Abstract
The efficacy of the Statens Serum Institut (SSI) enteric medium for isolation and direct identification of enteric pathogens was evaluated. Six different biochemical reactions can be read by using the SSI enteric medium, allowing direct identification of a range of enteric pathogens. All 248 gram-negative bacterial species that were tested grew on the SSI enteric medium. Only 10 of 248 bacteria (4%) showed discrepant results in the biochemical reactions, and none of these were enteric pathogens. Forty-three of 47 enteric pathogens (92%) produced identical rates of semiquantitative growth on the SSI enteric medium and 5% blood agar, whereas three Vibrio spp. and one Aeromonas spp. showed reduced growth. Gram-positive bacteria did not grow on the SSI enteric medium. Most enteric pathogens had a detection limit of 50 bacteria per ml of feces, but higher numbers of Vibrio spp. and some Shigella spp. were required for detection. The growth rates of 125 enteric pathogens and 12 Yersinia spp. on the SSI enteric medium, xylose lysine deoxycholate (XLD), Hektoen enteric (HE), Salmonella-Shigella (SS), and cefsulodin-irgasan-novobiocin (CIN) agar were compared. Detection rates after application of 200 CFU were 99% for SSI enteric medium, 92% for XLD, 88% for HE, and 82% for SS agar. The 12 Yersinia spp. grew excellently on both the SSI enteric medium and CIN agar. We conclude that the performance of the SSI enteric medium compares favorably to those of other media tested. Its ability to detect Yersinia spp. may limit the number of media needed in the typical laboratory. The direct identification of enteric pathogens on the medium may also provide a more rapid diagnosis.
Identification of organisms of the family Enterobacteriaceae constitutes a considerable proportion of the workload in many clinical microbiology laboratories. The primary goal in a diagnostic microbiologiy laboratory is the rapid identification of a pathogen. The traditional methodology for routine detection of enteric pathogens nearly always employs a combination of at least three different media in order to increase the sensitivity and the specificity of the detection and identification method. This is due to the fact that if one medium is highly inhibitory to some members of the Enterobacteriaceae, e.g., Escherichia coli, there is a simultaneous loss of sensitivity for fastidious pathogens such as the Shigella spp. (21). At the Statens Serum Institut (SSI; Copenhagen, Denmark), a medium that combines differentiating and selective properties without loss of the ability to detect has been developed. For more than 20 years, the SSI enteric medium alone has been used for detection of enteric pathogens (with the exception of Campylobacter spp. and anaerobic bacteria) in our reference laboratory, where 75,000 to 80,000 stool samples are investigated each year. Although the SSI enteric medium has been used succesfully for such a long time in typical clinical microbiology laboratories in Denmark, it has never been evaluated in tests designed for that purpose. The aims of this study were to evaluate the SSI enteric medium and to compare its ability to detect and identify enteric pathogens to those of other standard enteric media. Semiquantitative growth, the ability of the medium to produce distinctive biochemical reactions, and the detection limits in stools were evaluated. Also, quantitative growth on SSI medium was compared with that on other enteric media.
(This work was presented at the 98th General Meeting of the American Society for Microbiology, Atlanta, Ga., 17 to 21 May 1998 [12a].)
MATERIALS AND METHODS
Bacterial strains.
A total of 275 different strains, representing 28 genera, were used for the evaluation of SSI enteric medium. The strains were isolated from clinical specimens at the SSI, and they were chosen to include the whole spectrum of gram-negative rods (n = 264) with the addition of a number of gram-positive bacteria (n = 11) as controls to examine the selectivity of the medium (Table 1). The bacteria were identified according to routine procedures. Only major pathogenic species were evaluated for their detection limits in stools and compared for their quantitative growth on the different enteric media. The 275 strains represented a pool from which the bacteria for each investigation were selected.
TABLE 1.
