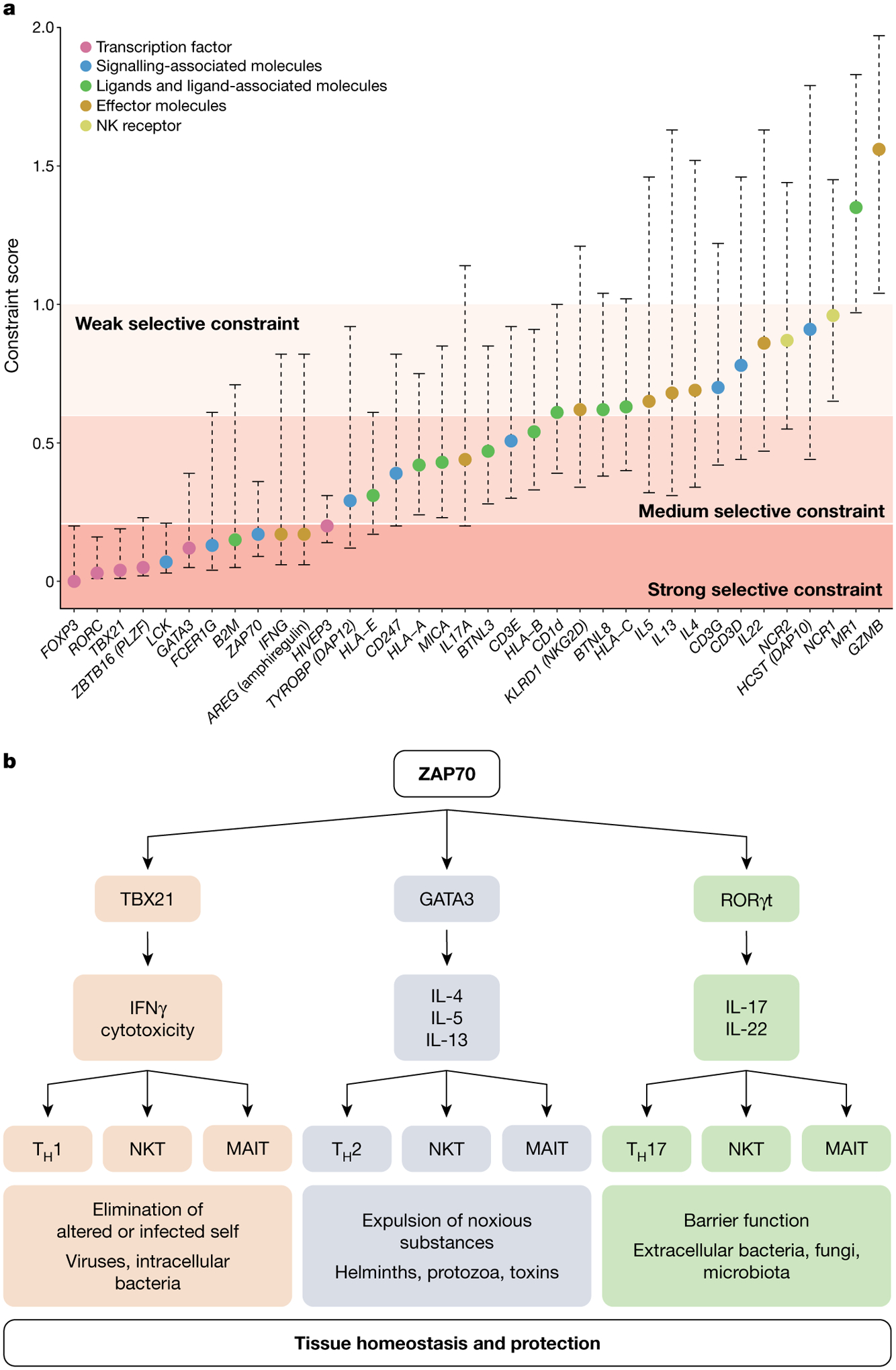
a, The selective constraint score shown (filled circles) is the ratio of the observed versus the expected (o/e) number of loss-of-function variants in that gene in the general population. The o/e metric comes with a 90% confidence interval, which is shown by the dashed lines. When a gene has a low o/e value, it is under stronger selection against loss-of-function mutations than a gene with a higher value. Genes were grouped into five major biological groups on the basis of their function and ranked from the most selectively constrained to the least constrained. The scores were obtained from the Genome Aggregation Database (gnomAD, v.2.1.1) and are based on sequencing data from 25,748 exome sequences and 15,708 whole-genome sequences from unrelated individuals. b, This figure shows that ZAP70 has a central role in the signalling hub of all T cells and, using MAIT and NKT cells as an example of unconventional T cell subsets, illustrates the multifaceted nature of the immune system that has evolved to have multiple conventional and unconventional T cell subsets that mediate the same key effector functions. Although there is redundancy at this functional level, these T cell subsets have different modes of recognition and are regulated by different stimuli, thereby increasing the robustness and resilience of the immune system. This property of the immune system also underlies the difficulty of showing a requirement for any given unconventional T cell subset. TH1, T helper 1 cell; TH2, T helper 2 cell; TH17, T helper 17 cell.
