Abstract
Background
(Val)ganciclovir resistance mutations in CMV UL97 (UL97-GCV-R) complicate anti-CMV therapy in recipients of solid organ and hematopoietic stem cell transplants, but comprehensive data on prevalence, emergence, and outcome are scarce.
Methods
Using next-generation sequencing (NGS; Illumina MiSeq platform), we analyzed UL97-GCV-R in patients with available plasma samples and refractory CMV replication/DNAemia (n = 87) containing viral loads ≥910 IU/mL. Twenty-one patients with CMV DNAemia resolving under antiviral therapy were analyzed as controls. Detected mutations were considered induced and of potential clinical significance if they increased by ≥10% compared with the first detected frequency or if they had a maximum frequency ≥25%.
Results
Nineteen of 87 (21.8%) with refractory CMV replication had ≥1 UL97-GCV-R detected by NGS, in comparison to 0/21 of the controls (P = .02). One-third of the recipients had 2 or more induced UL97-GCV-R mutations. The most frequently induced mutations affected codons 595 (42% [8/19]), 594 (32% [6/19]), and 603 (32% [6/19]). C592G was present in all episodes of both cases and controls at frequencies <15%, but never induced. UL97-GCV-R tended to be more frequent in donor/recipient CMV immunoglobulin G mismatch or following failure to complete primary prophylaxis, and many developed invasive CMV disease.
Conclusions
UL97-GCV-R is common among transplant patients with refractory CMV replication. Early testing by NGS allows for identification of major mutations at codons 595, 594, and 603 and excludes a major role of C592G in ganciclovir resistance. Large prospective studies on UL97-GCV-R are warranted.
Keywords: cytomegalovirus, ganciclovir resistance, hematopoietic stem cell transplantation, next-generation sequencing, solid organ transplantation
The currently available treatment options for post-transplant cytomegalovirus (CMV) DNAemia are generally effective, though their use is limited by toxic side effects and drug resistance [1–4]. Consequently, CMV DNAemia remains a challenge following the course of solid organ transplantation (SOT) and hematopoietic stem cell transplantation (HSCT) [5–8]. Of particular concern is the intrapatient emergence of CMV variants refractory or resistant to antiviral drugs, such as the first-line drug ganciclovir (GCV) and its oral prodrug valganciclovir [9–11]. (Val)ganciclovir resistance mutations are frequently localized in the CMV UL97 phosphotransferase gene (UL97-GCV-R); some of the most frequent UL97-GCV-R mutations confer up to a 15-fold increase in the IC50 and have been demonstrated to be associated with increased morbidity and mortality [9, 12]. Risk factors for development of UL97-GCV-R include, among others, prolonged exposure to (val)ganciclovir, CMV immunoglobulin G (IgG) mismatch of the donor (D) and recipient (R; ie, D+/R- for SOTs and D-/R+ for HSCTs), graft-vs-host disease (GVHD) in HSCTs, recurrent CMV DNAemia, failure to clear CMV DNAemia despite adequate dosages of therapy, and subtherapeutic levels of (val)ganciclovir [4, 9, 10]. The clinical picture in the UL97-GCV-R-infected patient may range from mild asymptomatic DNAemia to severe or fatal CMV organ disease [11], and it often necessitates a change of treatment to the second-line drugs foscarnet or, more rarely, cidofovir. Despite being equally as effective as (val)ganciclovir, foscarnet is considerably more nephrotoxic and only available as an intravenous formulation [3]. Similar limitations are even more pronounced with cidofovir, which is rarely used due to significant nephrotoxicity [13]. Maribavir is currently under investigation for treatment of refractory and resistant CMV DNAemia and disease and is showing promising results [14]. Whether the more recently available terminase inhibitor letermovir will play a major role in the treatment of refractory resistant CMV DNAemia is currently under investigation for SOT and HSCT recipients (ClinicalTrials.gov: NCT03728426).
In general, studies focusing on CMV resistance are limited by low power, making data on the prevalence and dynamics of UL97-GCV-R scarce. Furthermore, the current golden standard for detection of UL97-GCV-R is by Sanger sequencing, a method that is unable to detect viral subpopulations <20% [4, 10, 15]. Consequently, the early induction of resistance mutations may be missed by Sanger sequencing. In comparison, newer technologies using next-generation sequencing (NGS) can detect viral subpopulations already at frequencies around 1%–3%, yet data from studies using this method remain sparse [16–18].
The main objective of this study was to describe the viral evolution of UL97-GCV-R in a large cohort of SOT and HSCT recipients with refractory CMV DNAemia using NGS. To further the understanding, key samples from a control group of transplant recipients with uncomplicated CMV DNAemia with adequate response to (val)ganciclovir were also analyzed for comparison.
METHODS
Study Population and CMV Definitions
All SOT (heart, kidney, lung, and liver) and HSCT (myeloablative conditioning [MAC], nonmyeloablative conditioning regimen [NMA], and umbilical cord blood [UCB]) patients transplanted at Rigshospitalet in Copenhagen, Denmark, and included in the MATCH programme [19, 20] between January 1, 2010, and July 11, 2018, with ≥1 positive (≥273 IU/mL) CMV DNAemia were evaluated for inclusion in the study. Clinical information on the infectious episodes was collected from the PERSIMUNE data warehouse (https://www.persimune.dk/).
