Abstract
Constrained, membrane-permeable peptides offer the possibility of engaging challenging intracellular targets. Structure-permeability relationships have been extensively studied in cyclic peptides whose backbones are cyclized from head to tail, like the membrane permeable and orally bioavailable natural product cyclosporine A. In contrast, the physicochemical properties of lariat peptides, which are cyclized from one of the termini onto a side chain, have received little attention. Many lariat peptide natural products exhibit interesting biological activities, and some, such as griselimycin and didemnin B, are membrane permeable and have intracellular targets. To investigate the structure-permeability relationships in the chemical space exemplified by these natural products, we generated a library of scaffolds using stable isotopes to encode stereochemistry and determined the passive membrane permeability of over 1000 novel lariat peptide scaffolds with molecular weights around 1000. Many lariats were surprisingly permeable, comparable to many known orally bioavailable drugs. Passive permeability was strongly dependent on N-methylation, stereochemistry, and ring topology. A variety of structure-permeability trends were observed including a relationship between alternating stereochemistry and high permeability, as well as a set of highly permeable consensus sequences. For the first time, robust structure-permeability relationships are established in synthetic lariat peptides exceeding 1000 compounds.
Keywords: Cyclic peptide, lariat, permeability, mass-encoded library
INTRODUCTION
Targeting protein-protein interactions (PPIs) with small molecules remains challenging. Typical drug-like small molecules are too small to bind large protein interfaces, while larger molecules suffer from poor membrane permeability and are thus have limited access to intracellular targets. Membrane permeable cyclic peptides hold great promise for targeting intracellular PPIs; the tendency of macrocycles to adopt disk and sphere-like conformations enables them to bind flat binding sites,1–2 and a growing number of cyclic peptides are surprisingly membrane permeable despite molecular weights well above 600.3–8 Cyclic peptide natural products such as the well-known 11-mer cyclosporine A (CSA) have inspired a variety of model systems designed to probe the factors that govern membrane permeability in larger, peptidic molecules.3–8 Although the majority of these systems are, like CSA, cyclized from head to tail, a survey of the Natural Products Atlas9 revealed that roughly 30% of cyclic peptide natural products are cyclized between a terminal residue and a side chain (Figure 1b). The majority of these “lariat” natural products are cyclized from the C-terminal carboxylic acid onto a side-chain hydroxyl group to form an ester (i.e., depsipeptide) linkage.
Figure 1.
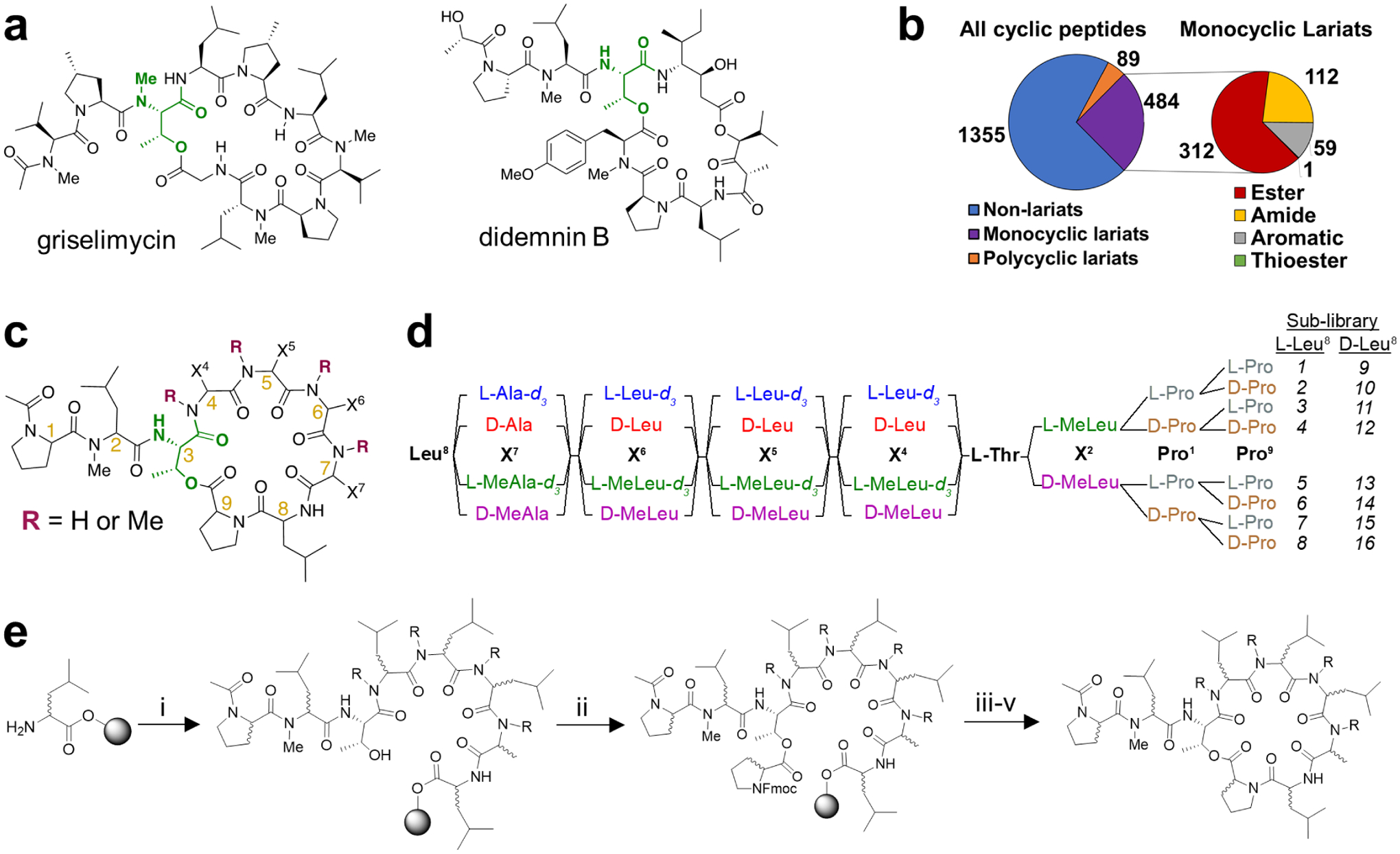
(a) Natural products inspiring the library design. (b) The portion of natural product cyclic peptides from the Natural Products Atlas comprised of lariat peptides and the portion of lariat peptides comprised of ester-cyclized lariats. (c) Design of Library 1. (d) Schematic of the split-pool library synthesis. (e) Synthesis of Library 1: (i) solid-phase peptide synthesis; (ii). Fmoc-Pro-OH, DIC, DMAP; (iii) piperidine, DBU; (iv) HFIP; (v) COMU, DIPEA.
Lariat depsipeptide natural products have a variety of ring sizes and tail lengths, and many contain non-peptidic polyketide elements in their backbones. Nonpolar members of this class are known to inhibit a variety of intracellular molecular targets. Griselimycin (Figure 1a), for example, is a lariat depsipeptide with an 8-residue macrocycle and 2-residue tail which potently inhibits the polymerase sliding clamp DnaN of Mycobacterium tuberculosis. The crystal structure of griselimycin bound to DnaN shows the cyclic portion of the molecule bound in one subsite with the lariat tail extended to engage a separate subsite.10 An optimized analogue of grislemycin exhibited an oral bioavailabity in mice of 89%.10 Didemnin B (Figure 1a) is a potent inhibitor of the eukaryotic translation elongation factor eEF-1A,11 and an analogue of didemnin B, plitidepsin (Aplidin), has been approved for multiple myeloma in Australia12 and is currently in clinical trials against the novel coronavirus SARS-CoV-2.13 Both griselimyicin and didemnin B have molecular weights of over 1000 Da, and although 3D structural data exist on both of griselimycin and didemnin B,11, 14 the relationship between structure and membrane permeability has not been established for either compound.
In this investigation we set out to determine the landscape of membrane permeability in structurally diverse lariat depsipeptides inspired by natural products, and to study relationships between structure and permeability for lariats with extensive variation in backbone N-methylation and stereochemistry. Previously, we generated split-pool libraries that permuted backbone elements to explore permeability in cyclic peptide scaffolds, using deconvolution by resynthesis to identify specific scaffolds of interest.15–16 Here we describe the synthesis and permeability data on a library of 4096 lariat peptides, in which stereochemistry was encoded using isotopic labels and tandem mass spectrometry (MS) used to sequence over 1000 of the library members. This library sequencing method was developed by Townsend et al.17 The data reveal specific patterns of stereochemistry and N-methylation that facilitate permeability and highlight the impact of the flexible tail on permeability across a wide range of loop geometries.