To formally quantify the levels of selective constraint in unconventional T cells, we analysed whether null mutations were allowed for ZBTB16 (also known as PLZF) and HIVEP3, which encode two critical regulators for NKT and MAIT cell differentiation78 and or expansion132; PLZF also has a role in the development of innate lymphoid cells133. Strikingly, in contrast to MR1 and CD1D, there is evidence for a strong selection constraint for PLZF and HIVEP3, suggesting that although NKT and MAIT cells may be dispensable individually, the loss of all PLZF- and HIVEP3-dependent unconventional T cell subsets may not be, as this would result in the loss of a functional niche that recognizes ‘microbial extended self’. Although we cannot formally eliminate that the lack of these transcription factors may have a critical non-immunological biological role, the strong selection constraint seen in humans and the observation that mice can reproduce and survive in the absence of these transcription factors provide evidence that contradict the notion that the unconventional T cell compartment is dispensable. Using approaches that can target multiple unconventional T cells simultaneously would help to ascertain to what extent redundancy exists within the adaptive and unconventional T cell compartments, and at what levels.
An unexpected observation is the constraint on loss-of-function mutations in HLA-E (Fig. 4a). Many viral infections result in the downregulation of MHC class I as a means to subvert conventional T cell responses109. In such settings, the leader peptide classically loaded on HLA-E can be substituted with virus-derived peptides to generate virus-specific T cells14 that provide the immune system with a solution to the riddle of the virus-mediated downregulation of MHC class I. This would be an example of a case in which HLA-E-restricted T cells are not redundant in the presence of conventional T cell immunity.
There is also some value in approaching the discussion of redundancy from the perspective of how many different unconventional T cells can recognize the same pathogen. An example is Mycobacterium tuberculosis, for which a variety of unconventional T cell subsets have been associated with the response, including CD1-restricted T cells45, HLA-E-restricted T cells134, Vγ9/Vδ2 γδ T cells135 and MAIT cells136. Given the broad array of unconventional T cells that are able to sense Mycobacterium tuberculosis—many with overlapping effector functions—it is unsurprising that in isolation each might not be essential for survival. Yet, the synergy between them is likely to provide the host with the best chance of fighting the infection, which is compatible with the high level of conservation of these molecules across species.
In summary, if the full spectrum of subsets were truly redundant it would be unlikely that such a diversity would have been conserved over time137. Moreover, there is an inherent advantage in being able to sample pathogens in many distinct ways and/or to survey and respond to internal changes, especially when considering that pathogens are also constantly evolving to subvert immunity.
Conclusion and future perspectives
Unconventional T cells can engage a broad spectrum of molecules that span peptides, lipid moieties, metabolites and phosphoantigens, and this recognition has been divided up across a multitude of cell subsets.