CMV PCR was performed on plasma samples acquired as part of routine clinical follow-up post-transplantation. The COBAS Amplicor Kit was used for CMV PCR analysis up until 2011; since then, the COBAS AmpliPrep/COBAS TaqMan has been used [21]. During the overlap period in 2011, the 2 kits were tested simultaneously, and a conversion factor of 1:1 between the 2 platforms was established. The reported DNAemia loads were converted from copies/mL to IU/mL using the conversion factor for COBAS AmpliPrep/COBAS TaqMan (1 copy/mL = 0.91 IU/mL).
CMV DNAemia was defined as 2 or more consecutive CMV PCRs with a CMV DNA load ≥273 IU/mL taken 2 weeks apart, or as ≥1 sample with viral load ≥2730 IU/mL. Solitary CMV DNAemia reads <2730 IU/mL were considered CMV blips [22]. The CMV DNAemia episodes were considered treatment responsive if there was an adequate response to treatment within 2 weeks after treatment initiation (viral load decrease ≥1 log) or if CMV DNAemia episodes cleared within 4 weeks on treatment. CMV DNAemia episodes with no decline in CMV DNAemia load (<1 log) despite 2 or more weeks on adequate treatment, or if the CMV DNAemia lasted more than 4 weeks despite adequate treatment, were considered refractory. For refractory cases, it was also noted whether CMV DNAemia occurred while (val)ganciclovir prophylaxis or treatment was administered. CMV-UL97-GCV-R was defined as a CMV DNAemia episode where genotypic resistance was demonstrated with NGS. Detected mutations were considered induced and of potential clinical significance if they increased by ≥10% compared with the first detected frequency or if they had a maximum frequency ≥25%. The GCV ratio (IC50 of mutant/IC50 of wild-type), as previously determined by others, is reported to illustrate the diminished effect of (val)ganciclovir in the presence of specific UL-97-GCV-R mutations [12].
Management of Post-transplant CMV Infection
The management of post-transplant CMV DNAemia in the MATCH programme at Rigshospitalet has been described previously [20, 23].
Most SOTs received 3 months of valganciclovir prophylaxis, with subsequent preemptive screening following prophylaxis discontinuation until 1 year post-transplantation. During the study period, primary prophylaxis consisted of valganciclovir 900 mg daily (adjusted for kidney function) for all SOTs except the kidney recipients, who received 450 mg every other day. The HSCTs were managed solely preemptively. In case of detection of CMV DNAemia <2730 IU/mL in an asymptomatic patient, the test was usually repeated, and if positive, preemptive treatment with valganciclovir was initiated. In cases of CMV DNAemia >2730 IU/mL or in a patient with possible CMV-related symptoms, preemptive valganciclovir treatment was commenced immediately.
Selection of Samples for NGS
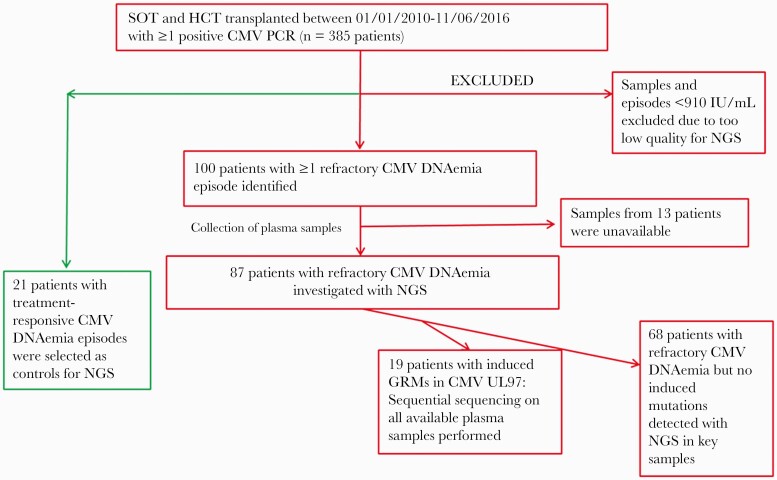
All CMV DNAemia episodes of the included transplant patients were retrospectively evaluated for refractory CMV replication. Cases with CMV DNAemia <910 IU/mL were excluded due to the risk of introducing variability during sequencing when using low-level DNAemia loads (Figure 1).
Figure 1.
Flowchart showing inclusion of 108 patients with either UL97-GCV-R or refractory CMV infection and control cases. Refractory CMV DNAemia was defined as no decline in CMV DNAemia load despite ≥2 weeks with adequate treatment or CMV infection ≥4 weeks despite treatment. Abbreviations: CMV, cytomegalovirus; HSCT, hematopoietic stem cell transplantation; GRM, ganciclovir resistance mutation; MAC, myeloablative conditioning regimen; NGS, next-generation sequencing; NMA, nonmyeloablative conditioning regimen; PCR, polymerase chain reaction; SOT, solid organ transplantation; UCB, umbilical cord blood.
The last positive available sample ≥910 IU/mL of all refractory episodes was selected for NGS analysis. In case of detection of UL97-GCV-R ≥10% in these episodes, all available samples ≥910 IU/mL for that patient were collected and analyzed with NGS. A selection of patients with treatment-responsive CMV episodes was also identified based on sample availability, and then matched by cohort characteristics (type of transplantation [SOT vs HSCT], CMV IgG serostatus, gender, and age). CMV PCR samples ≥910 IU/mL from these patients were then selected as controls for NGS analysis (Figure 1).
