Abstract
Protein arginine methyltransferase 5 (PRMT5) was discovered two decades ago. The first decade focused on the biochemical characterization of PRMT5 as a regulator of many cellular processes in a healthy organism. However, over the past decade, evidence has accumulated to suggest that PRMT5 may function as an oncogene in multiple cancers via both epigenetic and non-epigenetic mechanisms. In this review, we focus on recent progress made in prostate cancer, including the role of PRMT5 in the androgen receptor (AR) expression and signaling and DNA damage response, particularly DNA double-strand break repair. We also discuss how PRMT5-interacting proteins that are considered PRMT5 cofactors may cooperate with PRMT5 to regulate PRMT5 activity and target gene expression, and how PRMT5 can interact with other epigenetic regulators implicated in prostate cancer development and progression. Finally, we suggest that targeting PRMT5 may be employed to develop multiple therapeutic approaches to enhance the treatment of prostate cancer.
Introduction
Protein arginine methyltransferase 5 (PRMT5) is a type II enzyme of the emerging family of protein arginine methyltransferases (PRMTs) that can methylate arginine residues of histones and non-histone substrates [1]. Arginine methylation is a ubiquitous posttranslational modification across species [2]. Nine arginine methyltransferases have been described to date, and their function has been reviewed elsewhere [1,3]. However, PRMT5 is the most widely studied type II enzyme and PRMT9 is the only other type II enzyme. Biochemical and structural studies suggest that PRMT5 forms an octameric complex with the methylosome protein 50 (MEP50) for its catalytic activity [4–6]. In general, PRMT5 is considered an epigenetic repressor of target gene transcription via symmetric dimethylation of histones H4R3 (H4R3me2s), H3R8 (H3R8me2s), and H2AR3 (H2AR3me2s) [4–6]. However, several studies suggest that symmetric dimethylation of histones may activate gene transcription via dimethylation of H3R2 (H3R2me2s), H3R8me2s, and H4R3me2s [7–9]. Other than epigenetic regulation of gene transcription, PRMT5 post-translationally methylates many signaling molecules such as NF-κB, EGFR, p53, and others [6]. As a result, PRMT5 is a critical regulator of cellular proliferation, differentiation, cell cycle progression, DNA damage response (DDR), and cell death [4–6]. As a critical regulatory protein, PRMT5 is overexpressed in multiple cancers [4]. Mechanistic studies have suggested that PRMT5 may function as an oncogene through (1) epigenetic repression of tumor suppressor genes or cell cycle regulators or (2) post-translational regulation of signaling molecules. In this review, we will focus on recent progress in prostate cancer. We present evidence that PRMT5 functions as an epigenetic activator to promote transcription of DDR genes and androgen receptor (AR) in prostate cancer cells. Further, we discuss the identification of pICln as a novel cofactor of PRMT5 to activate transcription of these target genes independent of MEP50. These novel findings suggest potential PRMT5-based targeting approaches for prostate cancer treatment.
PRMT5-driven regulation of AR signaling in prostate cancer
Androgen/AR signaling is the major driver of the normal prostate function and prostate cancer growth and progression [10]. Due to the critical role of AR signaling in prostate cancer, AR remains the primary therapeutic focus for this disease. Indeed, androgen deprivation therapy (ADT) suppresses the production of androgens or inhibits AR signaling and is the standard of care for metastatic prostate cancer [11]. However, AR signaling plasticity leads to the emergence of the therapeutic resistance and the development of castration-resistant prostate cancer (CRPC) via multiple mechanisms of AR reactivation, including emergence of gain-of-function mutations, AR gene amplification, and expression of ligand-independent splice variants [12]. Thus, understanding the regulatory mechanisms of AR expression and activity is necessary to develop novel approaches for prostate cancer treatment.
Epigenetic regulation of AR transcription.
Epigenetic mechanisms mediate both positive and negative regulation of AR transcription [13]. As early as 2000, it was demonstrated that the level of AR promoter DNA methylation negatively correlates with AR expression [14]. Since then, multiple mechanisms such as histone methylation and expression of non-coding RNAs were identified to contribute to AR transcription [13]. One of the first reports indicating the potential implication of histone methylation on AR transcription was published in 2012 [15]. In this study, treatment of LNCaP cells with the inhibitor of multiple methyltransferases adenosine dialdehyde caused downregulation of AR expression, decrease of H3K9 methylation, and inhibition of cell growth. However, that study did not address the effect of general methyltransferases inhibition on other methylation marks.
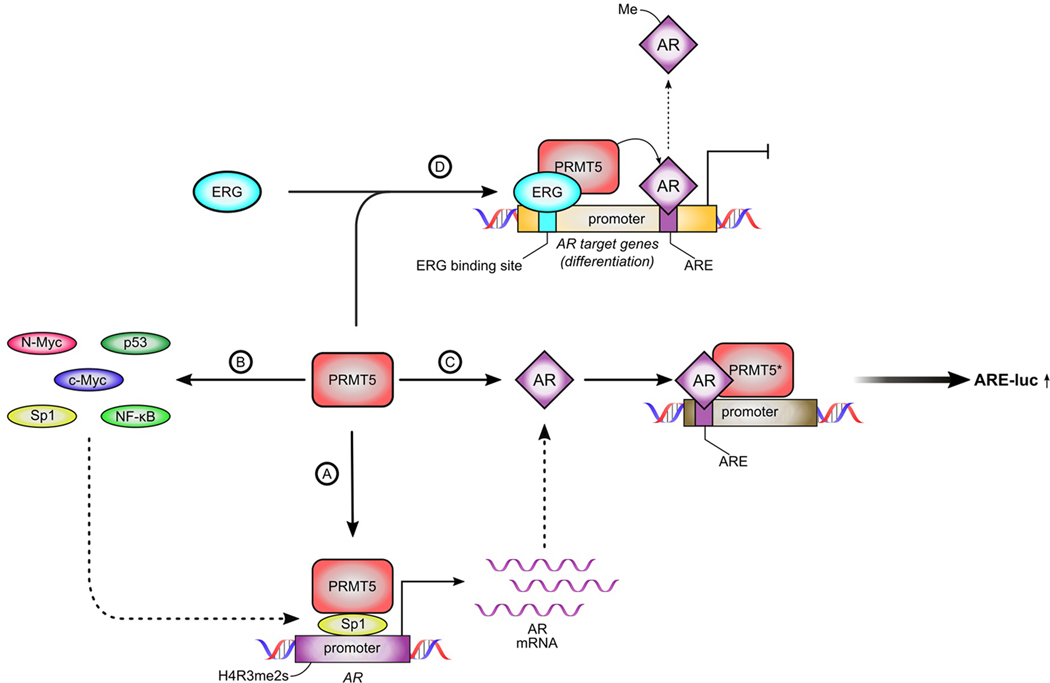
In 2017, it was shown that PRMT5 binds to the AR promoter and symmetrically dimethylates H4R3 at the AR promoter in hormone-naïve prostate cancer cell line LNCaP [16] (Fig. 1A). Targeting PRMT5 via either pharmacological inhibition or short hairpin RNA (shRNA)-mediated knockdown caused decrease of AR mRNA and protein expression accompanied by the decrease of cell proliferation in both cell culture and in LNCaP xenograft model. It was demonstrated that transcription factor Sp1 recruits PRMT5 to the AR promoter as PRMT5 does not have a DNA binding domain. Furthermore, later it was shown that PRMT5 similarly promotes AR transcription in CRPC cells in pICln-dependent manner [17]. Targeting PRMT5 in CRPC cells with different mechanisms of AR reactivation decreased cell proliferation and downregulated AR, AR splice variants, and AR target genes. As PRMT5-driven methylation of histones can also promote deposition of transcription activation marks such as H3K4me3 [7,9,18], the potential interplay between arginine methylation and other chromatin modification marks in the context of AR transcription regulation requires further investigation.
Fig. 1: Mechanisms of PRMT5-driven regulation of AR signaling in prostate cancer.
PRMT5 is implicated in the regulation of AR signaling at multiple steps: (A) PRMT5 is recruited to the AR promoter by Sp1 to symmetrically dimethylate H4R3 thus promoting AR transcription. (B) PRMT5 can modulate activity of transcriptions factors that regulate AR expression. (C) PRMT5 can function as an AR co-activator independently of its methyltransferase activity to enhance activation of AR target gene expression. (D) PRMT5 can methylate AR in an ERG-dependent manner leading to decreased recruitment of AR to AR target gene promoters of differentiation-promoting genes and increased cell proliferation. *mechanism (C) is independent of PRMT5 enzymatic activity.