Many questions remain when thinking about individual unconventional T cell subsets. Why did we evolve so many? Why did mammals develop a T cell subset that can recognize vitamin B2 metabolites in particular, and what other metabolite-sensing T cell subsets exist? Why was the adaptive immune T cell response selected on the basis of peptide recognition and not that of lipids or sugars? Was it because the key driving force was the need for protection against viruses? There is a clear case for unconventional T cells regulating the response to bacteria through the recognition of canonical antigens. Is this system in place to enable tissues to sense alterations in the intestinal or skin microbiota? Is there such a system in place to target fungi, protozoa, parasites and helminths, or to recognize toxins and allergens? Finally, does displacement of unconventional T cell subsets by chronic inflammation contribute to dysfunctional protective and anti-tumoral immune responses in tissues?
Most importantly, we should not think of unconventional T cells as a rudimentary attempt at adaptive immunity. Instead, they offer unique additions to the mosaic that is the immune system. Whereas conventional T cells have a critical role in sterilizing immunity, unconventional T cell subsets are well-suited for local responses, as they seed and mature with tissues from early in life and are more adapted to promote homeostasis and tissue healing. It is also more cost-effective for the host to rely as much as possible on preset defences rather than having to call on de novo conventional T cell responses constantly. Their role may be especially critical early in life, under periods of extended stress such as nutrient deprivation, and during chronic inflammation and ongoing infection to protect from additional insults. It also remains unclear how all the different arms of T cell immunity work together during the course of an infection and in different tissues. Future studies may want to consider looking at common hubs and targeting several unconventional T cell subsets simultaneously, through the deletion of PLZF, for example. Overall, a more systems-based approach that considers redundancy and the kinetics of immune responses, as well as niche formation and stability, will be necessary to truly appreciate the role and complexity of unconventional T cells and their relationship to adaptive immunity. Redundancy, rather than pointing to the trifling nature of these cells, may actually point to their importance in the defence and preservation of tissues.
Acknowledgements
We thank D. Littler for generating Fig. 1; A. Bendelac for sharing his insights on innate-like lymphocytes; D. Guy-Grand for sharing her insights on intraepithelial lymphocytes over many years; and M. Kronenberg and K. Sangani for discussions. This Review was supported by grants to B.J. from the National Institutes of Health (NIH: R01 DK67180 and R01 DK098435) and the Digestive Diseases Research Core Center at the University of Chicago (P30 DK42086); to J.R. from the Australian ARC Laureate Fellowship; and to L.B.B. from NIH: R01 GM134376.
Footnotes
Competing interests The authors declare no competing interests.
References
- 1.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ & Van Kaer L NKT cells: what’s in a name? Nat. Rev. Immunol 4, 231–237 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Lu L, Werneck MBF & Cantor H The immunoregulatory effects of Qa-1. Immunol. Rev 212, 51–59 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Colmone A & Wang C-R H2-M3-restricted T cell response to infection. Microbes Infect 8, 2277–2283 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Bendelac A, Savage PB & Teyton L The biology of NKT cells. Annu. Rev. Immunol 25, 297–336 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Cohen NR, Garg S & Brenner MB Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv. Immunol 102, 1–94 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Cheroutre H, Lambolez F & Mucida D The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol 11, 445–456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H-J & Cantor H Regulation of self-tolerance by Qa-1-restricted CD8+ regulatory T cells. Semin. Immunol 23, 446–452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J & Moody DB The burgeoning family of unconventional T cells. Nat. Immunol 16, 1114–1123 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Mayassi T & Jabri B Human intraepithelial lymphocytes. Mucosal Immunol 11, 1281–1289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogg G, Cerundolo V & McMichael AJ Capturing the antigen landscape: HLA-E, CD1 and MR1. Curr. Opin. Immunol 59, 121–129 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Legoux F, Salou M & Lantz O MAIT cell development and functions: the microbial connection. Immunity 53, 710–723 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Vantourout P & Hayday A Six-of-the-best: unique contributions of γδ T cells to immunology. Nat. Rev. Immunol 13, 88–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen MM, Witherden DA & Havran WL γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat. Rev. Immunol 17, 733–745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jouand N et al. HCMV triggers frequent and persistent UL40-specific unconventional HLA-E-restricted CD8 T-cell responses with potential autologous and allogeneic peptide recognition. PLoS Pathog 14, e1007041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bian Y et al. MHC Ib molecule Qa-1 presents Mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PLoS Pathog 13, e1006384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bendelac A et al. CD1 recognition by mouse NK1+ T lymphocytes. Science 268, 863–865 (1995). [DOI] [PubMed] [Google Scholar]; This paper demonstrates that NKT cells recognize CD1d.
