Summary
The specification of inhibitory neurons has been described for the mouse and human brain, and many studies have shown that pluripotent stem cells (PSCs) can be used to create interneurons in vitro. It is unclear whether in vitro methods to produce human interneurons generate all the subtypes found in brain, and how similar in vitro and in vivo interneurons are. We applied single-nuclei and single-cell transcriptomics to model interneuron development from human cortex and interneurons derived from PSCs. We provide a direct comparison of various in vitro interneuron derivation methods to determine the homogeneity achieved. We find that PSC-derived interneurons capture stages of development prior to mid-gestation, and represent a minority of potential subtypes found in brain. Comparison with those found in fetal or adult brain highlighted decreased expression of synapse-related genes. These analyses highlight the potential to tailor the method of generation to drive formation of particular subtypes.
Keywords: pluripotent stem cell, neuronal specification, human brain interneuron, transcriptional factor programming, single nuclei transcriptomics
Highlights
-
•
Comparison of interneurons derived from human pluripotent cells by various methods
-
•
Single-cell analyses define heterogeneity of in vitro-derived interneurons
-
•
Direct comparison of in vitro- and in vivo-derived interneurons
-
•
Identification of transcriptional modules that developmentally define interneurons
Plath, Lowry and colleagues profile interneurons generated from human pluripotent stem cells by various methods to understand the heterogeneity and cellular state of interneuron cultures in vitro. Using single-cell analyses, the authors define the homogeneity and maturity achieved with each in vitro method. By directly comparing these interneurons with those born in the human brain, the authors highlight distinctions particularly in synaptic genes and transcription factor modules that distinguish in vitro- and in vivo-derived neurons.
Introduction
Cortical inhibitory neurons (interneurons) are a critical component of the nervous system and act to regulate the degree of electrical excitation through direct interaction with excitatory and inhibitory neurons (Gelman et al., 2011, 2012; Gelman and Marin, 2010; Lim et al., 2018; Wonders and Anderson, 2006). Born from progenitor cells in the embryonic subpallidum, interneurons develop over a protracted period, undergoing important milestones of specification, migration, and maturation toward their ultimate function of cortical modulation (Gelman et al., 2012; Gelman and Marin, 2010). Many reports have elucidated the diversity and maturation of interneurons in mouse models (Boldog et al., 2018; Butt et al., 2017; Darmanis et al., 2015; Ghanem et al., 2008; Lim et al., 2018; Mayer et al., 2018; Wamsley and Fishell, 2017). Much less is known about the mechanisms through which interneurons acquire a mature state in the human context, and it has proven to be difficult to generate mature interneurons in vitro from human pluripotent stem cells (PSCs) as measured by their ability to generate repeated fast-spiking action potentials (Maroof et al., 2013).
Proper interneuron function is crucial for the appropriate establishment and activity of neural networks in the central nervous system (CNS) (Cobos et al., 2005). This is most apparent through the knowledge that defective interneurons are implicated in diverse human conditions such as intellectual disability (ID) (Chao et al., 2010, 2020; Ito-Ishida et al., 2015; Patra and Turner, 2014; Turner et al., 2002a, 2002b; Ure et al., 2016), Alzheimer disease (Martinez-Losa et al., 2018), and epilepsy (Cobos et al., 2005). With the advent of human PSCs, it is possible to generate interneurons in vitro, which have been utilized to model such diseases (Blair et al., 2018; Ohashi et al., 2018). This approach, however, has been stymied by the finding that PSC-derived cells, as in the developing brain, develop over an extended time, making applications such as disease modeling or cell therapy challenging (Nicholas et al., 2013). However, it is formally possible that PSCs do produce a small number of mature cells that are difficult to detect among a culture nearly full of immature neurons.
To begin to uncover the diversity and maturation mechanisms of cortical interneurons within the human context through development, single-cell approaches have proven to be useful. Over the past several years, advances in high-throughput single-cell RNA sequencing (scRNA-seq) techniques have allowed for a detailed picture of the transcriptional state of individual cells for the purpose of identifying cell types from a wide variety of tissues and in a variety of species (Macosko et al., 2015; Saunders et al., 2018; Zeisel et al., 2015). The human brain was until recently a more difficult target for these studies due to the availability of tissue and complications of dissociation of this tissue to single-cell suspension. To circumvent these issues, it is now possible to access human post mortem frozen brain, isolate nuclei, and profile the RNA from individual nuclei (Habib et al., 2017; Lake et al., 2016). Importantly, it has been established that sequencing specifically neuronal nuclei generates comparable transcriptomics data to whole-cell sequencing despite typically yielding fewer reads per cell (Lake et al., 2016).
Here, we used single-cell and single-nuclei transcriptomics to compare in vitro-derived interneurons with those in the fetal and adult human brain. We integrated multiple routinely used differentiation schemes to acquire a thorough understanding of human PSC-derived interneuron variability. Together, our work yields an integrated view of the transcriptional programs that distinguish individual subtypes of interneurons across different stages of development and defines programs specific to the mature state.
Results
ASCL1 and DLX2 overexpression generates PSC-derived GABAergic interneurons comparable with conventional differentiation methods
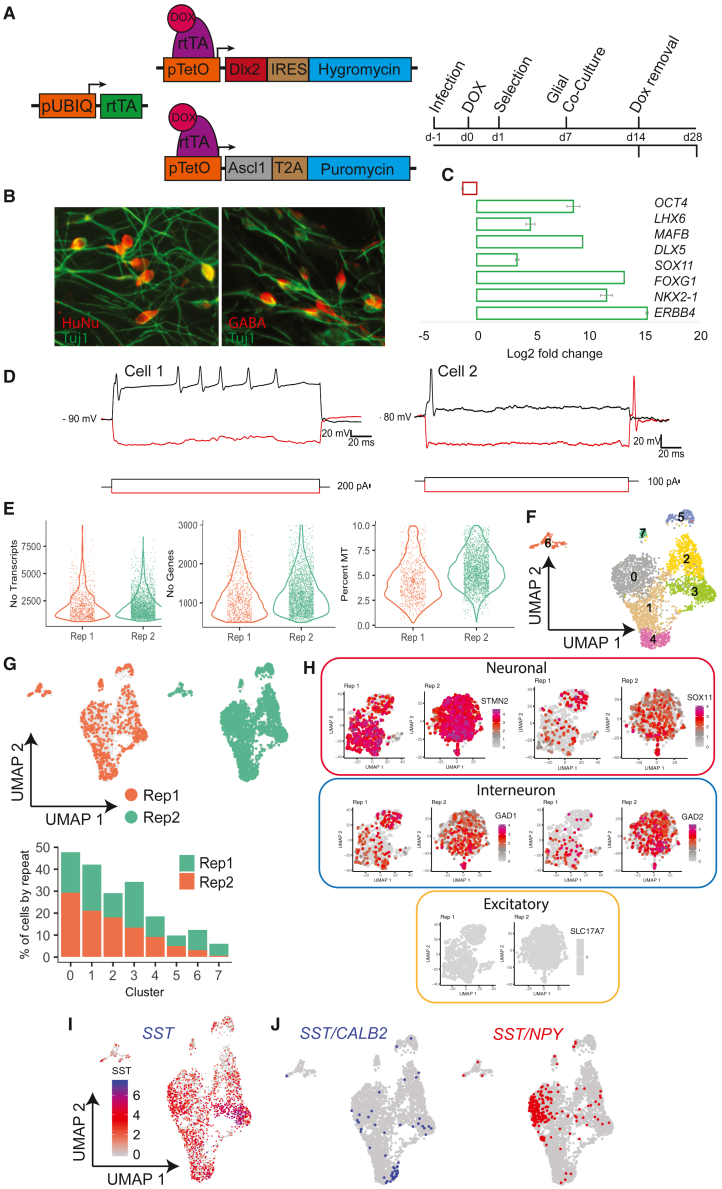
One method that was described to robustly generate interneurons from human PSCs is the overexpression of two transcription factors (TFs) that were previously shown to be important for interneuron fate in a variety of settings (Yang et al., 2017). To characterize the diversity of interneurons induced from human PSCs by TF-induced programming, we infected the H9 human embryonic stem cell (ESC) line with individual lentivirus particles containing the coding sequence of the TFs ASCL1 and DLX2, respectively (referred to here as the AD method), under the control of the tet-inducible promoter (Figure 1A). Infected cells were maintained in DOX/mTesr culture medium for 24 h before being switched to DOX/neuronal medium. After 6 days of treatment with puromycin and hygromycin to select for cells integrated with ASCL1 and DLX1 lentiviral backbone, induced interneurons (iINs) were dissociated and seeded onto mouse glial cultures, which are known to promote the viability of human neurons (Ahmed et al., 1983; Conde Guerri et al., 1989; Sugiyama et al., 1989). DOX was maintained for a further 7 days and subsequently removed (Figure 1A).