Regulation of transcription factor-mediated AR transcription.
In addition to direct regulation of AR transcription by methylation of histones at the proximal AR promoter, PRMT5 may also control AR transcription indirectly via modulation of AR-regulating transcription factors (Fig. 1B). For example, Sp1 is a major transcription factor to activate expression of AR [19]. In acute myeloid leukemia, PRMT5 downregulation causes downregulation of Sp1 expression likely via de-repression of miRNA miR-29b [8]. Interestingly, Sp1 recruits PRMT5 to the AR proximal promoter region to activate AR transcription in prostate cancer cells [16]. Given that expression of miR-29b is significantly lower in prostate cancer compared to normal tissues [20], future studies are needed to investigate if this positive feedback loop plays an essential role in the regulation of AR expression in prostate cancer cells.
Like Sp1, c-Myc is another positive regulator of AR transcription and a prominent oncogene in prostate cancer [21]. c-Myc can also recruit PRMT5 to its target genes in glioblastoma [22]. Interestingly, c-Myc has been shown to upregulate PRMT5 transcription [23], and, vice versa, PRMT5 has been shown to upregulate c-Myc expression in lymphoma [24], suggesting another potential positive feedback loop mechanism. N-Myc, another transcription factor of Myc family, was implicated in progression of prostate adenocarcinoma to neuroendocrine prostate cancer (NEPC) [25]. Importantly, N-Myc simultaneously interacts with AR and EZH2 to promote transcriptional repression of AR target genes during prostate cancer neuroendocrine differentiation [26]. In neuroblastoma, PRMT5 functions as a key regulator of N-Myc protein stability [27]. With the prominent role of N-Myc in NEPC, further studies are needed to investigate if PRMT5 also regulates N-Myc stability in prostate cancer and if targeting PRMT5 can suppress the growth of NEPC via down-regulation of N-Myc expression.
Another transcription factor implicated in regulation of AR transcription in prostate cancer is NF-κB. It was shown to be capable of both transcriptional activation and repression of AR, indicating the context-dependent role of this protein [28,29]. In several models, NF-κB is activated by PRMT5 methylation enhancing NF-κB binding to target genes [30,31]. Thus, it is likely that in prostate cancer with PRMT5 overexpression [16], PRMT5 promotes AR expression via NF-κB activation. Interestingly, p53 is implicated in regulation of AR expression as transcriptional repressor [32], while PRMT5-mediated methylation inactivates p53 in lymphoma model [33]. However, several studies show that PRMT5 activates p53 in the context of DNA damage which we discuss below. In summary, PRMT5 is involved in regulation of AR expression at several levels, including direct transcription activation via association with AR promoter and modulation of AR-regulating transcription factors.
Regulation of AR transcriptional activity and target gene expression.
Apart from regulating AR expression, PRMT5 can regulate AR activity directly by interacting with AR protein and modulating AR function as a transcription factor (Fig. 1C, D). The study by Hosohata et al. [34] suggested that PRMT5 may function as an AR co-activator independently of PRMT5 methyltransferase activity. Overexpression of either wild-type PRMT5 or the catalytically inactive PRMT5(R368A) mutant in PC3 cells enhanced luciferase activity of an androgen-responsive element (ARE)-containing luciferase reporter (Fig. 1C). However, later report by Mounir et al. [35] indicated that in VCaP cells, PRMT5-mediated methylation of AR attenuated AR binding to a subset of AR target genes (Fig. 1D). This methylation led to the repression of genes associated with prostatic epithelium differentiation and promoted VCaP cell proliferation. The interaction of PRMT5 and AR was mediated by ERG and only occurred in TMPRSS2-ERG-positive cell lines such as VCaP but not TMRSS2-ERG-negative cell lines such as 22Rv1. However, PRMT5 may also interact with AR in TMRSS2-ERG-negative PC3 via the PRMT5-interacting protein MEP50, which was also reported to act as AR co-activator [36,37]. Taken together, these studies demonstrate that PRMT5 can regulate AR activity via non-epigenetic mechanisms in a context-dependent manner.
PRMT5-mediated regulation of the DDR in prostate cancer
The genome is constantly exposed to both endogenous and environmental stresses that cause damage to DNA. DDR is an evolutionarily conserved cellular response which maintains genome integrity through coordinated regulation of cell cycle arrest, DNA damage repair, and apoptosis [38]. DNA double-stranded breaks (DSBs) are the most cytotoxic DNA lesions. Incorrect or incomplete repair of DSBs promotes accumulation of mutations and cancer development. Conversely, cancer therapies such as radiation therapy (RT) and chemotherapy kill cancer cells by inducing extensive DSBs and apoptosis. Thus, mutations in DDR proteins contribute to increased incidence of cancer whereas therapeutic agents targeting DDR regulators are used as anti-cancer agents alone or in combination with chemotherapy and RT.
Regulation of the cell cycle.
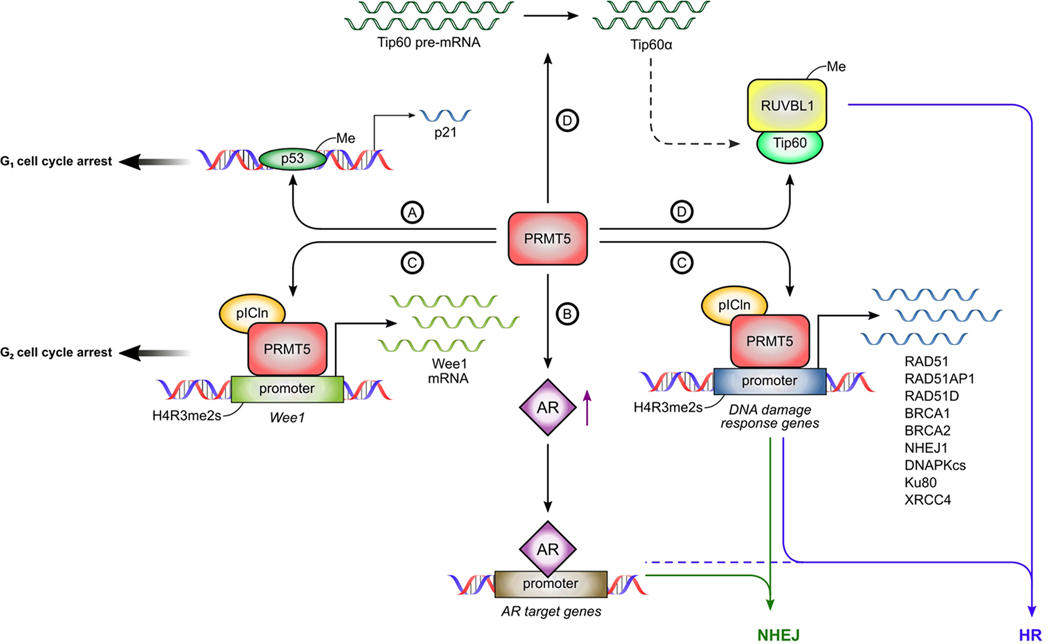
p53 is a critical transcription factor of cell cycle arrest and apoptosis in response to DNA damage. However, how p53 can selectively activate these cellular responses remained a central question in the field. In 2008, Jansson et al demonstrated that PRMT5 can methylate p53 at arginine residues 333, 335 and 337 to activate transcription of p21 and promote G1 cell cycle arrest in response to DNA damaging agent etoposide (Fig. 2A). This finding represented the first report to demonstrate a direct role for PRMT5 in DDR [39]. Since then, multiple studies have demonstrated that PRMT5 plays a pivotal role in DDR (Fig. 2A-D). In most cancer cells, including prostate cancer, PRMT5 knockdown induces G1 arrest, suggesting a positive regulatory role of G1 progression by PRMT5.
Fig. 2: Role of PRMT5 in DNA damage response in prostate cancer.
(A) PRMT5 can regulate cell cycle progression via methylation of p53 or (C) epigenetic activation of Wee1 transcription. (B) PRMT5 activates AR expression which can promote both HR and NHEJ. (C) PRMT5 functions as a master epigenetic regulator for a number of DSBs repair genes promoting both HR and NHEJ. (D) PRMT5 to regulate DSB repair pathway choice and promote HR through symmetrically demethylating RUVBL1 or through modulating splicing of Tip60.