- 17.Mattner J et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434, 525–529 (2005). [DOI] [PubMed] [Google Scholar]; This paper and the following reference show that NKT cells promote defence against bacterial infections by recognizing either self or microbial glycolipids.
- 18.Kinjo Y et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434, 520–525 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Kjer-Nielsen L et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). [DOI] [PubMed] [Google Scholar]; This paper demonstrates that MR1 presents vitamin B metabolites to MAIT cells.
- 20.Corbett AJ et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Shawar SM, Rodgers JR, Cook RG & Rich RR Specialized function of the nonclassical MHC class I molecule Hmt: a specific receptor for N-formylated peptides. Immunol. Res 10, 365–375 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Lenz LL, Dere B & Bevan MJ Identification of an H2-M3-restricted Listeria epitope: implications for antigen presentation by M3. Immunity 5, 63–72 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that the non-classical MHC molecule H2-M3 presents bacterial formylated peptides.
- 23.Tanaka Y et al. Natural and synthetic non-peptide antigens recognized by human gamma delta T cells. Nature 375, 155–158 (1995). [DOI] [PubMed] [Google Scholar]
- 24.de Jong A et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat. Immunol 15, 177–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birkinshaw RW et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat. Immunol 16, 258–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; The crystal structures in this paper show the breaking of the co-recognition paradigm as defined by the requirement for a TCR to simultaneously recognize an antigen and the antigen-presenting molecule.
- 26.Dimova T et al. Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc. Natl Acad. Sci. USA 112, E556–E565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasec P et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Braud V, Jones EY & McMichael A The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol 27, 1164–1169 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Jabri B et al. TCR specificity dictates CD94/NKG2A expression by human CTL. Immunity 17, 487–499 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Braud VM et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Oliveira CC et al. The nonpolymorphic MHC Qa-1b mediates CD8+ T cell surveillance of antigen-processing defects. J. Exp. Med 207, 207–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakaguchi S, Yamaguchi T, Nomura T & Ono M Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Barbee SD et al. Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc. Natl Acad. Sci. USA 108, 3330–3335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies SKINT-1 as the selecting ligand for DETCs.
- 34.Di Marco Barros R et al. Epithelia use butyrophilin-like molecules to shape organ-specific γδ t cell compartments. Cell 167, 203–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies BTNL molecules as the selecting ligands for gut-specific γδ T cells.
- 35.Mayassi T et al. Chronic inflammation permanently reshapes tissue-resident immunity in celiac disease. Cell 176, 967–981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper characterizes the functional and transcriptional program of the gut innate-like human BTNL-specific γδ T cells and shows how chronic inflammation permanently reshapes these niches.
- 36.Groh V, Steinle A, Bauer S & Spies T Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science 279, 1737–1740 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Kong Y et al. The NKG2D ligand ULBP4 binds to TCRγ9/δ2 and induces cytotoxicity to tumor cells through both TCRγδ and NKG2D. Blood 114, 310–317 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Crowley MP, Reich Z, Mavaddat N, Altman JD & Chien Y The recognition of the nonclassical major histocompatibility complex (MHC) class I molecule, T10, by the γδ T cell, G8. J. Exp. Med 185, 1223–1230 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin S et al. Antigen recognition determinants of γδ T cell receptors. Science 308, 252–255 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Lantz O & Bendelac A An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J. Exp. Med 180, 1097–1106 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawano T et al. CD1d-restricted and TCR-mediated activation of vα14 NKT cells by glycosylceramides. Science 278, 1626–1629 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Tilloy F et al. An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. J. Exp. Med 189, 1907–1921 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porcelli S, Yockey CE, Brenner MB & Balk SP Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4−8− α/β T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J. Exp. Med 178, 1–16 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treiner E et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Van Rhijn I et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat. Immunol 14, 706–713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gras S et al. T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat. Commun 7, 13257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melandri D et al. The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat. Immunol 19, 1352–1365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castro CD, Luoma AM & Adams EJ Coevolution of T-cell receptors with MHC and non-MHC ligands. Immunol. Rev 267, 30–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linehan JL et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates the role of H2-M3-restricted T cell responses in regulating the microbiota and wound healing in the skin.