Amplicon Generation and Sequencing
Total nucleic acids from 200 µL of plasma were extracted using Biomerieux easyMag. CMV UL97 amplicons were generated with Roche High Fidelity PCR Master (Roche, Penzberg, Germany). The amplicon starts at codon position 439 (TGGCCGACGCTATCAAATTT) and ends at position 619 (AGGCGCCGTAGCTCATTT) of UL97 (GenBank: X17403.1), giving a total length of 543 nucleotides. The PCR products were purified using Ampure XP (Agencourt) PCR purification according to the manufacturer’s instructions. The UL97 amplicons were pooled for each sample before library preparation. Libraries were prepared using a Nextera XT (Illumina, Carlsbad, CA, USA) sample preparation kit. We used a modified protocol, where input DNA and reagent volumes were halved, except for library normalization, in which 1.5 times the volume of magnetic normalization beads was used, compared with the manufacturer’s instructions. DNA libraries were sequenced on an Illumina MiSeq machine using a MiSeq 150-cycle V3 reagent kit (Illumina, Carlsbad, CA, USA), producing 75 base-pair paired-end reads.
Sequence Data Analysis
Raw reads were quality controlled by filtering low-quality reads and adapter and human sequence contamination. Trimmomatic.036 [24] was used for quality and adapter trimming, and bowtie2 and the human genome (GRCh38) were used for removal of human sequence reads [25]. The remaining reads were mapped to CMV (acc.no.X17403) using BWA-MEM [26] with the following parameters: bwa mem -t 4 -M -k 16 -r 1.0. Variants were called using LoFreq* [27]. Functional annotation of the variants was done with Annovar [28]. Frequencies of UL97-GCV-R detected with Lofreq* in any sample were calculated based on samtools mpileup results [29]. Depths of less than 1000 were considered insufficient data. Resistance variants were classified as dominant (ie, the most frequently detected variant at a specific codon/amino acid) or minority (less frequently detected at a specific codon/amino acid). Heat maps were generated using R and the package gplots (R package, version 3.0.1; https://CRAN.R-project.org/package=gplots).
Statistical Analysis
Patient demographics and characteristics were compared with standard descriptive statistics using SAS statistical software (Cary, NC, USA). Ninety-five percent CIs were calculated when relevant.
Patient Consent
All applicable regulatory and ethical approvals related to the project were obtained in accordance with the Danish national legislation. Collection of clinical data from the PERSIMUNE data warehouse was approved by the Danish National Board of Health and the Data Protection Agency. The clinical biobank containing plasma samples from CMV DNAemia episodes was approved by the Data Protection Agency. Furthermore, the current study was approved by Danish Data Protection and the National Committee on Health Research Ethics. Exemption from informed consent for the NGS analysis of plasma samples was sought and granted by the National Committee on Health Research Ethics, as the study was retrospective and utilized stored samples in a clinical biobank.
RESULTS
Patients and Samples Selected for NGS
There were 385 transplant recipients with ≥1 positive CMV PCR sample available for inclusion; out of these, 100/385 patients (26%) had at least 1 episode that was classified as refractory CMV DNAemia (corresponding to 303 samples) (Figure 1). Of these, plasma samples from 13 patients (89 samples) were unavailable. Of the remaining 87 patients (54 SOT and 33 HSCT), 212 samples were available for further analysis. The last available sample ≥910 IU/mL of each episode was selected for NGS analysis, and in case of detected UL97-GCV-R at a frequency ≥10%, all available plasma samples ≥910 IU/mL of that patient were selected for NGS analysis (n = 22), but for 3 cases (1 kidney recipient and 2 recipients of HSCT) additional data were unavailable, and thus induced resistance could not be proven. Thus, in total, 19 patients (6 HSCT and 13 SOT) with refractory CMV DNAemia had proven induced UL97-GCV-R mutations explaining the refractory course, whereas the remaining 68 patients (27 HSCT and 41 SOT) with refractory CMV DNAemia had no confirmed UL97-GCV-R to explain the refractory clinical course (Figure 1).
Twenty-one patients with CMV DNAemia episodes that resolved during (val)ganciclovir treatment were identified, and available plasma samples with CMV viral loads ≥910 IU/mL were analyzed by NGS as controls (Figure 1).
Age, gender, type of transplantation, and CMV IgG D/R status, stratified for type of transplantation and clinical presentation (refractory with UL97-GCV-R [from now on referred to as “resistant”], refractory without detected resistance [from now referred to as “refractory”], or control group), for the included recipients are listed in Table 1.
Table 1.