Figure 1.
Defining diversity of interneurons generated via TF-induced programming of human PSCs
(A) Schematic of the ASCL1/DLX2 expression constructs and procedure used to generate interneurons (iINs) with the AD method.
(B) Immunostaining of iINs after 2 weeks for HuNu, TUJ1, and GABA.
(C) qPCR for pluripotency- and interneuron-specific transcripts in iINs. Error bars are representative of three technical repeats.
(D) Representative patch clamp analysis of two iINs 2 weeks after induction, measuring action potential frequency. Membrane voltage traces (top) show the effects of depolarizing (black) and hyperpolarizing (red) current injections. The traces of the injected currents (duration 200 ms) are shown in bottom panels. Note the different modes of firing in the two cells: (left) repetitive firing and (right) only one action potential fired upon depolarization and a rebound action potential following the hyperpolarizing pulse.
(E) Quality analysis of scRNA-seq data for the two biological replicates of iINs at 2 weeks of induction (after dox withdrawal), measuring the number (No) of transcripts and genes, and percentage of mitochondrial reads per cell across.
(F) UMAP clustering of single iINs for the two repeats in (E).
(G) Quality control showing even distribution of cells from both replicates among the eight clusters from (F).
(H) Normalized expression levels of indicated genes superimposed onto the UMAPs from each replicate. Colored according to expression levels by cell.
(I) As in (H), except for the interneuron subtype marker, SST.
(J) As in (I), except for cells that express both SST and CALB2 (left, in blue) or SST and NPY (right, in red).
After 2 weeks of DOX removal, we characterized iINs to ensure proper specification of the GABAergic interneuron fate. We used an antibody against human nuclear antigen (HuNu) to identify human neurons against the co-cultured glial mouse cells. iIN cultures showed strong expression of the pan-neuronal marker TUJ1 and of the interneuron-specific neurotransmitter gamma-aminobutyric acid (GABA) (Figure 1B). qPCR for OCT4, a marker of PSCs, and for TFs involved in the development of GABAergic interneurons, including LHX6, MAFB, DLX5, SOX11, FOXG1, and NKX2-1, as well as for ERBB4, a marker specific for migrating interneurons (Fregnan et al., 2014; Rakic et al., 2015; Villar-Cervino et al., 2015), confirmed appropriate gene expression profiles for interneuron specification (Figure 1C).
Next, to validate the functional specification of these iINs, we performed electrophysiological recordings from five batches of cultured cells. We recorded from 29 fluorescently labeled iINs; 48% of them showed either multiple action potential firing (Figure 1D, left) or single action potential firing upon depolarizing current injections (Figure 1D, right) at their resting membrane potential. These results were not surprising given the requirement for many months in culture or for transplantation into the mouse brain to fully mature (Marin, 2013; Maroof et al., 2013).
To characterize heterogeneity of iINs, we performed droplet-based RNA sequencing (Drop-seq) (Macosko et al., 2015) of single iINs at 2 weeks of induction (post-DOX withdrawal). Cells were filtered to remove mouse cells, cells with low-quality reads (<500 genes and mitochondrial genes >10%) and cells with aberrantly high gene counts, indicative of doublets (>3,000 genes), resulting in transcriptomic data from a total of 3,363 cells across two repeats (replicate 1 [Rep 1] = 766 cells, Rep 2 = 2,597 cells) (Figure 1E), with a mean of 780 genes detected per cell. The Uniform Manifold Approximation and Projection (UMAP)-based embedding shows that iINs could be split into eight distinct clusters (Figure 1F), with similar distributions from both replicates (Figure 1G). The cells robustly expressed the general neuronal markers STMN2 and SOX11, as well as the interneuron-specific markers GAD1 and GAD2, and lacked expression of the excitatory marker SLC17A7, confirming interneuron specification (Figure 1H). Exploring the expression of broad interneuron subtype markers, we found that the vast majority of iINs expressed SST, reminiscent of interneurons from the medial ganglionic eminence (MGE) (Anderson et al., 2001; Fishell, 2007; Marin et al., 2000) (Figure 1I). Smaller sub-populations co-expressed SST/CALB2 and SST/NPY (Figure 1J); however, we did not find evidence of PVALB-expressing cells, which are not born in the brain until much later in development and are difficult to generate from human PSCs (Maroof et al., 2013). Furthermore, very few cells expressed caudal ganglionic eminence (CGE) subtype markers, including CALB2 (without SST), RELN and VIP (not shown). Thus, the expression of ASCL1/DLX2 generated a high proportion of GABAergic interneurons with a bias toward SST+ cells (Figure 1J).