- 50.Grant EJ et al. The unconventional role of HLA-E: the road less traveled. Mol. Immunol 120, 101–112 (2020). [DOI] [PubMed] [Google Scholar]
- 51.McDonald BD, Bunker JJ, Ishizuka IE, Jabri B & Bendelac A Elevated T cell receptor signaling identifies a thymic precursor to the TCRαβ+CD4−CD8β− intraepithelial lymphocyte lineage. Immunity 41, 219–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayans S et al. αβT cell receptors expressed by CD4−CD8αβ− intraepithelial T cells drive their fate into a unique lineage with unusual MHC reactivities. Immunity 41, 207–218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park SH et al. Selection and expansion of CD8α/α1 T cell receptor α/β1 intestinal intraepithelial lymphocytes in the absence of both classical major histocompatibility complex class I and nonclassical CD1 molecules. J. Exp. Med 190, 885–890 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gherardin NA, McCluskey J, Rossjohn J & Godfrey DI The diverse family of MR1-restricted T cells. J. Immunol 201, 2862–2871 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Uldrich AP et al. CD1d-lipid antigen recognition by the γδ TCR. Nat. Immunol 14, 1137–1145 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Luoma AM et al. Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells. Immunity 39, 1032–1042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Nours J et al. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science 366, 1522–1527 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Garcia KC et al. An αβ T cell receptor structure at 2.5 A and its orientation in the TCR–MHC complex. Science 274, 209–219 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Garboczi DN et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996). [DOI] [PubMed] [Google Scholar]
- 60.Rossjohn J et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Beringer DX et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat. Immunol 16, 1153–1161 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Gras S et al. Reversed T cell receptor docking on a major histocompatibility class I complex limits involvement in the immune response. Immunity 45, 749–760 (2016). [DOI] [PubMed] [Google Scholar]
- 63.La Gruta NL, Gras S, Daley SR, Thomas PG & Rossjohn J Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol 18, 467–478 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Adams EJ & Luoma AM The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol 31, 529–561 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Van Rhijn I, Godfrey DI, Rossjohn J & Moody DB Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol 15, 643–654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saunders PM et al. A bird’s eye view of NK cell receptor interactions with their MHC class I ligands. Immunol. Rev 267, 148–166 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Hoare HL et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol 7, 256–264 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Borg NA et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 (2007). [DOI] [PubMed] [Google Scholar]; This paper provides the first insights into how unconventional TCR recognition differs from that of conventional TCRs in the form of type I NKT TCR–CD1d–lipid complex.
- 69.Rossjohn J, Pellicci DG, Patel O, Gapin L & Godfrey DI Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol 12, 845–857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel O et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun 4, 2142 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Eckle SBG et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med 211, 1585–1600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patel O et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat. Immunol 13, 857–863 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Girardi E et al. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat. Immunol 13, 851–856 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gherardin NA et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 44, 32–45 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Wun KS et al. T cell autoreactivity directed toward CD1c itself rather than toward carried self lipids. Nat. Immunol 19, 397–406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bendelac A Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med 182, 2091–2096 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seach N et al. Double-positive thymocytes select mucosal-associated invariant T cells. J. Immunol 191, 6002–6009 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Savage AK et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates the critical role of PLZF in the effector programming of unconventional T cells such as NKT cells and MAIT cells.
