Rice (Oryza sativa L.) is a facultative short-day plant and provides staple food for more than half of the human population. Heading date (also known as flowering time) is one of the most important agronomic traits of rice as it determines the regional and seasonal adaptability of rice varieties and has a significant influence on the grain yield (Zhou et al. 2021). To maximize rice production, it is crucial to breed rice cultivars with optimum heading date suitable for the cropping areas where the cultivars are to be used. Domestication and breeding activities had created diverse natural variations of flowering time and artificially modulated the flowering regulatory pathways of rice, which helped significant expansion of rice cultivating areas (Goretti et al. 2017; Itoh et al. 2018; Hu et al. 2019). The genetic pathways regulating rice flowering time have been extensively investigated and a number of flowering time genes have been cloned and used to modify rice heading date using traditional and state-of-art molecular methods, such as cross-based introgression of key heading date gene(s) with the assistance of molecular markers and molecular modulation of the expression levels of heading date genes by gene editing (Zhou et al. 2021).
Rice has a complex genetic network regulating flowering time (Hori et al. 2016; Zhou et al. 2021). One of the major components of the network is phytochromes, including PHYA, PHYB, and PHYC. They are regulators of Ghd7, a key floral repressor in rice. PHYA alone or a combination of PHYB and PHYC can induce the expression level of Ghd7 (Osugi et al. 2011). The function of PHYC depends on the PHYB protein which participates in the regulation of PHYC expression level and in photomorphogenesis via PHYB/PHYC heterodimer (Osugi et al. 2011; Xie et al. 2014). While, under long-day (LD) conditions, phyA single mutation hardly affects flowering time of rice, phyB or phyC single mutant flowers ~ 12 days earlier than the wild-type (Takano et al. 2005, 2009).
Nanjing46 (NJ46 for short), ranked as one of the top varieties for its palatability by consumers in the triangle region of the Yangtze River, is a low amylose content rice variety suitable for planting in southern Jiangsu, China. Northward expansion of NJ46 requires shortening its long growth duration by promoting early flowering without yield penalty. In a previous study, we generated early-maturing japonica rice lines (on the Nanjing9108 genetic background) by CRISPR/Cas9-mediated editing of Hd2, Hd4, and Hd5 genes, but the lines flowered too early to be suitable for planting in Jiangsu (Li et al. 2017). We were thus seeking alternative genes for fine tuning the heading date and considered PHYC as a suitable candidate based on its moderate function in regulating flowering time (Takano et al. 2005; Osugi et al. 2011).
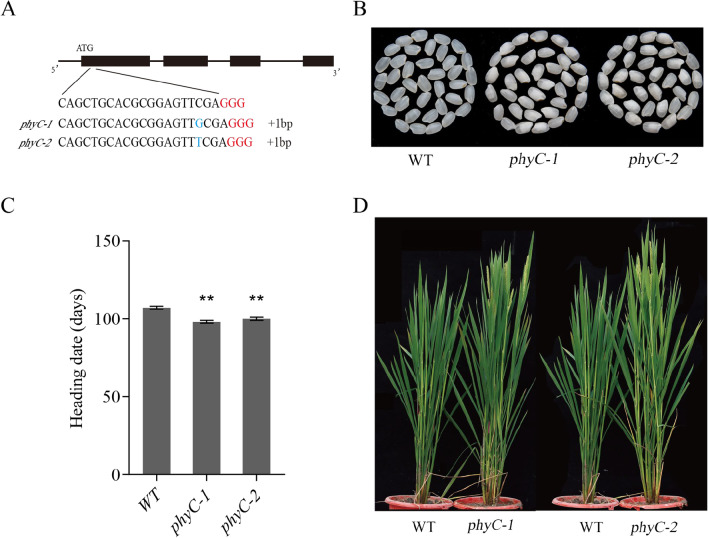
In this study, we used the CRISPR/Cas9 gene editing system to knock out PHYC in NJ46. To generate loss-of-function PHYC mutants, we designed guide RNA targeting the first exon of PHYC (Fig. 1A). We generated 20 independent T0 plants using Agrobacterium-mediated transformation approach. Positive T0 transgenic plants were identified by PCR amplification of a fragment of the Hyg gene that was used as a selection marker. The target genomic region of PHYC was amplified by a pair of primers (PHYC-F/R; Additional file 1: Table S1) flanking the target site and sequenced. The sequencing results were decoded by the Degenerate Sequence Decoding (DSD) method (Liu et al. 2015) (Fig. 1A, Additional file 1: Fig. S1A). Based on screening of the absence of Hyg and Cas9 and the presence of mutation in the target site using PCR in T1 generation, we found two transgene-free homozygous gene editing lines (both with a single base insertion that caused frame shift and pre-mature stop codon; Additional file 1: Fig. S1B) and named them phyC-1 and phyC-2. We analyzed the phenotypes of homozygous T1 and T2 populations, together with the wild-type NJ46, by recording heading date and other agronomic traits and evaluating rice grain quality under natural conditions (same as LD conditions).