Genera and number of strains used to evaluate the SSI enteric medium
| Species | n |
|---|---|
| Gram-negative bacteria | |
| Salmonella spp. | |
| Salmonella typhi | 2 |
| Salmonella paratyphi | 3 |
| Other Salmonella spp. | 65 |
| Shigella spp. | |
| Shigella dysenteriae | 2 |
| Other Shigella spp. | 29 |
| Vibrio spp. | |
| Vibrio cholerae | 7 |
| Other Vibrio spp. | 2 |
| Yersinia spp. | |
| Yersinia enterocolitica | 21 |
| Other Yersinia spp. | 7 |
| Citrobacter spp. | 3 |
| Escherichia coli | 16 |
| Klebsiella spp. | 12 |
| Providencia spp. | 11 |
| Proteus spp. | 20 |
| Enterobacter spp. | 17 |
| Serratia spp. | 16 |
| Erwinia spp. | 1 |
| Alcaligenes spp. | 4 |
| Aeromonas spp. | 7 |
| Burkholderia spp. | 1 |
| Agrobacterium spp. | 2 |
| Cedecea spp. | 1 |
| Acinetobacter spp. | 4 |
| Flavobacterium spp. | 1 |
| Moraxella spp. | 1 |
| Pseudomonas spp. | 5 |
| Shewanella spp. | 1 |
| Plesiomonas spp. | 1 |
| Flavimonas spp. | 1 |
| Xanthomonas spp. | 1 |
| Subtotal | 264 |
| Gram-positive bacteria | |
| Staphylococcus spp. | 4 |
| Enterococcus spp. | 2 |
| Streptococcus spp. | 5 |
| Subtotal | 11 |
| Total | 275 |
SSI enteric medium.
The SSI enteric medium is available both as agar plates and granulated powder. The selective action on SSI enteric medium is caused by the combination of a detergent (sodium dodecyl benzene sulfonate) and sodium deoxycholate, on which gram-positive bacteria, gram-negative cocci, and certain gram-negative rods (e.g., Pasteurella and Haemophilus) are excluded from growth. Also, the detergent is an efficient inhibitor of the swarming of nearly all Proteus strains. Upon inoculation, the medium produces four differentiating reactions.
(i) H2S reaction.
Due to the presence of sodium thiosulfate and ferric citrate in the agar, H2S-positive bacteria produce a distinct ferrosulfide precipitate located centrally and deeply in the more anaerobic parts of the colony.
(ii) Lactose fermentation.
Fermentation is manifested by acid production made visible by using as an indicator the combination of deoxycholate and neutral red. The acid production is confined to the colony and its immediate surroundings due to the reaction with deoxycholate, which yields deoxycholic acid. Deoxycholic acid precipitates and becomes moderately colored by neutral red. The confinement of the reaction to the colony makes recognition of adjacent lactose-negative colonies possible.
(iii) Phenylalanine deaminase reaction.
Phenylalanine deaminase-active bacteria are recognized. With ferric citrate in an acid environment, the conversion of l-phenylalanine to phenylpyruvate is visible as a brown, diffusible pigment in the SSI enteric medium.
(iv) Indole reaction.
This reaction is included as a differentiating characteristic in a pure subculture from a suspect colony on the primary medium. Deamination of tryptophan produces a volatile chemical, indole, which may be detected by placing a piece of filter paper inside the lid of the inverted petri dish that has been wetted with the reagents of Ehrlich’s indole test, i.e., p-dimethylaminobenzaldehyde, ethyl alcohol, and hydrogen chloride.
Table 2 provides more detailed information on how different species are identified on the SSI enteric medium (see also Fig. 1).
TABLE 2.
Differentiating reactions produced by the SSI enteric medium and possible identity of the pathogenic bacteria
| Reaction
|
Possible identity | |||
|---|---|---|---|---|
| H2S | Lactose | PDa | Indole | |
| + | − | − | − | Salmonella spp. (including Salmonella arizonae), lactose-negative Citrobacter freundii, Shewanella putrefaciens |
| + | − | − | + | E. coli (of H2S-positive variety), Edwardsiella tarda |
| + | + | − | − | Citrobacter freundii, lactose-positive Salmonella spp. |
| + | + | − | + | E. coli (of H2S-positive variety) |
| + | − | + | − | Proteus mirabilis |
| + | − | + | + | Proteus vulgaris |
| − | − | + | + | Morganella morganii, Providencia spp. |
| − | − | − | − | Shigella spp., Yersinia spp., Salmonella paratyphi A |
| − | − | − | + | Shigella spp., Vibrio spp., Aeromonas spp., E. coli, Plesiomonas spp. |
| − | + | − | − | Klebsiella pneumoniae, Enterobacter spp. |
| − | + | − | + | E. coli, Aeromonas spp., Klebsiella oxytoca |
Phenylalanine deaminase.
