Abstract
Metastasis is the leading cause of cancer-related deaths. Understanding the pathogenesis of metastasis at the molecular levels is of great significance for cancer research. However, the molecular diagnosis or treatment of cancer metastasis is limited. Accumulating and growing evidence shows that epigenetic changes are present in all human cancers, and epigenetic regulation is an indispensable factor to promote tumor metastasis. With the deepening of research and the advancement of technology, the function and mechanism of epigenetic regulation, including DNA methylation, histone/RNA modification, and precursor messenger RNA alternative splicing and noncoding RNAs, has become more increasingly clear. At present, the application of epigenetic therapies in tumor treatment is becoming a feasible therapeutic route. In this review, we looked for the key molecules in epigenetic regulation and discuss their relative regulating mechanisms in cancer metastasis. Furthermore, we highlight promising therapeutic strategies, including monitoring serum DNA for diagnostic purposes and early phase clinical trial therapies that target DNA and histone methylation. This may also be beneficial in finding new targets for further prognosis and diagnosis of cancer metastasis.
1. Introduction
Cancer is one of the major public health problems which has become a dominant cause of death in the global world, while the stage of metastasis is responsible for approximately 90% of deaths [1]. In the early phases, uncontrolled multiplication of tumor cells leads to the formation of expanded tumor mass that affects normal tissue through physical stress. With the gradual growth of the tumor, metastasis then occurs step by step. The primary tumor cells invade the adjacent tissues, migrate to new sites, survive and adapt to the microenvironment of distant tissues, and form secondary tumors (colonization) [2, 3], which mean going to the most fearsome stage of cancer [2]. Therefore, understanding the molecular mechanism of metastasis is of great significance for the diagnosis of advanced cancer, delaying tumor progression, and finding new therapeutic targets. It is widely accepted that mutations in key genes drive cancer formation at the genetic level [4]. However, only the genetic alterations cannot explain the mechanism of cancer metastasis [5]. A massive amount of evidence suggests epigenetic changes closely related to the process of metastasis [6–8].
Epigenetic regulation refers to the heritable alterations in gene expression that are not due to the changes from the DNA sequences themselves. Since its discovery in 1983 [9], the role of epigenetic modification in cancer has become increasingly prominent with the in-depth knowledge of epigenetic mechanisms. In the past 40 years, the roles of DNA hypo- and hypermethylation in cancer have been well established, along with the reported discovery that DNA methylation at a CpG (nonmethylated cytosine/guanine base pairs) abnormality is strongly associated with cancer development [9, 10]. And in the last 30 years, imprinted genes have been found to influence cancer by chromatin modifications [11]. In the last two decades, the study of posttranscription regulation in cancer has been rapidly developed, accompanied with the application of new technologies. For example, the widespread use of next generation sequencing and the development of bioinformatics analysis have been beneficial in studying the role of different subtypes of the same gene in cancer. Here, we review the functional roles and mechanisms of epigenetic regulation in cancer metastasis (Figure 1). It may be beneficial in providing new insights into the diagnosis, prognosis, and prevention of cancer research.
Figure 1.
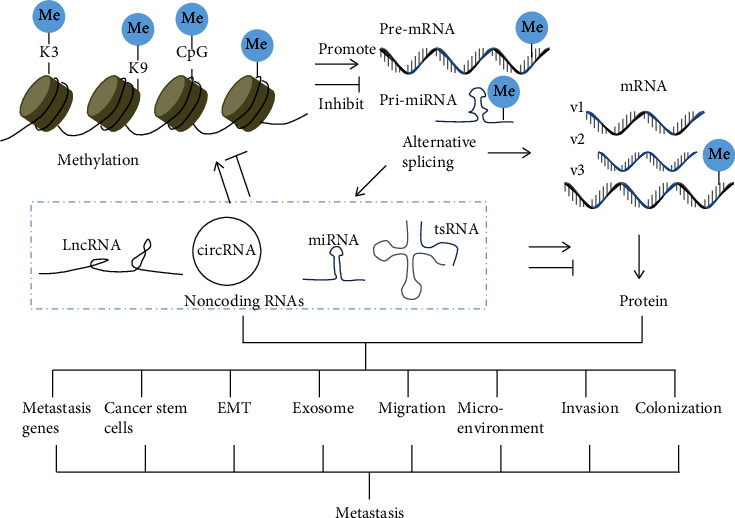
Schematic diagram of epigenetic modifications in cancer metastasis. DNA/histone methylation and RNA methylation regulate the transcription and translation of target genes, respectively. Methylation also affects alternative splicing, which increases the diversity of proteins and noncoding RNAs. Methylation, alternative splicing, and noncoding RNAs may act alone or in combination in cancer metastasis.
1.1. Methylation Influences Metastasis-Associated Gene Expression
Methylation is one of the well-known modifications in DNA, RNA, and histone, which are catalyzed through methyltransferases (writers) with thorough mechanisms about how to write, read, and erase the covalent epigenetic marks [12]. Dynamic changes in methylation of DNA, RNA, and histone are essential for cellular functions. As the effect of methylation on carcinogenesis is addressed elsewhere, we pay attention to the alteration of methylation in cancer metastasis.
1.1.1. DNA Methylation
5-Methylcytosine (5mC) is one of the most studied modifications of DNA methylation written by the family of DNA methyltransferase 1-3 (DNMT1-3) [13]. 5mC was erased through ten-eleven translocation (TET) dioxygenase-mediated oxidation to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) [14]. In order to identify whether its function is to activate or suppress gene expression, the position of DNA methylation throughout the genome needs to be considered [15]. Whole-genome profiling of 5mC has shown that 5mC dynamically exists in enhancers, gene bodies, and extended transcriptionally inactive partially methylated domains, implicating its important function involved in disease progression, including cancer [16]. Various DNA methylation pathways in cancer metastasis have been reported and verified in tissue and cells at pancancer or specific cancers. Here, we just picked a small part of them from different cancers and cells as examples, which are listed in Table 1. Moreover, DNA modifications may serve as potential prognostic biomarkers and future therapeutic targets. For example, in breast cancer patients, serum deprivation response (SDPR) expression was silenced by promoter DNA methylation, and its expression were negatively correlated with metastasis and relapse-free survival [17]. The discovery of novel methylation of genes specifically on the metastasis of certain cancers may help reveal the underlying mechanisms and progression of specific cancer metastasis. In addition, DNA methylation could be shaped by circulating tumor cells (CTCs), which are related to the formation of metastasis seeding and promote cancer stemness and metastasis [18]. Lastly, monitoring serum DNA methylation might be used as diagnostic biomarkers to predict survival outcomes for metastatic cancers. Because aberrant DNA can be released into the bloodstream by cancer cells, it is easy to detect common tissue changes from the blood using quantitative multiplex assays, especially for cell-free DNA methylation [19]. Studies such as TBCRC 005 might provide the first potential clinical tool for measuring the serum level of methylation, which was conducive to the detection of metastatic breast cancer [20]. In another example, IRX1 promoter hypomethylation detected in serum DNA of patients with osteosarcoma might be a potential biomarker for predicting lung metastasis in osteosarcoma [21]. Compared with tissue-based biomarkers, blood DNA biomarkers were easier for repetitive monitoring and might be more informative and specific than protein biomarkers. The use of serum DNA methylation analysis might be beneficial for exploiting personalized medicine and monitoring of therapeutic effects [19].