Comparison of iINs induced from PSCs by different methods
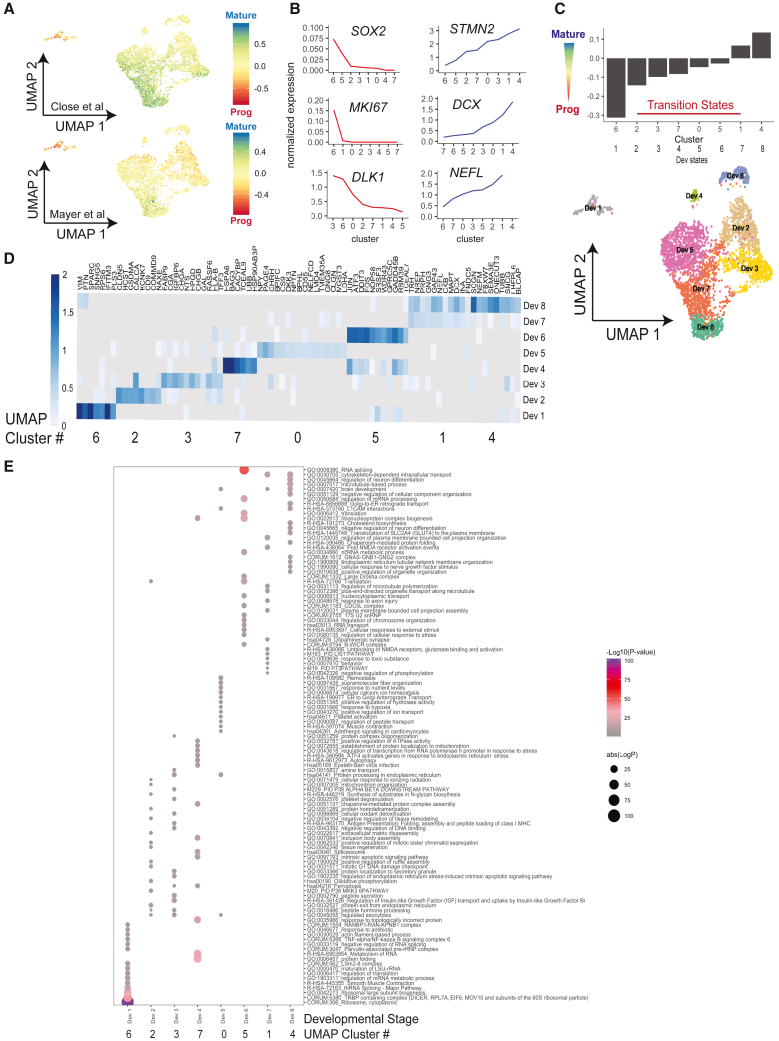
We next investigated the relative maturity of the iINs generated by forced expression of ASCL1 and DLX2 in PSCs. We assigned all cells a score based on both neural progenitor and maturing interneuron gene sets described by two separate scRNA-seq datasets on maturing interneurons generated from PSCs in defined interneuron induction culture medium or on in vivo murine development (Close et al., 2017; Mayer et al., 2018; Schuman et al., 2019; Wamsley and Fishell, 2017) (Figure 2A). Using the Close et al. signatures from PSC-derived INs (Figure 2A), we found progenitor-like cells in cluster 6 and the most mature cells in cluster 4 (clusters as described in Figure 1F), indicating that the overexpression of AD can generate similar maturation states, but in only 2 to 4 weeks as opposed to 7 weeks necessary for typical directed differentiation toward interneurons. Accordingly, transcripts for progenitor markers such as SOX2, MKI67, and DLK1 were enriched in cluster 6, whereas migration and axonal markers DCX, STMN2, and NEFL were most expressed in cluster 4 (Figure 2B). Using the maturity scores derived from the in vivo gene sets from Mayer et al. (2018) (Figure 2A), a similar distribution was apparent. However, the most mature cluster 4 did not reach full maturation and most iINs showed a strong progenitor score, aligning with previous reports that human PSC-derived interneurons do not fully mature in vitro.
Figure 2.
Characterization of specification and maturation of iINs
(A) UMAP of scRNA-seq data from iINs (as defined in Figure 1F) colored according to maturity score, from red, immature progenitors, to blue, mature neurons, based on the maturation signatures for in vitro differentiation (Close et al., 2017) and for in vivo data (Mayer et al., 2018). Scores were computed by averaging each cell's normalized expression of maturation genes and subtracting from the average of the normalized expression of progenitor genes.
(B) Normalized transcript levels of progenitor (SOX2, MKI67, DLK1) and neuron (STMN2, DCX, NEFL) marker genes across each of the eight iIN clusters from Figure 1F. Clusters are organized by increasing expression levels.
(C) Bar plot showing the average maturation score from (A) across clusters 0–7 from Figure 1F, ordered from most progenitor-like to most mature. Based the increasing maturation signature axis, clusters 0–7 were redefined as developmental transition states (Dev1–8).
(D) Top 20 differentially expressed genes (above a threshold of 1 for average log2 fold change) between the iIN clusters, ordered by developmental group.
(E) GO analysis of all differentially expressed genes above a threshold of 0.5 average log2 fold change for each cluster, ordered by Dev state. The original cluster number is given below. Dots are colored according to each GO term’s p value, and the size of dots represent the number of genes identified within each GO term.
Using the expression status of progenitor and maturation genes, we generated a trajectory and inferred a developmental state for each of the eight clusters of AD-induced iINs (Figure 2C) and defined gene expression signatures for each developmental state (Dev1–8), from progenitor, neuron-specified, to postmitotic iINs (Figure 2D). The most primitive developmental stage (Dev1, cluster 6) showed a strong enrichment for the splicing and regulation of mRNA, which has been previously described as an enriched feature of developing tissues, including the CNS (Figure 2E) (Lopez, 1998; Voineagu et al., 2011; Wamsley et al., 2018). The Dev 2–4 states demonstrated a shift in the metabolism of cells to oxidative phosphorylation, and activation of the p38-MAPK pathway and insulin growth factor (IGF) signaling (Figure 2E). The p38-MAPK pathway has been associated with cell-cycle arrest and may signify the exit of the cell cycle typical of progenitors becoming neurons (Brancho et al., 2003). The Dev 5 state captured a third transition and associated genes are linked to peptide and ion transport and calcium homeostasis, important features for the stabilization of neuronal membrane potential (Figure 2E). Genes characterizing the final Dev 6–8 states yielded terms related to microtubule polymerization, synaptic regulation, brain development, and cholesterol biosynthesis. Taken together, the forced expression of ASCL1 and DLX2 generated GABAergic iIN populations as previously reported; however, the single-cell analysis provided here highlighted developmental heterogeneity within these cultures.
Heterogeneity and maturity of interneurons created in organoid cultures
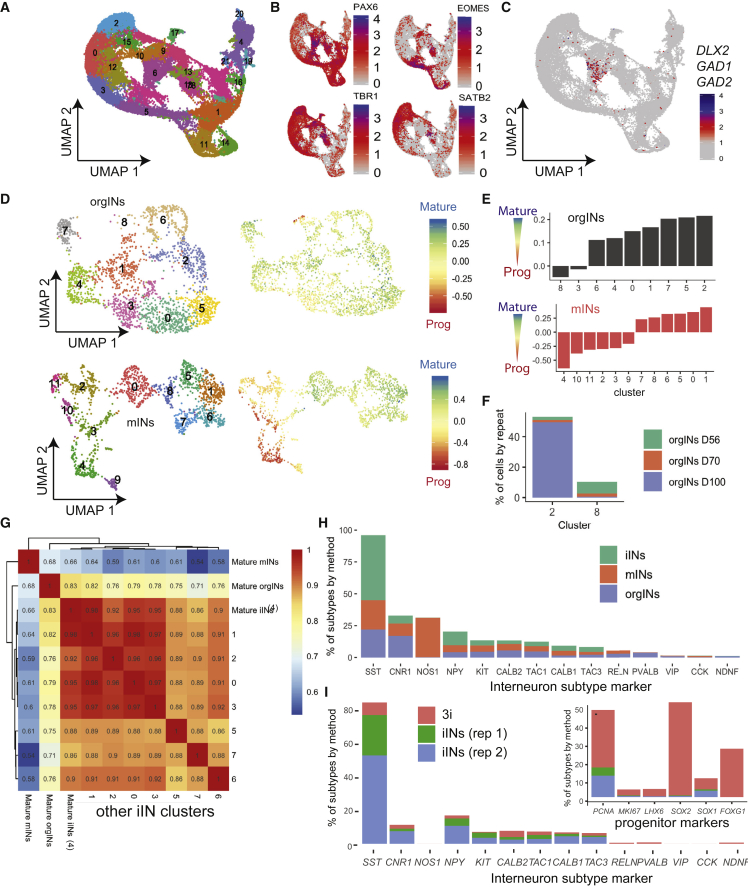
To better understand how the use of TF-induced programming of iINs compares with conventional directed differentiation methods for GABAergic interneuron generation, we took advantage of the same scRNA-seq dataset from monolayer differentiation exploited in Figure 2 (Close et al., 2017) (monolayer interneurons [mINs]) and a scRNA-seq dataset generated from unfused or fused MGE-cortex organoids after 56, 70, and 100 (days of induction from PSCs (organoid interneurons [orgINs]) (Samarasinghe et al., 2021). We detected 22 clusters in the organoid dataset (Figure 3A), including clusters of basal radial glia (marked by PAX6 transcripts), intermediate progenitors (EOMES), deep cortical plate (TBR1), superficial cortical layers (SATB2), and interneurons (DLX2, GAD1, and GAD2, cluster 6) (Figures 3B and 3C). Re-clustering of the interneuron cluster generated nine interneuron-specific clusters (Figure 3D). From the mIN dataset, interneurons separated into 12 distinct clusters (Figure 3D).