Baseline Demographics of 108 Included Transplant Recipients, Stratified by Clinical Presentation and Overall Type of Transplantation
| Demographics | Resistant Cases (n = 19) | Refractory Cases (n = 68) | Control Cases (n = 21) | Total (n = 108) | ||||
|---|---|---|---|---|---|---|---|---|
| SOT (n = 13) | HSCT (n = 6) | SOT (n = 41) | HSCT (n = 27) | SOT (n = 14) | HSCT (n = 7) | SOT (n = 68) | HSCT (n = 40) | |
| Age and gender | ||||||||
| Median (IQR) age at transplantation, y | 47.0 | 42.5 | 53.0 | 48.0 | 55.0 | 15.0 | 51.5 | 43.0 |
| (38.0–51.0) | (4.0–45.0) | (42.0–64.0) | (25.0–62.0) | (43.0–63.0) | (8.0–40.0) | (40.5–61.0) | (16–54.5) | |
| Proportion male | 9/13 | 5/6 | 24/41 | 13/27 | 8/14 | 3/7 | 41/68 | 21/40 |
| Type of transplantation | ||||||||
| SOT | ||||||||
| Kidney | 8/13 | NA | 24/41 | NA | 6/14 | NA | 38/68 | NA |
| Liver | 3/13 | NA | 7/41 | NA | 4/14 | NA | 14/68 | NA |
| Lung | 2/13 | NA | 7/41 | NA | 3/14 | NA | 12/68 | NA |
| Heart | 0/13 | NA | 3/41 | NA | 1/14 | NA | 4/68 | NA |
| HSCT | ||||||||
| MAC | NA | 5/6 | NA | 13/27 | NA | 5/7 | NA | 23/40 |
| NMA | NA | 1/6 | NA | 11/27 | NA | 2/7 | NA | 14/40 |
| UCB | NA | 0/6 | NA | 3/27 | NA | 0/7 | NA | 3/40 |
| Donor/recipient CMV IgG | ||||||||
| D+/R+ | 1/13 | 0/6 | 10/41 | 12/27 | 7/14 | 3/7 | 18/68 | 15/40 |
| D+/R- | 12/13 | 0/6 | 25/41 | 1/27 | 6/14 | 0/7 | 43/68 | 1/40 |
| D-/R+ | 0/13 | 4/6 | 3/41 | 13/27 | 0/14 | 4/7 | 3/68 | 21/40 |
| D-/R- | 0/13 | 1/6 | 1/41 | 0/27 | 0/14 | 0/7 | 1/68 | 1/40 |
| Unknown | 0/13 | 1/6 | 2/41 | 1/27 | 1/14 | 0/7 | 3/68 | 2/40 |
Abbreviations: CMV, cytomegalovirus; D, donor; HSCT, hematopoietic stem cell transplantation; IgG, immunoglobulin G; IQR, interquartile range; MAC, myeloablative conditioning regimen; NMA, nonmyeloablative conditioning regimen; R, recipient; SOT, solid organ transplantation; UCB, umbilical cord blood.
Clinical Features of CMV DNAemia in Resistant Episodes
Details on clinical and genotypic features of the 19 recipients with induced UL97-GCV-R are shown in Table 2.
Table 2.
Details on Clinical and Genotypic Features of 19 Transplant Recipients With Induced Ganciclovir Resistance Mutations in the CMV UL97 Gene
| Patient | Tx Type | Pre-tx Donor/Recipient CMV IgG | Clinical Presentation | Codon in Amino Acid | No. of Induced GRMs/Patient | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 460 I/V | 520 Q | 594 V/G | 595 S/F | 596 G | 603 W | 607 Y/F | |||||
| 1 | MAC | D-/R+ | CMV colitis, retinitis, and suspected pneumonia | ++a | 2 | ||||||
| 2 | MAC | D-/R+ | CMV colitis | ++ | 1 | ||||||
| 3 | MACb | D-/R- | CMV retinitis | ++ | ++ | 2 | |||||
| 4 | MACb | D-/R+ | CMV colitis | ++ | ++ | 2 | |||||
| 5 | MAC | D-/R+ | None proven | ++ | 1 | ||||||
| 6 | NMA | D-/R+ | CMV colitis | ++ | 1 | ||||||
| 7 | Liver | D+/R- | CMV pneumonia | ++ | ++ | 2 | |||||
| 8 | Liver | D+/R- | Asymptomatic | ++ | 1 | ||||||
| 9 | Liver | D+/R- | Asymptomatic | ++ | 1 | ||||||
| 10 | Lung | D+/R- | CMV pneumonia | ++ | 1 | ||||||
| 11 | Lung | D+/R- | CMV colitis | ++ | 1 | ||||||
| 12 | Kidney | D+/R- | Asymptomatic | ++ | ++ | ++ | 3 | ||||
| 13 | Kidney | D+/R- | Asymptomatic | ++ | 1 | ||||||
| 14 | Kidney | D+/R- | Asymptomatic | ++ | ++ | ++ | 3 | ||||
| 15 | Kidney | D+/R- | CMV pneumonia | ++a | ++ | 3 | |||||
| 16 | Kidney | D+/R- | Asymptomatic | ++ | 1 | ||||||
| 17 | Kidney | D+/R+ | Asymptomatic | ++ | 1 | ||||||
| 18 | Kidney | D+/R- | Asymptomatic | ++ | 1 | ||||||
| 19 | Kidney | D+/R- | Asymptomatic | ++ | 1 | ||||||
| No. of patients with mutation | 2/19 | 2/19 | 6/19 | 8/19 | 1/19 | 6/19 | 2/19 | ||||
| % with mutation induced | 10.5 | 10.5 | 31.6 | 42.1 | 5.3 | 31.6 | 10.5 |
Patient number corresponds to numbering in Figure 3A.
Abbreviations: CMV, cytomegalovirus; D, donor; GRMs, ganciclovir resistance mutations; HSCT, hematopoietic stem cell transplantation; MAC, myeloablative conditioning regimen; NMA, nonmyeloablative conditioning regimen; R, recipient; SOT, solid organ transplantation; Tx, transplantation; UCB, umbilical cord blood.
aBoth mutation in same codon.
bPediatric patients.