Table 1.
Examples of DNA methylation with cancer metastasis.
| Gene name | DNA methylation location/level | Cancer types | Samples | Refs |
|---|---|---|---|---|
| POPDC1, POP1 | Promoter, hypermethylation | Colorectal carcinoma (CRC), adenomatous polyps | Human tissues, CRC cell lines, athymic mice | [64] |
|
| ||||
| Akr1b1, Hoxb4, Rasgrf2, Rassf1, Hist1h3c, Tm6sf1 | High cumulative methylation index | Breast cancer | Serum, 182 women | [20] |
|
| ||||
| IRX1 | Hypomethylation | Osteosarcoma | 2 cell lines, serum from 67 primary osteosarcoma | [21] |
|
| ||||
| Genome-scale analyses of DNA methylation | High consistency of hypermethylation across metastases | Lethal metastatic prostate cancer | Tissue, 71 specimens from each of 13 subjects | [65] |
|
| ||||
| 2481 differentially methylated regions | CpG island, flanking regions, and CpG sparse promoters | Prostate cancer | 17 tissues, 6 cell lines | [66] |
|
| ||||
| HOP homeobox HOPX | Promoter, hypermethylation | Nasopharyngeal carcinoma | 443 formalin-fixed paraffin-embedded (FFPE) NPC tissues, 5 human NPC cell lines | [67] |
|
| ||||
| 47 genes (RDBP), 48 CpGs significant association | Hypermethylation | Pheochromocytoma, paraganglioma | 310 tumors were obtained from patients, 67 with metastases | [68] |
1.1.2. RNA m6A Methylation
It has been more than 40 years since N6-methyladenosine (m6A) was identified [22]. m6A, one of the most abundant modifications in mRNAs, similar to DNA methylation, exists dynamically in biological effects and is influenced by “writer” (METTL3, METTL14, WTAP, and KIAA1429), “eraser” (FTO and ALKBH5), and “reader” (YTH and HNRNP) proteins [23]. RNA m6A modification is abundant in the near stop codons and 3′ untranslated terminal regions (3′UTRs), which affect the expression of a transcriptome [24]. Increasing evidence shows that the diversity of m6A methylation is associated with multiple biological functions in mammals, such as mRNA splicing procession [25], mRNA stability [26], protein translation efficiency [27], and cell differentiation [28] (Figure 2(a)). Along with the recent breakthrough findings of two mammalian RNA demethylases, FTO (the fat mass and obesity-associated protein) [29] and ALKBH5 (alkylation repair homolog protein 5) [30], the dynamic and reversible modification of m6A methylation in physiological processes has begun to be appreciated [31, 32]. Disturbing m6A RNA methylation might be associated with cancer metastasis. Abnormal expression of the writer, eraser, and reader proteins of m6A was strikingly linked with cancer advance. For instance, in vitro and in vivo experiments have shown that METTL14 (methyltransferase-like 14) inhibition significantly promoted tumor metastasis by modulating the primary microRNA 126 process in an m6A-dependent manner [33]. And loss of function of METTL3 decreased protein abundance for several m6A-containing mRNAs but had minimal impact on mRNA levels. METTL3 expression was upregulated in lung adenocarcinoma, and using its knockdown and overexpression studies demonstrated that METTL3 promoted invasion of human lung cancer cells [34]. Furthermore, expression of metastasis-associated genes shaped by m6A methylation was one of the key factors for cancer metastasis. As an example, TRIM7 expression was influenced by m6A modification and regulated osteosarcoma cell migration and invasion [35]. With the application of more and more sensitive detection techniques, researchers will discover the distribution of m6A methylation in transcriptomics and their associated fundamental properties in cancer [36]. Epigenetic modification of RNA is a rapidly developing field in biology, and the knowledge of its modification mainly focuses on m6A, which gives us a promising future therapeutic strategy in cancer. Clarifying the role of m6A modification in genes and their classical mechanisms involved in tumor metastasis will probably be the focus of future research.
Figure 2.
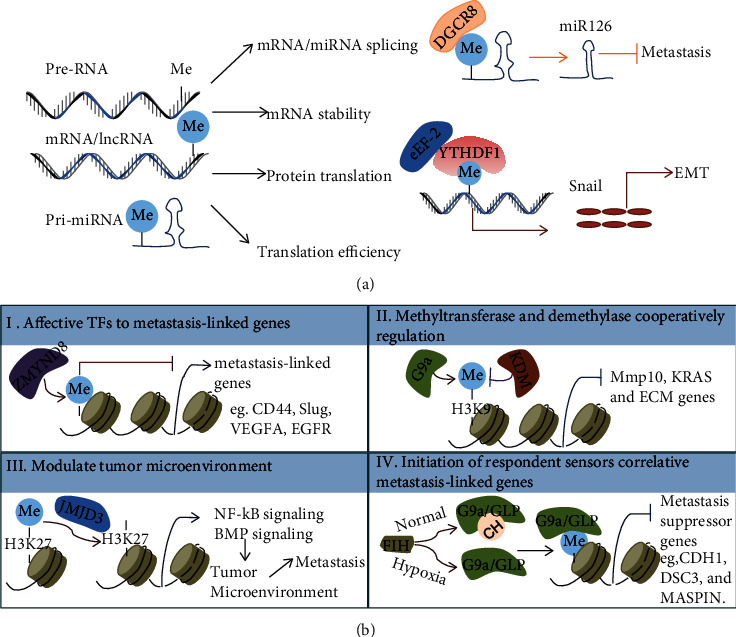
Diverse mechanisms of RNA/histone methylation in metastasis. (a) m6A methylation influenced mRNA/noncoding-RNA splicing and stabilized as well as mRNA translation. (b) Histone methylation directly or indirectly influenced the expression of target genes through transcription factors binding in their promoter areas, such as histone methylation inhibiting recruitment of the transcriptional cofactors (TFs). Histone methylation was also involved in the regulation of tumor microenvironment.