In the case of DNA damage, PRMT5 plays a different role in the regulation of the cell cycle. In prostate cancer cells, PRMT5 is required for DNA damage-induced G2 arrest due to epigenetic activation of WEE1 [40]. In other cancer cells, PRMT5 regulates G2 arrest via methylation of RAD9 [41] and accumulation of KLF4 [42]. These mechanisms may contribute to G2 arrest in prostate cancer cells as well. Future studies will further define the role of PRMT5 in cell cycle arrest in prostate cancer cells.
Epigenetic regulation of DSB repair gene expression.
One major discovery regarding the role of PRMT5 in DDR is that PRMT5 functions as a master epigenetic activator of DSB repair genes in prostate cancer cells (Fig. 2C) [40]. PRMT5 is required for the repair of ionizing radiation (IR)- and etoposide-induced DSBs and activation of both homologous recombination (HR) and non-homologous enjoining (NHEJ). Interestingly, knockdown of PRMT5 increased spontaneous DSBs independent of external DNA damage inducers, indicating that PRMT5 is required to prevent or repair endogenous DSBs. Subsequent analysis identified several PRMT5 target genes involved in the DDR, including genes involved in HR (RAD51, RAD51D, RAD51AP1, BRCA1, and BRCA2) and NHEJ (NHEJ1/XLF and DNAPKcs/PRKDC). Upon DNA damage, PRMT5 was upregulated and recruited to the promoters of DDR genes to promote transcription. Surprisingly, PRMT5-mediated activation of these genes was independent of the canonical cofactor MEP50 but dependent on pICln. Upon recognition of DSBs, repair proteins (such as RAD51, BRCA1, and BRCA2) are transiently upregulated to facilitate repair through HR or NHEJ. Although this transient upregulation is required for cell survival following genotoxic stresses, little is known how proteins are quickly upregulated to promote repair of DNA damage [43]. Upregulated PRMT5 expression by DNA damage and subsequent activation of DSB repair genes likely contribute significantly to DSB repair at the protein level as well.
PRMT5-regulated DSB repair genes appear to be conserved in multiple cancer and normal human cells [40]. However, PRMT5 may be particularly critical to DSB repair in prostate cancer cells given its regulatory role in transcription of AR [16], due to the fact that AR transcriptionally regulates expression of several target genes involved in NHEJ (Fig. 2B) [44–46]. Targeting PRMT5 in prostate cancer cells decreased expression of AR and AR-target genes involved in NHEJ (Ku80/XRCC5, XRCC4, and DNAPKcs/PRKDC). Furthermore, PRMT5-mediated regulation of Ku80/XRCC5, XRCC4, and DNAPKcs/PRKDC was stronger in AR-expressing prostate cancer cells compared to cells of other origins (such as breast cancer or glioblastoma). Therefore, in prostate cancer cells, PRMT5 can regulate the expression of DDR genes directly as well as indirectly through regulating AR expression. Indeed, the correlation between PRMT5 and AR target genes involved in NHEJ was stronger in prostate cancer compared to most other cancers [40]. Interestingly, DNAPKcs/PRKDC is both an AR and PRMT5 target gene in prostate cancer cells [40]. As PRMT5 does not contain any DNA binding domain, it remains to be determined whether AR also recruits PRMT5 to the promoter of DNAPKcs/PRKDC.
Splicing regulation of DDR genes.
PRMT5 methylates several spliceosome proteins and is a critical regulator of alternative splicing [47]. In 2017, Braun et al. demonstrated that, at least in malignant glioma cells, PRMT5 could modulate gene expression by regulating the splicing out of detained-introns (DIs) [48]. Additionally, in 2019 Tan et al. reported that in hematopoietic stem cells, PRMT5 maintains appropriate splicing activity to prevent aberrant intron retention and exon skipping [49]. Further, genes annotated as “DNA repair” were enriched when functional enrichment analysis was performed on alternative splicing events upon PRMT5 knockdown in hematopoietic stem cells [49]. Although there was little overlap between these differentially spliced genes in hematopoietic stem cells [49] and the validated PRMT5 target genes involved in DDR in prostate cancer cells (RAD51, RAD51AP1, RAD51D, BRCA1, BRCA2, NHEJ1, DNAPKcs, and WEE1) [40], RNA-seq analysis also identified many other genes involved in DDR in prostate cancer cells. Future research is needed to determine if PRMT5 may also regulate the expression of DDR genes in prostate cancer cells via splicing.
Regulation of DSB repair pathways.
DSBs can be efficiently repaired regardless of pathway choice. However, pathway choice affects the speed and accuracy of the repair [50]. HR and NHEJ are the two major pathways for DSB repair. Acetylation of H4K16 (H4K16Ac) catalyzed by Tip60 can relax local chromatin and facilitate the displacement of 53BP1 from DSBs, enabling BRCA1 binding, RPA filament formation, and subsequent DSBs repair via HR.
Recently, it was discovered that in osteosarcoma cells, PRMT5 might promote the DSBs repair via promoting HR over NHEJ by methylating R205 on RUVBL1, a member of the Tip60 complex (Fig. 2D). Mechanistically, the methylation of RUVBL1increases the Tip60’s acetylase activity resulting in increased H4K16ac. Additionally, PRMT5 is required for appropriate splicing of Tip60, which promotes Tip60 acetyltransferase activity, demotes the binding of 53BP1 to DSBs, and promotes HR [51]. PRMT5-deficient hematopoietic progenitor cells have reduced levels of full-length Tip60, reduced Tip60 acetyltransferase activity, and defective HR.
Although it remains to be determined if PRMT5 also regulates the appropriate splicing of Tip60 or methylates RUVBL1in prostate cancer cells, PRMT5 may likely have distinct roles in DSB repair choice in a cell type-dependent manner. For example, knockdown of RUVBL1 had no effect on IR-induced 53BP1 foci in hematopoietic cells [51], suggesting that the role of PRMT5 in DSB repair choice may be different in various cell types. In prostate cancer cells, PRMT5 activates transcription of RAD51, and targeting PRMT5 causes downregulation of RAD51 protein [40]. However, in osteosarcoma cells, depletion of PRMT5 impaired HR without affecting the expression of RAD51 [52]. Future studies may elucidate if PRMT5 regulates DSB repair choice via methylation of RUVBL1 or Tip60 splicing in prostate cancer cells.
In summary, it is clear that PRMT5 is a critical regulator of DDR in prostate cancer. PRMT5 regulates the function of other proteins associated with DDR (p53 [39,53], E2F1 [54,55], FEN1 [56], RAD9 [41], KLF4 [42], and TDP1 [57]). Future characterization of these PRMT5 substrates and identification of additional substrates and target genes will shed new light on both the epigenetic and non-epigenetic roles of PRMT5 in the DDR in prostate cancer cells.
Regulation of PRMT5 activity and target gene expression by interacting proteins
PRMT5 can interact with several proteins in addition to MEP50, and these interacting proteins may regulate the enzymatic activity and substrate specificity [58–62]. Studies utilizing purified recombinant proteins suggest that MEP50 is an obligate cofactor of PRMT5 required for methyltransferase activity while proteins such as RioK1 and pICln alter substrate specificity [59,62]. However, in the context of prostate cancer, pICln [17,40] but not MEP50 [16] mediates PRMT5 activity towards histones at the promoters of AR and DDR genes.
Recently, we demonstrated that PRMT5 and pICln bind to the promoters of multiple DNA damage response genes to symmetrically dimethylate histone H4R3, and this recruitment of PRMT5/pICln and histone methylation is enhanced upon DNA damage [40]. Mechanistically, while overall expression of pICln and MEP50 did not change upon DNA damage, subcellular distribution of pICln and MEP50 changed oppositely: pICln accumulated in the nucleus while MEP50 localized in the cytoplasm. As pICln and MEP50 can both enhance PRMT5 activity and H4R3 methylation is involved in both activation and repression of target gene expression [4–6,40], the selection of its interacting proteins may likely determine the activation or repression of PRMT5 target gene expression. Indeed, MEP50, pICln, and PRMT5 all bound to the promoter of PRMT5-repressed gene involucrin [63] and mediated repression of involucrin transcription. The transcriptional activation of target gene expression by PRMT5/pICln was also demonstrated in activation of AR transcription in CRPC cells [17]. It is also interesting to note that whereas only H4R3me2s was observed at the proximal promoters of AR and DDR genes, all H4R3me2s, H3R2me2s, H3R8me2s and H2AR3me2s were all detected at the proximal promoter of IVL [17, 40]. Thus, the composition of PRMT5-centered complexes and the types of histone arginine methylations will likely determine transcriptional activation versus repression of target genes. Genome-wide analysis of PRMT5 target genes and their relationship with its cofactors as well as histone modifications is needed in future studies.