RESULTS AND DISCUSSION
Library Design
The design of Library 1 was inspired by the structures of griselimycin and didemnin B, which feature a linkage between the C-terminus and an internal Thr residue, and an N-acylated, two-residue tail (Figure 1c). Orally bioavailable compounds exceeding MW 1000 are quite rare.18 Library 1 samples a chemical space extending slightly beyond this limit. Lipophilicity was kept within a range characteristic of orally bioavailable macrocycles, while the number of hydrogen bond donors was varied.19 The library was designed to sample diverse lariat peptide backbone geometries by permuting stereochemistry as well as the number and pattern of amide N-methyl groups within the macrocycle. For Library 1, we included only simple aliphatic side chains to focus specifically on the effect of backbone geometry on membrane permeability. All compounds with the same number of N-methyl groups have the same molecular weight and predicted lipophilicities (as captured by the calculated octanol-water partition coefficient AlogP) (Table S2); therefore, variation in permeability within an isomeric series must be the result of conformational differences associated with stereochemistry and N-methyl position. Leucine was selected as the predominant amino acid due to its prevalence among passively permeable natural products, including lariats, with a single Ala residue included to adjust lipophilicity to a range optimal for permeability.16, 20 Two Pro residues were included, one in the tail and one in the macrocycle, similar to didemnin B and griselimycin. The average molecular weight of the library was 1016, slightly smaller than griselimycin and didemnin B but significantly larger than model systems previously reported in studies of passive permeability.
The 4096-member library was synthesized as 16 sub-libraries, each theoretically containing a mixture of 256 members (Figure 1d). The number of compounds in each sub-library was limited by the necessity of chromatographic separation of compounds with the same parent mass during analysis as required for optimal MS2-based sequencing using CycLS, a program developed previously in our lab.21 The stereochemistry of residues 1, 2, 8, and 9 are specific to each sub-library. The stereochemistry of residues 4–7 were encoded using stable isotope labelling to allow for identification by LC-MS2. The number of N-methyl groups in Library 1 ranges from one to five and the number of hydrogen-bond donors from two to six (Table S2). The N-methyl group on the lariat tail is constant for all library members while the degree of N-methylation among the mass-encoded residues ranges from zero to four.
Synthetic strategy
The synthesis of Library 1 was devised to avoid potential challenges associated with lariat peptides (Figure 1e). Cyclization efficiency often determines the overall efficiency of cyclic peptide synthesis; therefore, we avoided macrolactonization as the cyclization step and instead opted to form the ester linkage on-resin during the linear synthesis. However, since continuing peptide synthesis on the Thr hydroxyl group could lead to diketopiperazine formation after attachment and deprotection of the second residue, we chose the Pro9-Leu8 connection as the site for cyclization. This strategy has been successful in several lariat depsipeptide natural product syntheses.22–24 Leu8 was not N-methylated to prevent premature cleavage from the 2-chlorotrityl resin caused by diketopiperazine formation.25 The Thr hydroxyl was not protected during the synthesis.22–23 Couplings with N-methylated amino acids are known to be challenging. We chose HATU as the coupling agent for library synthesis due to its established use in the coupling of N-methylated amino acids.26 For the formation of the ester linkage, we adapted conditions applied previously in the total synthesis of theonellapeptolide 1d.22
Overall permeability of the library
Permeability was acquired using the parallel artificial membrane permeability assay (PAMPA), a high-throughput method which correlates well to cell-based passive permeability methods and even oral absorption.27 Permeabilities among library members varied widely, ranging from below 0.01 × 10−6 to over 10 × 10−6 cm/s. To benchmark the permeability of the lariat peptides, 1NMe3 (Figure S1), a membrane permeable cyclic hexapeptide developed previously,28 was added to each sub-library. The average permeability of 1NMe3 in sub-libraries 1–16 was 8.1 × 10−6 cm/s (Table S5). In total, 29 lariats in Library 1 exceeded this permeability and may be considered highly permeable.
Effect of N-methylation
On average, compounds with more N-Me groups were more permeable (Figure 2b), as previously reported in a variety of head-to-tail cyclized peptides.6, 15, 28–29 Most of the compounds with no N-Me groups in the macrocycle had negligible permeability. Although the most permeable compounds with only one N-Me group in the macrocycle were among the most permeable in the library, the proportion of impermeable compounds and the variance of logPapp decreased with increasing degree of N-methylation (Table S6).
Figure 2.
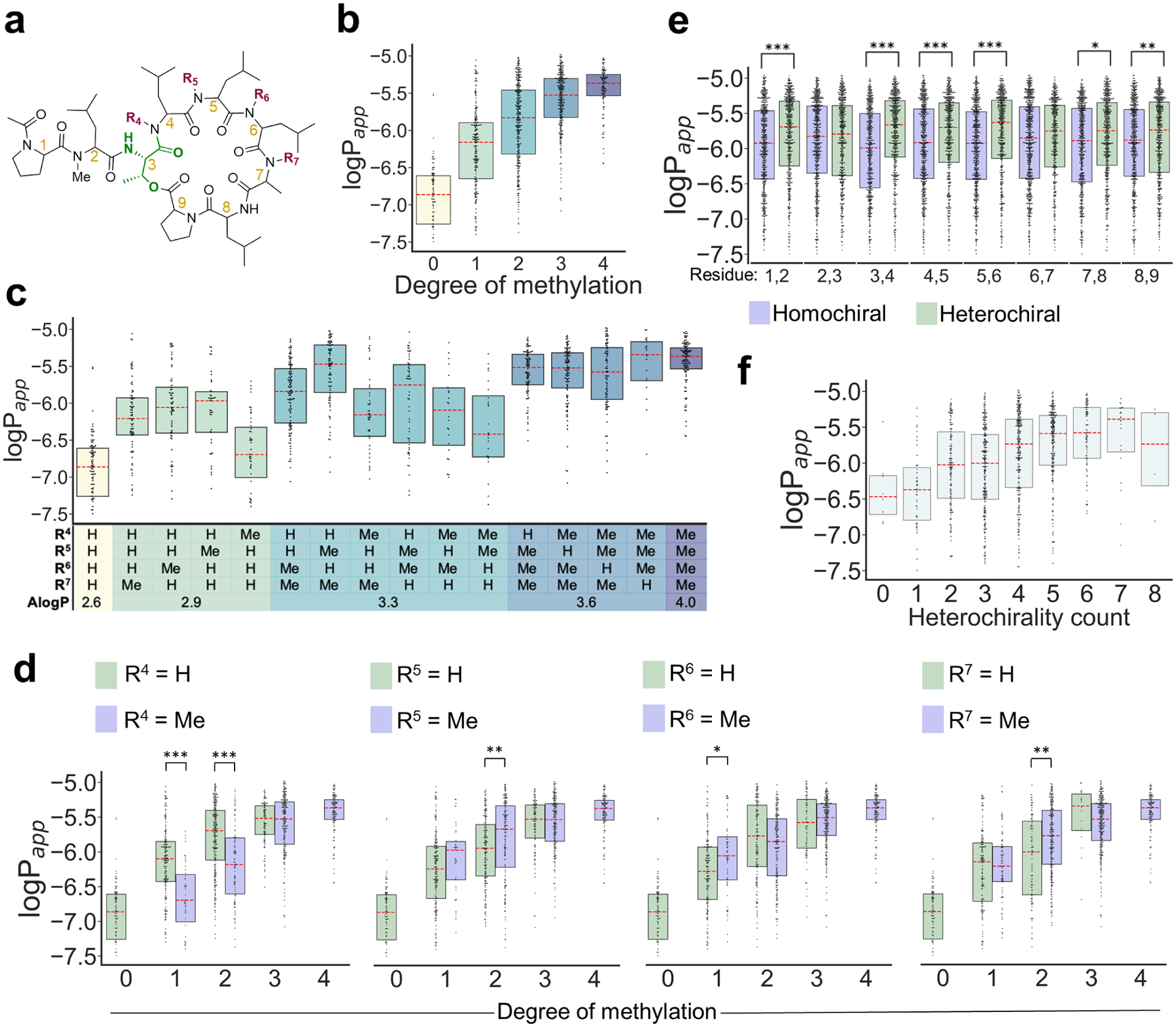
(a) Skeletal structure of Library 1. (b) Effect of number of N-methyl groups on permeability. (c) Effect of N-methyl position on permeability. (d) Effect of methylation on permeability for each variable position. (e) Higher number of heterochiral relationships between adjacent residues is associated with higher permeability. (f) Effect of relative stereochemistry between adjacent residues on permeability. The red dashed lines represent medians and the boxes represent quartiles. Statistics are as follows: Mann-Whitney U test; ***P < 0.0001, **P < 0.001, *P < 0.01.