Fig. 1.
Characterization of early-maturing rice generated by CRISPR/Cas9-mediated editing of PHYC. A Schematic diagram of the PHYC gene and the position of the editing target site. Black boxes and lines in between represent exons and introns, respectively. The target sequence is shown in black and the PAM sequence (CGG) in red. Underneath the target sequence is the sequence alignment to show the 1-bp insertion (highlighted in blue) induced by gene editing in the two independent transgenic lines (phyC-1 and phyC-2). B Comparison of the morphology of the milled rice grains from T2 plants. C Comparison of heading date between the two mutants and the wild-type. D Plant morphology of the PHYC mutants (phyC-1 and phyC-2; T2 generation) and NJ46 at the flowering stage
The heading date of phyC-1 and phyC-2 was about 7 days earlier than the wild-type (Fig. 1C, D). It has been reported that in addition to the regulation of heading date, PHYC is also involved in the regulation of chlorophyll content and leaf angle in rice seedlings, plant height, panicle architecture, and grain size (Li et al. 2019). We thus compared these traits between the mutants and wild-type in 2019 and 2020. In both T1 and T2 generations, no significant difference in plant height and panicle length was observed between the mutants and the wild-type (Additional file 1: Fig. S2). Compared to the wild-type, the mutants showed a slight but significant increase of grain length and 1000-grain weight, but no difference in grain width and grain thickness (Additional file 1: Fig. S3). The obviously noticeable difference was the appearance quality of polished rice grains after shelling (Fig. 1B). The possible reason for this phenomenon was that the transparency of mutants of the semi-glutinous variety NJ46 was decreased due to the higher level of chalkiness degree and the low water content (Additional file 1: Fig. S4A, B). Gel consistency and amylose content are two important traits for evaluating the eating quality of rice. There was no difference in gel consistency between the mutants and the wild-type (Additional file 1: Fig. S4C). However, the amylose content of phyC-1 and phyC-2 decreased by 13.6% and 12.7%, compared with that of the wild-type (Additional file 1: Fig. S4D). Decrease of appearance quality and amylose content in phyC-1 and phyC-2 might be a result of cooccurrences of high temperature in the early stage of grain-filling due to the advanced heading stage (Nevame et al. 2018; Hirano and Sano 1998; Ahmed, et al. 2015) (Additional file 1: Fig. S5).
These results indicated that knockout PHYC by CRISPR/Cas9-mediated gene editing could moderately shorten the heading date of NJ46, making it suitable for cultivation in northern Jiangsu, although probably would have some negative influences on grain quality. But the slightly adverse effects of loss-of-function of PHYC on the appearance quality of rice caryopses may be eliminated if the novel germplasm is planted in the regional with more suitable light and temperature conditions to avoid high temperatures during grain filling stage. We will address this issue in the follow-up studies.