1.1.3. Histone Methylation
Histone modification, as a key characteristic of epigenetic regulation, has an important role in cancer progression. A diverse set of identified histone modifications known in cancer are deregulated, such as methylation, acetylation, phosphorylation, sumoylation, adenosine diphosphate (ADP) ribosylation, deimination, and crotonylation [37]. Misregulation of these modifications can lead to inappropriate activation of oncogenes or tumor suppressors [5]. Currently, methylation modification of histone has been considered a potential target for cancer treatment, and clinical trials have shown promising results [38]. It has been well documented that how histone methylation was modified relative to writers, erasers, and readers dynamically regulated methylation in association with cancer [12]. It is widely accepted that dysregulation of histone methylation may be a driver in diverse types of cancer. Here, we focus on the latest reports on histone methylation in cancer metastasis. There are at least four mechanisms that explain methylation's role in metastasis (Figure 2(b)). Firstly, histone modification affects the recruitment of the transcriptional factor, which is linked with dysregulated expression of metastasis-linked genes [39]. It was reported that H3K9 histone methyltransferase G9a enhanced the level of H3K9 dimethylation, which contributed to the recruitment of the transcription cofactors HP1, DNMT1, and HDAC1 into the promoter region of the cell adhesion molecule Ep-CAM, thus inhibiting the expression of Ep-CAM and promoting the invasion and metastasis of lung cancer [40]. Secondly, histone methyltransferase and demethylase coordinate to regulate the expression of prometastatic genes [41, 42]. Thirdly, histone modification takes part in modulating the tumor microenvironment through molecular signaling pathways to promote cancer progression and metastasis [43]. Fourthly, the altered microenvironment conditions in cancer initiate respondent sensor reactions by controlling the histone methyltransferase activity, leading to dysregulated expression of metastasis genes. For instance, under the hypoxia condition of ovarian cancer, FIH, as an oxygen sensor, drove the expression of metastasis-related genes through histone lysine methyltransferases G9a and GLP [44]. Multiple known mechanisms can regulate the expression of metastasis-related genes in a synergistic or interdependent manner, and they can be combined with sensor reaction in the abnormal microenvironment to participate in tumor metastasis [45]. In summary, histone modification provides a critical regulatory role in cancer metastasis-linked gene transcription and microenvironment-induced cancer progression. As a form of epigenetic therapy, some inhibitors of histone modification have shown efficacy in preclinical trials. Inhibitors of EZH2 (enhancer of zeste homolog 2, H3K27 methyltransferase [46]), DOT1L (H3K79 methyltransferase [47]), PRMT1 (arginine N-methyltransferases [48]), and PRMT5 have now entered clinical trials [12]. The data from these studies will benefit a better understanding of the role of histone modifications in cancer progression and give new hope for cancer treatment.
1.2. Alternative Splicing as a Regulator of Cancer Metastasis
Alternative splicing acts on the precursor messenger RNA (pre-mRNA) of one gene to generate diverse isoforms of RNA and protein [49]. In humans, up to 94% of genes undergo alternative splicing [50]. These different isoforms of mRNA can affect their own stability, localization, or translation [51]. And relevant protein isoforms may have related, distinct, or even opposing functions that vary from tissue to tissue [50]. No matter whether it is in normal tissues or in cancer progression, there are at least five main alternative splicing patterns (Figure 3(a)): cassette exons, intron retention, alternative 5′ splice sites, alternative 3′ splice sites, and mutually exclusive exons [52]. Histone modifications or DNA methylation has been reported to have a direct effect on alternative splicing [53] (Figure 3(b)). For example, several studies showed significant aggregation of the H3K36me3 (histone H3 lysine 36 trimethyl) signals on exons, and these histone signals might be associated with lower levels in alternatively spliced exons compared to constitutive exons [54]. In addition, with technological advances in single-cell sequencing, researchers have used scM&T-seq (single-cell methylation and transcriptome sequencing) [55] to discover the correlation and variation between transcription and methylation. It also has been found that local DNA methylation profiles influence splicing variation across cells. And DNA methylation information might accurately predict different splicing patterns of individual cassette exons during cell differentiation [56]. DNA methylation promoted the exclusion of weak upstream exons by inhibiting DNA-binding protein function in genes, such as CCCTC-binding factor- (CTCF-) recognized CD45 exon 5 inhibited by DNA methylation 5mC, which mediated local RNA polymerase II pausing and linked to alternative splicing [57]. Alternative splicing is not only an important mechanism for cell development, differentiation, and regulation of tissue-specific function [51], but it has also been found in a variety of pathological processes, including tumorigenesis and cancer metastasis [58]. And alternative splicing has been used as a relevant therapeutic target for cancer treatment [59]. Alternative splicing is involved in most stages of metastasis, including EMT (epithelial-mesenchymal transition) [60], stemness feature [60], invasion [61], surviving in circulation [62], extravasation [61], and colonization in new sites [63] (Figure 3(c)). Here, epithelial-restricted splicing regulator (ESRP) is a salient example of how alternative splicing promotes cancer metastasis. Inhibition of the expression of ESRP is enough to make some changes in morphology in epithelial cells and contribution to EMT [69]. ESRP also mediates the splicing event of EMT-related genes in their pre-mRNA to produce different isoforms, such as p120 and NUMB [70, 71]. Although an enormous work has been done on the roles of alternatively splicing in cancer metastasis, it is still important to identify the key molecules and epigenetic modifications that control the abnormal subtypes of proteins, which contribute to disease progression in certain types of cancer.
Figure 3.
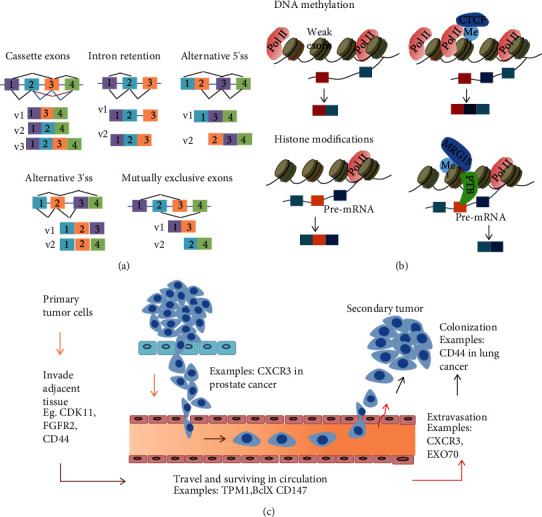
Schematic diagram of an alternative splicing process and its relative function in metastasis. (a) The five different alternative splicing types. (b) DNA methylation or histone methylation influenced the production of alternative splicing. (c) Alternative splicing is involved in every step of metastasis, from primary tumor cells invading adjacent tissues to moving to new sites, surviving in the circulation, and adapting to distant tissues to form secondary tumor (colonization).