To identify features that could be associated with increased permeability when comparing isomeric compounds, we investigated the relationship between the pattern of N-methylation in the macrocycle and permeability. The positions of the N-methyl groups had a marked effect on permeability (Figure 2c–d). Lariats with a single N-methyl group in the macrocycle at R5, R6, or R7 had higher permeability than unmethylated lariats, while N-methylation at R4 did not significantly increase permeability (Figure 2c–d). Similarly, for compounds with two N-methyl groups in the macrocycle, those with N-methylation at R4 were less permeable than those with the N-methyl groups elsewhere in the ring (Figure 2d). No compounds with N-methylation at Leu4 appear among the 25 most permeable compounds that contain a single macrocyclic N-methyl group. Therefore, although both the number and position of N-methyl groups give rise to strong permeability trends within the library, the large variation in permeability among closely related sequences highlights the subtle dependence of conformation (and therefore, permeability) on backbone geometry.
Effect of stereochemistry
While the degree and position of N-methylation had a profound effect on permeability, we also observed strong stereochemical effects. We found that the number of heterochiral residue junctions, defined as adjacent residues having opposite stereochemical configurations, correlated strongly with permeability (Figure 2f). Most adjacent residue pairs in the macrocycle contributed to this effect, except for MeLeu2/Thr3 and Leu6/Ala7 (Figure 2e).
The general association between heterochirality and permeability occurs for 1–3 degrees of N-methylation but appears strongest for lariats with two N-methyl groups in the macrocycle (Figure S6). The effect appears absent 4 degrees of N-methylation, although the low representation of compounds with 4 degrees of N-methylation and low heterochirality may obscure the effect. For each pair of adjacent stereocenters, the effect of heterochirality is mostly consistent for each degree of N-methylation (Table S13). The stereochemistry at Leu8 had a clear effect on permeability (Figure S7), with higher permeability observed for compounds featuring L stereochemistry at Leu8. This increase in permeability only occurred when either Ala7 or Pro9 had D stereochemistry (Figure S8) and was highest when both Ala7 and Pro9 had D stereochemistry (Figure S9). The impact of individual stereocenters on permeability was generally consistent for all degrees of N-methylation and does not appear related to variation in representation among the different classes (Tables S11 and S12). Even in the context of extensive variation in both stereochemistry and N-methylation, certain stereochemical patterns favor permeability, suggesting that some structural elements are consistently instrumental in producing favorable membrane-associated conformations.
Permeability validation
To validate the permeability results from this library, compounds were individually synthesized and evaluated in PAMPA (Figure 3, Table S25). Compounds were selected at random from the entire dataset and represented a range of permeabilities and N-methyl group counts. Of 11 resynthesized compounds, 9 had retention times matching the corresponding library member, indicating that two of the 11 compounds were mis-sequenced. The permeability trends were reproduced overall, although Papp values were roughly four times higher in the library. This effect is most likely a result of the higher total peptide concentration when assaying the library versus individual compounds. In the case of individual compounds, adhesion of compound to the walls of the PAMPA plate and dissolution in the lipid layer may have disproportionately affected the results, whereas in the library these sinks were saturated. We found that the average recovery in the library was 67% while average recovery for the nine individual compounds was 42%, providing some support for this hypothesis. We also assayed these compounds in the MDCK cell-based assay (Figure 3). The MDCK assay results confirm the trend in permeabilities. Interestingly, the Papp values obtained in the MDCK assay are generally higher than those obtained in the library PAMPA, in contrast to the Papp values from the individual PAMPA.
Figure 3.
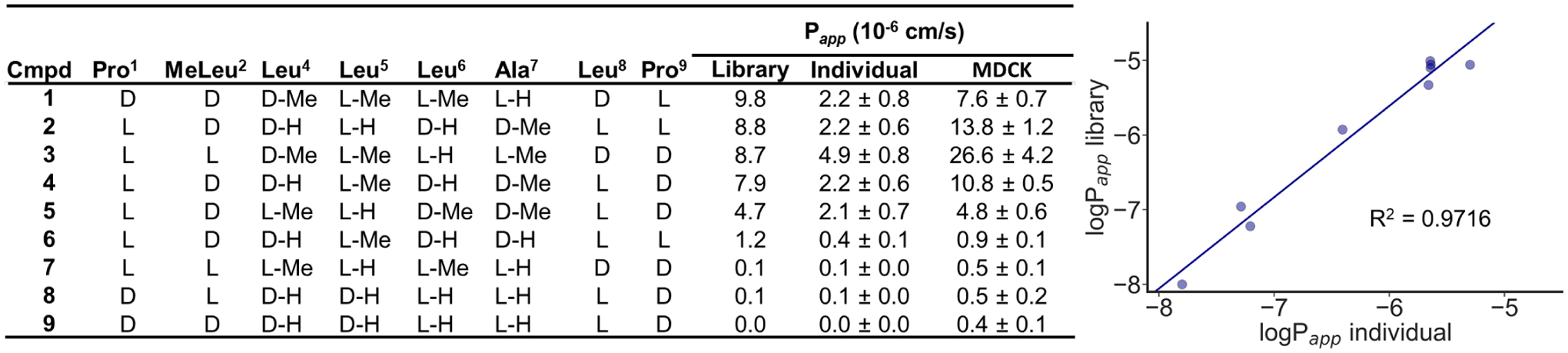
Resynthesized compound permeability. These compounds were individually synthesized and tested for PAMPA permeabilities. Permeabilities of compounds assayed individually in PAMPA are plotted against permeabilities from the library. The relationship is linear (R2 = 0.9716).
Sequencing and Representation
The final Library 1 dataset included 1099 unique entries that were sequenced with high confidence using a conservative scoring function based on the original application of CycLS in the identification of cyclic hexa- and heptapeptides. Although the library design describes a 4096-member library, the final data set contains 27% of the theoretical number. This prompted us to determine to what extent the observed structure-property trends were influenced by differences in representation within the dataset among different stereochemical and N-methylation patterns. We examined representation in the library with respect to all the features from which we derived structure-permeability relationships including number of N-methyl groups, N-methylation pattern, and stereochemistry at individual stereocenters and between adjacent residues.
Chromatographic overlap of compounds with the same m/z impeded data recovery and decreased the representation of lariats in the dataset (Table S3, selected ion chromatograms are provided in the SI). Thus, higher heterochirality (specifically among the mass-encoded positions 4–7) and intermediate numbers of N-methyl groups were associated with decreased representation due to increased mass redundancy (Figure S2, Table S14). A notable exception occurs for compounds with 4 N-methyl groups in the macrocycle. Here, the sequences with no heterochirality between positions 4–7 were poorly represented in the data. This is most likely due to low synthetic efficiency. Surprisingly, the dataset is biased in favor of lariats with higher degrees of N-methylation (Table S4), despite the presumed decreased synthetic efficiency of incorporating N-methylated residues. This is explained by differences in average retention time and retention time variance between each degree of N-methylation (Table S19). Retention times vary more for higher levels of N-methylation, resulting in reduced peak overlap. Thus, disparities in representation are related to the chromatographic gradient employed. The gradient was designed to place the bulk of the peaks in the middle of the gradient while ensuring the most lipophilic lariats eluted within the run (conditions provided in Table S1). Given the known relationship between reversed-phase retention time and permeability, the data could be biased towards exclusion of less permeable compounds, although the library chromatograms show the highest peak density towards the middle of the run. The number of compounds in the dataset with each N-methylation pattern varied widely (Table S7) as did the number of compounds identified from each sub-library (Table S8). However, average permeability did not correlate with the number of compounds with a given N-methylation pattern or in each sub-library (Figure S3).