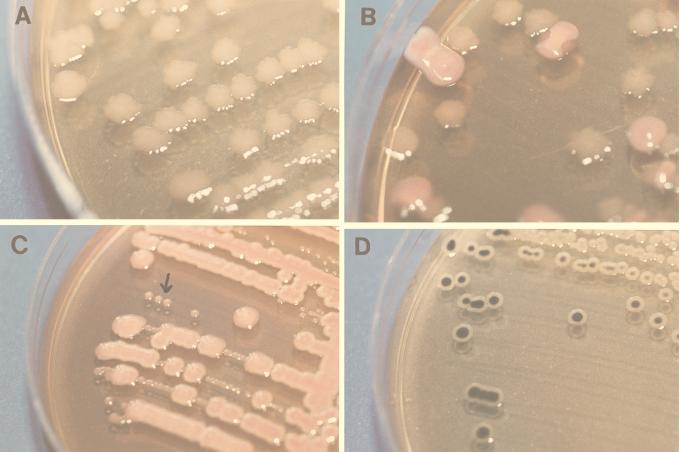
FIG. 1.
Colony appearance of four different genera of the Enterobacteriaceae on SSI enteric medium. SSI enteric medium yields six differentiating biochemical reactions that allow direct identification of a range of enteric pathogens (see the text). In this figure, only the four major reactions are illustrated, with different bacteria. (A) Shigella sonnei, with the characteristic rough transformations, causes the surfaces and the edges of the colonies to become serrated. (B) E. coli and Shigella sonnei. E. coli organisms appear as red colonies due to lactose production, and there is no rough transformation. (C) E. coli and Y. enterocolitica. Yersinia spp. appear as tiny round translucent colonies which resemble pearls on a string (indicated by the arrow). They are easily recognized compared to the much larger, red colonies of E. coli. (D) H2S-positive Salmonella spp. H2-positive organisms appear with a black precipitate located centrally and deeply in the anaerobic parts of the colony.
Two other reactions visible with the SSI enteric medium are useful for the identification of Salmonella spp. and Shigella sonnei; however, both require a certain degree of practical experience before routine use.
(i) Metallic sheen reaction.
This reaction is characterized by a metallic sheen on top of the black center in H2S-producing Salmonella (except in Salmonella typhi). The cause of this metallic sheen is largely unknown.
(ii) Rough transformation.
Due to rather high concentrations of divalent ions (Mg++ and Ca++), a rough transformation is regularly induced in Shigella sonnei, affecting the surfaces and the edges of the colonies. The resulting increased spreading of the colonies makes recognition possible also when they are located among colonies of other bacteria.
Other media. (i) XLD agar.
Xylose-lysine-desoxycholate (XLD) agar (Oxoid, Basingstoke, United Kingdom) was originally formulated by Taylor for the isolation and identification of Shigella spp. from stool specimens (17). It has since been found also to be a satisfactory medium for Salmonella spp. (8). XLD agar contains deoxycholate as the selective agent and identifies the bacteria according to lactose, sucrose, and xylose fermentation; H2S production; and lysine decarboxylation.
(ii) HE agar.
Hektoen enteric (HE) agar (Oxoid) was developed by King and Metzger (10) to grow shigellae as readily as other pathogens while inhibiting normal intestinal flora. HE agar contains bile salts and may contain novobiocin as selective agent; it identifies the bacteria according to lactose, sucrose, and salicin fermentation and H2S production.
(iii) SS agar.
Salmonella-Shigella (SS) agar (Becton Dickinson, Basel, Switzerland) is a differential selective medium for the isolation of Salmonella and some Shigella species from clinical speciments. It contains brilliant green and bile salts as selective agents and identifies bacteria according to lactose fermentation and H2S production.
(iv) CIN agar.
Cefsulodin-irgasan-novobiocin (CIN) agar (Oxoid) was developed by Schiemann (15) for the isolation of Yersinia enterocolitica. It contains bile salts and antibiotics as selective agents and identifies the bacteria by mannitol fermentation.
All the media were prepared according to the manufacturers’ instructions at the Media Department at the SSI. XLD, HE, SS, and CIN agar have been described, evaluated, and compared by others (1, 3–11, 14–21).