1.3. Noncoding RNAs That Play an Increasingly Important Role in Tumor Metastasis
The effects of noncoding RNAs on physiological and pathological processes are increasingly becoming clear. Now we know that noncoding RNAs interact with DNA, protein, or RNA to execute their function [71]. For example, noncoding RNAs have been documented to change protein contents by regulating gene expression at the transcriptional level [72] or by altering mRNA translational efficiency [73]. Noncoding RNAs can also change the location of proteins by turning their signaling pathways [74], or act as working partners of proteins [75]. The discovery of these novel noncoding RNAs has sparked a new round of exploration and further research into cancer. Here, we present recent advances and explore how noncoding RNAs are involved in cancer metastasis. The well-known noncoding RNAs, including tRNAs and rRNAs, are in abundance and have well-defined regulatory roles in translation. Novel classes of noncoding RNAs emerged more than two decades ago, for example, mircroRNAs (miRNAs), circular RNAs (circRNAs), and long noncoding RNAs (lncRNAs). Accumulating evidence suggests that noncoding RNAs have essential roles in regulating gene expression and cancer metastasis.
1.3.1. tRNA-Derived ncRNAs
tRNA-derived ncRNAs (tdRNAs) have been discovered in a variety of organisms and species [76]. tdRNAs, as a novel class of small noncoding RNAs, are generated from specific cleavages of mature or precursor tRNAs. According to the cleavage position, tsRNAs can be currently categorized into at least three types [77]. tRNA-derived small RNAs (tsRNAs) and tRNA-derived fragments (tRFs or tDRs) are generated during the maturation of tRNA precursors and maturation sequences, respectively. The third type is the tRNA halve (tiRNA), which is generated from the production of mature tRNAs that undergo cleavage in the anticodon loops. tdRNAs are believed to play important roles in diverse aspects of biological processes, for instance, cell proliferation [78], regulation of gene expression [79], stress response [80], control of retrotransposon [81], and tumor suppression [82]. tdRNAs also have functions in cancer metastasis (Figure 4(a)). Here, we summarize the known mechanisms of tdRNAs in promoting the cancer process. First of all, tdRNAs competed with the mRNAs of metastasis-linked genes at binding sites for functional proteins, such as RNA-binding proteins. For example, endogenous tdRNAs, like tRFs, tRFAsp, tRFGly, tRFGlu, and tRFTyr, competed with multiple metastasis-relative transcriptions for their binding sites in RNA-binding protein YBX1. The competitive binding to YBX1 resulted in the destabilization of metastasis-relative transcriptions that suppressed cancer cell invasion and metastasis [82]. In addition, tdRNAs are functionally similar to miRNAs in that they downregulate targeted genes and interfere with their related biological processes. For instance, tRF/miR-1280, derived from both tRNALeu and pre-microRNA, was significantly decreased in human colorectal cancer (CRC) tissues. And tRF/miR-1280 inhibited the Notch signaling pathway and relative metastatic features by directly downregulating JAG2 expression, which leads to the cancer stem cell-like phenotype [83]. Furthermore, tdRNAs were specifically generated by cleavage within tRNA anticodon loops using ribonuclease angiogenin under stress-activated conditions. And the incision production of 5′-tiRNAs, not 3′-tiRNAs, inhibited translation and protein synthesis by competing to displace eIF4G/eIF4A from mRNA-binding sites [84]. Over the past 40 years, the understanding of tdRNAs has undergone dramatic changes, from meaningless degradation products of tRNA to key players in orchestrating gene regulation in physiological or pathological processes. However, the multifaceted regulatory potential of tdRNAs in cancer still requires deeper validation. It is a challenge to find out key tdRNAs as biomarkers of cancer processes and new approaches to targeted tdRNAs for cancer diagnosis and treatment.
Figure 4.
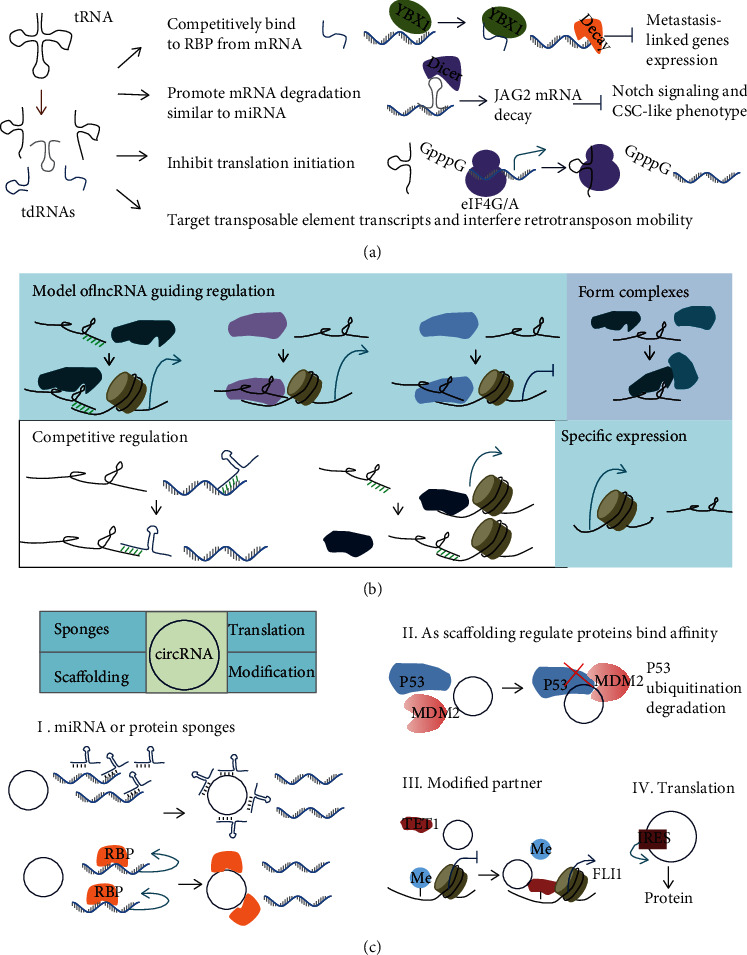
Diverse mechanisms of noncoding RNAs in metastasis. (a) tRNA-derived ncRNAs (tdRNAs) inhibited the mRNA expression of metastasis-linked genes by competing with its work-partner RBP, promoting mRNA degradation, inhibiting its translation, or interfering with retrotransposon mobility. (b) Schematic shows long noncoding RNAs (lncRNAs) working in cancer progression through a variety of molecular mechanisms. lncRNAs guided proteins, such as transcription factors, to specific sites and further affected the expression of downstream genes by complementary base pairing or complex formation. lncRNAs also competed with miRNAs or proteins to bind to mRNA, thereby affecting its stability or translation initiation. The specific expression of lncRNAs were also found in metastasis. (c) circRNAs worked in metastasis. circRNAs acted as competing endogenous RNAs (ceRNA) of proteins or miRNAs, as modified partners, or disturbed metastasis-linked gene transcription or translation.