In this study, we demonstrated that the CRISPR/Cas9-mediated gene editing approach is an effective tool for manipulating heading date in rice, consistent with the results by Cui et al. (2019), who investigated the usage of gene editing in alteration of flowering time by targeting 10 heading time genes. Given that rice yield is positively correlated with growth duration, or days to heading, it is important to balance the trade-off between yield and early flowering. We envision that the balance can be achieved by choosing suitable genes and appropriate approaches for manipulating their functions. While a number of genes with a known function in regulating rice flowering time have been or are being investigated using the gene editing technology, more genes, single or in different combinations, with micro-effects on heading date changes, should be investigated in future studies in order to breed novel elite varieties suitable for different cultivation areas. Base editing and prime editing can be used to precisely alter gene sequence (Komor et al. 2016; Anzalone et al. 2019) and have been demonstrated to be promising in crop improvement, such as fine-tuning of amylose content (Xu et al. 2021; Li et al. 2020). Application of precise editing technology in the right genes will finally help us achieve the goal of fine-tuning the heading date of rice without penalty in yield and quality.
Supplementary Information
Additional file 1. Materials, methods and data.
Acknowledgements
This study was supported by Jiangsu Agricultural Science and Technology Innovation Fund CX(19)1002 and the Key Research and Development Program of Jiangsu Province (Modern Agriculture, BE2018381-2).
Abbreviations
- PHYA
Phytochrome A
- PHYB
Phytochrome B
- PHYC
Phytochrome C
- Ghd7
Grain number, plant height and heading date 7
- LD
Long-day
- Hd2
Heading date 2
- Hd4
Heading date 4
- Hd5
Heading date 5
- WT
Wild type
- CRISPR
Clustered regulatory interspaced short palindromic repeat
- sgRNA
Single guide RNA
- CTAB
Cetyltrimethylammonium Bromide
- Hyg
Hygromycin
Authors' Contributions
JY, XD and YYF designed and supervised the research. FQW, YX, ZHC, FJF and JW performed most experiments. BL, WQL, YJJ, and YJT analyzed date. BL, XL and QHZ wrote the paper. All authors read and commented on the manuscript.
Funding
This work was funded by Jiangsu Agricultural Science and Technology Innovation Fund CX(19)1002, and the Key Research and Development Program of Jiangsu Province(Modern Agriculture, BE2018381-2).
Availability of Data and Materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bin Li, Email: 1904227922@qq.com.
Xi Du, Email: 1139088579@qq.com.
Yunyan Fei, Email: wlclekd@126.com.
Fangquan Wang, Email: wangfangquan@jaas.ac.cn.
Yang Xu, Email: xuyang@jaas.ac.cn.
Xia LI, Email: jspplx@jaas.ac.cn.
Wenqi Li, Email: 58190059@qq.com.
Zhihui Chen, Email: 987762831@qq.com.
Fangjun Fan, Email: 15026501@qq.com.
Jun Wang, Email: wangjunjaas@aliyun.com.
Yajun Tao, Email: ricetao@163.com.
Yanjie Jiang, Email: 99485140@qq.com.
Qian-Hao Zhu, Email: Qianhao.Zhu@csiro.au.
Jie Yang, Email: yangjie168@aliyun.com.