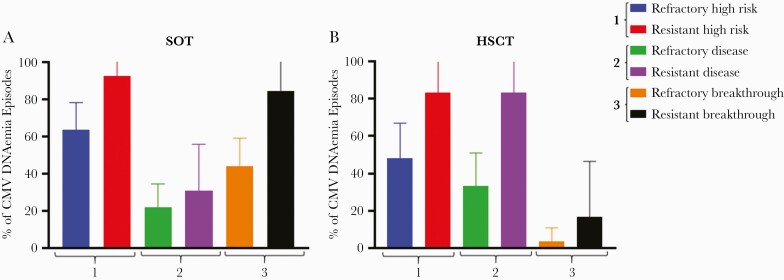
CMV IgG D/R mismatch was found in 5/6 of the HSCT and 12/13 of the SOT recipients with UL97-GCV-R (Table 2). Most HSCTs had received an MAC regimen before transplantation, and 2 were pediatric patients. CMV disease was a dominant feature, with 4 cases of gastrointestinal disease and 1 case of CMV retinitis. The remaining case was a patient with a complex cycle of severe intestinal GVHD necessitating intensification of immunosuppressive treatment, followed by multiple episodes of recurrent CMV DNAemia. CMV disease was suspected but never confirmed. Most SOTs were kidney recipients, and only 1 of these had CMV disease. Furthermore, prophylaxis breakthrough was common amongs the SOTs (Figure 2).
Figure 2.
Comparison of clinical factors of CMV DNAemia between included refractory and resistant cases. Comparison between the proportion of cases with (1) high-risk serostatus, (2) CMV disease, and (3) breakthrough CMV replication on antiviral treatment, stratified by refractory and resistant CMV DNAemia in SOT (A) and HSCT (B). High-risk serostatus in SOT is associated with pretransplant CMV IgG D+/R- in SOT and D-/R+ in HSCT. Disease refers to CMV end organ disease in HSCT and CMV end organ disease and/or CMV syndrome in SOT. “Breakthrough” refers to prior treatment failure. Abbreviations: CMV, cytomegalovirus; D, donor; HSCT, hematopoietic stem cell transplantation; IgG, immunoglobulin G; R, recipient; SOT, solid organ transplantation.
There was a trend toward higher frequencies of high-risk CMV IgG D/R mismatch among UL97-GCV-R cases for both SOT and HSCT (Figure 2), even though the 95% CIs were overlapping for both types of transplantations. For HSCTs, there was also a trend toward higher frequencies of CMV disease among cases with UL97-GCV-R (33.3%; 95% CI, 15.5%–51.1%; vs 83.3%; 95% CI, 53.4%–113.1%; for refractory and resistant cases, respectively), a trend that was less pronounced among the SOTs (Figure 2).
Prevalence of Induced UL97-GCV-R
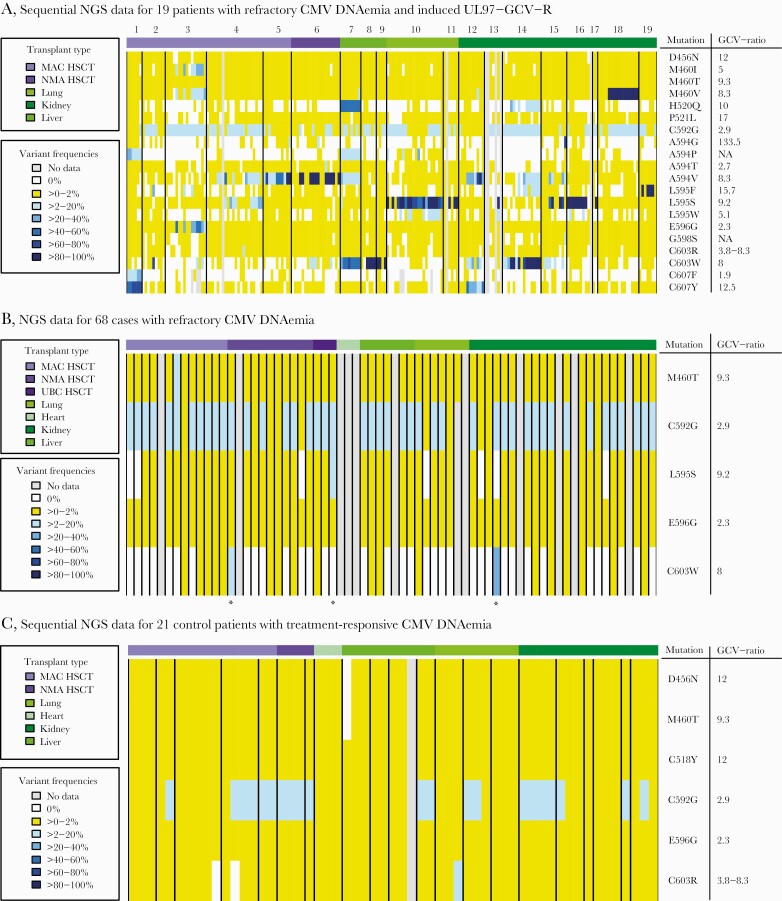
Of the 87 recipients with refractory CMV replication, 19 (22%) had ≥1 induced UL97-GCV-R detected by NGS. In comparison, 0/21 (0%) of the controls had any induced UL97-GCV-R mutations (P = .02). The majority of the 19 resistant recipients (63%) only had 1 dominant UL97-GCV-R induced, whereas 2 induced UL97-GCV-R were detected in 21%. In the remaining 16%, 3 different induced UL97-GCV-R were detected (Table 2).
Separate heat maps of the detected UL97-GCV-R mutations stratified for type of transplantation were constructed for the 19 cases with confirmed induced mutations, as well as for the 68 cases with refractory/breakthrough CMV DNAemia and the 21 controls with treatment-responsive CMV DNAemia (Figures 3A–C respectively). The diversity of detected mutations was more pronounced among the 19 cases with induced UL97-GCV-R, with 20 different amino acid substitutions detected in this group, compared with 5 among the 68 refractory cases and 6 among the 21 control cases (Figures 3A–C). The C592G mutation, conferring low-level resistance against (val)ganciclovir, was present in low frequencies, but never induced in any of the 3 patient populations (Figures 3A–C).