Interplay of PRMT5 with epigenetic regulators in prostate cancer cells
PRMT5 and other arginine methyltransferases.
Out of nine protein arginine methyltransferases, four were shown to monomethylate and asymmetrically dimethylate the same histone residues as PRMT5: H2AR3 by PRMT7, H3R2 by PRMT6, H3R8 by PRMT2, H4R3 by PRMT1, PRMT6, and PRMT7.
While PRMT1 and PRMT5 deposit different types of methylation marks (asymmetrical vs symmetrical dimethylation) and possibly act in the opposite ways [64,65], inhibition of both PRMT5 and PRMT1 had synergistic effect in lung and pancreatic cells [66]. However, their potential competition was not explored in the context of prostate cancer. It remains to be established if inhibition of both enzymes is a better therapeutic approach, especially in methylthioadenosine phosphorylase (MTAP)-deficient prostate cancer [67]. There is no direct evidence suggesting interplay between PRMT2 and PRMT5, and their relationship remains to be investigated. PRMT6 was suggested to be an oncogene in prostate cancer via activation of PI3K/Akt pathway, possibly by increasing asymmetrical dimethylation of H3R2 on the target gene promoters [68]. Interestingly, PRMT6 knockdown increased AR expression in prostate cancer cells, though whether PRMT6 directly regulates AR transcription via asymmetrical dimethylation of H3R2 or H4R3 remains to be determined [16]. However, PRMT5-driven symmetrical dimethylation of H3R2 enhanced the binding of H3R2 by the epigenetic reader WDR5 [18,69], while asymmetrical dimethylation of H3R2 (mark that is catalyzed by PRMT6) prevented the binding of WDR5 in biochemical assay [70]. These observations suggest that PRMT5 and PRMT6 may play opposite roles via regulation of target gene expression by depositing different types of arginine methylation, at least on H3R2 in prostate cancer cells.
Role of PRMT7 was not explored in the context of prostate cancer. However, PRMT7 expression is detected in prostate cancer (Protein Atlas), and PRMT7-mediated H4R17 monomethylation can allosterically promote PRMT5-mediated H4R3 dimethylation [71]. Additionally, multiple evidences suggest at least partial overlap of PRMT7 with PRMT5 function [18] while retaining unique substrates [72]. Future research elucidating genome-wide differential binding of PRMT5 and PRMT7 and their substrates in prostate cancer cells may establish whether co-targeting PRMT5 and PRMT7 is a better approach than targeting single enzyme.
Apart from functional overlap in the epigenetic regulation, other arginine methyltransferases might interact with PRMT5 via post-translational modification of non-histone substrates. PRMT4 (also known as co-activator-associated arginine methyltransferase 1, CARM1) is an established co-activator of androgen receptor [73]. A 2006 study demonstrated that in hormone-naïve LNCaP cells, PRMT4 binding increased AR transcriptional activity and promoted cell proliferation and survival. Since PRMT5 may also function as AR co-activator, as discussed above [34], it will be beneficial to explore the possible combinational effect of PRMT4 and PRMT5 targeting on AR signaling.
PRMT5 and lysine methylation.
Histone lysine methylation is a histone post-translational modification that has been linked to a variety of cellular processes such as transcription regulation, DNA replication, and DNA repair [74]. In prostate cancer, polycomb repressive complex 2 (PRC2) and its enzymatic component EZH2 is an established oncogene [75]. Importantly, EZH2 functions as an oncogene in prostate cancer both as a part of PRC2 and in PRC2-independent manner. Notably, independently of PRC2, EZH2 may function as a co-activator of AR-mediated transcription in LNCaP-abl model [76] and as an epigenetic activator of AR transcription in LNCaP cells [77] while in NEPC models EZH2 suppressed AR expression and signaling [78]. Thus, targeting EZH2 could be an effective approach to either repress AR signaling in AR-dependent prostate cancer or restore AR signaling in AR-independent prostate cancer. In fact, dual targeting of AR and EZH2 was explored in CRPC cell models [79]. While direct PRMT5/PRC2 or PRMT5/EZH2 interaction was not investigated in prostate cancer, in leukemia model, PRMT5 colocalized with PRC2 at the promoters of tumor suppressors RBL2 and ST7, and PRMT5/PRC2 interaction was mediated by BRD7. This interaction was associated with transcriptional repression of RBL2 and ST7 [80]. Additionally, in lymphoma cells PRMT5 promotes expression of PRC2 through epigenetic repression of RBL2 [81], which possibly can lead to even further suppression of genes co-regulated by PRMT5 and PRC2. Based on in vitro evidence, it was suggested that SUZ12 directly interacts with MEP50 to recruit PRMT5 to histone H2A substrate [82]. However, contrary to the evidence that PRMT5 cooperates with PRC2 to repress gene transcription, observation in AML cells demonstrated that PRMT5-driven histone H3R8 symmetric dimethylation prevented methylation of H3K27 by PRC2 and activated multiple gene expression [83]. Taken together, these observations suggest that interaction of PRMT5 and PRC2, and transcriptional outcome of this interaction is highly context-dependent. Given the importance of PRC2 and EZH2 in prostate cancer, it is imperative to investigate this interaction in detail. EZH2 also functions as an AR co-activator outside of PRC2 [84], thus exploring the effect of co-targeting PRMT5 and EZH2 on AR signaling is warranted.
In addition to research on PRC2/PRMT5 axis, significant research effort was devoted to investigating the interplay of WDR5 and PRMT5. WDR5 recruits H3K4 methyltransferase complexes MLL1–4 to chromatin and functions as an oncogene in prostate cancer to promote AR recruitment to its target genes [85]. Despite well-characterized functional interaction of PRMT5 and WDR5, this interaction has not been investigated in prostate cancer. It was demonstrated that PRMT5-driven methylation of H3R2 in vitro [70], in lymphoma [18], lung cancer [9], and ovarian cancer [86] cells enhances binding of WDR5 to promote H3K4 trimethylation and activation of target gene transcription. On the contrary, in erythroleukemia cells and bone marrow, PRMT5-driven recruitment of WDR5 lead to transcriptional repression of γ-globin gene expression [69] again suggesting context-dependent outcome of PRMT5-driven histone methylation. Elucidation of composition of PRMT5-containing protein complexes at the regulatory elements of target genes may shed light on this discrepancy.
PRMT5 and lysine acetylation.
The connection between histone acetylation and gene expression modulation was discovered as early as 1964, and since then, a variety of histone acetyltransferases and histone deacetylases was described [87]. p300/CBP is a well-known co-activator of AR transcriptional activity and was shown to be a potential driver of prostate cancer progression [88]. In prostate cancer cells, PRMT5 was present in the same complex with p300/CBP and nucleolin, and this interaction was facilitated by p300/CBP-interacting transactivator with E/D-rich carboxy-terminal domain-2 (CITED2) [89]. The formation of the complex promoted methylation and acetylation of nucleolin by PRMT5 and p300/CBP, respectively, and subsequent nucleolin nuclear export to promote AKT-mediated EMT and prostate cancer metastasis via increased translation of AKT mRNA. Interestingly, PRMT5- and p300/CBP-containing complex also included both RioK1 and MEP50, though their functional involvement remains to be determined.
Another lysine acetyltransferase demonstrated to interact with and be regulated by PRMT5 is TIP60, as discussed above. Notably, both TIP60 and p300/CBP acetylate H4K5. In vitro, acetylation of H4K5 enhances methylation of H4R3 by PRMT5/MEP50 complex [90]. However, another study suggested that non-acetylated histone H4 was methylated by purified PRMT5 in complex with Brg1 or hBrm more efficiently than H4K5-acetylated H4 [91]. The same study also found that PRMT5 was present in a complex containing c-Myc, histone deacetylase (HDAC) 2, and Brg1 on the two Myc target gene promoters to repress gene transcription, demonstrating the interplay between PRMT5-mediated histone methylation, histone acetylation/deacetylation, and chromatin remodeling. As Brg1 and PRMT5 bind to the AR promoter and catalyze H4R3 dimethylation to activate AR transcription [16], the status of H4K5 acetylation on the AR promoter requires further investigation.