1.3.2. lncRNAs
lncRNAs are identified by their transcription length of more than 200 nucleotides, without or with partly protein-coding potential, and many of them are uniquely expressed in tissues and cells. lncRNAs influence a number of cellular functions in pathological and physiological processes; for example, lncRNAs have a variety of functions in tumorigenesis and metastasis by regulating the expression of targeted genes [85]. It has been reviewed that lncRNAs are involved in cancer phenotypes, like proliferation and invasion, through interactions with DNA, protein, or RNA [70, 86]. Because of lack of common characteristics, the existing lncRNAs can be classified according to their sequence features, biochemical pathways, subcellular location, or functions [87]. And the sequences of lncRNAs are poorly conserved; however, their action modes are similar. Here, we segregated their action models into four categories in cancer metastasis (Figure 4(b)). First, lncRNAs act as guides for downstream molecules to a special location. For example, upregulation of lncRNA HOTAIR indicated an increased risk of tumor invasion and metastasis in primary breast tumors. And the molecular mechanism was HOTAIR dysregulated downstream metastasis-linked gene expression by guiding Polycomb Repressive Complex 2 (PRC2) to histone H3 altering lysine 27 methylation [88]. Furthermore, lncRNA may be competitive with other molecules. For instance, lncRNA-ATB competitively binds with the miR-200 family from their targeted ZEB1 and ZEB2 gene, which indirectly contributes to the expression of the oncogene ZEB1/2 and promotes EMT and invasion in hepatocellular carcinoma [89]. Third, lncRNAs act as scaffolds to facilitate the molecules in executing their functions. lncRNA-mPvt1, as an example, is bound to NOP2 protein and enhances its stability, which is beneficial to cell proliferation, cell cycling, and the acquisition of stem cell-like properties in hepatocellular carcinoma (HCC) cells [90]. Lastly, lncRNAs may be specifically expressed in tissues or cells [91]. With more and more evidence showing that lncRNAs are emerging as important actors involved in cancer metastasis, a better understanding of the regulation and regulatory mechanism of lncRNA will shed light on the hope for novel clinical treatment.
1.3.3. miRNAs
MicroRNAs (miRNAs) are noncoding RNAs with a length of about 18~22 nucleotides that regulate gene expression by binding to the mRNA 3′UTR of targeted genes, leading to translational repression or mRNA decay [92]. It has been well studied how miRNAs are involved in the initiation, progression, and inhibition of cancer [93]. And miRNAs have been shown potential roles in the diagnosis, prognosis, and treatment in cancer clinical applications [94]. About 30% of human genes are regulated by miRNAs, and miRNA expression profiles might be associated with cancer types and their stages [95]. In general, miRNA expression is always downregulated in cancer compared with normal tissues [95]. Since miRNAs mediating metastasis were reported in 2007, various mechanisms about how miRNAs work in metastasis have been found [3]. Now, a growing body of evidence demonstrated that miRNA mediated metastasis gene expression, cancer stem cell-like phenotype, epithelial-to-mesenchymal transition (EMT), invasion, and migration, remodeling the tumor tissue microenvironment by exosomes [96]. Here, we selected 3 examples for each of these five aspects, as shown in Table 2. The key miRNAs related to metastasis specifically in cancer could be used as biomarkers for diagnosis and therapeutic target.
Table 2.
List of examples of miRNA-mediated pathways in cancer metastasis.
| Relative phenotypes | Pathways | Cancer types | Refs |
|---|---|---|---|
| Metastasis gene expression | mir-9-E-cadherin | Breast cancer | [97] |
| miR-1269a-Smad7 and HOXD10 | Colorectal cancer | [98] | |
| miR-122-ADAM17 | Hepatocellular carcinoma | [99] | |
|
| |||
| Cancer stem cell-like phenotype | miR-199a-FOXP2 | Breast cancer | [100] |
| miR-34a-CD44 | Prostate cancer | [101] | |
| miR-23a-metastasis suppressor 1 (MTSS1) | Colorectal cancer | [102] | |
|
| |||
| EMT | miR-30a-Snai1 | Non-small-cell lung cancer | [103] |
| miR-200 family/miR-205-ZEB1/SIP1 | Breast cancer | [104] | |
| miR-1296-SRPK1 | Hepatocellular carcinoma | [105] | |
|
| |||
| Invasion and migration | miR-21-programmed cell death 4/maspin | Breast cancer | [106] |
| miR-132-ZEB2 | Colorectal cancer | [107] | |
| miR-940-migration and invasion enhancer 1 | Prostate cancer | [108] | |
|
| |||
| Remodeling microenvironment by exosomal miRNA | miR-10b-HOXD10 and KLF4 | Breast cancer | [109] |
| miR-17, miR-19a, miR-19b, miR-20a, and miR-92-PTEN | Brain metastasis | [110] | |
| miR-103-VE-cadherin, p120-catenin, and zonula occludens 1 | Hepatocellular carcinoma | [111] | |
|
| |||
| Metastasis biomarker | miR-203 | Colorectal cancer | [112] |
| miR-1246 and miR-1290 | Non-small-cell lung cancer | [113] | |
| miR-200b | Breast cancer | [114] | |
1.3.4. circRNAs
Since the discovery of the first circRNA in 1976 [115], circRNAs have gone from being considered as “junk” generated by nonsense splicing events to a large class of noncoding RNAs exerting important biological functions in physiological or pathological processes [116]. circRNAs lack poly(A) tails and 5′ termini capping, and most of them come from known protein-coding genes by a back-splicing event [117]. And their expression have tissue- and cell-specific patterns [118, 119]. Studies have explored circRNAs involved in cancer with a steadily increasing pace, and there are at least four potential ways to elucidate the mechanisms of circRNAs in cancer (Figure 4(c)). Firstly, circRNAs act as miRNAs or protein sponges that indirectly regulate the functions of their downstream target genes. For example, circTP63 function as a competitive endogenous RNA (ceRNA) of miR-873-3p, thereby inhibiting miR-873-3p function on FOXM1, which finally leads to upregulating FOXM1 expression and cell cycle progression in lung squamous cell carcinoma [120]. In another example, HuR is a well-studied RNA-binding protein (RBP) regulating a range of RNA expression. The binding site of HuR and PABPN1 mRNA was blocked by circPABPN1, and the competition decreased the translation of PABPN1 [121]. Secondly, circRNAs work as protein scaffolds to regulate binding affinity. circ-Foxo3, as a case in point, promoted MDM2-induced p53 ubiquitination but prevented Foxo3 ubiquitination and affected subsequent degradation [122]. In addition, circRNAs recruit proteins to specific genomic locations and cooperate with epigenetic modifications to regulate the expression of downstream genes; for example, circular RNA FECR1 recruited TET1, a DNA demethylase, to the FLI1 promoter and activated FLI1 expression [123]. Furthermore, a number of recent studies have shown that circRNA own an open reading frame (ORF), and an internal ribosome entry site (IRES) can be translated into functional proteins [124]. Lastly, a subset of circRNAs that is dominantly localized in the nucleus may interact with the Pol II transcription complex to enhance transcription of their parent coding genes [125]. circRNAs used in translational and precision medicine researches has become a hotspot.