We also examined the effect of differences in representation of stereochemical features on permeability trends. Overall, we did not find a meaningful correlation between representation of heterochiral compounds relative to homochiral compounds and permeability for any of the degrees of N-methylation (Figure S4). Considering the known effect of N-methylation on permeability, we assessed the effects of stereochemistry on permeability separately for each degree of N-methylation (Tables S11 and S13). We noted four representation disparities (exceeding 20% difference), affecting Ala7, Pro9, and relative stereochemistry at Leu5/Leu6, and Leu6/Ala7 (Tables S9, S10, S12, S14). The overrepresentation of L-Ala7 in the library is strongest at lower numbers of N-methyl groups, implying that our observation of higher permeability for D-Ala7 may be an artifact of representation bias (Table S12). The particularly large disparity in representation for Ala7 stereochemistry combined with that for Leu6/Ala7 prompted us to examine the effect of disparate representation of individually impactful stereocenters (Leu4, Ala7, and Leu8) on the heterochirality results for adjacent residue pairs that contain those stereocenters. For Leu6/Ala7, Ala7/Leu8, and Leu8/Pro9, the more permeable of the two heterochiral configurations was underrepresented (Tables S15 and S16). This pattern did not occur for the homochiral configurations. If the permeability trends observed here are consistent with the theoretical library space, then this disparity will cause the permeability enhancement associated with heterochirality to be artificially suppressed. To some degree, this would account for the weaker heterochirality effect observed for these three pairs compared to the other three pairs in the cyclic portion of the lariat structure. Moreover, this disparity could significantly skew the average permeability of the entire library downwards. Alternatively, the underrepresentation of the most permeable diastereomers could reflect an absence of the impermeable library members bearing those diastereomers. This appears to be the case for Leu6/Ala7 and Ala7/Leu8, in which the higher degrees of N-methylation were overrepresented for the most permeable diastereomer, skewing the perceived permeability of the affected diastereomers upwards (Table S17). To better understand the extent of this effect, we determined the permeability of all stereochemistries for Leu6/Ala7, Ala7/Leu8, and Leu8/Pro9. The effect of stereochemistry on permeability is consistent for each degree of N-methylation for Ala7/Leu8 and Leu8/Pro9, while the high permeability of L-Leu6/D-Ala7 only occurred for 1 and 2 degrees of N-methylation (Table S18). We conclude that the structure-permeability relationships discovered in Library 1 remain valid in spite of differences in representation among structural sub-classes.
The occurrence of mis-sequenced entries in the data set is a potential source of error. Of 11 compounds resynthesized individually, two (18%) were mis-sequenced. This is similar to our previous study, in which the mis-sequencing rate was 23%.21 At worst, patterns of mis-sequencing specifically affecting permeable compounds could result in false conclusions regarding the effect of molecular features on membrane permeability. However, in the absence of evidence to the contrary, mis-sequencing most likely adds noise to the data, weakening observed structure-permeability relationships. Thus, the observed structure-permeability relationships likely emerged in spite of mis-sequencing rather than because of it. Future efforts will be directed toward determining the extent of systematic bias resulting from mis-sequencing, using a much larger set of individually synthesized compounds.
Consensus stereochemistry of highly permeable compounds
Polar contacts between macrocycles and their targets are dominated by interactions involving the amide backbone of the macrocycle.1 Therefore, we were interested in further investigating the factors associated with high permeability among compounds with fewer N-methyl groups. Stereochemical consensus among highly permeable compounds with specific N-methylation patterns indicates which stereocenters are critical for allowing permeable conformations and which may be varied without compromising permeability. A cursory glance revealed that the three most permeable compounds with one N-methyl group in the macrocycle were N-methylated at Ala7 and only varied with respect to two stereocenters. To identify more N-methylation and stereochemical patterns associated with high permeability, we examined the 25 most permeable compounds with one or two N-methyl groups in the macrocycle. Among the 25 most permeable compounds with one macrocyclic N-methyl group, 12 were N-methylated at Leu6 and showed strong stereochemical consensus at Leu4, Leu5, Leu8, and Pro9 (Figure 4b, Scaffold A). On the other hand, 17 of the 25 most permeable lariats with two N-methyl groups in the macrocycle were N-methylated at Leu5 and Ala7, although there was no stereochemical consensus among these compounds (Figure 4b, Scaffold B). This N-methylation pattern is clearly exceptional in its ability to accommodate stereochemical variation and thus support diverse permeable 3D geometries.
Figure 4.
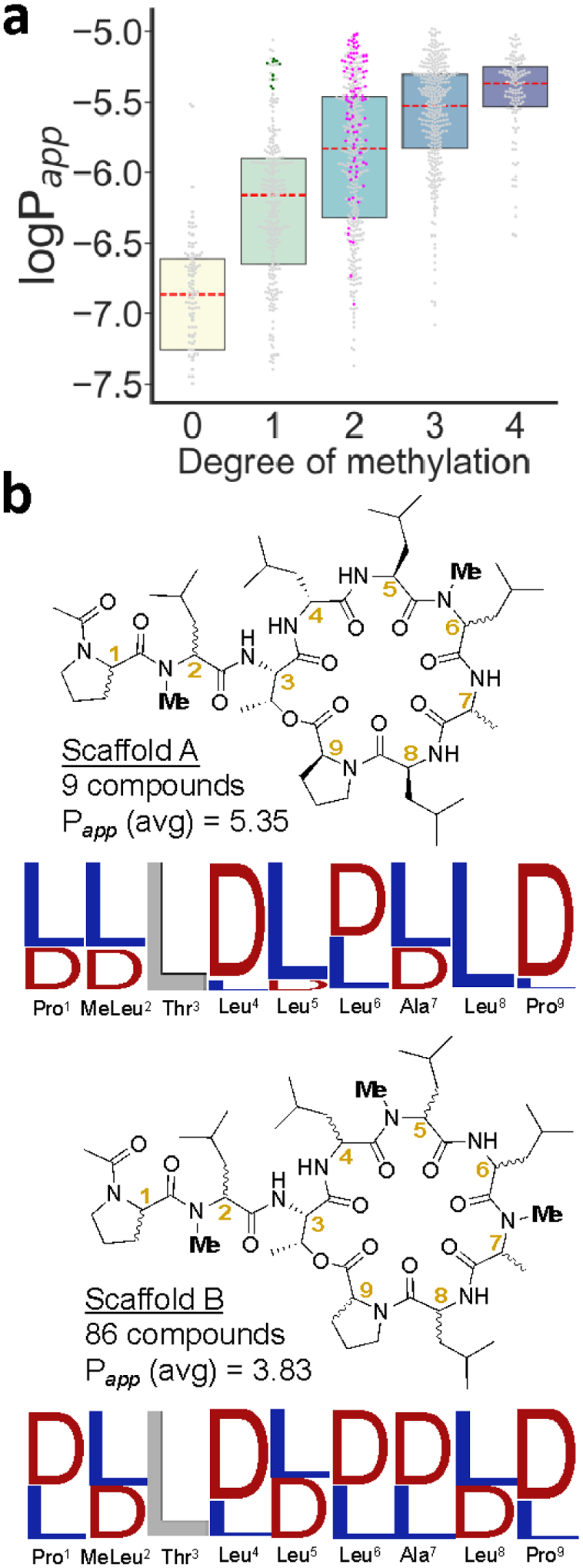
Scaffolds derived from the stereochemical consensus of permeable compounds with specific N-methylation patterns. (a) The compounds of scaffolds A (green) and B (magenta) relative to the rest of the library. (b) The structures of the scaffolds with sequence logos. The relative height of the letter symbolizes the representation of that stereochemistry among compounds with specific N-methylation patterns from among the top 25 most permeable compounds with that degree of N-methylation. The number of compounds contributing to the consensus is listed along with the average permeability in units of 10−6 cm/s.
Structural study of compound 2
To better understand the low dielectric conformations facilitating high permeability among lariats with multiple hydrogen bond donors (HBDs), we used NMR and molecular modelling to study the solution conformation of compound 2, the most permeable library member with only one macrocyclic N-methyl amide. The compound has several features associated with high permeability: high heterochirality, D stereochemistry at Leu4, non-N-methylated Leu4, and L stereochemistry at Leu8. We used ROESY-derived interproton distance restraints (Table S22) and dihedral (ϕ-angle) restraints derived from 3J vicinal coupling constants between Hα-HN protons (Table S24). These restraints were provided as input to ForceGen, an algorithm designed to model macrocycles using NMR-derived restraints.30–31 Our implementation of this method is described in the Supporting Information.
A very strong ROESY crosspeak between the Hα atoms of D-Leu6 and D-MeAla7 provided convincing evidence that this amide bond adopts the cis conformation (ω = 0°). Strong ROESY crosspeaks between the δ protons of Pro9 and the α-protons of Leu8, and between the δ protons of Pro1 and the terminal acetyl protons, along with an absence of Hα-Hα crosspeaks, indicate trans geometry for these amides. The strong ROESY crosspeak between the N-methyl protons of D-MeLeu2 and the α-proton of Pro1, along with no Hα-Hα crosspeak, indicate a trans geometry for this amide as well. These backbone amide geometries were therefore used as ω torsional restraints in the ForceGen structure calculations.