Support for the interplay between histone methylation and acetylation also comes from a recent study demonstrating that in lymphoma, PRMT5-mediated H3R8 symmetric dimethylation is coupled with HDAC2- or 3-mediated deacetylation at H2BK12, H3K9, H3K14, and H4K8 on the promoter regions of three target miRNAs [24], which regulate expression of cyclin D1 and c-Myc. Interestingly, methylation of H4R3 by PRMT5/MEP50 may also impact the acetylation of H4K5 by TIP60, at least in vitro [92]. In short, interplay between PRMT5-mediated methylation and other marks has been explored extensively; future research will likely provide more insight into complex epigenetic regulation. However, biochemical evidence from in vitro studies must be cautiously taken as epigenetic regulation is heavily dependent on chromatic status and protein complex composition in cells.
PRMT5 and DNA methylation.
DNA methylation, mediated by DNA methyltransferases (DNMTs), is essential for the maintenance of various cellular processes such as DNA repair, recombination, replication, and gene expression [93]. In cancer, including prostate cancer, alterations of DNA methylation often lead to promoter hypo- and hypermethylation at oncogenes and tumor suppressor genes, respectively [93]. Although changes of DNA methylation status were well described in prostate cancer, and accumulating evidence suggests connection between PRMT5 activity and such alterations, this relationship has not been explored in prostate cancer specifically.
The direct relationship between PRMT5-driven histone methylation and DNA methylation was first observed in erythroid cells [94]. In this study, H4R3 methylation by PRMT5 at the γ-globin promoter caused recruitment of DNMT3a to the same region via interaction of H4R3me2s with DNMT3a. Methylation of CpG islands in this region was PRMT5-dependent, a phenomenon also observed in gastric cancer [95]. However, it was not elucidated whether DNMT3a interacted with methylated histones or directly with PRMT5. In addition to potentially recruiting DNA methylases to its target regions, PRMT5 can also be recruited to already methylated DNA regions. In breast cancer cells, PRMT5/MEP50 was recruited to methylated DNA via interaction with methyl CpG binding domain 2 (MBD2) protein and coincided with increased methylation of histone H4R3 [96]. If both mechanisms are active in prostate cancer, it suggests a possible DNA methylation – histone H4R3 methylation positive feedback loop. Future research needs to investigate the relationship between other histone methylation marks mediated by PRMT5 and DNA methylation status.
Therapeutic potential of PRMT5 targeting for prostate cancer treatment
Although prostate cancer is well managed with a 10-year survival of over 90% [97], localized high-risk prostate cancer and metastatic prostatic cancer remain the major clinical challenge. The gold standard treatment for localized high-risk prostate cancer is RT in combination with ADT whereas metastatic hormone-naïve prostate cancer (HNPC) patients are treated with the first line ADT, and CRPC patients are treated with the second generation of AR signaling inhibitors (ASI) abiraterone and enzalutamide [97]. Unfortunately, 30–50% of high-risk prostate cancers recur and can eventually progress to metastatic disease [97]. Despite the initial positive response of most HNPC to ADT, almost all HNPC cases progress to CRPC within 2 years. Further, evidence has emerged that nearly 20% of CRPC progress to NEPC after ASI [98]. NEPC is an aggressive cancer with a median survival of one year. Currently, NEPC is considered the end-stage of the disease with no effective treatment option. Additionally, significant adverse effects are associated with RT and ADT/ASI, diminishing the quality of life. Thus, there is an urgent need to develop novel treatment strategies or improve the existing treatment efficacy. Given the importance of PRMT5 in regulation of radiation-induced DSB repair and AR signaling in prostate cancer cells, targeting PRMT5 may be explored as a novel AR targeting approach for treatment of metastatic prostate cancer or as a radiosensitization approach for treatment of localized prostate cancer.
Targeting PRMT5 as a monotherapy for prostate cancer treatment.
AR is the major driver of prostate cancer development and progression, and targeting AR remains the mainstay of treatment for metastatic prostate cancer. Recent findings that PRMT5 regulates AR signaling at multiple levels discussed above suggest that targeting PRMT5 may be used as a novel approach to treat prostate cancer. Several inhibitors for PRMT5 have been developed by multiple labs [99–101]. Currently, five inhibitors are in clinical phase I trials in the US: GSK3326595, JNJ-64619178, PF06939999, PRT543, and PRT811 (clinicaltrials.gov). GSK3326595 and JNJ-6461917 are catalytic inhibitors of PRMT5, with GSK3326595 competing with peptide substrates in the substrate binding pocket and JNJ-64619178 binding competitively with both the SAM and substrate binding pockets. These trials enroll patients with hematological malignancies and solid tumors and are expected to be complete in 2021 – 2023.
Given that knockdown of PRMT5 significantly inhibited prostate cancer cell growth and suppressed xenograft tumor growth in mice [16,17], it is possible that catalytic PRMT5 inhibitors may phenocopy the effect of PRMT5 knockdown in preclinical models. As PRMT5 not only regulates AR transcription but also regulates AR activity and target gene expression [34,35], targeting PRMT5 with these inhibitors may likely offer additivity or synergy to current ADT and ASI therapies. As HNPC/CRPC growth and survival are dependent on AR signaling, targeting PRMT5 may similarly suppress the growth of CRPC cells via down-regulation of the AR signaling. Interestingly, MTAP deletion, which renders cells heavily dependent on PRMT5, may represent a particularly useful patient stratum for PRMT5-specific therapy [102]. As 10–70% of prostate cancer tissues have decreased MTAP expression [67], this subset of patients may be effectively treated with PRMT5 inhibitors alone. Future preclinical evaluation of these PRMT5 inhibitors should include prostate cancer cells with and without MTAP deletion. If these preclinical studies validate and confirm the therapeutic potential of PRMT5 inhibition, future clinical trials will enable evaluation of the therapeutic effect of these PRMT5 inhibitors to treat metastatic prostate cancer.
PRMT5 targeting as a radiosensitization approach.
RT is used to treat more than 50% of human cancers, and the clinical efficacy is limited by adverse toxicity to surrounding tissues. Co-administration of an effective radiosensitizer would decrease the IR dose required for the same therapeutic effect. For prostate cancer, ADT has been used as such a radiosensitizer, and several large clinical studies confirmed the clinical benefit of RT in combination with adjuvant or neoadjuvant ADT for localized high-risk disease when compared with RT or ADT alone [97]. Although the mechanism was initially unclear, recent studies suggest that AR transcriptionally activates several genes involved in NHEJ and HR [44–46].
Because PRMT5 epigenetically activates AR transcription in prostate cancer cells [16], Owens et al. recently evaluated whether targeting PRMT5 can radiosensitize prostate cancer cells and discovered that PRMT5 functions as a master epigenetic activator of multiple DDR genes in an AR-independent manner as discussed above [40]. Significantly, targeting PRMT5 by either knockdown or inhibition with their PRMT5 inhibitor BLL3.3 increased radiation-induced DSBs and increased cell death. Further, PRMT5 expression correlates positively with most of its DDR target genes in prostate and other cancers. Because PRMT5 regulates expression of DSB repair genes (HR, NHEJ, G2 arrest) and the expression of AR, PRMT5 may be a more effective radiosensitization target as opposed to ADT for treatment of localized high-risk prostate cancer patients. Preclinical studies utilizing mouse models to mimic localized, high-risk prostate cancer could assess PRMT5 inhibition as a radiosensitization approach. If successful, PRMT5 targeting would represent a novel mode of action for control of this disease stage.
Concluding remarks and future perspectives
The cloning of the human PRMT5 gene was reported in 1999 [103]. However, the role of PRMT5 in human cancers has only been reported in recent years. Results from our lab and several others have clearly suggested that PRMT5 plays a critical role in DDR and the AR signaling in prostate cancer. Notably, PRMT5 expression is significantly higher in prostate cancer tissues compared to benign hyperplasia, and expression of PRMT5 highly correlated with AR at both protein and mRNA level [16]. While PRMT5 was generally considered an epigenetic repressor of tumor suppressors and cell cycle regulators in human cancers, we have provided compelling evidence that PRMT5 also functions as an epigenetic activator of DDR genes and AR [16,17,40]. Consistent with our findings, several recent RNA-seq studies have also identified comparable number of genes that are activated or repressed by PRMT5 knockdown [83,104], although whether these genes are direct targets of PRMT5 remains to be determined. Another surprising finding is identification of pICln, but not MEP50, as a potential cofactor of PRMT5 to transcriptionally activate DDR gene expression.