Figure 3.
A, Sequential NGS data for 19 patients with CMV DNAemia with known induced UL97-GCV-R. In case of detection of UL97-GCV-R at variant frequencies ≥10%, all available samples ≥910 IU/mL for that patient were collected and analyzed with NGS in a second round of analysis. The colors in the heat map indicate the frequencies of the detected known UL97-GCV-R mutations. The numbers above the heat map refer to patient number and correspond to patient numbers in Table 2. B, NGS data for 68 cases with refractory CMV DNAemia. In cases with refractory CMV replication without known UL97-GCV-R, the last available sample of each episode ≥910 IU/mL was selected for NGS analysis. The colors in the heat map indicate the frequencies of the detected known UL97-GCV-R mutations. *Three patients in whom NGS detected resistance mutations ≥10% but <25%, but where no additional data were available. C, Sequential NGS data for 21 control cases with treatment-responsive CMV DNAemia. A selection of patients with treatment-responsive CMV episodes were identified, and available CMV PCR samples ≥910 IU/mL from these patients were selected as controls for NGS analysis. The colors in the heat map indicate the frequencies of the detected known UL97-GCV-R mutations. The width of each patient column varies depending on how many sequential samples matching the inclusion criteria were available for sequencing with NGS for each individual patient. Abbreviations: GCV-ratio, ganciclovir ratio; IC50 of mutant, IC50 of wild-type; HSCT, hematopoietic stem cell transplantation; MAC, myeloablative conditioning regimen; NGS, next-generation sequencing; NMA, nonmyeloablative conditioning regimen; SOT, solid organ transplantation; UCB, umbilical cord blood.
Dynamics of Induced UL97-GCV-R
In 14/19 of the resistant cases, plasma samples were available for NGS within 4 weeks from onset of CMV DNAemia (corresponding to a total of 20 induced mutations). In these earliest samples, the UL97-GCV-R detected later in the clinical course for each patient was either not detected (n = 11), detected as a minority variant (n = 7), or detected as a dominating variant (n = 2).
The most frequent UL97-GCV-R affected codons 595 (42%), 594 (32%), or 603 (32%). The proportion of C592G was low in all episodes (<15%) during the whole course (Figures 3A–C, Table 2).
DISCUSSION
In this study, we systematically explored the emergence of GCV resistance using NGS in 87 transplant patients with refractory CMV DNAemia. Our results show that drug resistance mutations documented to confer a 3–10-fold increase in IC50 are emerging in >20% of the recipients with refractory CMV DNAemia. In line with previous observations, important clinical factors such as CMV IgG D/R mismatch and failure of prior treatment tended to be more common among these cases [9, 30–35].
We have previously reported a higher rate of CMV breakthrough during primary prophylaxis among kidney recipients in our center due to the administration of (val)ganciclovir prophylaxis of 450 mg every other day [34], and this lower dosage may very well account for some of the UL97-GCV-R observed among the kidney recipients in the current study. The results of these two studies have prompted an increase in the administered dosage of valganciclovir prophylaxis for the kidney recipients at our center and underline the importance of optimal dosing of primary prophylaxis.
The development of UL97-GCV-R was heterogeneous in this cohort; 55% of the induced UL97-GCV-Rs were not detected with NGS within the first 4 weeks of replication. The remaining cases were either found as minority variants or only in a few cases detected as the dominating mutant within 4 weeks of infection offset. This observation implies a clinical challenge in the timing and interpretation of NGS technology for the diagnosis of UL97-GCV-R and highlights the highly complex and dynamic interactions between the immunocompromised host and CMV [36, 37]. To properly address this research question, larger, prospective, and better powered studies on UL97-GCV-R are warranted.
Whether the use of NGS provides a significant clinical advantage over Sanger sequencing cannot be addressed using the results of this study. It may be that as NGS becomes cheaper and more accessible, it could be more integrated into clinical practice. Alternatively, the results generated using NGS may provide crucial information on key mutations leading to refractory CMV DNAemia and disease, and these can be assessed in the clinical setting using more traditional technologies. Regardless, more studies are needed to better understand the impact of these lower allele frequency mutations on the clinical course of refractory CMV DNAemia and disease. Furthermore, future studies need to focus on molecular validation and external quality assurance programs in order to enhance the specificity of allele calling, something the present study is lacking. This may not only enhance our management of CMV DNAemia and disease based on (val)ganciclovir, but also provide important insights into and lessons in the context of newer antiviral agents such as maribavir and letermovir.
In the CMV UL97 phosphotransferase gene, there are certain hot-spots for mutations that have been documented in the literature [4, 11, 35]. These mutations are usually referred to as the 7 canonical mutations of UL97, and they are believed to account for most of the observed clinical cases of UL97-GCV-R, as is also seen in our data. Interestingly, one of the most common of the canonical mutations, C592G, which confers low-grade resistance toward (val)ganciclovir, was present in all selected episodes in low frequencies (<15%), regardless of whether the patient was a control (ie, treatment responding), refractory, or verified resistant case (Figures 3A–C). However, C592G was never induced in any of the patients, suggesting that this mutation may be hitchhiking during the selective sweep of the virus during treatment with (val)ganciclovir. Thus, if this mutation is detected in a patient, it should potentially be considered a normal variation and not associated with treatment resistance.