Consistent with its membrane permeability, the NMR solution conformation of 2 in chloroform is characterized by an extensive intramolecular hydrogen bonding (IMHB) network involving all five HBDs (Figure 5). The type VI β-turn centered about the cis-amide between D-Leu6 and D-MeAla7 facilitates two transannular hydrogen bonds between Leu5 and Leu8, flanked on one side by an inverse γ-turn centered around Leu5. The NH group of Leu4 points towards the center of the macrocycle to form hydrogen bonds with the Thr3 sidechain and Leu8 carbonyl. In the lariat tail, the NH group of Thr3 is sequestered from solvent by a γ-turn centered around D-MeLeu2. While the central ring motif is well conserved among the lowest energy conformers, the tail is quite mobile. We used variable temperature 1H NMR to determine the extent of solvent exposure of each NH group. The low temperature shift coefficients (<4 ppb/K) are consistent with the NMR structure showing exclusion of all five NH groups from solvent (Table S23).
Figure 5.
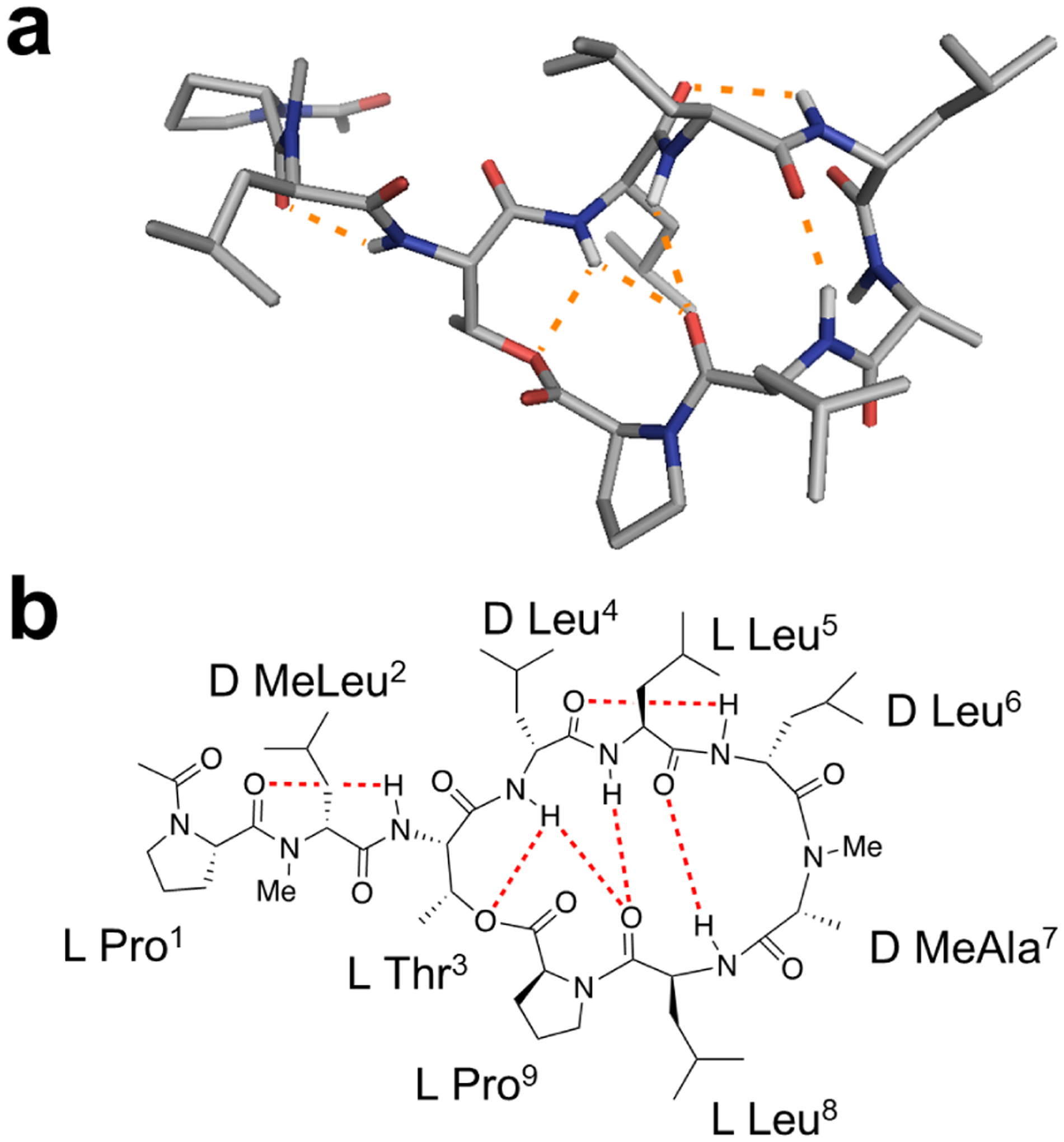
(a) 3D representation of the low-dielectric solution structure of compound 2. (b) Schematic structure of 2 showing hydrogen bonding observed in low-dielectric solution structure.
Some backbone features associated with permeability may be understood in the context of this 3D solution structure. Heterochirality in the region between Thr3 and Leu6 likely supports IMHB networks by providing a torsional space that favors turns and allows transannular orientation of HBDs.32–34 For a transannular orientation of HBDs to be permitted, bulky leucine sidechains must face opposite directions when adjacent, requiring heterochiral relationships. Additional solution structures of compounds from this library are likely to reveal low-dielectric conformations that may provide additional insight into the observed structure-permeability relationships.
Role of lariat topology
We investigated the effect of lariat topology on permeability by creating additional libraries with the Thr residue transposed, resulting in varied tail length and ring size (Figure 6, Scheme S1). For Libraries 2 and 3, the Thr was transposed towards the N-terminus, resulting in lariats containing 8 residues in the macrocycle and one residue in the tail. Library 3 lacks an N-methyl group on the residue adjacent to the Thr to control for the deleterious effect of N-methylation at this position that was observed in the initial library. For Library 4, the Thr residue was situated at the N-terminus, resulting in a 9-residue macrocycle that lacked a lariat tail. Compounds with the same number of N-methyl groups are isomeric, allowing direct comparison of permeability without lipophilicity (ALogP) and molecular weight as confounding factors. Libraries 2–4 were prepared as single sub-libraries of 256 compounds each. The stereochemistry of sub-library 6 from Library 1 (“Library 1.6”) was used for Libraries 2–4 as this was the most permeable sub-library.
Figure 6.