Confirmatory reactions.
H2S production was investigated by the addition of several colonies into a ferrocloride-peptone-gelatin medium (SSI). After incubation at 22°C for a maximum of 4 days, H2S-positive bacteria were black or grey along the stab line (12). We tested for phenylalanine deaminase by the addition of several colonies into a culture medium containing l-phenylalanine (SSI) followed by incubation at 35°C for 2 to 4 h and the addition of a few drops of reagent (FeCl3 · 6H2O and 5 N HCl). Phenylalanine deaminase-active bacteria give an immediate and strong dark green coloring of the culture medium (2). Lactose and indole fermentation was confirmed by using standard procedures. The indole reaction was performed after xylene extraction.
Growth and reactivity of gram-negative rods on the SSI enteric medium.
Bacteria were grown overnight on 5% blood agar (SSI). Several colonies were streaked on the SSI enteric medium and incubated overnight at 35°C. The plates were examined for the colony characteristics, size, and color of the test organisms, and the number of colonies on the medium was determined. Isolates showing unexpected reactions on the SSI enteric medium were retested further in confirmatory reactions by using the standard procedures described earlier.
Semiquantitative growth.
A total of 59 bacterial strains were tested; 48 represented the relevant gram-negative organisms, and 11 were gram-positive controls. Bacteria of each strain were suspended in saline and were adjusted to approximately 2 × 108 CFU/ml as measured by optical density (540 nm) using a colorimeter (Ciba-Corning Colorimeter 254; Ciba Corning Diagnostics Ltd., Suffolk, United Kingdom). The bacterial suspensions were diluted in saline to produce 10−1 to 10−7 suspensions, and 200 μl of each (including the undiluted bacterial suspension) was plated onto 5% blood agar (SSI) and the SSI enteric medium. The bacteria were categorized as having the same growth on both media, less growth on SSI enteric medium than on 5% blood agar, or no growth. Same growth indicates that the number of colonies on the SSI enteric medium was between 10 and 120% of the number on the 5% blood agar, whereas less growth was between 0.1 and 10%.
Detection limit for enteric pathogens in simulated stool cultures.
The most common enteric pathogens (a total of 28 strains) were selected for this experiment. The bacteria in question were suspended in saline, adjusted to approximately 2 × 108 CFU/ml and diluted in saline from 10−1 to 10−7. The diluted bacteria were mixed with normal feces (e.g., fecal samples with no growth of enteric pathogens) by suspending 10 μl of feces into 2 ml of phosphate-buffered saline (pH 7.38) and adding 5 μl of each dilution of bacteria. The suspension was mixed, and 10 μl was plated on SSI enteric medium and incubated at 35°C overnight. The highest dilution resulting in growth on the medium was determined.
Comparison with other enteric media.
A total of 125 strains belonging to the major pathogenic species were tested on XLD, CIN, HE, and SS agar, and the results were compared with those obtained with SSI enteric medium. The bacteria in question were suspended and diluted so that an inoculum of ∼200 CFU was applied. Each of the five media were inoculated on the same day by the same technician. After incubation overnight at 35°C the number of colonies was counted.
RESULTS
Growth and reactivity of gram-negative rods on the SSI enteric medium.
All tested bacteria (n = 248) grew on SSI enteric medium and very few (10 of 248 [4%]) showed discrepant biochemical reactions (H2S production, lactose fermentation, phenylalanine deaminase production, and indole production) (Table 3). Five strains (Klebsiella pneumoniae, Providencia rettgeri, and three Enterobacter cloacae strains) gave a false-negative reaction for lactose fermentation. Three other strains (Citrobacter freundii, Proteus vulgaris, and Proteus mirabilis) did not produce H2S, and one strain (Alcaligenes piechaudii) did not produce phenylalanine deaminase. However, all of these strains expressed the expected reactions in confirmatory tests.
TABLE 3.