Figure 3.
Comparison of iINs, mINs, and orgINs
(A) Unsupervised UMAP clustering of 36,076 single cells derived from day 56 (D56) unfused, day 70 (D70) and day 100 (D100) fused cortex and MGE organoids.
(B) UMAP from (A) colored by the normalized expression of indicated genes in individual cells.
(C) As in (B), except that the plot shows the co-expression of indicated interneuron marker genes, indicating that cluster 6 is expressing these markers together.
(D) (Top) Cells from interneuron cluster 6 of the organoid scRNA-seq data in (A) (orgINs) were divided into nine interneuron clusters (0–8) and plotted on a UMAP (left). In the same UMAP to the right, individual cells are colored by the maturation score from Close et al. (2017). (Bottom) UMAP of the scRNA-seq data from the mIN dataset of 2D cultures of interneurons made by directed differentiation (Close et al., 2017), separating interneurons into 12 distinct clusters (0–11) (left) and colored by the maturation score from (Close et al., 2017) (right).
(E) Maturation score for the clusters in (D), ordered from least to most mature.
(F) The contribution of cells from different organoid harvest time points to the orgINs clusters with highest (cluster 2) and lowest (cluster 8) maturation state.
(G) Pearson correlation analysis for the most mature orgIN cluster (cluster 2 from (E)), the most mature mIN cluster (cluster 1 from (E)), and all iIN clusters (AD-method-derived interneurons) from Figures 1 and 2 (with cluster 4 containing the most mature cells).
(H) A survey of the subtypes of interneurons found in iINs, mINs, and orgINs.
(I) A similar analysis as in (H), but for individual experiments for directed differentiation (3i) of human induced PSCs or the two biological replicates of iINs from hESCs.
Next, we attempted to understand whether the mINs and orgINs showed a similar type of progressive developmental trajectory as found for iINs. Using the gene signatures from Figure 2A, we found that, while there was a wide spectrum of developmental states in the mIN culture, this was not the case for orgINs (Figures 3D and 3E). Aside from a small number of cells with a strong progenitor-like signature (cluster 8), orgINs produced mostly interneurons that appear to be most similar to a postmitotic, fully specified fate (Figures 3D and 3E). In orgINs, clusters 8 and 2 were the most progenitor- and mature-like, respectively (Figure 3E). Cells from day 56 predominated in cluster 8, whereas cells from day 100 predominated in cluster 2, indicating a maturation with different time course and/or organoid fusion (Figure 3F) (Samarasinghe et al., 2021).
Compared with orgINs, mINs showed a more progressive developmental transition with a slower drop in progenitor marker levels and less dramatic acquisition of mature markers (Figures 3D and 3E). This observation was more similar to that for iINs (AD method), indicating that the 3D structure and network generation of multiple cell types within organoids results in a more uniform maturation of interneurons than monolayer-based methods. On the other hand, this distinction could also be a product of the fact that the interneurons profiled from the organoids were profiled after 7–14 weeks, while the iINs were profiled after just 2–3 weeks. However, neither in vitro method generated significant numbers of fully mature interneurons as defined by the maturation signature (defined by data from the Close and Fishell maturation signatures, signified by blue color) (Figure 3D). To further compare these in vitro methods of interneuron generation, we correlated the most fully specified clusters of interneurons from mINs and orgINs with all iIN clusters and found highest similarity to the most mature iIN cluster 4 (Figure 3G), followed by cluster 1, which was the only other more mature cluster of iINs (Figure 2C).
We also explored whether each of the different methods generated different proportions of interneuron subtypes (Figure 3H). Using established markers (Darmanis et al., 2015; Habib et al., 2017), we found that the majority of interneurons from all in vitro methods were SST+, arguing that the modeling of SST-related disorders would be most informative in vitro. Smaller proportions of interneurons defined by expression of KIT, CALB2, TAC1, CALB1, and TAC3 were generated in all the methods (Figure 3H). orgINs were not substantially more diverse than monolayer-based methods, but contained a small number of PVALB+ interneurons (Figure 3H). To determine whether each individual human PSC line or each derivation showed consistent results, we also plotted cell type proportions from the replicates of AD-enforced cultures and our own directed differentiation cultures (without TF overexpression, 3i; Maroof et al., 2013) generated from distinct human PSC lines, and came to similar conclusions (Figure 3I). However, iIN cultures contained fewer progenitors than interneuron cultures generated by directed differentiation (Figure 3I, inset).
Identifying hallmarks of maturation in interneurons in the adult human brain
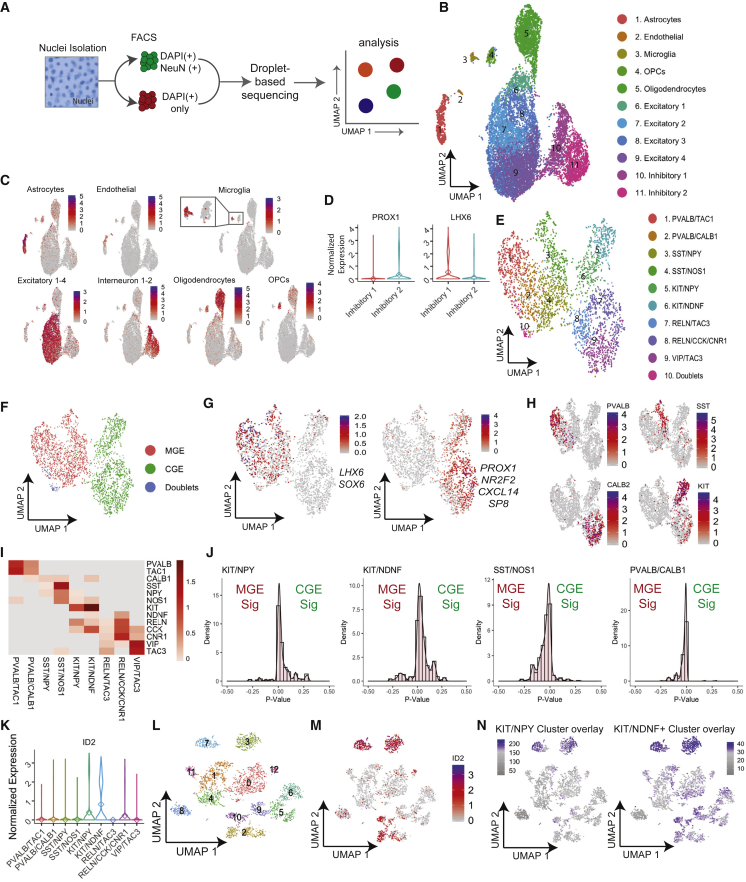
We next set out to compare the PSC-derived interneurons generated by the AD-method with bona fide interneurons from the human brain. We took advantage of methods to analyze RNA from individual nuclei of frozen post mortem human frontal cortex, a reliable method to generate informative transcriptomes from frozen samples (Habib et al., 2017). To improve RNA capture, we slightly modified the Drop-seq approach for scRNA-seq, whereby droplet size was decreased and lysis buffer concentration increased to improve nuclear membrane lysis (Habib et al., 2017). We isolated and fluorescence-activated cell sorted individual nuclei based on DAPI staining from six healthy adult frontal cortex tissue samples. From the same donors, we also sorted nuclei based on DAPI and NeuN staining to enrich neuronal cells, as interneurons represent only a small proportion of total cells within the adult brain (Figure 4A). We merged the data from all six brains and, using UMAP-based clustering, identified 11 clusters that corresponded to previously established cell types, including astrocytes, endothelial cells, microglia, oligodendrocyte precursor cells (OPCs), oligodendrocytes, and excitatory and inhibitory neurons, identified using established markers of each cell type respectively (Figures 4B and 4C). Within the clusters that showed markers of inhibitory neurons, cells were broadly split into two groups of distinct developmental origin, such that inhibitory interneuron cluster 1 highly expresses LHX6, indicative of MGE-derived interneurons, whereas inhibitory interneuron cluster 2 expresses PROX1, indicative of CGE-derived interneurons (Figure 4D).