As activation of AR transcription by PRMT5 requires the participation of Sp1 and Brg1 [16,17], PRMT5 may likely form higher-order complexes with other transcriptional regulators such as transcription factors and chromatin remodelers to determine the transcriptional output, and in a locus-specific manner. Additionally, the interplay between PRMT5 and other epigenetic regulators (e.g., other PRMTs, HDACs, KMTs, DNMTs, Fig. 3) should have a similarly important role in controlling the transcription of PRMT5 target genes. Future research focusing on the analysis of PRMT5-centered and locus-specific complexes and the functional relationship with other transcriptional and epigenetic regulators will shed new light on the epigenetic role and mechanisms of PRMT5 in prostate cancer. Additionally, it remains to be investigated whether and how PRMT5 promotes prostate cancer growth and progression by regulating key prostate cancer modulators (Rb, p53, N-Myc, and EZH2 [4–6]) in the nucleus or through regulating other unidentified signaling molecules in the cytoplasm in prostate cancer [105]. Importantly, these mechanistic studies will likely offer new avenues for development of novel therapeutics. This may be particularly important given that targeting PRMT5 with the catalytic inhibitors may cause some unwanted side effects due to its many normal cellular roles [4–6]. Although targeted delivery of PRMT5 inhibitors could potentially circumvent this problem, identification of pICln as a novel cofactor of PRMT5 to activate transcription of AR and DDR genes suggests that targeting the interaction of PRMT5 with pICln may offer a specific and unique approach to treat prostate cancer as a monotherapy or as a radiosensitizer.
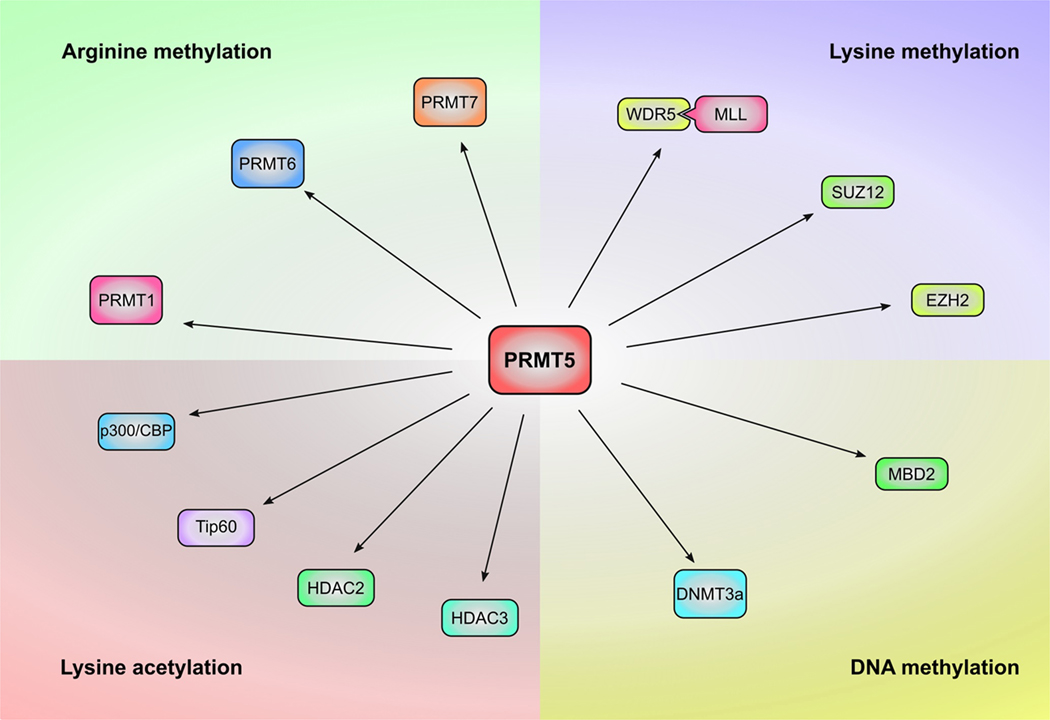
Fig. 3: Potential interplay of PRMT5 with other epigenetic regulators.
PRMT5 can interact with and regulate multiple epigenetic regulatory molecules leading to various cellular effects in various types of cancer cells. Investigation of these interplays in the context of prostate cancer will shed new light on the roles and mechanisms of PRMT5 in prostate cancer.
As our understanding of prostate cancer biology and pathology continues to evolve and new therapies are developed, exploration of PRMT5’s role in the context of prostate cancer development and progression is warranted. One emerging clinical challenge is the development of ASI-induced NEPC. As most NEPC cells lose the expression or activity of AR [98], it would be essential to determine if PRMT5 targeting may promote NEPC development via AR down-regulation. Conversely, targeting PRMT5 in combination with the current platinum chemotherapy may offer a better treatment approach for NEPC. Likewise, co-targeting AR expression (PRMT5 inhibitors) and AR activity (e.g., abiraterone, enzalutamide, and darolutamide) may offer additive or even synergistic effect on the suppression of CRPC and prevention of progression to NEPC.
In conclusion, the role of PRMT5 in prostate cancer is beginning to emerge. The evidence discussed above clearly suggests that targeting PRMT5 may be explored as a novel AR targeting approach and as a radiosensitization approach. Future effort in elucidating the role and the underlying mechanism of PRMT5 in the context of prostate cancer development, progression, and therapeutic intervention will likely lead to development of novel and specific therapeutic approaches for prostate cancer management.
Acknowledgements
This work was supported by following grants: U.S. Army Medical Research Acquisition Activity, Prostate Cancer Research Program (PC11190, PC120512, PC150697), NCI R01CA212403, Purdue University Center for Cancer Research Small Grants, and Purdue Research Foundation Research Grant. J. L. Owens was supported by the Indiana Clinical and Translational Sciences Institute (CTSI) Pre-Doctoral Fellowship, which was made possible with partial support from Grant Numbers TL1 TR001107, TL1 TR002531, UL1 TR001108, and UL1 TR002529 (A. Shekhar, PI) from the NIH, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. A. M. Asberry was supported by NIH T32 Grant NIH T32GM125620.
Footnotes
Competing Interests
The authors declare that they have no conflict of interest.