Percentages of agreement with expected biochemical reactions of 248 gram-negative rods on the SSI enteric medium
| Bacterial type | No. of strains whose reaction was in agreement with the expected reaction/no. tested (% in agreement) | Result for strain whose reaction was not in agreement with the expected reaction
|
|
|---|---|---|---|
| n | Reaction | ||
| Salmonella spp. | 61/61 (100) | ||
| Shigella spp. | 28/28 (100) | ||
| Vibrio spp. | 5/5 (100) | ||
| Yersinia spp. | 24/24 (100) | ||
| Citrobacter spp. | 12/13 (92) | 1 | H2S negative |
| Escherichia coli | 12/12 (100) | ||
| Klebsiella spp. | 11/12 (92) | 1 | Lactose negative |
| Providencia spp. | 10/11 (90) | 1 | Lactose negative |
| Proteus spp. | 18/20 (90) | 2 | H2S negative |
| Enterobacter spp. | 14/17 (82) | 3 | Lactose negative |
| Serratia spp. | 15/16 (94) | 1a | Indole positive |
| Erwinia spp. | 1/1 (100) | ||
| Alcaligenes spp. | 3/4 (75) | 1 | PDb |
| Other gram-negative rodsc | 24/24 (100) | ||
Judged positive for a negative strain.
Phenylalanine deaminase production.
Other gram-negative rods include Aeromonas spp. (n = 4), Burkholderia spp. (n = 1), Agrobacterium spp. (n = 2), Cedecea spp. (n = 1), Acinetobacter spp. (n = 4), Flavobacterium spp. (n = 1), Moraxella spp. (n = 1), Pseudomonas spp. (n = 7), Shewanella spp. (n = 1), Plesiomonas spp. (n = 1), and Flavimonas spp. (n = 1).
Semiquantitative growth.
Most gram-negative rods (42 of 47 tested [92%]) grew with little or no inhibition on SSI enteric medium compared to growth on 5% blood agar (Table 4). Three Vibrio spp. (one each of V. vulnificus, V. cholera O139, and V. parahaemolyticus) and an Aeromonas sobria strain showed reduced growth (10- to 1,000-fold). The gram-positive bacteria did not grow on SSI enteric medium.
TABLE 4.
Semiquantitative growth of 59 bacterial strains on the SSI enteric medium compared to growth on 5% blood agara
| Bacterial type | No. of strains whose growth rate on SSI enteric medium wasb:
|
||
|---|---|---|---|
| The same as that on 5% blood agar | Lower than that on 5% blood agar | Not observed (no growth) | |
| Salmonella spp. | 9 | ||
| Shigella spp. | 5 | ||
| Vibrio spp. | 2 | 3 | |
| Yersinia spp. | 4 | ||
| Citrobacter spp. | 2 | ||
| Escherichia coli | 6 | ||
| Klebsiella spp. | 2 | ||
| Proteus spp. | 4 | ||
| Enterobacter spp. | 2 | ||
| Serratia spp. | 3 | ||
| Other gram-negative rodsc | 5 | 1 | |
| Gram-positive bacteriad | 11e | ||
The original (100) and 10-fold dilutions of 10−1 to 10−7 were plated.
Relative growth rates were calculated as follows: (CFU per milliliter on SSI enteric medium/CFU per milliliter on 5% blood agar) × 100. Growth rates were deemed the same if growth on SSI enteric medium was 10 to 120% of that on blood agar and were deemed lower if growth on SSI enteric medium was ≤0.1 to <10% of that on 5% blood agar.
Other gram-negative rods include Aeromonas spp. (n = 3), Acinetobacter spp. (n = 1), Pseudomonas (n = 1), and Xanthomonas spp. (n = 1).
Gram-positive bacteria include Staphylococcus spp. (n = 4), Enterococcus spp. (n = 2), and Streptococcus spp. (n = 5).
Four strains grew undiluted (∼108 CFU/ml).
Detection limit for enteric pathogens in simulated stool cultures.
Most enteropathogens (16 of 28 [57%]) had a detection limit of 50 bacteria/ml of feces except for Vibrio spp. and some Shigella spp. (Table 5). Of the five Vibrio spp., one was not detected (V. cholera non-O1), two had a detection limit of 5,000 bacteria/ml of feces (V. vulnificus and V. cholera O139), and two had a detection limit of 50,000 CFU/ml of feces (V. parahaemolyticus and V. cholera O1). The strains with the high detection limits were the same as the ones showing reduced growth in the semiquantitative growth experiment. Of the four Shigella spp. tested, one had a detection limit of only 5 CFU/ml of feces (S. sonnei) and three had a detection limit of 500 CFU/ml of feces (two S. flexneri and S. boydii).