2. Conclusions and Future Perspective
In tumor progression, finding the key epigenetic molecules that control metastasis is of great clinical significance and will provide a new way to search for effective therapeutic drugs and molecular markers for diagnosis. Here, we present the available evidence from four parts on how epigenetics plays a role in cancer metastasis. First, DNA methylation or histone methylation, either alone or in combination with other epigenetic molecules (such as noncoding RNA), regulated the expression of metastasis-linked genes at the transcriptional level. Second, RNA methylation also took part in translation initiation, translation efficiency, and mRNA stability of metastasis-linked genes. Third, alternative splicing increased the diversity of mRNA and noncoding RNA that played important roles in tumor metastasis, and these processes were partly influenced by methylation modification of DNA or RNA. Fourth, noncoding RNAs were almost involved in the whole process of metastasis-linked genes from transcription initiation to protein production to biological function. It has been reported that some early clinical trials targeted epigenetics, for instance, by designing drugs that specifically target DNA methylation or histone methylation for cancer therapeutics (see the review in [12]). Epigenetic drugs' combination strategies, such as DNMT inhibitors combined with HDAC inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia, have also been investigated in clinical trials. [126]. While many researches have explored the role of epigenetics as a biomarker and have addressed key pathways driving disease processes, there are still some major challenges that exist in their application in broader clinical medicine. For example, how to ensure the accuracy and repeatability of the measurements, how to distinguish transient changes in disease from the true biomarkers, and how to translate the results into the precise medication for the individual to make sure maximum benefit for patients [127]. Epigenetic therapies present opportunities and challenges, and further research is still needed to translate these relatively significant findings into clinical applications.
Acknowledgments
This study is supported partially by the National Natural Science Foundation of China (grant number 82001159) and the Natural Science Foundation of Jiangxi Province (20202BCD42011).
Contributor Information
Jianping Xiong, Email: xiong_jp_ncu@126.com.
Mei Ma, Email: mamei1223@163.com.
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Bin Yu, Mei Ma, and Xin Yu retrieved the data and drafted the manuscript. Jianping Xiong supervised the study. Bin Yu and Xin Yu contributed equally to this work.
References
- 1.Chaffer C. L., Weinberg R. A. A perspective on cancer cell metastasis. Science . 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Fidler I. J. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nature Reviews. Cancer . 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 3.Ma L., Teruya-Feldstein J., Weinberg R. A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature . 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee A., Rodger E. J., Eccles M. R. Epigenetic drivers of tumourigenesis and cancer metastasis. Seminars in Cancer Biology . 2018;51:149–159. doi: 10.1016/j.semcancer.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B., Papadopoulos N., Velculescu V. E., Zhou S., Diaz L. A., Jr., Kinzler K. W. Cancer genome landscapes. Science . 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lujambio A., Calin G. A., Villanueva A., et al. A microRNA DNA methylation signature for human cancer metastasis. Proceedings of the National Academy of Sciences . 2008;105(36):13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu X., Kuang Y., Li L., et al. Epigenomic and functional characterization of junctophilin 3 (JPH3) as a novel tumor suppressor being frequently inactivated by promoter CpG methylation in digestive cancers. Theranostics . 2017;7(7):2150–2163. doi: 10.7150/thno.18185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuo W., Liu Y., Li S., et al. Long Noncoding RNA _GMAN_ , Up-regulated in Gastric Cancer Tissues, Is Associated With Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding _GMAN-AS_. Gastroenterology . 2019;156(3):676–691.e11. doi: 10.1053/j.gastro.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg A. P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature . 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 10.Goelz S. E., Vogelstein B., Hamilton S. R., Feinberg A. P. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science . 1985;228(4696):187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 11.Rainier S., Johnson L. A., Dobry C. J., Ping A. J., Grundy P. E., Feinberg A. P. Relaxation of imprinted genes in human cancer. Nature . 1993;362(6422):747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 12.Michalak E. M., Burr M. L., Bannister A. J., Dawson M. A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nature Reviews. Molecular Cell Biology . 2019;20(10):573–589. doi: 10.1038/s41580-019-0143-1. [DOI] [PubMed] [Google Scholar]
- 13.Bestor T. H. The DNA methyltransferases of mammals. Human Molecular Genetics . 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 14.Kohli R. M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature . 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews. Genetics . 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 16.Lister R., Pelizzola M., Dowen R. H., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature . 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozturk S., Papageorgis P., Wong C. K., et al. SDPR functions as a metastasis suppressor in breast cancer by promoting apoptosis. Proceedings of the National Academy of Sciences of the United States of America . 2016;113(3):638–643. doi: 10.1073/pnas.1514663113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gkountela S., Castro-Giner F., Szczerba B. M., et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell . 2019;176(1-2):98–112.e14. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzenbach H., Hoon D. S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews. Cancer . 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 20.Visvanathan K., Fackler M. S., Zhang Z., et al. Monitoring of serum DNA methylation as an early independent marker of response and survival in metastatic breast cancer: TBCRC 005 prospective biomarker study. Journal of Clinical Oncology . 2017;35(7):751–758. doi: 10.1200/JCO.2015.66.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J., Song G., Tang Q., et al. IRX1 hypomethylation promotes osteosarcoma metastasis via induction of CXCL14/NF-κB signaling. The Journal of Clinical Investigation . 2015;125(5):1839–1856. doi: 10.1172/JCI78437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conkie D., Affara N., Harrison P. R., Paul J., Jones K. In situ localization of globin messenger RNA FORMATION. The Journal of Cell Biology . 1974;63(2):414–419. doi: 10.1083/jcb.63.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B. S., Roundtree I. A., He C. Post-transcriptional gene regulation by mRNA modifications. Nature Reviews. Molecular Cell Biology . 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell . 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X., Yang Y., Sun B. F., et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Research . 2014;24(12):1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., Lu Z., Gomez A., et al. _N_ 6-methyladenosine-dependent regulation of messenger RNA stability. Nature . 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Zhao B. S., Roundtree I. A., et al. _N_ _6_ -methyladenosine Modulates Messenger RNA Translation Efficiency. Cell . 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geula S., Moshitch-Moshkovitz S., Dominissini D., et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science . 2015;347(6225):1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 29.Jia G., Fu Y., Zhao X., et al. _N_ 6-Methyladenosine in nuclear RNA is a major substrate of the obesity- associated FTO. Nature Chemical Biology . 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng G., Dahl J. A., Niu Y., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell . 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemkine G. F., Raji A., Alfama G., et al. Adult neural stem cell cycling in vivo requires thyroid hormone and its alpha receptor. The FASEB Journal . 2005;19(7):1–17. doi: 10.1096/fj.04-2916fje. [DOI] [PubMed] [Google Scholar]