- 79.Koay H-F et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol 4, eaay6039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Griewank K et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27, 751–762 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Urdahl KB, Sun JC & Bevan MJ Positive selection of MHC class Ib-restricted CD8+ T cells on hematopoietic cells. Nat. Immunol 3, 772–779 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bediako Y et al. SAP is required for the development of innate phenotype in H2-M3-restricted CD8+ T cells. J. Immunol 189, 4787–4796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDonald BD, Jabri B & Bendelac A Diverse developmental pathways of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol 18, 514–525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei DG et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J. Exp. Med 202, 239–248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guy-Grand D et al. Origin, trafficking, and intraepithelial fate of gut-tropic T cells. J. Exp. Med 210, 1839–1854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Constantinides MG et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that there is a window in early life for the colonization of tissues by MAIT cells and stresses the role of MAIT cells in wound repair.
- 87.Mazzarino P et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur. J. Immunol 35, 3240–3247 (2005). [DOI] [PubMed] [Google Scholar]
- 88.Doorduijn EM et al. T cells engaging the conserved MHC class Ib molecule Qa-1b with TAP-independent peptides are semi-invariant lymphocytes. Front. Immunol 9, 60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Havran WL & Allison JP Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature 344, 68–70 (1990). [DOI] [PubMed] [Google Scholar]
- 90.Benlagha K, Weiss A, Beavis A, Teyton L & Bendelac A In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med 191, 1895–1904 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dusseaux M et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011). [DOI] [PubMed] [Google Scholar]
- 92.Mueller SN & Mackay LK Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol 16, 79–89 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Masopust D & Soerens AG Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol 37, 521–546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olszak T et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chan AC et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4+ and CD4− subsets. Clin. Exp. Immunol 172, 129–137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Loh L et al. Human mucosal-associated invariant T cells in older individuals display expanded TCRαβ clonotypes with potent antimicrobial responses. J. Immunol 1950, 1119–1133 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Zaid A et al. Persistence of skin-resident memory T cells within an epidermal niche. Proc. Natl Acad. Sci. USA 111, 5307–5312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hayday A, Theodoridis E, Ramsburg E & Shires J Intraepithelial lymphocytes: exploring the Third Way in immunology. Nat. Immunol 2, 997–1003 (2001). [DOI] [PubMed] [Google Scholar]
- 99.Hinks TSC et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep 28, 3249–3262.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Legoux F, Salou M & Lantz O Unconventional or preset αβ T cells: evolutionarily conserved tissue-resident T cells recognizing nonpeptidic ligands. Annu. Rev. Cell Dev. Biol 33, 511–535 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Guy-Grand D, Cuénod-Jabri B, Malassis-Seris M, Selz F & Vassalli P Complexity of the mouse gut T cell immune system: identification of two distinct natural killer T cell intraepithelial lineages. Eur. J. Immunol 26, 2248–2256 (1996). [DOI] [PubMed] [Google Scholar]; This paper provides the first evidence that intestinal tissue-resident unconventional intraepithelial T lymphocytes express NK receptors and mediate NK-cell-like killing.
- 102.Toulon A et al. A role for human skin-resident T cells in wound healing. J. Exp. Med 206, 743–750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Groh V et al. Broad tumor-associated expression and recognition by tumor-derived γδ T cells of MICA and MICB. Proc. Natl Acad. Sci. USA 96, 6879–6884 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pamer EG, Bevan MJ & Lindahl KF Do nonclassical, class Ib MHC molecules present bacterial antigens to T cells? Trends Microbiol 1, 35–38 (1993). [DOI] [PubMed] [Google Scholar]
- 105.Le Bourhis L et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol 11, 701–708 (2010). [DOI] [PubMed] [Google Scholar]
- 106.Gaya M et al. Initiation of antiviral B cell immunity relies on innate signals from spatially positioned NKT cells. Cell 172, 517–533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boismenu R & Havran WL Modulation of epithelial cell growth by intraepithelial γδ T cells. Science 266, 1253–1255 (1994). [DOI] [PubMed] [Google Scholar]; This paper was the first study to propose that TCRγδ T cells have a role in wound healing.