TABLE 5.
Detection limits of 28 enteric pathogens with SSI enteric medium
| Bacterial strain | n | Detection limita |
|---|---|---|
| Salmonella spp. | 7 | 50 |
| Salmonella typhimurium | 1 | 500 |
| Salmonella virchow | 1 | 500 |
| Shigella spp. | 3 | 500 |
| Shigella sonnei | 1 | 5 |
| Vibrio spp. | 2 | 5,000 |
| Vibrio parahaemolyticus | 1 | 50,000 |
| Vibrio cholerae O1 | 1 | 50,000 |
| Vibrio cholerae non-O1 | 1 | No detection |
| Yersinia spp. | 3 | 50 |
| Yersinia sp. strain O139 | 1 | 500 |
| Escherichia coli | 6 | 50 |
Detection limit is defined as the lowest number of detectable CFU per gram of feces. Tenfold dilutions of bacteria were mixed with fecal samples and plated, and CFU per gram of feces were determined.
Growth on SSI enteric medium compared to that on standard enteric media.
Growth on SSI enteric medium was as good as or better than that on other standard enteric media (Table 6). This was particularly evident for Shigella spp. and enteropathogenic E. coli. Possible exceptions were Vibrio spp., which grew slightly better on HE agar than on all other media. The SSI enteric medium also detected Yersinia spp. When an inoculum of ∼200 CFU was applied to all five media and the quantitative growth rates were compared, SSI enteric medium showed >50 CFU in 75% of the tested bacteria compared to 48% for XLD medium, 75% for HE medium, and 71% for SS agar (Table 6).
TABLE 6.
Comparison of growth of 125 bacteria strains on the SSI enteric medium to growth on other media used for detection of enteropathogens
| Medium type and strain (no. of strains tested) | No. (%) of strains exhibiting growth | No. (%) of strains whose growth wasa:
|
|
|---|---|---|---|
| >10 and <50 CFU | >50 CFU | ||
| SSI enteric medium | |||
| Salmonella spp. (58) | 58 (100) | 9 (16) | 49 (84) |
| Shigella spp. (28) | 28 (100) | 10 (36) | 18 (64) |
| Yersinia spp. (12) | 12 (100) | None | 12 (100) |
| Vibrio spp. (8) | 7 (87) | 6 (86) | 1 (14) |
| E. coli (16) | 16 (100) | 4 (25) | 12 (75) |
| Aeromonas spp. (3) | 3 (100) | 2 (67) | 1 (33) |
| Total (125) | 124 (99) | 31 (25) | 93 (75) |
| XLD plate | |||
| Salmonella spp. (58) | 58 (100) | 12 (21) | 46 (79) |
| Shigella spp. (28) | 26 (92) | 26 (100) | NAb |
| Yersinia spp. (12) | 12 (100) | 8 (67) | 4 (33) |
| Vibrio spp. (8) | 7 (87) | 4 (57) | 3 (43) |
| E. coli (16) | 10 (62) | 8 (80) | 2 (20) |
| Aeromonas spp. (3) | 3 (100) | 2 (67) | 1 (33) |
| Total (125) | 116 (93) | 60 (52) | 56 (48) |
| HE medium | |||
| Salmonella spp. (58) | 56 (96) | 3 (5) | 53 (95) |
| Shigella spp. (28) | 23 (82) | 12 (52) | 11 (48) |
| Yersinia spp. (12) | 12 (100) | 2 (17) | 10 (83) |
| Vibrio spp. (8) | 8 (100) | 3 (38) | 5 (62) |
| E. coli (16) | 8 (50) | 7 (88) | 1 (12) |
| Aeromonas spp. (3) | 3 (100) | None | 3 (100) |
| Total (125) | 110 (88) | 27 (25) | 83 (75) |
| SS agar plate | |||
| Salmonella spp. (58) | 58 (100) | 6 (10) | 52 (90) |
| Shigella spp. (28) | 23 (82) | 18 (78) | 5 (22) |
| Yersinia spp. (12) | 12 (100) | 1 (8) | 11 (92) |
| Vibrio spp. (8) | 6 (75) | 3 (50) | 3 (50) |
| E. coli (16) | 1 (6) | 1 (100) | NA |
| Aeromonas spp. (3) | 3 (100) | 1 (33) | 2 (67) |
| Total (125) | 103 (82) | 30 (29) | 73 (71) |
| CIN agar | |||
| Yersinia spp. (12) | 12 (100) | None | 12 (100) |
An inoculum of ∼200 CFU was applied for all 125 bacteria.