Interestingly, more than 70% of the refractory episodes did not have induced UL97-GCV-R in this study; however, there are a couple of caveats to consider in relation to this observation. First, it should be noted that because CMV DNA measured in plasma is unencapsulated and thus subject to DNase breakdown, plasma measurements of DNA may not be entirely representative [38]. Second, although the data in our biobank were collected prospectively, they were analyzed retrospectively in the current study. We were not able to retrospectively analyze all available plasma samples of the refractory infections with NGS, but used the last available sample ≥910 IU/mL of these episodes as a proxy for resistance (assuming the refractory nature would be most likely to be more reflected toward the end of the episode) and only selected those episodes with a mutation frequency ≥10% in this sample. However, it is plausible that some of the refractory episodes with mutation frequency <10% were indeed the result of UL97-GCV-R. Furthermore, some patients had missing data, preventing full NGS on samples that were suspected to contain UL97-GCV-R. Thus, it is possible that the prevalence of UL97-GCV-R among the refractory CMV DNAemia episodes is in fact underestimated in this study. Finally, at the time, we were unable to sequence the DNA polymerase gene UL54 and can consequently not make any assumptions about the development and presence of cross-resistance among the presented cases. However, a thorough investigation of UL54 mutations is underway and will be published in due time.
There are several strengths of the present study. The sequenced data are generated from a large, standardized, and prospectively collected biobank, with detailed corresponding clinical data. Furthermore, the longitudinal structure of the data allowed analysis of the dynamics of UL97-GCV-R development. The use of NGS enabled investigation of both dominant mutants and minor variants. However, the present study, as well as most other studies on CMV resistance mutations, is hampered by the low number of actual resistant cases to study [18, 36, 37]. Despite having a relatively large sample size, UL97-GCV-R is a rare event, emphasizing the need for larger multicenter studies. In order to increase the studied population, this study includes both recipients of SOT and HSCT. However, it is important to remember that SOT and HSCT represent two very heterogenous groups of patients, and caution should be taken before drawing overall conclusions based on these data. It should also be noted that the definition used for refractory CMV infection in this study differs slightly from the recently published definitions by Chemaly et al. [11], the main difference being that we did not differentiate between “probable refractory CMV DNAemia” and “refractory CMV DNAemia,” thus losing granularity and applicability to other cohorts using this newer definition. Future studies are encouraged to use the published definitions on refractory CMV DNAemia for wider applicability.
In conclusion, the use of NGS technology provided detailed insights into the heterogenous and dynamic development of UL97-GCV-R among transplant recipients. Resistant CMV DNAemia remains a serious challenge and is common among refractory CMV replication, and the need for novel, less toxic anti-CMV drugs for primary prophylaxis and treatment of refractory CMV DNAemia is evident. In order to properly address this issue, large prospective and multicenter studies on UL97-GCV-R are warranted.
Acknowledgments
Financial support. This work was supported by the Danish National Research Foundation (grant number DNRF126).
Potential conflicts of interest. F.G. has received personal fees from Pfeizer, Astra-Zeneca, Boehringer-Ingelheim, and Novartis outside the framework of the submitted work. None of the other authors have any conflicts of interest to disclose. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Ljungman P. Prophylaxis against herpesvirus infections in transplant recipients. Drugs 2001; 61:187–96. [DOI] [PubMed] [Google Scholar]
- 2. Goodrich JM, Mori M, Gleaves CA, et al. Early treatment with ganciclovir to prevent cytomegalovirus disease after allogeneic bone marrow transplantation. N Engl J Med 1991; 325:1601–7. [DOI] [PubMed] [Google Scholar]
- 3. Avery RK, Arav-Boger R, Marr KA, et al. Outcomes in transplant recipients treated with foscarnet for ganciclovir-resistant or refractory cytomegalovirus infection. Transplantation 2016; 100:e74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lurain NS, Chou S. Antiviral drug resistance of human cytomegalovirus. Clin Microbiol Rev 2010; 23:689–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harvala H, Stewart C, Muller K, et al. High risk of cytomegalovirus infection following solid organ transplantation despite prophylactic therapy. J Med Virol 2013; 85:893–8. [DOI] [PubMed] [Google Scholar]
- 6. Stern M, Hirsch H, Cusini A, et al. ; Members of Swiss Transplant Cohort Study. Cytomegalovirus serology and replication remain associated with solid organ graft rejection and graft loss in the era of prophylactic treatment. Transplantation 2014; 98:1013–8. [DOI] [PubMed] [Google Scholar]