- 32.Batista P. J., Molinie B., Wang J., et al. m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell . 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma J. Z., Yang F., Zhou C. C., et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulatingN6‐methyladenosine‐dependent primary microRNA processing. Hepatology . 2017;65(2):529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 34.Lin S., Choe J., Du P., Triboulet R., Gregory R. I. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Molecular Cell . 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou C., Zhang Z., Zhu X., et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. eBioMedicine . 2020;59:p. 102955. doi: 10.1016/j.ebiom.2020.102955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linder B., Grozhik A. V., Olarerin-George A. O., Meydan C., Mason C. E., Jaffrey S. R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nature Methods . 2015;12(8):767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y., Garcia B. A. Comprehensive catalog of currently documented histone modifications. Cold Spring Harbor Perspectives in Biology . 2015;7(9, article a025064) doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Experimental & Molecular Medicine . 2017;49(4) doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li N., Li Y., Lv J., et al. ZMYND8 reads the dual histone mark H3K4me1-H3K14ac to antagonize the expression of metastasis-linked genes. Molecular Cell . 2016;63(3):470–484. doi: 10.1016/j.molcel.2016.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M. W., Hua K. T., Kao H. J., et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Research . 2010;70(20):7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 41.Kim J. H., Sharma A., Dhar S. S., et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Research . 2014;74(6):1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowbotham S. P., Li F., Dost A. F. M., et al. H3K9 methyltransferases and demethylases control lung tumor-propagating cells and lung cancer progression. Nature Communications . 2018;9(1):p. 4559. doi: 10.1038/s41467-018-07077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park W. Y., Hong B. J., Lee J., Choi C., Kim M. Y. H3K27 demethylase JMJD3 employs the NF-κB and BMP signaling pathways to modulate the tumor microenvironment and promote melanoma progression and metastasis. Cancer Research . 2016;76(1):161–170. doi: 10.1158/0008-5472.CAN-15-0536. [DOI] [PubMed] [Google Scholar]
- 44.Kang J., Shin S. H., Yoon H., et al. FIH is an oxygen sensor in ovarian cancer for G9a/GLP-driven epigenetic regulation of metastasis-related genes. Cancer Research . 2018;78(5):1184–1199. doi: 10.1158/0008-5472.CAN-17-2506. [DOI] [PubMed] [Google Scholar]
- 45.Joyce J. A., Pollard J. W. Microenvironmental regulation of metastasis. Nature Reviews. Cancer . 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piunti A., Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science . 2016;352(6290, article aad9780) doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 47.Stein E. M., Garcia-Manero G., Rizzieri D. A., et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood . 2018;131(24):2661–2669. doi: 10.1182/blood-2017-12-818948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klose R. J., Kallin E. M., Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature Reviews. Genetics . 2006;7(9):715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 49.Yang X., Coulombe-Huntington J., Kang S., et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell . 2016;164(4):805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang E. T., Sandberg R., Luo S., et al. Alternative isoform regulation in human tissue transcriptomes. Nature . 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baralle F. E., Giudice J. Alternative splicing as a regulator of development and tissue identity. Nature Reviews. Molecular Cell Biology . 2017;18(7):437–451. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J., Manley J. L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discovery . 2013;3(11):1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luco R. F., Pan Q., Tominaga K., Blencowe B. J., Pereira-Smith O. M., Misteli T. Regulation of alternative splicing by histone modifications. Science . 2010;327(5968):996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X. S., Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genetics . 2009;41(3):376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angermueller C., Clark S. J., Lee H. J., et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature Methods . 2016;13(3):229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linker S. M., Urban L., Clark S. J., et al. Combined single-cell profiling of expression and DNA methylation reveals splicing regulation and heterogeneity. Genome Biology . 2019;20(1) doi: 10.1186/s13059-019-1644-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shukla S., Kavak E., Gregory M., et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature . 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Climente-Gonzalez H., Porta-Pardo E., Godzik A., Eyras E. The functional impact of alternative splicing in cancer. Cell Reports . 2017;20(9):2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 59.Lee S. C., Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nature Medicine . 2016;22(9):976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pradella D., Naro C., Sette C., Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Molecular Cancer . 2017;16(1) doi: 10.1186/s12943-016-0579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Q., Dhir R., Wells A. Altered CXCR3 isoform expression regulates prostate cancer cell migration and invasion. Molecular Cancer . 2012;11(1) doi: 10.1186/1476-4598-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oltean S., Bates D. O. Hallmarks of alternative splicing in cancer. Oncogene . 2014;33(46):5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 63.Yae T., Tsuchihashi K., Ishimoto T., et al. Alternative splicing of _CD44_ mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nature Communications . 2012;3(1):p. 883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 64.Williams C. S., Zhang B., Smith J. J., et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. The Journal of Clinical Investigation . 2011;121(10):4056–4069. doi: 10.1172/JCI44228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aryee M. J., Liu W., Engelmann J. C., et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Science Translational Medicine . 2013;5(169, article 169ra110) doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J. H., Dhanasekaran S. M., Prensner J. R., et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Research . 2011;21(7):1028–1041. doi: 10.1101/gr.119347.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ren X., Yang X., Cheng B., et al. _HOPX_ hypermethylation promotes metastasis via activating _SNAIL_ transcription in nasopharyngeal carcinoma. Nature Communications . 2017;8(1) doi: 10.1038/ncomms14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Cubas A. A., Korpershoek E., Inglada-Perez L., et al. DNA methylation profiling in pheochromocytoma and paraganglioma reveals diagnostic and prognostic markers. Clinical Cancer Research . 2015;21(13):3020–3030. doi: 10.1158/1078-0432.CCR-14-2804. [DOI] [PubMed] [Google Scholar]
- 69.Warzecha C. C., Jiang P., Amirikian K., et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. The EMBO Journal . 2010;29(19):3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ishiyama N., Lee S. H., Liu S., et al. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell . 2010;141(1):117–128. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 71.Schmitt A. M., Chang H. Y. Long noncoding RNAs in cancer pathways. Cancer Cell . 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caiment F., Gaj S., Claessen S., Kleinjans J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Research . 2015;43(5):2525–2534. doi: 10.1093/nar/gkv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell . 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 74.Yang Q., Du W. W., Wu N., et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death and Differentiation . 2017;24(9):1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du W. W., Zhang C., Yang W., Yong T., Awan F. M., Yang B. B. Identifying and characterizing circRNA-protein interaction. Theranostics . 2017;7(17):4183–4191. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar P., Kuscu C., Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends in Biochemical Sciences . 2016;41(8):679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balatti V., Nigita G., Veneziano D., et al. tsRNA signatures in cancer. Proceedings of the National Academy of Sciences of the United States of America . 2017;114(30):8071–8076. doi: 10.1073/pnas.1706908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Honda S., Loher P., Shigematsu M., et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proceedings of the National Academy of Sciences of the United States of America . 2015;112(29):E3816–E3825. doi: 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A. Z., Kay M. A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA . 2010;16(4):673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saikia M., Hatzoglou M. The Many Virtues of tRNA-derived Stress-induced RNAs (tiRNAs): Discovering Novel Mechanisms of Stress Response and Effect on Human Health∗. The Journal of Biological Chemistry . 2015;290(50):29761–29768. doi: 10.1074/jbc.R115.694661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schorn A. J., Gutbrod M. J., LeBlanc C., Martienssen R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell . 2017;170(1):61–71.e11. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodarzi H., Liu X., Nguyen H. C., Zhang S., Fish L., Tavazoie S. F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell . 2015;161(4):790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang B., Yang H., Cheng X., et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Research . 2017;77(12):3194–3206. doi: 10.1158/0008-5472.CAN-16-3146. [DOI] [PubMed] [Google Scholar]