Figure 4.
Profiling interneurons from human adult brains with snRNA-seq
(A) Schematic of the workflow for snRNA-seq from frozen archived adult brain cortex.
(B) UMAP of 17,879 nuclei from six adult cortexes. Cell identities were assigned to clusters based on marker gene expression.
(C) Simultaneous expression of several genes known to define clusters in (B).
(D) Violin plots for the normalized expression of PROX1 and LHX6 in the two interneuron clusters 10 and 11 in (B), called Inhibitory 1 and 2, to decipher MGE- versus CGE-derived interneurons.
(E)The UMAP of the cells from interneurons from clusters 10/11 in (B), revealing 10 interneuron sub-clusters (1–10) of the adult brain, labeled based on key marker expression.
(F) UMAP from (E) split into MGE- versus CGE-derived interneurons based on simultaneous expression of region-specific markers.
(G) UMAP as in (E) showing the simultaneous expression of established markers of either MGE (LHX6, SOX6) or CGE (PROX1, NRF2, CXCL14, SP8).
(H) Normalized expression of indicated interneuron subtype marker on the UMAP from (E).
(I) Heatmap of normalized expression of selected interneuron markers by clusters from (E).
(J) Histograms to demonstrate the developmental origin of indicated KIT+ clusters and SST/NOS1, PVALB/CALB1 clusters as controls. MGE and CGE gene signatures were generated by taking the top 20 most differentially expressed genes between MGE- and CGE-derived interneurons. Individual cells were assigned MGE and CGE scores based on their individual expression of each MGE or CGE gene, using the Wilcoxon test.
(K) Normalized expression for ID2 in interneuron sub-clusters from in (E).
(L) UMAP of all interneuron subtypes in a human brain dataset from the Allen Brain Database.
(M) As in (L), with an overlay of the normalized expression of ID2.
(N) Simultaneous expression of KIT/NPY and KIT/NDNF cluster signature genes (genes unique to KIT/NPY or KIT/NDNF clusters from E with an average log2 fold-change cutoff of 0.5), overlayed on the UMAP in (L).
To ascertain whether we captured the interneuron diversity of the adult brain in our in vivo dataset, we re-clustered the inhibitory clusters 1 and 2, yielding 10 clusters broadly separable according to developmental origin, including clusters enriched for either MGE markers LHX6 and SOX6, or CGE markers, PROX1, NR2F2, SP8, and CXCL14 (Figures 4E–4G). One cluster was consistent with doublets of cells, and was not considered in subsequent analyses. To assign subtypes, we cross-validated the gene expression of the remaining nine interneuron clusters against a published dataset of single-cell profiling of human cortex (Habib et al., 2017) and were able to assign the four overarching interneuron types PVALB, SST, CALB2, and KIT (Figures 4H and 4I).
Interestingly, KIT+ inhibitory neurons were recently described in mouse cortex (Fishell, 2007; Mayer et al., 2018; Wamsley and Fishell, 2017). Therefore, our data suggest that they also exist in the adult human brain. To determine the developmental origin the KIT+ interneuron subsets, we looked for expression of gene signatures representative of MGE- and CGE-derived cells (Figure 4J),and found that both KIT+ populations appeared to be closer to CGE, as opposed to SST or PVALB populations, which looked more similar to MGE. Both our KIT+ clusters also showed strong expression of ID2, characteristic of a subtype recently described in the mouse and human cortex (https://portal.brain-map.org/atlases-and-data/rnaseq) (Figure 4K). An overlay of the enriched genes from the KIT/NPY and KIT/NDNF clusters onto the Allen Brain Database of single-nuclei RNA sequencing (snRNA-seq) data from the human cortex (Figures 4L–4N) showed a very strong similarity with two of the ID2+ clusters in the Allen dataset (Figure 4N), confirming the presence of these cells in our independent dataset. These data indicate that our method to derive and characterize interneurons from adult brain encompassed most known subtypes of interneurons.
Comparing iINs with interneurons during human gestation
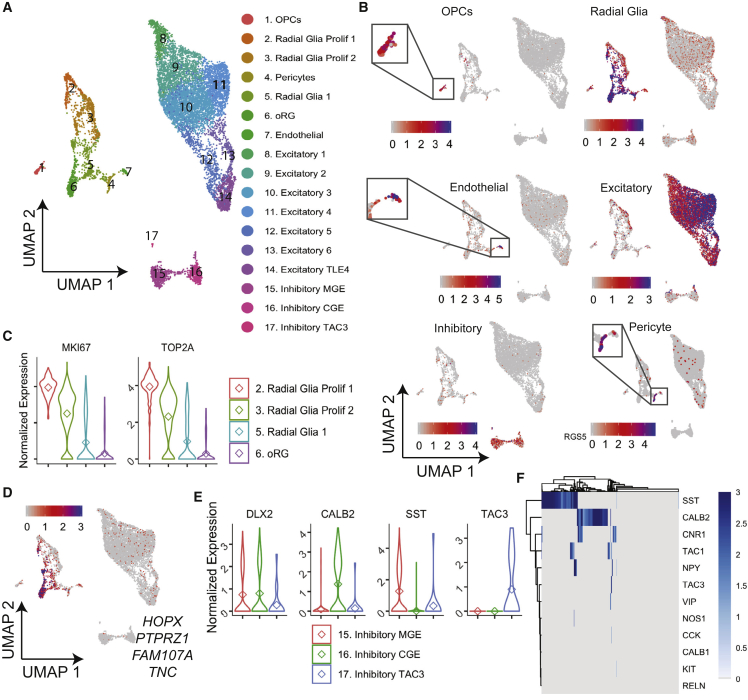
To further determine genes that define the adult state of human interneurons in vivo, we reasoned that the comparison with an earlier developmental state would also be informative, and integrated our Drop-seq-generated single-cell transcriptomes from interneurons in fetal development (Polioudakis et al., 2019). We used profiles of 10,101 cells from two samples at 15 postconceptional weeks (pcw), which formed 17 clusters upon dimensionality reduction (Figure 5A). Analyzing the expression signatures of these clusters, we identified similar groups of cell types as in the adult human brain, including OPCs, endothelial cells, excitatory and inhibitory neurons, as well as cell types not present in our adult brain dataset, namely, radial glia and pericytes (Figure 5B). Within the radial glia, we found clusters that were separated based on cell proliferation and cell identity, with clusters 2 and 3 showing an enrichment for proliferation markers (MKI67 and TOP2A) and cluster 6 for oRG markers (HOPX, PTPRZ1, FAM107A, and TNC) (Figures 5A–5D), which are known to contribute to new cell formation in upper layers specifically in the developing human brain.
Figure 5.
Single-cell profiling of fetal brain interneurons
(A) Unsupervised clustering using UMAP analysis of 11,796 cells from fetal cortex (Polioudakis et al., 2019).