- 108.Jabri B & Abadie V IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol 15, 771–783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tortorella D, Gewurz BE, Furman MH, Schust DJ & Ploegh HL Viral subversion of the immune system. Annu. Rev. Immunol 18, 861–926 (2000). [DOI] [PubMed] [Google Scholar]
- 110.Turtle CJ et al. Innate signals overcome acquired TCR signaling pathway regulation and govern the fate of human CD161hi CD8α+ semi-invariant T cells. Blood 118, 2752–2762 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Slichter CK et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight 1, e86292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wesley JD, Tessmer MS, Chaukos D & Brossay L NK cell-like behavior of Vα14i NK T cells during MCMV infection. PLoS Pathog 4, e1000106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tyznik AJ et al. Cutting edge: the mechanism of invariant NKT cell responses to viral danger signals. J. Immunol 181, 4452–4456 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Strid J et al. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nat. Immunol 9, 146–154 (2008). [DOI] [PubMed] [Google Scholar]
- 115.Strid J, Sobolev O, Zafirova B, Polic B & Hayday A The intraepithelial T cell response to NKG2D-ligands links lymphoid stress surveillance to atopy. Science 334, 1293–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu Y et al. An innate-like Vδ1+ γδ T cell compartment in the human breast is associated with remission in triple-negative breast cancer. Sci. Transl. Med 11, eaax9364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carnaud C et al. Cutting edge: cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J. Immunol 163, 4647–4650 (1999). [PubMed] [Google Scholar]
- 118.Godfrey DI, Le Nours J, Andrews DM, Uldrich AP & Rossjohn J Unconventional T cell targets for cancer immunotherapy. Immunity 48, 453–473 (2018). [DOI] [PubMed] [Google Scholar]
- 119.Crowther MD et al. Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol 21, 178–185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Edholm E-S, Banach M & Robert J Evolution of innate-like T cells and their selection by MHC class I-like molecules. Immunogenetics 68, 525–536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mondot S, Boudinot P & Lantz O MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics 68, 537–548 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Boudinot P et al. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc. Natl Acad. Sci. USA 113, E2983–E2992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Flajnik MF & Kasahara M Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet 11, 47–59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rodgers JR & Cook RG MHC class Ib molecules bridge innate and acquired immunity. Nat. Rev. Immunol 5, 459–471 (2005). [DOI] [PubMed] [Google Scholar]
- 125.Van Rhijn I, Ly D & Moody DB CD1a, CD1b, and CD1c in immunity against mycobacteria. Adv. Exp. Med. Biol 783, 181–197 (2013). [DOI] [PubMed] [Google Scholar]
- 126.van Wilgenburg B et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat. Commun 9, 4706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Legoux F et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366, 494–499 (2019). [DOI] [PubMed] [Google Scholar]
- 128.Fischer A Severe combined immunodeficiencies. Immunodefic. Rev 3, 83–100 (1992). [PubMed] [Google Scholar]
- 129.Barreiro LB et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet 5, e1000562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barreiro LB & Quintana-Murci L From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet 11, 17–30 (2010). [DOI] [PubMed] [Google Scholar]
- 131.Howson LJ et al. Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1. Sci. Immunol 5, eabc9492 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Harsha Krovi S et al. Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat. Commun 11, 6238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Constantinides MG, McDonald BD, Verhoef PA & Bendelac A A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Heinzel AS et al. HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med 196, 1473–1481 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kabelitz D et al. The primary response of human γ/δ+ T cells to Mycobacterium tuberculosis is restricted to Vγ9-bearing cells. J. Exp. Med 173, 1331–1338 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gold MC et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol 8, e1000407 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nish S & Medzhitov R Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity 34, 629–636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Borbulevych OY, Santhanagopolan SM, Hossain M & Baker BM TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J. Immunol 187, 2453–2463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wun KS et al. Human and mouse type I natural killer T cell antigen receptors exhibit different fine specificities for CD1d-antigen complex. J. Biol. Chem 287, 39139–39148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]