References
- 1.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer [Internet]. 2013;13:37–50. [DOI] [PubMed] [Google Scholar]
- 2.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. [DOI] [PubMed] [Google Scholar]
- 3.Jahan S, Davie JR. Protein arginine methyltransferases (PRMTs): Role in chromatin organization. Adv Biol Regul. 2015;57:173–84. [DOI] [PubMed] [Google Scholar]
- 4.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci [Internet]. 2015;72:2041–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci. 2011;36:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shailesh H, Zakaria ZZ, Baiocchi R, Sif S. Protein arginine methyltransferase 5 (PRMT5) dysregulation in cancer. Oncotarget [Internet]. 2018;9:36705–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Z, Kong X, Xia J, Wu X, Li H, Xu H, et al. The arginine methyltransferase PRMT5 regulates CIITA-dependent MHC II transcription. Biochim Biophys Acta - Gene Regul Mech. 2016;1859:687–96. [DOI] [PubMed] [Google Scholar]
- 8.Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia [Internet]. 2015;30:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Lorton B, Gupta V, Shechter D. A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene. 2017;36:373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.La Vignera S, Condorelli RA, Russo GI, Morgia G, Calogero AE. Endocrine control of benign prostatic hyperplasia. Andrology. 2016;4:404–11. [DOI] [PubMed] [Google Scholar]
- 11.Snow O, Lallous N, Singh K, Lack N, Rennie P, Cherkasov A. Androgen receptor plasticity and its implications for prostate cancer therapy. Cancer Treat Rev. 2019;81:101871. [DOI] [PubMed] [Google Scholar]
- 12.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cucchiara V, Yang JC, Mirone V, Gao AC, Rosenfeld MG, Evans CP. Epigenomic regulation of androgen receptor signaling: Potential role in prostate cancer therapy. Cancers (Basel). 2017;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama T, Watanabe M, Suzuki H, Toyota M, Sekita N, Hirokawa Y, et al. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab Investig. 2000;80:1789–96. [DOI] [PubMed] [Google Scholar]
- 15.Shiota M, Takeuchi A, Yokomizo A, Kashiwagi E, Tatsugami K, Naito S. Methyltransferase inhibitor adenosine dialdehyde suppresses androgen receptor expression and prostate cancer growth. J Urol. 2012;188:300–6. [DOI] [PubMed] [Google Scholar]
- 16.Deng X, Shao G, Zhang H-T, Li C, Zhang D, Cheng L, et al. Protein arginine methyltransferase 5 functions as an epigenetic activator of the androgen receptor to promote prostate cancer cell growth. Oncogene [Internet]. 2016;36:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beketova E, Fang S, Owens JL, Liu S, Chen X, Zhang Q, et al. Protein Arginine Methyltransferase 5 Promotes pICln-Dependent Androgen Receptor Transcription in Castration-Resistant Prostate Cancer. Cancer Res [Internet]. 2020;80:4904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Migliori V, Müller J, Phalke S, Low D, Bezzi M, Mok WC, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–45. [DOI] [PubMed] [Google Scholar]
- 19.Faber PW, Van Rooij HCJ, Schipper HJ, Brinkmann AO, Trapman J. Two different, overlapping pathways of transcription initiation are active on the TATA-less human androgen receptor promoter. The role of Sp1. J Biol Chem. 1993;268:9296–301. [PubMed] [Google Scholar]
- 20.Zhu C, Hou X, Zhu J, Jiang C, Wei W. Expression of miR-30c and miR-29b in prostate cancer and its diagnostic significance. Oncol Lett [Internet]. 2018;16:3140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai S, Cao S, Jin L, Kobelski M, Schouest B, Wang X, et al. A positive role of c-Myc in regulating androgen receptor and its splice variants in prostate cancer. Oncogene. 2019;38:4977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mongiardi MP, Savino M, Bartoli L, Beji S, Nanni S, Scagnoli F, et al. Myc and Omomyc functionally associate with the Protein Arginine Methyltransferase 5 (PRMT5) in glioblastoma cells. Sci Rep. 2015;5:15494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh CM, Bezzi M, Low DHP, Ang WX, Teo SX, Gay FPH, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature [Internet]. 2015;523:96–100. [DOI] [PubMed] [Google Scholar]
- 24.Karkhanis V, Alinari L, Ozer HG, Chung J, Zhang X, Sif S, et al. Protein arginine methyltransferase 5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1 and c-MYC expression in aggressive B-cell lymphoma. J Biol Chem. 2020;295:1165–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger A, Brady NJ, Bareja R, Robinson B, Conteduca V, Augello MA, et al. N-Myc-mediated epigenetic reprogramming drives lineage plasticity in advanced prostate cancer. J Clin Invest. 2019;129:3924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, et al. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell [Internet]. 2016;30:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JH, Szemes M, Vieira GC, Melegh Z, Malik S, Heesom KJ, et al. Protein arginine methyltransferase 5 is a key regulator of the MYCN oncoprotein in neuroblastoma cells. Mol Oncol [Internet]. 2015;9:617–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Altuwaijri S, Deng F, Chen L, Lal P, Bhanot UK, et al. NF-κB regulates androgen receptor expression and prostate cancer growth. Am J Pathol. 2009;175:489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas-Jardin SE, Dahl H, Nawas AF, Bautista M, Delk NA. NF-κB signaling promotes castration-resistant prostate cancer initiation and progression. Pharmacol Ther. 2020;19:107538. [DOI] [PubMed] [Google Scholar]
- 30.Harris DP, Chandrasekharan UM, Bandyopadhyay S, Willard B, DiCorleto PE. PRMT5-mediated methylation of NF-κB p65 at Arg174 is required for endothelial CXCL11 gene induction in response to TNF-α and IFN-γ costimulation. PLoS One [Internet]. 2016;11:e0148905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei H, Wang B, Miyagi M, She Y, Gopalan B, Bin Huang D, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc Natl Acad Sci U S A. 2013;110:13516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alimirah F, Panchanathany R, Cheny J, Zhang X, Ho SM, Choubey D. Expression of androgen receptor is negatively regulated by p53. Neoplasia. 2007;9:1152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Chitnis N, Nakagawa H, Kita Y, Natsugoe S, Yang Y, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5:288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosohata K, Li P, Hosohata Y, Qin J, Roeder RG, Wang Z. Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription. Mol Cell Biol [Internet]. 2003;23:7019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mounir Z, Korn JM, Westerling T, Lin F, Kirby CA, Schirle M, et al. ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the androgen receptor. Elife [Internet]. 2016;5:e13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, Wu H, Lee P, Wang Z. Roles of the androgen receptor cofactor p44 in the growth of prostate epithelial cells. J Mol Endocrinol [Internet]. 2006;37:283–300. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Kumari S, Hu Q, Senapati D, Venkadakrishnan VB, Wang D, et al. A comprehensive analysis of coregulator recruitment, androgen receptor function and gene expression in prostate cancer. Elife. 2017;6:e28482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein M, Kastan MB. The DNA Damage Response: Implications for Tumor Responses to Radiation and Chemotherapy. Annu Rev Med [Internet]. 2015;66:129–43. [DOI] [PubMed] [Google Scholar]
- 39.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–9. [DOI] [PubMed] [Google Scholar]
- 40.Owens JL, Beketova E, Liu S, Tinsley SL, Asberry AM, Deng X, et al. PRMT5 Cooperates with pICln to Function as a Master Epigenetic Activator of DNA Double-Strand Break Repair Genes. iScience. 2020;23:100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He W, Ma X, Yang X, Zhao Y, Qiu J, Hang H. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res [Internet]. 2011;39:4719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu D, Gur M, Zhou Z, Gamper A, Hung MC, Fujita N, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun. 2015;6:8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rieger KE, Chu G. Portrait of transcriptional responses to ultraviolet and ionizing radiation in human cells. Nucleic Acids Res [Internet]. 2004;32:4786–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3:1254–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asim M, Tarish F, Zecchini HI, Sanjiv K, Gelali E, Massie CE, et al. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat Commun. 2017;8:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radzisheuskaya A, Shliaha PV, Grinev V, Lorenzini E, Kovalchuk S, Shlyueva D, et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat Struct Mol Biol. 2019;26:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braun CJ, Stanciu M, Boutz PL, Patterson JC, Calligaris D, Higuchi F, et al. Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma. Cancer Cell. 2017;32:411–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan DQ, Li Y, Yang C, Li J, Tan SH, Chin DWL, et al. PRMT5 Modulates Splicing for Genome Integrity and Preserves Proteostasis of Hematopoietic Stem Cells. Cell Rep [Internet]. 2019;26:2316–28. [DOI] [PubMed] [Google Scholar]