(B) UMAP as in (A) colored by the simultaneous expression of marker gene panels previously established as unique to broad CNS cell types in the fetal brain (Polioudakis et al., 2019). For pericytes, the normalized expression level of the pericyte marker RGS5 is given.
(C) Violin plots of normalized expression counts in single cells from all radial glial clusters from (A), for genes associated with cell proliferation.
(D) UMAP as in (A) overlayed to show the simultaneous expression indicated for outer radial glia markers.
(E) Violin plots of normalized expression counts for indicated interneuron-specific markers, in single cells of interneuron clusters in (A).
(F) Heatmap of the normalized expression of indicated interneuron subtype markers with optimal leaf hierarchical ordering.
Probing for inhibitory neuron markers within the fetal dataset, we defined three interneuron clusters, as shown through the expression of DLX2, that, similarly to the adult dataset, separated based on developmental origin of MGE (SST+ interneurons) versus CGE (CALB2+ interneurons) (Figure 5E). Interneuron cluster 17 showed a strong enrichment for TAC3, a marker often found expressed alongside PVALB in mature interneurons (Figure 5E). This cluster may represent interneurons destined to become PVALB+ later in development as previously suggested (Polioudakis et al., 2019). Re-clustering of the interneurons did not reveal any further separation of the broad subtypes (not shown). We also found that few fetal interneurons expressed the diverse subtype markers that we found within the adult brain, with SST and CALB2 being the predominant markers, identical to our findings of in vitro-derived interneurons (Figure 5F) but distinct from adult brain data (Figure 4). These observations demonstrate that, in this developmental time frame, interneurons have a vastly reduced heterogeneity compared with the adult brain, leading to the conclusion that the acquisition of subtype diversity correlates with interneuron maturation.
Identifying expression patterns that distinguish in vitro- from in vivo-derived interneurons
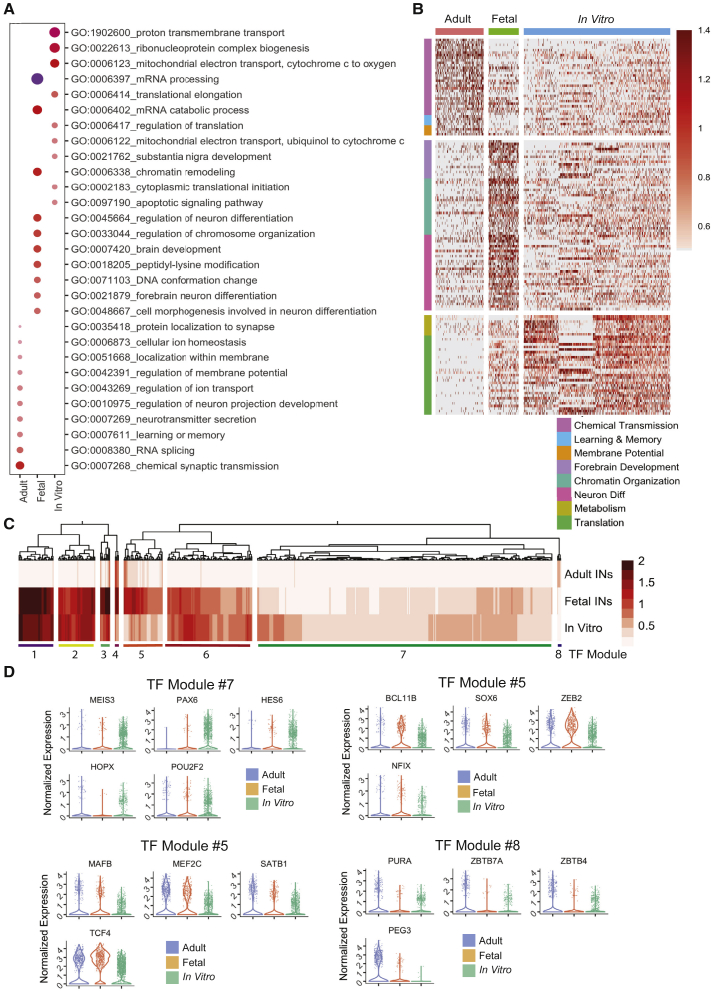
Next, we were interested in understanding the dynamic expression patterns of genes across our in vitro-derived (iINs, mINs, and orgINs) and in vivo-generated datasets for the fetal and adult brain. Due to the power of sc/snRNA-seq, we were able to perform these analyses in a subtype-specific manner, which is of importance due to the different functional behaviors of interneuron subtypes. First, we merged SST+ interneurons from our in vitro iINs, mINs, and orgINs datasets and compared them with fetal and adult SST+ interneurons through differential gene expression analysis. Using Gene Ontology (GO) analysis to understand the biological context of gene expression changes, we found that each set of SST+ interneurons exhibited unique signatures, such that adult interneurons enriched for synaptic terms, learning, memory, and neurotransmission, fetal interneurons for chromatin organization, forebrain development, and neuron differentiation, and in vitro-derived cells for metabolism and protein translation (Figures 6A and 6B).
Figure 6.
Defining transcriptional modules that distinguish SST+ interneurons born in vitro versus in vivo
(A) GO analysis of differentially expressed genes (unique genes with an average log2 fold change above 0.5) across SST+ interneurons from adult, fetal, and in vitro samples. In vitro samples consist of SST+ interneurons from iINs, mINs, and orgINs. Dot color represents the log10 p value for each term and the size represents the number of genes present per GO term.
(B) Heatmap of normalized counts in individual cells of the top differentially expressed genes between SST+ adult, fetal, and in vitro interneurons from (A), clustered according to their functional properties as defined by GO terms.
(C) TF module analysis, comparing differential expression of TFs and chromatin regulators across adult, fetal, and in vitro-derived SST+ interneurons. Normalized expression data are plotted and optimal leaf hierarchical clustering was used to generate modules of highly correlating regulators.
(D) The normalized expression of indicated TFs/chromatin regulators from different modules in (C) is depicted in violin plots.
We next turned our attention more specifically to differentially expressed TFs to gain an insight into the regulation of differences in cell states between in SST+ interneurons, as these are known to be critical to specify and maintain cell fate. We built networks of co-expressed TFs and chromatin regulators across the developmental stages through a correlation analysis and divided them into eight modules (Figure 6C). Some of these modules were expressed specifically in either the adult, fetal, or in vitro interneurons, whereas other modules were common to all three samples (Figure 6C). This also implied that fetal SST interneurons at 15 pcw were developmentally progressed compared with their in vitro counterpart.
Many of the TFs within the modules are known to play important roles during neurogenesis and are typically found early during development (Figure 6D). For instance, module 7 captured TFs most enriched in PSC-derived cells, including MEIS3, PAX6, HES6, HOPX, and POU2F2. Within module 5, we identified TFs that showed strongest enrichment in fetal interneurons. These TFs were typical of developing interneurons, including BCL11B, SOX6, ZEB2, and NFIX. Within this same module, we also found the enrichment of some MGE-specific TFs in both fetal and adult cells, including MAFB, MEF2C, SATB1, and TCF4. Finally, in module 8, we found TFs specific to adult cells, including PURA, ZBTB7A, ZBTB4, and PEG3. Both ZBTB4 and ZBTB7A regulate chromatin organization, and mutations in other types of genes with roles in regulation of gene expression in this module are often found in ID syndromes, such as Angelman syndrome (UBE3A) (Kishino et al., 1997; Matsuura et al., 1997) and ICF syndrome (DNMT3B) (Huang et al., 2014; Moarefi and Chedin, 2011; Sagie et al., 2014).