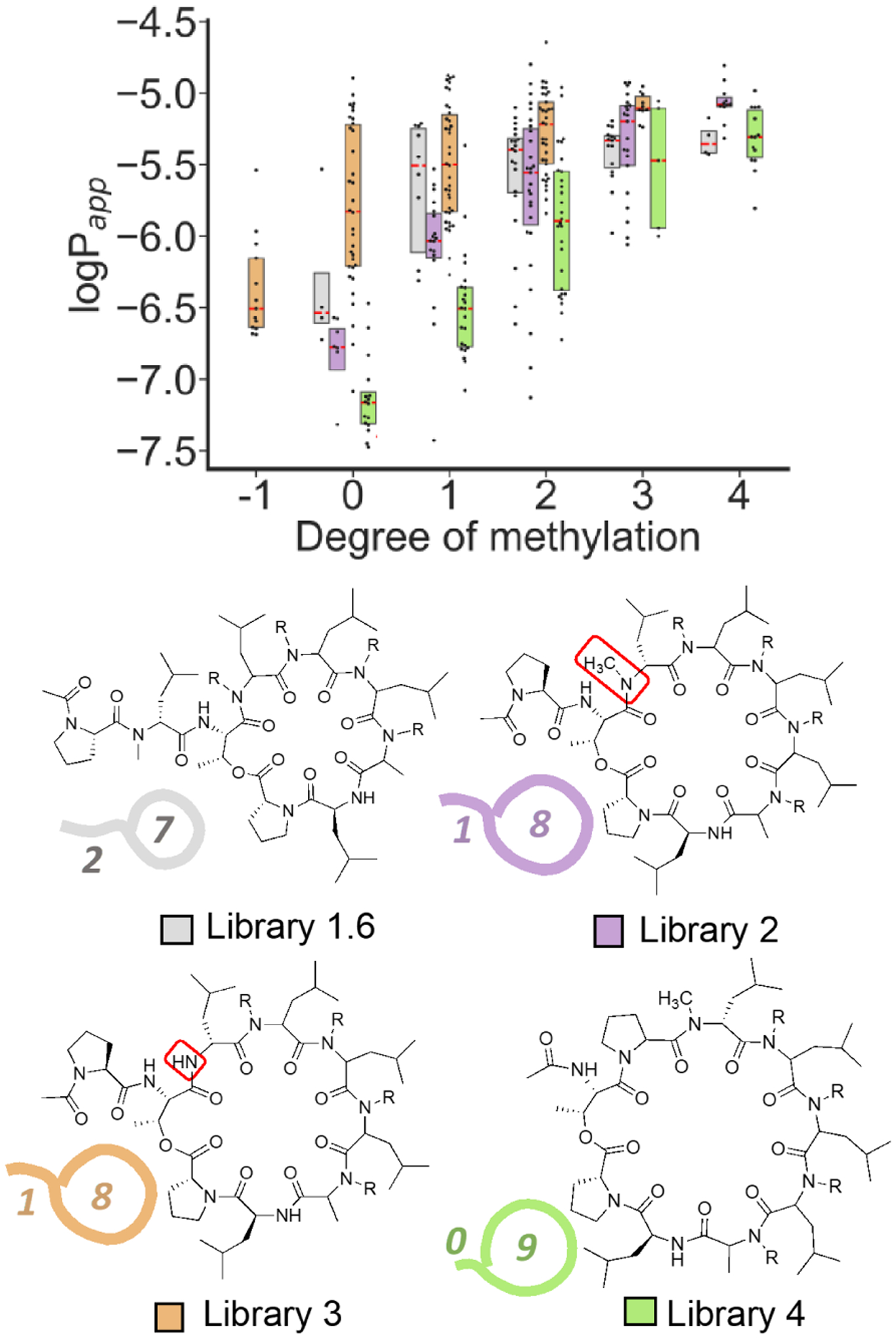
Effect of threonine position on permeability. The molecular weight and AlogP are constant among these libraries. Library 3 has one fewer N-methyl group than the other three libraries. To maintain consistency with library 1, degree of methylation refers to the total number of N-methyl groups minus one. Within each degree of methylation, all compounds are isomeric. The red dashed lines represent medians and the boxes represent quartiles.
Library 3 with the 8–1 macrocycle-tail topology was slightly more permeable, on average, than the parent Library 1.6, indicating that the favorable permeability observed for Library 1.6 was not specific to its 7–2 macrocycle-tail topology. Interestingly, the deleterious effect of N-methylation at the residue neighboring the Thr which was observed in Library 1 was also observed in Library 2 with the 8–1 lariat topology. The large difference in permeability between Libraries 2 and 3 further establishes that N-methyl group location is as important as the type of linkage in determining permeability. Library 4, bearing the “9–0” linkage, was significantly less permeable than any of the other libraries, indicating a role for at least one residue in the tail in facilitating permeability across a broad range of macrocycle geometries. Although a single residue lariat tail is theoretically sufficient to promote a γ-turn involving the Thr NH, as seen in the solution structure of 2, the significantly diminished permeability of Library 4 may alternatively be due to the addition of a second Pro residue to the macrocycle, a rigidifying element that could disfavor permeable conformations.
Sidechain variation
Sidechain substitution is an ideal approach to combinatorial drug discovery. An effect of sidechain identity on the permeability of cyclic peptides has been observed in previous studies.16, 35 We designed Library 5 to investigate the effect of sidechain variation on the permeability of 2. We chose 2 as the scaffold on which to perform sidechain substitution because it is highly permeable in spite of having five HBD, and its relatively low ALogP allowed for substitution with bulkier side chains while keeping much of the library below the solubility threshold. Keeping stereochemistry and N-methylation constant, we varied positions 4–6 among eight nonpolar amino acids, producing a theoretical diversity of 512 library members (Figure 7). At each position, all eight amino acids had unique masses to allow for identification using CycLS.21 Amino acids without side-chain hydrogen bond donors were selected to avoid scaffold perturbation caused by potential side-chain-to-backbone hydrogen bonding. The amino acids, ranging in size from Abu to 1-Nal, were chosen at random from Fmoc amino acids already available in the lab. We limited the scope of this library to amino acids without β-branching.
Figure 7.
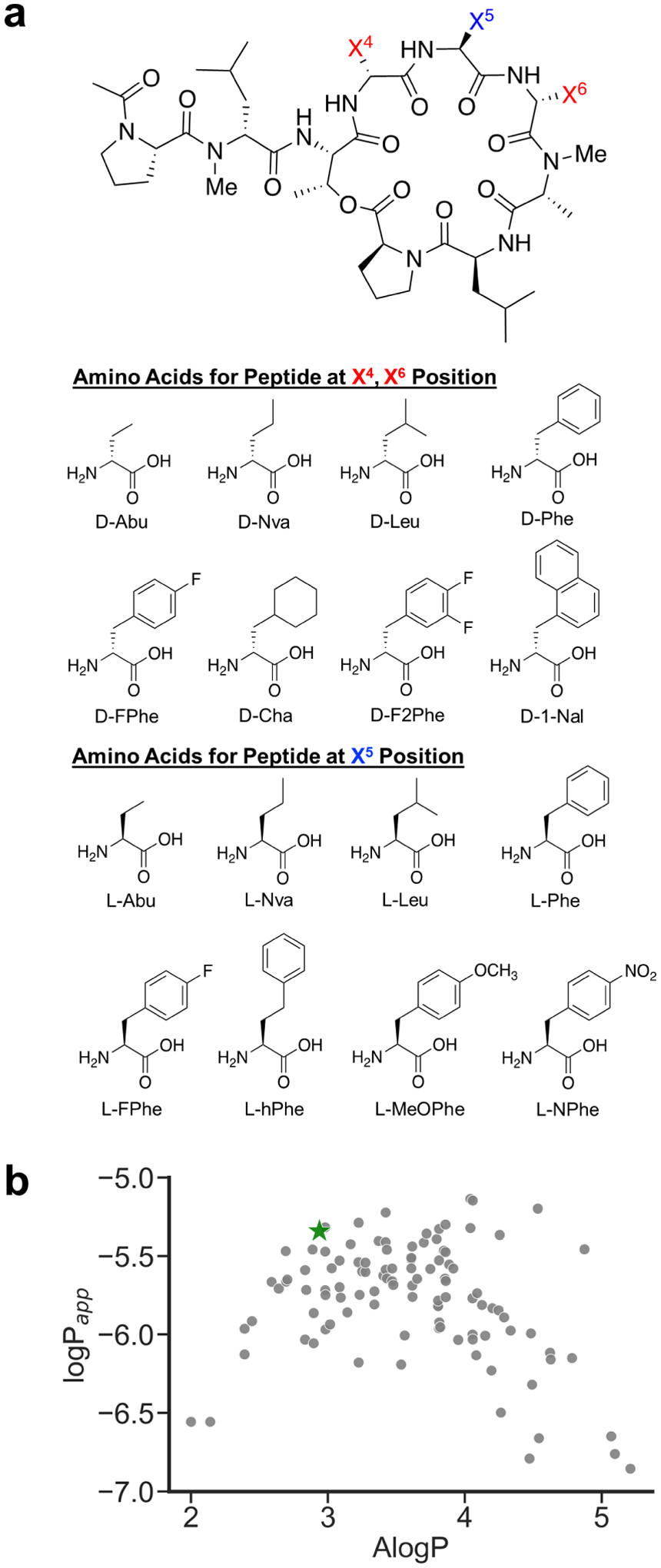
(a) Design of Library 5. (b) logPapp values plotted against AlogP values. Compound 2 is indicated by a green star.
Due to the poor overall aqueous solubility observed for this library, we adapted the “sink PAMPA” conditions that have been described for the analysis of highly lipophilic compounds.36 Encouragingly, 2 was permeable under these conditions with Papp = 4.6 × 10−6 cm/s. After data processing, the dataset contained 121 library members from a theoretical diversity of 512.
Of the 121 compounds identified in Library 5, 84 had Papp values above 1 × 10−6 cm/s. As seen in previous studies in both peptidic and non-peptidic systems, the relationship between AlogP and permeability followed a bell-shaped curve,6, 16, 37 indicative of the mutually opposing effects of lipophilicity and solubility on permeabililty along the polarity continuum.20 Permeable library members were most abundant between AlogP of 3 and 4. Of the 57 Library 5 members with AlogP values in this range, 52 had Papp values above 1 × 10−6 cm/s. Although 2 was one of the more permeable compounds in Library 5, side chain substitution did not abrogate permeability. Substitution with Abu did not prevent permeability at any of the three positions, indicating that steric shielding of HBDs is not vital for allowing permeability. This result indicates that mass-encoded libraries probing stereochemistry and N-methylation may be useful for the development of permeable libraries with diversity provided by side chain variation, at least among those with nonpolar funtionality.