NA, not applicable.
DISCUSSION
The SSI enteric medium evaluated in this study provided the most effective universal medium for the recovery of enteric pathogens. Since we could not evaluate all of the many different types of media available for the isolation of enteric pathogens, we decided to include in this study only those widely used in typical clinical bacteriology laboratories.
XLD agar performed second best, although it had a lower sensitivity towards Shigella spp. and E. coli than did the SSI enteric medium. Like Taylor and Schelhart (21), Altwegg et al. (1), and Desmond and Janda (3), we found XLD agar to be superior to HE agar and SS agar, although HE agar supported growth of Vibrio spp. best of all tested media. This may explain why some investigators have reported HE agar to be equal to or slightly superior to XLD agar (3, 4, 8). Even though this study showed reduced growth and high detection limits of some Vibrio spp. on the SSI enteric medium, the medium detected V. cholerae O1-positive stools in a 1-year prospective household-based diarrhea study in Guinea-Bissau (13). The SSI enteric medium also proved its utility during a V. cholerae epidemic in September 1987 in Guinea-Bissau, although the sensitivity of detecting V. cholerae was increased with the additional use of thiosulfate-citrate-bile-sucrose (TCBS) agar (12b). All cases of cholera diagnosed in the Department of Gastrointestinal Infection at the SSI during the past 20 years have been diagnosed as fast as or faster on the SSI enteric medium than on the traditional TCBS medium after preenrichment in peptone broth. This is probably due to the extremely high numbers of organisms present in stools of patients with cholera.
Some investigators have questioned the efficiency of CIN agar for its use in detection of Yersinia enterocolitica (6, 7, 9). As the SSI enteric medium allows growth of all enteric pathogens, including Yersinia spp., CIN agar could be removed from the list of standard plating agars in the typical laboratory and thereby diminish workload and cost to the laboratory. Yersinia spp. grow as easily recognizable tiny round translucent colonies which resemble pearls on a string on SSI enteric medium.
The performance of any medium is dependent upon the skill and training of the technician and his or her familiarity with the medium. To fully benefit from the capacity to directly identify enteric pathogens on SSI enteric medium, the recognition of the metallic-sheen reaction and the rough transformation reaction in particular requires practical experience.
In conclusion, SSI enteric medium is selective for gram-negative bacteria and allows growth of all aerobic enteric pathogens. The medium compares favorably to similar media, and its ability to detect Yersinia spp. would limit the number of media needed in a typical laboratory. Also, the direct identification of enteric pathogens on the SSI enteric medium may provide a more rapid diagnosis.
REFERENCES
- 1.Altwegg M, Buser J, von Graevenitz A. Stool cultures for Shigella spp.: improved specificity by using MacConkey agar with xylose. Diagn Microbiol Infect Dis. 1996;24:121–124. doi: 10.1016/0732-8893(96)00021-1. [DOI] [PubMed] [Google Scholar]
- 2.Bøvre K, Fuglesang J E, Henriksen S D, Lapage S P, Lautrop H, Snell J J S. Studies on a collection of gram-negative bacterial strains showing resemblance to Moraxellae: examination by conventional bacteriological methods. Int J Syst Bacteriol. 1974;24:438–446. [Google Scholar]
