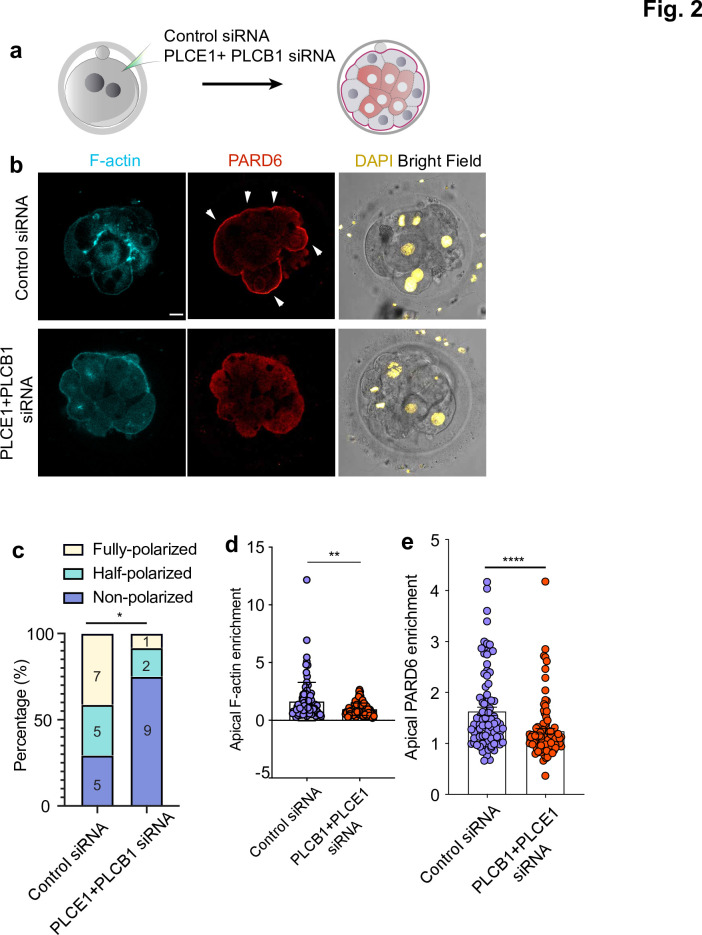
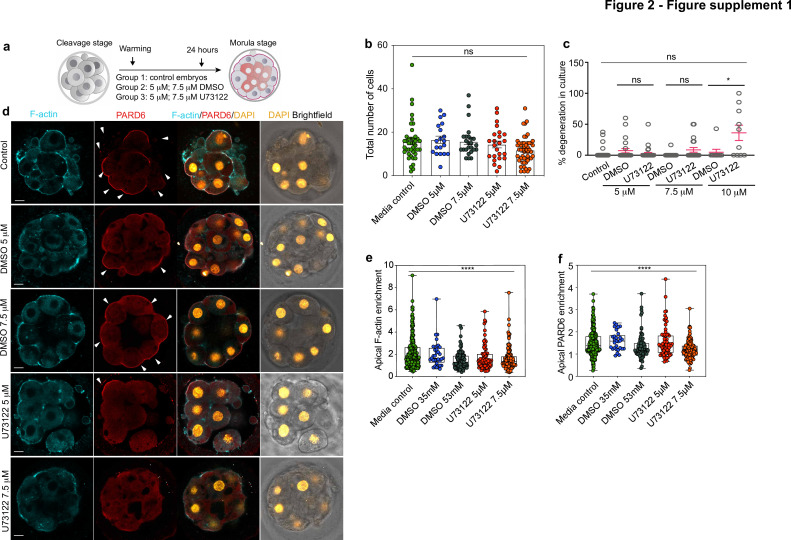
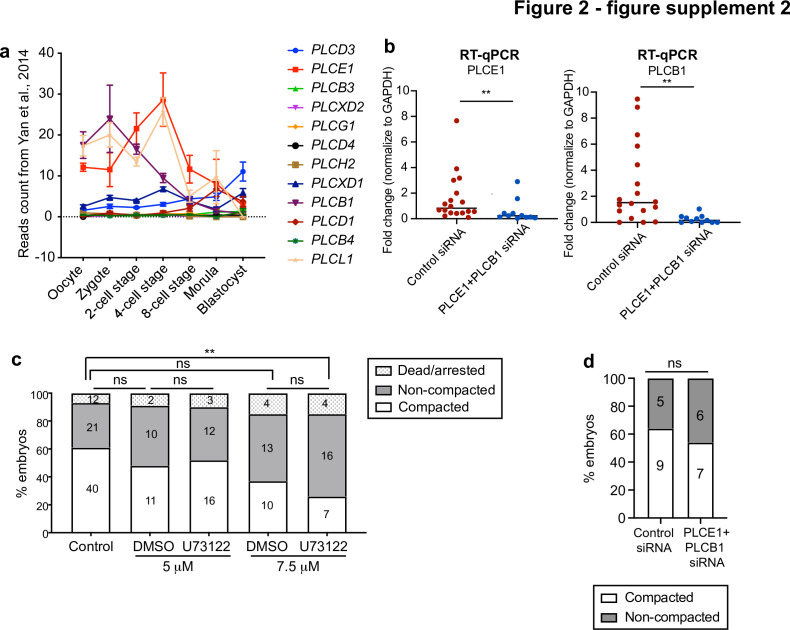
Figure 2. PLC activity regulates cell polarization in the human embryo.
(a) Scheme of the PLCE1/PLCB1 siRNA injections. (b), Representative images of embryos injected with control siRNA or PLCE1/PLCB1 siRNA and cultured until embryonic day four to reveal the localization of F-actin, PARD6, and DAPI. (c) Quantification of the percentage of embryos showing PARD6 polarized cells in embryos from panel b. The number in each bar indicates the number of embryos analyzed. N = 17 embryos for control siRNA injected group; N = 12 for PLCB1+ PLCE1 siRNA injected group. * p < 0.05. Fisher’s exact test. Three independent experiments. (d) Quantification of apical F-actin fluorescence intensity in embryos from panel b. N = 92 control siRNA cells and N = 100 PLCB1+ PLCB1 siRNA cells. **p < 0.0001, Mann-Whitney test. (e) Quantification of apical PARD6 fluorescence intensity in embryos from panel f. N = 89 control siRNA cells and N = 100 PLCB1+ PLCB1 siRNA cells. ****p < 0.0001, Mann-Whitney test. Scale bars, 15 µm.