CONCLUSION
We set out to investigate the permeability landscape of lipophilic lariat peptides inspired by natural products. Using a mass-encoded library with varied stereochemistry and N-methylation but invariant, aliphatic side chains, we obtained the permeabilities of over 1000 lariat peptides with diverse backbone geometries. The striking variation in passive permeability observed among isomeric compounds that differ only in N-methyl position and stereochemistry highlight the key role of conformation in determining passive membrane permeability in this chemical space. Although we identified intriguing structure-permeability relationships from the data without using computational tools, automated data mining techniques would undoubtedly reveal deeper relationships. In addition, machine learning approaches may yield models with more predictive power that could be applied to a larger set of scaffolds. Nonetheless, the surprisingly high number of permeable scaffolds in this dataset indicates that the landscape of passive permeability in lariat depsipeptides extends well beyond that defined by existing lariat peptide natural products.
Although the vast majority of orally bioavailable macrocycles are below MW 1000,18 permeability above MW 1000 has been reported for synthetic macrocycles. A designed permeable cyclic decapeptide scaffold has been reported previously,38 and further studied with side-chain substitution and peptide-peptoid substitution.39 However, the results reported here reveal numerous scaffolds supporting drug-like permeability above MW 1000. The permeability of many compounds in this library, even some with as many as 5 HBDs, as well as the maintenance of permeability upon side chain variation of compound 2, indicates a potentially important role for lariat peptides in future discovery efforts. The crystal structure of the DNA sliding clamp from M. smegmatis in complex with griselimycin, in which the cyclic part of the molecule binds one subsite and the lariat tail extends into an adjacent subsite,10 illustrates the unique potential of lariats to target biomolecular interactions.
Our results reveal that, as with cyclic peptides, the permeability of lariat peptides is sensitive to stereochemistry as well as the number and location of N-methyl groups. The emergence of consensus features and general trends among permeable compounds illustrates how this data can be applied to the design of compounds or libraries biased towards membrane permeability in this relatively uncharted chemical space.
EXPERIMENTAL SECTION
Most of the experimental procedures are in the supporting information.
General procedure for manual solid-phase peptide coupling.
To a solution of the Fmoc amino acid (2 eq, 0.5 M in DMF) was added HATU (1.9 eq, 0.5 M in DMF) followed by DIPEA (2.5eq). The resultant solution was swirled and allowed to stand for 5 minutes, then added to the drained resin. The resin was heated to 50°C for 2 h. After coupling, the resin was washed with DMF (3x) and DCM (3x). For Fmoc deprotection, the resin was treated with a solution containing 2% 1,8-diazabicycloundec-7-ene (DBU) and 2% piperidine in DMF for 15 min at room temperature. The resin was then washed with DMF (3x) and DCM (3x).
Ester formation using DIC.
Fmoc-proline (10 eq) was dissolved in DMF/DCM (1:9, roughly 3mL/g Fmoc-proline). DMAP was added (0.25 eq) followed by DIC (10 eq). The solution was swirled rapidly until a precipitate formed. The mixture was added to the resin and the SPE tube capped. The reaction was shaken at room temperature for 3 h, then drained. Another portion of reactants (10 eq Fmoc-proline, 0.25 eq DMAP, 10 eq DIC) was immediately added without washing the resin and the resin was allowed to react overnight at room temperature. The resin was drained and washed with DMF until the thick precipitate which formed during the reaction was removed, then washed with DCM (3x).
Supplementary Material
ACKNOWLEDGMENT
We thank J. Lee for his assistance with NMR spectroscopic experiments. We thank Akihiro Furukawa and Daichi-Sankyo Company, Ltd. for providing MDCK data.
Funding Sources
This work was supported by the US National Institutes of Health (GM131135).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Detailed experimental procedures and analytical data (PDF)
Library PAMPA data (Excel)
Library chromatograms (ZIP)
REFERENCES
- 1.Villar EA; Beglov D; Chennamadhavuni S; Porco JA Jr.; Kozakov D; Vajda S; Whitty A, How proteins bind macrocycles. Nat Chem Biol 2014, 10 (9), 723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doak BC; Zheng J; Dobritzsch D; Kihlberg J, How Beyond Rule of 5 Drugs and Clinical Candidates Bind to Their Targets. Journal of Medicinal Chemistry 2016, 59 (6), 2312–2327. [DOI] [PubMed] [Google Scholar]
- 3.Rezai T; Yu B; Millhauser GL; Jacobson MP; Lokey RS, Testing the Conformational Hypothesis of Passive Membrane Permeability Using Synthetic Cyclic Peptide Diastereomers. Journal of the American Chemical Society 2006, 128 (8), 2510–2511. [DOI] [PubMed] [Google Scholar]
- 4.Pye CR; Hewitt WM; Schwochert J; Haddad TD; Townsend CE; Etienne L; Lao Y; Limberakis C; Furukawa A; Mathiowetz AM; Price DA; Liras S; Lokey RS, Nonclassical Size Dependence of Permeation Defines Bounds for Passive Adsorption of Large Drug Molecules. J Med Chem 2017, 60 (5), 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen DS; Hoang HN; Lohman RJ; Hill TA; Lucke AJ; Craik DJ; Edmonds DJ; Griffith DA; Rotter CJ; Ruggeri RB; Price DA; Liras S; Fairlie DP, Improving on nature: making a cyclic heptapeptide orally bioavailable. Angew Chem Int Ed Engl 2014, 53 (45), 12059–63. [DOI] [PubMed] [Google Scholar]
- 6.Wang CK; Northfield SE; Swedberg JE; Colless B; Chaousis S; Price DA; Liras S; Craik DJ, Exploring experimental and computational markers of cyclic peptides: Charting islands of permeability. European Journal of Medicinal Chemistry 2015, 97, 202–213. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen DS; Shepherd NE; Xu W; Lucke AJ; Stoermer MJ; Fairlie DP, Orally Absorbed Cyclic Peptides. Chemical Reviews 2017, 117 (12), 8094–8128. [DOI] [PubMed] [Google Scholar]
- 8.Dougherty PG; Sahni A; Pei D, Understanding Cell Penetration of Cyclic Peptides. Chemical Reviews 2019, 119 (17), 10241–10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Santen JA J. G; Leen Singh A; Aniebok V; Balunas MJ; Bunsko D; Carnevale Neto F; Castaño-Espriu L; Chang C; Clark TN; Cleary Little JL; Delgadillo DA; Dorrestein PC; Duncan KR; Egan JM; Galey MM; Haeckl FPJ; Hua A; Hughes AH; Iskakova D; Khadilkar A; Lee J-H; Lee S; LeGrow N; Liu DY; Macho JM; McCaughey CS; Medema MH; Neupane RP; O’Donnell TJ; Paula JS; Sanchez LM; Shaikh AF; Soldatou S; Terlouw BR; Tran TA; Valentine M; van der Hooft JJJ; Vo DA; Wang M; Wilson D; Zink KE; Linington RG, The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Central Science 2019, 5 (11), 1824–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kling A; Lukat P; Almeida DV; Bauer A; Fontaine E; Sordello S; Zaburannyi N; Herrmann J; Wenzel SC; Konig C; Ammerman NC; Barrio MB; Borchers K; Bordon-Pallier F; Bronstrup M; Courtemanche G; Gerlitz M; Geslin M; Hammann P; Heinz DW; Hoffmann H; Klieber S; Kohlmann M; Kurz M; Lair C; Matter H; Nuermberger E; Tyagi S; Fraisse L; Grosset JH; Lagrange S; Muller R, Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 2015, 348 (6239), 1106–1112. [DOI] [PubMed] [Google Scholar]
- 11.Marco E; Martín-Santamaría S; Cuevas C; Gago F, Structural Basis for the Binding of Didemnins to Human Elongation Factor eEF1A and Rationale for the Potent Antitumor Activity of These Marine Natural Products. Journal of Medicinal Chemistry 2004, 47 (18), 4439–4452. [DOI] [PubMed] [Google Scholar]
- 12.PharmaMar, PharmaMar announces the approval of Aplidin in Australia for the treatment of multiple myeloma. 2018.