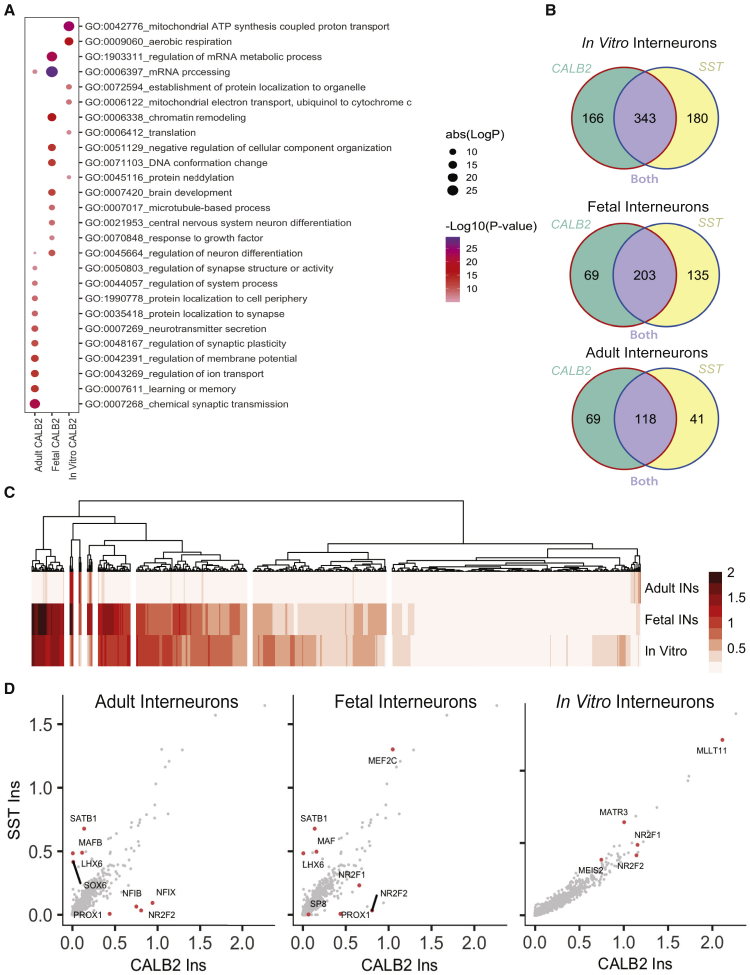
Using the same form of analysis, we interrogated how interneurons that expressed CALB2 changed over development and between in vivo and in vitro conditions. Surprisingly, we found that PSC-derived, fetal, and adult CALB2+ interneurons showed highly similar changes to SST interneurons from the same states. Consequently, we found similar GO terms and a large overlap for differentially expressed genes for SST and CALB2 interneuron states (Figures 7A and 7B). Pearson correlation analysis again highlighted eight clusters of co-expressed TFs that distinguish in vitro CALB+ interneurons from fetal and adult CALB+ interneurons (Figure 7C). We also found that many of the TFs that define in vitro, fetal, and adult SST+ interneurons were also enriched in CALB2+ interneurons from the respective stage (Figure 7D). It was apparent that the differences between SST+ and CALB2+ interneurons from both the adult and fetal samples were characterized by TFs that are known to play a role in subtype specification, including the MGE-specific TFs (SATB1, MAF, LHX6) for SST+ interneurons and the CGE-specific TFs (NFIX, NR2F2, and PROX1) for CALB2+ interneurons (Figure 7D). We conclude, therefore, that the process of maturation is conserved between subtypes, leading to the notion that the functional differences observed between subtypes may be established early in development.
Figure 7.
CALB2+ interneurons display maturation signatures similar to that of SST+ interneurons
(A) GO analysis of differentially expressed genes (average log2 fold change above 0.5) across CALB2+ interneurons from the adult, fetal, and in vitro samples. In vitro samples consist of iINs, mINs, and orgINs. Dot color represents the log10 p value for each term and the size represents the number of genes present per GO term.
(B) Venn diagrams to show the overlap in vitro-, fetal-, or adult-specific genes in SST+ and CALB2+ interneurons.
(C) TF module analysis, comparing differential expression of TFs/chromatin regulators across adult, fetal, and in vitro-derived CALB2+ interneurons. Normalized expression data were plotted and optimal leaf hierarchical clustering was used to generate modules of highly correlating TFs, and the results were similar to those found in Figure 6D.
(D) Scatterplots comparing the average normalized expression of all TFs of cells from adult (left), fetal (middle) and in vitro-derived (right) CALB2+ and SST+ interneurons.
Discussion
Interneurons made from human PSCs could be useful to ameliorate patient symptoms not only because of their importance in a variety of diseases, such as epilepsy, but also because they can be derived easily and appear to survive in transplantation models. However, our work highlights the molecular and physiological immaturity of interneurons derived from human PSCs relative to both mid-gestation and adult INs. We elaborated on molecular distinctions between particular subtypes of interneurons as they proceed through maturation as a resource to both understand their development and also provide clues as to how to promote their maturity. We provided a single-cell characterization and comparison of various subtypes of interneurons derived in vitro and in vivo. It is not clear at this point whether excitatory neurons employ the same types of molecular tools to advance their maturation as do interneurons, but they are known to do so at a much faster rate. As a result, similar studies will need to be carried out to identify equivalent patterns of gene expression that drive maturation in the excitatory lineages.
The in vitro work described here demonstrates several advantages and disadvantages of distinct culture methods. First, all three methods described here only generated a limited diversity of subtypes of interneurons. iINs were highly homogeneous and were generated quickly, while directed differentiation via culture conditions was slow. orgINs were more diverse than the other methods, but took significant effort and time to produce. None of these methods produced mature, post-natal-like interneurons as defined functionally (electrophysiology) or transcriptionally. Future work will focus on forced expression of additional TFs that could drive maturation beyond that observed with ASCL1 and DLX1.
This work also sheds light on the processes by which different subtypes of neurons achieve maturation. A priori, it was possible that each subtype of interneuron takes advantage of distinct mechanisms to drive maturation. Our data suggest that in vitro-derived SST+ and CALB2+ interneurons each show similar patterns of expression that distinguish them from their brain-derived counterparts. This is not to say that the genes that distinguished the in vitro and brain-derived interneurons were identical, but instead that the functional categories of genes, including TFs, were highly similar. This suggests that different subtypes of interneurons use the same physiological processes to achieve maturation. Going forward, we expect that studies such as this will lead to improved methods for generating mature interneurons for in vitro modeling of various neurological disorders that are linked in this lineage.
Experimental procedures
Generation of interneurons in organoid culture
The generation of brain organoids was performed as described (Samarasinghe et al., 2021).
Tissue samples
Anonymous fetal tissue samples were obtained from the University of California, Los Angeles (UCLA), gene and cell therapy core according to institutional review board guidelines (Table S1). Anonymized discarded adult brain tissue was acquired from the NIH BioBrainBank (Table S1). NeuroBioBank (Sepulveda repository, Los Angeles, CA) for BioBrainBank. This study was performed according to the legal and institutional ethical regulations of the UCLA Office of Human Research Protection. Full informed consent was obtained from all parent donors.
Additional materials and methods can be found in the supplemental information.
Data availability
All datasets generated or used in this study and are deposited in NIH GEO (GSE180132 and GSE181715) and are summarized in Table S1.
Author contributions
Data collection and analysis, T.A., J.L., A.L., M.O.G., D.P., X.W., I.L., J.H., and R.S.; data analysis, T.A., S.S., J.L., and D.P.; project design and support, I.C., K.P., D.G., I.M., B.N.N., and W.E.L.; manuscript preparation, T.A., K.P., W.E.L.