References
- Ahmed N, Tetlow IJ, Nawaz S, Iqbal A, Mubin M, Nawaz ul Rehman MS, Butt A, Lightfoot DA, Maekawa M. Effect of high temperature on grain filling period, yield, amylose content and activity of starch biosynthesis enzymes in endosperm of basmati rice. J Sci Food Agric. 2015;95:2237–2243. doi: 10.1002/jsfa.6941. [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zhu M, Xu Z, et al. Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor Appl Genet. 2019;132(6):1887–1896. doi: 10.1007/s00122-019-03324-1. [DOI] [PubMed] [Google Scholar]
- Goretti D, Martignago D, Landini M, Brambilla V, Gómez-Ariza J, Gnesutta N, Galbiati F, Collani S, Takagi H, Ryohei T, Mantovani R, Fornara F. Transcriptional and post-transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLOS Genet. 2017 doi: 10.1371/journal.pgen.1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, Sano Y. Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. Plant Cell Physiol. 1998;39(8):807–812. doi: 10.1093/oxfordjournals.pcp.a029438. [DOI] [Google Scholar]
- Hori K, Matsubara K, Yano M. Genetic control of flowering time in rice: integration of mendelian genetics and genomics. Theor Appl Genet. 2016;129(12):2241–2252. doi: 10.1007/s00122-016-2773-4. [DOI] [PubMed] [Google Scholar]
- Hu Y, Li S, Xing Y. Lessons from natural variations: artificially induced heading date variations for improvement of regional adaptation in rice. Theor Appl Genet. 2019;132:383–394. doi: 10.1007/s00122-018-3225-0. [DOI] [PubMed] [Google Scholar]
- Itoh H, Wada KC, Sakai H, Shibasaki K, Fukuoka S, Wu J, Yonemaru J-I, Yano M, Izawa T. Genomic adaptation of flowering-time genes during the expansion of rice cultivation area. Plant J. 2018;94:895–909. doi: 10.1111/tpj.13906. [DOI] [PubMed] [Google Scholar]
- Komor A, Kim Y, Packer M, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhou W, Ren Y, Tian X, Lv T, Wang Z, Fang J, Chu C, Yang J, Bu Q. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J Genet Genomics. 2017;44(3):175–178. doi: 10.1016/j.jgg.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng C, Zhang Z, Zhou J, Zhang H, Xie X. Characterization of phytochrome C functions in the control of de-etiolation and agronomic traits in rice. Plant Physiol Biochem. 2019;142:117–124. doi: 10.1016/j.plaphy.2019.07.004. [DOI] [PubMed] [Google Scholar]
- Li J, Li H, Chen J, Yan L, Xia L. Toward precision genome editing in crop plants. Mol Plant. 2020;13:811–813. doi: 10.1016/j.molp.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Liu W, Xie X, Ma X, Li J, Chen J, Liu Y-G. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant. 2015;8(9):1431–1433. doi: 10.1016/j.molp.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Nevame AYM, Emon RM, Malek MA, Hasan MM, Amirul Alam Md, Muharam FM, Aslani F, Rafii MY, Ismail MR. Relationship between high temperature and formation of chalkiness and their effects on quality of rice. Biomed Res Int. 2018 doi: 10.1155/2018/1653721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osugi A, Itoh H, Ikeda-Kawakatsu K, Takano M, Izawa T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 2011;157(3):1128–1137. doi: 10.1104/pp.111.181792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, et al. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell. 2005;17:3311–3325. doi: 10.1105/tpc.105.035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Inagaki N, Xie X, Kiyota S, Baba-Kasai A, Tanabata T, Shinomura T. Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. PNAS. 2009;106(34):14705–14710. doi: 10.1073/pnas.0907378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kagawa T, Takano M. The phytochrome B/phytochrome C heterodimer is necessary for phytochrome C-mediated responses in rice seedlings. PLoS ONE. 2014;9(5):e97264. doi: 10.1371/journal.pone.0097264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lin Q, Li X, Wang F, Chen Z, Wang J, Li W, Fan F, Tao Y, Jiang Y, Wei X, Zhang R, Zhu QH, Bu Q, Yang J, Gao C. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol J. 2021;19(1):11–13. doi: 10.1111/pbi.13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Zhu S, Cui S, Hou H, Wu H, Hao B, Cai L, Xu Z, Liu L, Jiang L, Wang H, Wan J. Transcriptional and post-transcriptional regulation of heading date in rice. New Phytol. 2021;230:943–956. doi: 10.1111/nph.17158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Materials, methods and data.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.