- 84.Ivanov P., Emara M. M., Villen J., Gygi S. P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Molecular Cell . 2011;43(4):613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhan A., Soleimani M., Mandal S. S. Long noncoding RNA and cancer: a new paradigm. Cancer Research . 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hanahan D., Weinberg R. A. The hallmarks of cancer. Cell . 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 87.St Laurent G., Wahlestedt C., Kapranov P. The landscape of long noncoding RNA classification. Trends in Genetics . 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta R. A., Shah N., Wang K. C., et al. Long non-coding RNA _HOTAIR_ reprograms chromatin state to promote cancer metastasis. Nature . 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan J. H., Yang F., Wang F., et al. A Long Noncoding RNA Activated by TGF-β Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer Cell . 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 90.Wang F., Yuan J. H., Wang S. B., et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology . 2014;60(4):1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 91.Ward M., McEwan C., Mills J. D., Janitz M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. Journal of Human Transcriptome . 2015;1(1):2–9. doi: 10.3109/23324015.2015.1077591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews. Genetics . 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 93.Garzon R., Calin G. A., Croce C. M. MicroRNAs in cancer. Annual Review of Medicine . 2009;60(1):167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 94.Brown B. D., Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nature Reviews. Genetics . 2009;10(8):578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 95.Lu J., Getz G., Miska E. A., et al. MicroRNA expression profiles classify human cancers. Nature . 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 96.Kim J., Yao F., Xiao Z., Sun Y., Ma L. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Reviews . 2018;37(1):5–15. doi: 10.1007/s10555-017-9712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma L., Young J., Prabhala H., et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biology . 2010;12(3):247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bu P., Wang L., Chen K. Y., et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nature Communications . 2015;6(1) doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsai W. C., Hsu P. W., Lai T. C., et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology . 2009;49(5):1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 100.Cuiffo B. G., Campagne A., Bell G. W., et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell . 2014;15(6):762–774. doi: 10.1016/j.stem.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 101.Liu C., Kelnar K., Liu B., et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature Medicine . 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jahid S., Sun J., Edwards R. A., et al. miR-23aPromotes the transition from indolent to invasive colorectal cancer. Cancer Discovery . 2012;2(6):540–553. doi: 10.1158/2159-8290.CD-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumarswamy R., Mudduluru G., Ceppi P., et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. International Journal of Cancer . 2012;130(9):2044–2053. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 104.Gregory P. A., Bert A. G., Paterson E. L., et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology . 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 105.Xu Q., Liu X., Liu Z., et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Molecular Cancer . 2017;16(1) doi: 10.1186/s12943-017-0675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y. Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Research . 2008;18(3):350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 107.Zheng Y. B., Luo H. P., Shi Q., et al. miR-132 inhibits colorectal cancer invasion and metastasisviadirectly targeting ZEB2. World Journal of Gastroenterology . 2014;20(21):6515–6522. doi: 10.3748/wjg.v20.i21.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rajendiran S., Parwani A. V., Hare R. J., Dasgupta S., Roby R. K., Vishwanatha J. K. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Molecular Cancer . 2014;13(1) doi: 10.1186/1476-4598-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Singh R., Pochampally R., Watabe K., Lu Z., Mo Y. Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Molecular Cancer . 2014;13(1) doi: 10.1186/1476-4598-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang L., Zhang S., Yao J., et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature . 2015;527(7576):100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fang J. H., Zhang Z. J., Shang L. R., et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology . 2018;68(4):1459–1475. doi: 10.1002/hep.29920. [DOI] [PubMed] [Google Scholar]
- 112.Hur K., Toiyama Y., Okugawa Y., et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut . 2017;66(4):654–665. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang W. C., Chin T. M., Yang H., et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nature Communications . 2016;7(1) doi: 10.1038/ncomms11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Madhavan D., Zucknick M., Wallwiener M., et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clinical Cancer Research . 2012;18(21):5972–5982. doi: 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 115.Sanger H. L., Klotz G., Riesner D., Gross H. J., Kleinschmidt A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proceedings of the National Academy of Sciences of the United States of America . 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kristensen L. S., Andersen M. S., Stagsted L. V. W., Ebbesen K. K., Hansen T. B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nature Reviews. Genetics . 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 117.Guo J. U., Agarwal V., Guo H., Bartel D. P. Expanded identification and characterization of mammalian circular RNAs. Genome Biology . 2014;15(7):p. 409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xia S., Feng J., Lei L., et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings in Bioinformatics . 2017;18(6):984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 119.Salzman J., Chen R. E., Olsen M. N., Wang P. L., Brown P. O. Cell-type specific features of circular RNA expression. PLoS Genetics . 2013;9(9, article e1003777) doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cheng Z., Yu C., Cui S., et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nature Communications . 2019;10(1):p. 3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Abdelmohsen K., Panda A. C., Munk R., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation byCircPABPN1. RNA Biology . 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Du W. W., Fang L., Yang W., et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death and Differentiation . 2017;24(2):357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen N., Zhao G., Yan X., et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biology . 2018;19(1) doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pamudurti N. R., Bartok O., Jens M., et al. Translation of circRNAs. Molecular Cell . 2017;66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li Z., Huang C., Bao C., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology . 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 126.Prebet T., Sun Z., Figueroa M. E., et al. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US Leukemia Intergroup Trial E1905. Journal of Clinical Oncology . 2014;32(12):1242–1248. doi: 10.1200/JCO.2013.50.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carter A. C., Chang H. Y., Church G., et al. Challenges and recommendations for epigenomics in precision health. Nature Biotechnology . 2017;35(12):1128–1132. doi: 10.1038/nbt.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]