- 50.Gupta A, Hunt CR, Chakraborty S, Pandita RK, Yordy J, Ramnarain DB, et al. Role of 53BP1 in the Regulation of DNA Double-Strand Break Repair Pathway Choice. Radiat Res. 2014;181:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamard PJ, Santiago GE, Liu F, Karl DL, Martinez C, Man N, et al. PRMT5 Regulates DNA Repair by Controlling the Alternative Splicing of Histone-Modifying Enzymes. Cell Rep. 2018;24:2643–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clarke TL, Sanchez-Bailon MP, Chiang K, Reynolds JJ, Herrero-Ruiz J, Bandeiras TM, et al. PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination. Mol Cell [Internet]. 2017;65:900–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res [Internet]. 2009;37:4965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho E-C, Zheng S, Munro S, Liu G, Carr SM, Moehlenbrink J, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J [Internet]. 2012;31:1785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu L, Yang X, Duan X, Cui L, Li G. Exogenous expression of marine lectins DlFBL and SpRBL induces cancer cell apoptosis possibly through PRMT5-E2F-1 pathway. Sci Rep. 2014;4:4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Z, Kanjanapangka J, Liu N, Liu S, Liu C, Wu Z, et al. Sequential Posttranslational Modifications Program FEN1 Degradation during Cell-Cycle Progression. Mol Cell. 2012;47:444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rehman I, Basu SM, Das SK, Bhattacharjee S, Ghosh A, Pommier Y, et al. PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes. Nucleic Acids Res [Internet]. 2018;46:5601–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol [Internet]. 2001;21:8289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guderian G, Peter C, Wiesner J, Sickmann A, Schulze-Osthoff K, Fischer U, et al. RioK1, a New Interactor of Protein Arginine Methyltransferase 5 (PRMT5), Competes with pICln for Binding and Modulates PRMT5 Complex Composition and Substrate Specificity. J Biol Chem [Internet]. 2011;286:1976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang M, Lin X, Segers F, Suganthan R, Hildrestrand GA, Rinholm JE, et al. OXR1A, a Coactivator of PRMT5 Regulating Histone Arginine Methylation. Cell Rep [Internet]. 2020;30:4165–78. [DOI] [PubMed] [Google Scholar]
- 61.Lacroix M, Messaoudi S El, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep [Internet]. 2008;9:452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pesiridis GS, Diamond E, Van Duyne GD. Role of pICLn in methylation of Sm proteins by PRMT5. J Biol Chem. 2009;284:21347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saha K, Adhikary G, Eckert RL. MEP50/PRMT5 reduces gene expression by histone arginine methylation and this is reversed by PKCd/p38d signaling. J Invest Dermatol [Internet]. 2016;136:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M, An W, Xu L, Lin Y, Su L, Liu X. The arginine methyltransferase PRMT5 and PRMT1 distinctly regulate the degradation of anti-apoptotic protein CFLARL in human lung cancer cells. J Exp Clin Cancer Res [Internet]. 2019;38:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Favia A, Salvatori L, Nanni S, Iwamoto-Stohl LK, Valente S, Mai A, et al. The Protein Arginine Methyltransferases 1 and 5 affect Myc properties in glioblastoma stem cells. Sci Rep [Internet]. 2019;9:15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao G, Zhang L, Villarreal OD, He W, Su D, Bedford E, et al. PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic Acids Res [Internet]. 2019;47:5038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collins CC, Volik SV, Lapuk AV, Wang Y, Gout PW, Wu C, et al. Next generation sequencing of prostate cancer from a patient identifies a deficiency of methylthioadenosine phosphorylase, an exploitable tumor target. Mol Cancer Ther. 2012;11:775–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Almeida-Rios D, Graça I, Vieira FQ, Ramalho-Carvalho J, Pereira-Silva E, Martins AT, et al. Histone methyltransferase PRMT6 plays an oncogenic role of in prostate cancer. Oncotarget [Internet]. 2016;7:53018–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Z, He Y, Ju J, Rank G, Cerruti L, Ma C, et al. The role of WDR5 in silencing human fetal globin gene expression. Haematologica. 2012;97:1632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lorton BM, Harijan RK, Burgos ES, Bonanno JB, Almo SC, Shechter D. A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5. Biochemistry [Internet]. 2020;e-pub ahead of print 31 March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain K, Jin CY, Clarke SG. Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc Natl Acad Sci U S A. 2017;114:10101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, et al. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev [Internet]. 2012;26:2749–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Majumder S, Liu Y, Ford OH, Mohler JL, Whang YE. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate [Internet]. 2006;66:1292–301. [DOI] [PubMed] [Google Scholar]
- 74.Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol. 2019;26:880–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deb G, Thakur VS, Gupta S. Multifaceted role of EZH2 in breast and prostate tumorigenesis: Epigenetics and beyond. Epigenetics. 2013;8:464–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 Oncogenic Activity in Castration-Resistant Prostate Cancer Cells Is Polycomb-Independent. Science (80- ) [Internet]. 2012;338:1465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J, Lee Y, Lu X, Song B, Fong KW, Cao Q, et al. Polycomb- and Methylation-Independent Roles of EZH2 as a Transcription Activator. Cell Rep [Internet]. 2018;25:2808–2820.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science (80- ) [Internet]. 2017;355:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shankar E, Franco D, Iqbal O, Moreton S, Kanwal R, Gupta S. Dual targeting of EZH2 and androgen receptor as a novel therapy for castration-resistant prostate cancer. Toxicol Appl Pharmacol. 2020;404:115200. [DOI] [PubMed] [Google Scholar]
- 80.Tae S, Karkhanis V, Velasco K, Yaneva M, Erdjument-Bromage H, Tempst P, et al. Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res [Internet]. 2011;39:5424–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung J, Karkhanis V, Tae S, Yan F, Smith P, Ayers LW, et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) Silencing. J Biol Chem. 2013;288:35534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Furuno K, Masatsugu T, Sonoda M, Sasazuki T, Yamamoto K. Association of Polycomb group SUZ12 with WD-repeat protein MEP50 that binds to histone H2A selectively in vitro. Biochem Biophys Res Commun [Internet]. 2006;345:1051–8. [DOI] [PubMed] [Google Scholar]
- 83.Liu F, Xu Y, Lu X, Hamard P-J, Karl DL, Man N, et al. PRMT5-mediated histone arginine methylation antagonizes transcriptional repression by polycomb complex PRC2. Nucleic Acids Res [Internet]. 2020;48:2956–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Q, Wang G, Li Q, Jiang W, Kim J-S, Wang R, et al. Polycomb group proteins EZH2 and EED directly regulate androgen receptor in advanced prostate cancer. Int J Cancer [Internet]. 2019;145:415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim JY, Banerjee T, Vinckevicius A, Luo Q, Parker JB, Baker MR, et al. A Role for WDR5 in Integrating Threonine 11 Phosphorylation to Lysine 4 Methylation on Histone H3 during Androgen Signaling and in Prostate Cancer. Mol Cell. 2014;54:613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cao L, Wu G, Zhu J, Tan Z, Shi D, Wu X, et al. Genotoxic stress-triggered β-catenin/JDP2/PRMT5 complex facilitates reestablishing glutathione homeostasis. Nat Commun [Internet]. 2019;10:3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Briefings Funct Genomics Proteomics [Internet]. 2006;5:209–21. [DOI] [PubMed] [Google Scholar]
- 88.Jin L, Garcia J, Chan E, De La Cruz C, Segal E, Merchant M, et al. Therapeutic targeting of the CBP/p300 bromodomain blocks the growth of castration-resistant prostate cancer. Cancer Res. 2017;77:5564–75. [DOI] [PubMed] [Google Scholar]
- 89.Shin SH, Lee GY, Lee M, Kang J, Shin HW, Chun YS, et al. Aberrant expression of CITED2 promotes prostate cancer metastasis by activating the nucleolin-AKT pathway. Nat Commun. 2018;9:4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng Y, Wang J, Asher S, Hoang L, Guardiani C, Ivanov I, et al. Histone H4 acetylation differentially modulates arginine methylation by an in cis mechanism. J Biol Chem. 2011;286:20323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol [Internet]. 2003;23:7475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scaglione A, Patzig J, Liang J, Frawley R, Bok J, Mela A, et al. PRMT5-mediated regulation of developmental myelination. Nat Commun [Internet]. 2018;9:2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joseph DB, Strand DW, Vezina CM. DNA methylation in development and disease: an overview for prostate researchers. Am J Clin Exp Urol [Internet]. 2018;6:197–218. [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu X, Zhang J, Liu L, Jiang Y, Ji J, Yan R, et al. Protein arginine methyltransferase 5-mediated epigenetic silencing of IRX1 contributes to tumorigenicity and metastasis of gastric cancer. Biochim Biophys Acta - Mol Basis Dis. 2018;1864:2835–44. [DOI] [PubMed] [Google Scholar]
- 96.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WAM, Cohen A, Lasonder E, et al. MBD2/NuRD and MBD3/NuRD, Two Distinct Complexes with Different Biochemical and Functional Properties. Mol Cell Biol. 2006;26:843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chang AJ, Autio KA, Roach M, Scher HI. High-risk prostate cancer-Classification and therapy. Nat Rev Clin Oncol. 2014;11:308–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas G V., et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J Clin Oncol [Internet]. 2018;36:2492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bonday ZQ, Cortez GS, Grogan MJ, Antonysamy S, Weichert K, Bocchinfuso WP, et al. LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity. ACS Med Chem Lett. 2018;9:612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol [Internet]. 2015;11:432–7. [DOI] [PubMed] [Google Scholar]
- 101.Gulla A, Hideshima T, Anderson KC. PRMT5 inhibitors on the (myeloma) road. Oncotarget [Internet]. 2018;9:36646–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep [Internet]. 2016;15:574–87. [DOI] [PubMed] [Google Scholar]
- 103.Pollack BP, Kotenko S V, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hs17p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem [Internet]. 1999;274:31531–42. [DOI] [PubMed] [Google Scholar]
- 104.Serio J, Ropa J, Chen W, Mysliwski M, Saha N, Chen L, et al. The PAF complex regulation of Prmt5 facilitates the progression and maintenance of MLL fusion leukemia. Oncogene [Internet]. 2018;37:450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gu Z, Li Y, Lee P, Liu T, Wan C, Wang Z. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS One. 2012;7:e44033. [DOI] [PMC free article] [PubMed] [Google Scholar]