- 3.Desmond E, Janda J M. Growth of Aeromonas species on enteric agars. J Clin Microbiol. 1986;23:1065–1067. doi: 10.1128/jcm.23.6.1065-1067.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn C, Martin W J. Comparison of media for isolation of Salmonellae and Shigellae from fecal specimens. Appl Microbiol. 1971;22:17–22. doi: 10.1128/am.22.1.17-22.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dusch H, Altwegg M. Evaluation of five new plating media for isolation of Salmonella species. J Clin Microbiol. 1995;33:802–804. doi: 10.1128/jcm.33.4.802-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukushima H. New selective agar medium for isolation of virulent Yersina enterocolitica. J Clin Microbiol. 1987;25:1068–1073. doi: 10.1128/jcm.25.6.1068-1073.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukushima H, Gomyoda M. Growth of Yersinia pseudotuberculosis and Yersinia enterocolitica biotype 3B serotype O3 inhibited on cefsulodin-irgasan-novobiocin agar. J Clin Microbiol. 1986;24:116–120. doi: 10.1128/jcm.24.1.116-120.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isenberg H D, Kominos S, Siegel M. Isolation of Salmonellae and Shigellae from an artificial mixture of fecal bacteria. Appl Microbiol. 1969;18:656–659. doi: 10.1128/am.18.4.656-659.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kachoris M, Rouff K L, Welch K, Kallas W, Ferraro M J. Routine culture of stool specimens for Yersinia enterocolitica is not a cost-effective procedure. J Clin Microbiol. 1988;26:582–583. doi: 10.1128/jcm.26.3.582-583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King S, Metzger W I. A new plating medium for the isolation of enteric pathogens. I. Hektoen enteric agar. Appl Microbiol. 1968;16:577–578. doi: 10.1128/am.16.4.577-578.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King S, Metzger W I. A new plating medium for the isolation of enteric pathogens. II. Comparison of Hektoen enteric agar with SS and EMB agar. Appl Microbiol. 1968;16:579–581. doi: 10.1128/am.16.4.579-581.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolmos H J, Schmidt J. Failure to detect hydrogen-sulfide production in lactose/sucrose-fermenting Enterobacteriaceae, using triple sugar iron agar. Acta Pathol Microbiol Immunol Scand Sect B. 1987;95:85–87. doi: 10.1111/j.1699-0463.1987.tb03092.x. [DOI] [PubMed] [Google Scholar]
- 12a.Meyer A A, Blom M, Frimodt-Møller N, Espersen F. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Evaluation of SS1 enteric plate for rapid detection of enteric pathogens, abstr. C-247; p. 172. [Google Scholar]
- 12b.Mølbak, K. Personal communication.
- 13.Mølbak K, Wested N, Hj̀lyng N, Scheutz F, Gottschau A, Aaby P, José da Silva A P. The etiology of early childhood diarrhea: a community study from Guinea-Bissau. J Infect Dis. 1994;169:581–587. doi: 10.1093/infdis/169.3.581. [DOI] [PubMed] [Google Scholar]
- 14.Morris G K, Koehler J A, Gangarosa E J, Sharrar R G. Comparison of media for direct isolation and transport of Shigellae from fecal specimens. Appl Microbiol. 1970;19:434–437. doi: 10.1128/am.19.3.434-437.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiemann D A. Synthesis of a selective agar medium for Yersinia enterocolitica. Can J Microbiol. 1979;25:1298–1304. doi: 10.1139/m79-205. [DOI] [PubMed] [Google Scholar]
- 16.Silva R M, Toledo M R F, Trabulsi L R. Biochemical and cultural characteristics of invasive Escherichia coli. J Clin Microbiol. 1980;11:441–444. doi: 10.1128/jcm.11.5.441-444.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor W I. Isolation of Shigellae. I. Xylose lysine agars: new media for isolation of enteric pathogens. Am J Clin Pathol. 1965;44:471–475. [PubMed] [Google Scholar]
- 18.Taylor W I, Harris B. Isolation of Shigellae. II. Comparison of plating media and enrichment broths. Am J Clin Pathol. 1965;44:476–479. [PubMed] [Google Scholar]
- 19.Taylor W I, Schelhart D. Isolation of Shigellae. IV. Comparison of plating media with stools. Am J Clin Pathol. 1967;48:356–362. [PubMed] [Google Scholar]
- 20.Taylor W I, Schelhart D. Isolation of Shigellae. VI. Performance of media with stool specimens. Appl Microbiol. 1968;16:1387–1393. doi: 10.1128/am.16.9.1387-1393.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor W I, Schelhart D. Isolation of Shigellae. VIII. Comparison of xylose lysine deoxycholate agar, Hektoen enteric agar, Salmonella-Shigella Agar, and eosin methylene blue agar with stool specimens. Appl Microbiol. 1971;21:32–37. doi: 10.1128/am.21.1.32-37.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]