- 13.Hodgson J, The pandemic pipeline. Nature biotechnology 20 March 2020. [DOI] [PubMed] [Google Scholar]
- 14.Fredersdorf M; Kurz M; Bauer A; Ebert MO; Rigling C; Lannes L; Thiele CM, Conformational Analysis of an Antibacterial Cyclodepsipeptide Active against Mycobacterium tuberculosis by a Combined ROE and RDC Analysis. Chemistry 2017, 23 (24), 5729–5735. [DOI] [PubMed] [Google Scholar]
- 15.Hewitt WM; Leung SS; Pye CR; Ponkey AR; Bednarek M; Jacobson MP; Lokey RS, Cell-permeable cyclic peptides from synthetic libraries inspired by natural products. J Am Chem Soc 2015, 137 (2), 715–21. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa A; Townsend CE; Schwochert J; Pye CR; Bednarek MA; Lokey RS, Passive Membrane Permeability in Cyclic Peptomer Scaffolds Is Robust to Extensive Variation in Side Chain Functionality and Backbone Geometry. J Med Chem 2016, 59 (20), 9503–9512. [DOI] [PubMed] [Google Scholar]
- 17.Townsend C; Jason E; Naylor MR; Pye CR; Schwochert J; Edmonson QD; Lokey RS, The Passive Permeability Landscape Around Geometrically Diverse Hexa- and Heptapeptide Macrocycles. 2020, ChemRxiv archive. https://chemrxiv.org/articles/preprint/The_Passive_Permeability_Landscape_Around_Geometrically_Diverse_Hexa-_and_Heptapeptide_Macrocycles/13335941 (accessed Dec 16, 2020).
- 18.Doak BC; Over B; Giordanetto F; Kihlberg J, Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem Biol 2014, 21 (9), 1115–42. [DOI] [PubMed] [Google Scholar]
- 19.Over B; Matsson P; Tyrchan C; Artursson P; Doak BC; Foley MA; Hilgendorf C; Johnston SE; Lee MD; Lewis RJ; McCarren P; Muncipinto G; Norinder U; Perry MWD; Duvall JR; Kihlberg J, Structural and conformational determinants of macrocycle cell permeability. Nature Chemical Biology 2016, 12 (12), 1065–1074. [DOI] [PubMed] [Google Scholar]
- 20.Naylor MR; Ly AM; Handford MJ; Ramos DP; Pye CR; Furukawa A; Klein VG; Noland RP; Edmondson QD; Turmon AC; Hewitt WM; Schwochert J; Townsend CE; Kelly CN; Blanco MJ; Lokey RS, Lipophilic Permeability Efficiency Reconciles the Opposing Roles of Lipophilicity in Membrane Permeability and Aqueous Solubility. J Med Chem 2018, 61 (24), 11169–11182. [DOI] [PubMed] [Google Scholar]
- 21.Townsend C; Furukawa A; Schwochert J; Pye CR; Edmondson Q; Lokey RS, CycLS: Accurate, whole-library sequencing of cyclic peptides using tandem mass spectrometry. Bioorg Med Chem 2018, 26 (6), 1232–1238. [DOI] [PubMed] [Google Scholar]
- 22.Kuranaga T; Enomoto A; Tan H; Fujita K; Wakimoto T, Total Synthesis of Theonellapeptolide Id. Org Lett 2017, 19 (6), 1366–1369. [DOI] [PubMed] [Google Scholar]
- 23.Yao G; Wang W; Ao L; Cheng Z; Wu C; Pan Z; Liu K; Li H; Su W; Fang L, Improved Total Synthesis and Biological Evaluation of Coibamide A Analogues. J Med Chem 2018, 61 (19), 8908–8916. [DOI] [PubMed] [Google Scholar]
- 24.Seo H; Lim D, Total Synthesis of Halicylindramide A. The Journal of Organic Chemistry 2009, 74 (2), 906–909. [DOI] [PubMed] [Google Scholar]
- 25.Teixidó M; Albericio F; Giralt E, Solid-phase synthesis and characterization of N-methyl-rich peptides. The Journal of Peptide Research 2008, 65 (2), 153–166. [DOI] [PubMed] [Google Scholar]
- 26.Angell Yvonne M., G.-E. C, Rich Daniel H., Comparative Studies of the Coupling of N-Methylated, Sterically Hindered Amino Acids During Solid-Phase Peptide Synthesis. Tetrahedron Letters 1994, 35, 5981–5984. [Google Scholar]
- 27.Kansy M; Senner F; Gubernator K, Physicochemical high throughput screening: parallel artificial membrane permeation assay in the description of passive absorption processes. J Med Chem 1998, 41 (7), 1007–10. [DOI] [PubMed] [Google Scholar]
- 28.White TR; Renzelman CM; Rand AC; Rezai T; McEwen CM; Gelev VM; Turner RA; Linington RG; Leung SS; Kalgutkar AS; Bauman JN; Zhang Y; Liras S; Price DA; Mathiowetz AM; Jacobson MP; Lokey RS, On-resin N-methylation of cyclic peptides for discovery of orally bioavailable scaffolds. Nat Chem Biol 2011, 7 (11), 810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang CK; Northfield SE; Colless B; Chaousis S; Hamernig I; Lohman R-J; Nielsen DS; Schroeder CI; Liras S; Price DA; Fairlie DP; Craik DJ, Rational design and synthesis of an orally bioavailable peptide guided by NMR amide temperature coefficients. Proceedings of the National Academy of Sciences 2014, 111 (49), 17504–17509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleves AE; Jain AN, ForceGen 3D structure and conformer generation: from small lead-like molecules to macrocyclic drugs. Journal of Computer-Aided Molecular Design 2017, 31 (5), 419–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain AN; Cleves AE; Gao Q; Wang X; Liu Y; Sherer EC; Reibarkh MY, Complex macrocycle exploration: parallel, heuristic, and constraint-based conformer generation using ForceGen. Journal of Computer-Aided Molecular Design 2019, 33 (6), 531–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haque TS, L. J, Gellman SH, Stereochemical Requirements for ?beta-Hairpin Formation: Model Studies with Four-Residue Peptides and Depsipeptides. Journal of the American Chemical Society 1996, 118 (29), 6975–6985. [Google Scholar]
- 33.Ghosh D; Lahiri P; Verma H; Mukherjee S; Chatterjee J, Engineering β-sheets employing N-methylated heterochiral amino acids. Chemical Science 2016, 7 (8), 5212–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoang HN; Hill TA; Ruiz-Gómez G; Diness F; Mason JM; Wu C; Abbenante G; Shepherd NE; Fairlie DP, Twists or turns: stabilising alpha vs. beta turns in tetrapeptides. Chemical Science 2019, 10 (45), 10595–10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rand AC; Leung SS; Eng H; Rotter CJ; Sharma R; Kalgutkar AS; Zhang Y; Varma MV; Farley KA; Khunte B; Limberakis C; Price DA; Liras S; Mathiowetz AM; Jacobson MP; Lokey RS, Optimizing PK properties of cyclic peptides: the effect of side chain substitutions on permeability and clearance(). Medchemcomm 2012, 3 (10), 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh MH; Lee HJ; Jo SH; Park BB; Park S-B; Kim E-Y; Zhou Y; Jeon YH; Lee K, Development of Cassette PAMPA for Permeability Screening. Biological & Pharmaceutical Bulletin 2017, 40 (4), 419–424. [DOI] [PubMed] [Google Scholar]
- 37.Sawada GA; Barsuhn CL; Lutzke BS; Houghton ME; Padbury GE; Ho NFH; Raub TJ, Increased Lipophilicity and Subsequent Cell Partitioning Decrease Passive Transcellular Diffusion of Novel, Highly Lipophilic Antioxidants. J Pharm Exp Ther 1999, 288 (3), 1317–1326. [PubMed] [Google Scholar]
- 38.Fouche M; Schafer M; Berghausen J; Desrayaud S; Blatter M; Piechon P; Dix I; Martin Garcia A; Roth HJ, Design and Development of a Cyclic Decapeptide Scaffold with Suitable Properties for Bioavailability and Oral Exposure. ChemMedChem 2016, 11 (10), 1048–59. [DOI] [PubMed] [Google Scholar]
- 39.Furukawa A; Schwochert J; Pye CR; Asano D; Edmondson QD; Turmon A; Klein V; Ono S; Okada O; Lokey RS, Drug‐like properties in macrocycles above MW 1000: Backbone rigidity vs. side‐chain lipophilicity. Angewandte Chemie International Edition 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