- 7. Green ML, Leisenring WM, Xie H, et al. CMV reactivation after allogeneic HCT and relapse risk: evidence for early protection in acute myeloid leukemia. Blood 2013; 122:1316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Green ML, Leisenring W, Xie H, et al. Cytomegalovirus viral load and mortality after haemopoietic stem cell transplantation in the era of pre-emptive therapy: a retrospective cohort study. Lancet Haematol 2016; 3:e119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fisher CE, Knudsen JL, Lease ED, et al. Risk factors and outcomes of ganciclovir-resistant cytomegalovirus infection in solid organ transplant recipients. Clin Infect Dis 2017; 65:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El Chaer F, Shah DP, Chemaly RF. How I treat resistant cytomegalovirus infection in hematopoietic cell transplantation recipients. Blood 2016; 128:2624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chemaly RF, Chou S, Einsele H, et al. Definitions of resistant and refractory cytomegalovirus infection and disease in transplant recipients for use in clinical trials. Clin Infect Dis 2019; 68:1420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Landry ML, Stanat S, Biron K, et al. A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates. Antimicrob Agents Chemother 2000; 44:688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mehta Steinke SA, Alfares M, Valsamakis A, et al. Outcomes of transplant recipients treated with cidofovir for resistant or refractory cytomegalovirus infection. Transpl Infect Dis 2021; 23:e13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Papanicolaou GA, Silveira FP, Langston AA, et al. Maribavir for refractory or resistant cytomegalovirus infections in hematopoietic-cell or solid-organ transplant recipients: a randomized, dose-ranging, double-blind, phase 2 study. Clin Infect Dis 2019; 68:1255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Göhring K, Hamprecht K, Jahn G. Antiviral drug- and multidrug resistance in cytomegalovirus infected SCT patients. Comput Struct Biotechnol J 2015; 13:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chou S, Ercolani RJ, Sahoo MK, et al. Improved detection of emerging drug-resistant mutant cytomegalovirus subpopulations by deep sequencing. Antimicrob Agents Chemother 2014; 58:4697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sahoo MK, Lefterova MI, Yamamoto F, et al. Detection of cytomegalovirus drug resistance mutations by next-generation sequencing. J Clin Microbiol 2013; 51:3700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garrigue I, Moulinas R, Recordon-Pinson P, et al. Contribution of next generation sequencing to early detection of cytomegalovirus UL97 emerging mutants and viral subpopulations analysis in kidney transplant recipients. J Clin Virol 2016; 80:74–81. [DOI] [PubMed] [Google Scholar]
- 19. Lodding IP, Sengeløv H, da Cunha-Bang C, et al. ; MATCH Programme Study Group. Clinical application of variation in replication kinetics during episodes of post-transplant cytomegalovirus infections. EBioMedicine 2015; 2:699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lodding IP, da Cunha Bang C, Sørensen SS, et al. Cytomegalovirus (CMV) disease despite weekly preemptive CMV strategy for recipients of solid organ and hematopoietic stem cell transplantation. Open Forum Infect Dis 2018; 5:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DiDomenico N, Link H, Knobel R, et al. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem 1996; 42:1915–23. [PubMed] [Google Scholar]
- 22. Lodding IP, Mocroft A, da Cunha Bang C, et al. Impact of CMV PCR blips in recipients of solid organ and hematopoietic stem cell transplantation. Transplant Direct 2018; 4:e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ekenberg C, da Cunha-Bang C, Lodding IP, et al. Evaluation of an electronic, patient-focused management system aimed at preventing cytomegalovirus disease following solid organ transplantation. Transpl Infect Dis 2020; 22:e13252. [DOI] [PubMed] [Google Scholar]
- 24. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilm A, Aw PP, Bertrand D, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 2012; 40:11189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. da Cunha-Bang C, Sørensen SS, Iversen M, et al. Factors associated with the development of cytomegalovirus infection following solid organ transplantation. Scand J Infect Dis 2011; 43:360–5. [DOI] [PubMed] [Google Scholar]
- 31. Kalra A, Williamson T, Daly A, et al. Impact of donor and recipient cytomegalovirus serostatus on outcomes of antithymocyte globulin-conditioned hematopoietic cell transplantation. Biol Blood Marrow Transplant 2016; 22:1654–63. [DOI] [PubMed] [Google Scholar]
- 32. Gor D, Sabin C, Prentice HG, et al. Longitudinal fluctuations in cytomegalovirus load in bone marrow transplant patients: relationship between peak virus load, donor/recipient serostatus, acute GVHD and CMV disease. Bone Marrow Transplant 1998; 21:597–605. [DOI] [PubMed] [Google Scholar]
- 33. Hassan-Walker AF, Kidd IM, Sabin C, et al. Quantity of human cytomegalovirus (CMV) DNAemia as a risk factor for CMV disease in renal allograft recipients: relationship with donor/recipient CMV serostatus, receipt of augmented methylprednisolone and antithymocyte globulin (ATG). J Med Virol 1999; 58:182–7. [PubMed] [Google Scholar]
- 34. Khurana MP, Lodding IP, Mocroft A, et al. Risk factors for failure of primary (val)ganciclovir prophylaxis against cytomegalovirus infection and disease in solid organ transplant recipients. Open Forum Infect Dis 2019; 6:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. López-Aladid R, Guiu A, Mosquera MM, et al. Improvement in detecting cytomegalovirus drug resistance mutations in solid organ transplant recipients with suspected resistance using next generation sequencing. PLoS One 2019; 14:e0219701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hage E, Wilkie GS, Linnenweber-Held S, et al. Characterization of human cytomegalovirus genome diversity in immunocompromised hosts by whole-genome sequencing directly from clinical specimens. J Infect Dis 2017; 215:1673–83. [DOI] [PubMed] [Google Scholar]
- 37. Guermouche H, Burrel S, Mercier-Darty M, et al. Characterization of the dynamics of human cytomegalovirus resistance to antiviral drugs by ultra-deep sequencing. Antiviral Res 2020; 173:104647. [DOI] [PubMed] [Google Scholar]
- 38. Naegele K, Lautenschlager I, Gosert R, et al. Cytomegalovirus sequence variability, amplicon length, and DNase-sensitive non-encapsidated genomes are obstacles to standardization and commutability of plasma viral load results. J Clin Virol 2018; 104:39–47. [DOI] [PubMed] [Google Scholar]