Conflict of interests
W.E.L. is a founder and president of Pelage Pharmaceuticals and Secretary of Sardona Therapeutics, but this work was not related to these companies. K.P. is on the Editorial Board of Stem Cell Reports.
Acknowledgments
We would like to thank Iris Dror for help with computational approaches, Andrew Elefanty for sharing the NKX2.1 GFP reporter hESC line, and Marius Wernig for sharing the ASCL1 and DLX2 plasmids. The Broad Center for Regenerative Medicine and Stem Cell Research (BSCRC) at UCLA supports core facilities used in this research and this project specifically. Several authors were supported by training grants (T.A.: BSCRC. S.S.: BSCRC Rose Hills Foundation Training Award. J.L.: Tumor Cell Biology Training Program [US HHS Ruth L. Kirschstein Institutional National Research Service Award #T32 CA009056]. R.A.S.: UCLA/NINDS Translational Neuroscience Training Grant R25NS065723, Research and Training Fellowship from the American Epilepsy Society, Taking Flight Award from CURE Epilepsy, and the BSCRC). This work was also supported by the Paul Allen Family Foundation (W.E.L. and K.P.), and the Steffy Family Foundation (W.E.L. and B.G.N.). W.E.L. is supported by NIH NINDS (NS103788). W.E.L. was also supported by a research award from the BSCRC. K.P. is supported by the David Geffen School of Medicine at UCLA, the UCLA BSCRC, the NIH (P01 GM099134), and a Faculty Scholar grant from the Howard Hughes Medical Institute. R.A.S. is supported by NIH NINDS (K08NS119747) and the Simons Foundation. B.G.N. is supported by the NIH (R01NS089817, R01DA051897 and P50HD103557), California Institute of Regenerative Medicine (DISC1-08819), and the BSCRC and UCLA Jonsson Comprehensive Cancer Center Ablon Scholars Program. I.C. is supported by the NIH (R01AG059848).
Published: September 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.08.006.
Contributor Information
Kathrin Plath, Email: kplath@mednet.ucla.edu.
William E. Lowry, Email: blowry@ucla.edu.
Supplemental information
References
- Ahmed Z., Walker P.S., Fellows R.E. Properties of neurons from dissociated fetal rat brain in serum-free culture. J. Neurosci. 1983;3:2448–2462. doi: 10.1523/JNEUROSCI.03-12-02448.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.A., Marin O., Horn C., Jennings K., Rubenstein J.L. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Blair J.D., Hockemeyer D., Bateup H.S. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat. Med. 2018;24:1568–1578. doi: 10.1038/s41591-018-0139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog E., Bakken T.E., Hodge R.D., Novotny M., Aevermann B.D., Baka J., Borde S., Close J.L., Diez-Fuertes F., Ding S.L. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 2018;21:1185–1195. doi: 10.1038/s41593-018-0205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancho D., Tanaka N., Jaeschke A., Ventura J.J., Kelkar N., Tanaka Y., Kyuuma M., Takeshita T., Flavell R.A., Davis R.J. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 2003;17:1969–1978. doi: 10.1101/gad.1107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt S.J., Stacey J.A., Teramoto Y., Vagnoni C. A role for GABAergic interneuron diversity in circuit development and plasticity of the neonatal cerebral cortex. Curr. Opin. Neurobiol. 2017;43:149–155. doi: 10.1016/j.conb.2017.03.011. [DOI] [PubMed] [Google Scholar]
- Chao H.T., Chen H., Samaco R.C., Xue M., Chahrour M., Yoo J., Neul J.L., Gong S., Lu H.C., Heintz N. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao O.Y., Marron Fernandez de Velasco E., Pathak S.S., Maitra S., Zhang H., Duvick L., Wickman K., Orr H.T., Hirai H., Yang Y.M. Targeting inhibitory cerebellar circuitry to alleviate behavioral deficits in a mouse model for studying idiopathic autism. Neuropsychopharmacology. 2020;45:1159–1170. doi: 10.1038/s41386-020-0656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close J.L., Yao Z., Levi B.P., Miller J.A., Bakken T.E., Menon V., Ting J.T., Wall A., Krostag A.R., Thomsen E.R. Single-cell profiling of an in vitro model of human interneuron development reveals temporal dynamics of cell type production and maturation. Neuron. 2017;96:949. doi: 10.1016/j.neuron.2017.10.024. [DOI] [PubMed] [Google Scholar]
- Cobos I., Calcagnotto M.E., Vilaythong A.J., Thwin M.T., Noebels J.L., Baraban S.C., Rubenstein J.L. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat. Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Conde Guerri B., Sinues Porta E., Arrazola Schlamilch M., Comunas Gonzalez F., Calatayud Maldonado V. Effects of glia-conditioned medium on primary cultures of central neurons. Histol. Histopathol. 1989;4:217–222. [PubMed] [Google Scholar]
- Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U S A. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G. Perspectives on the developmental origins of cortical interneuron diversity. Novartis Found. Symp. 2007;288:21–35. discussion 35-44, 96-28. [PubMed] [Google Scholar]
- Fregnan F., Gnavi S., Macri L., Perroteau I., Gambarotta G. The four isoforms of the tyrosine kinase receptor ErbB4 provide neural progenitor cells with an adhesion preference for the transmembrane type III isoform of the ligand neuregulin 1. Neuroreport. 2014;25:233–241. doi: 10.1097/WNR.0000000000000073. [DOI] [PubMed] [Google Scholar]
- Gelman D., Griveau A., Dehorter N., Teissier A., Varela C., Pla R., Pierani A., Marin O. A wide diversity of cortical GABAergic interneurons derives from the embryonic preoptic area. J. Neurosci. 2011;31:16570–16580. doi: 10.1523/JNEUROSCI.4068-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman D.M., Marin O. Generation of interneuron diversity in the mouse cerebral cortex. Eur. J. Neurosci. 2010;31:2136–2141. doi: 10.1111/j.1460-9568.2010.07267.x. [DOI] [PubMed] [Google Scholar]
- Gelman D.M., Marin O., Rubenstein J.L.R. In: Jasper's Basic Mechanisms of the Epilepsies, Fourth Edition. Noebels J.L., Avoli M., Rogawski M.A., Olsen R.W., Delgado-Escueta A.V., editors. (Oxford University Press); 2012. The generation of cortical interneurons. [Google Scholar]
- Ghanem N., Yu M., Poitras L., Rubenstein J.L., Ekker M. Characterization of a distinct subpopulation of striatal projection neurons expressing the Dlx genes in the basal ganglia through the activity of the I56ii enhancer. Dev. Biol. 2008;322:415–424. doi: 10.1016/j.ydbio.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S.R., Aguet F., Gelfand E., Ardlie K. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Wu Z., Liu Z., Hu G., Yu J., Chang K.H., Kim K.P., Le T., Faull K.F., Rao N. Selective demethylation and altered gene expression are associated with ICF syndrome in human-induced pluripotent stem cells and mesenchymal stem cells. Hum. Mol. Genet. 2014;23:6448–6457. doi: 10.1093/hmg/ddu365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Ishida A., Ure K., Chen H., Swann J.W., Zoghbi H.Y. Loss of MeCP2 in parvalbumin-and somatostatin-expressing neurons in mice leads to distinct Rett syndrome-like phenotypes. Neuron. 2015;88:651–658. doi: 10.1016/j.neuron.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T., Lalande M., Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Lake B.B., Ai R., Kaeser G.E., Salathia N.S., Yung Y.C., Liu R., Wildberg A., Gao D., Fung H.L., Chen S. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L., Mi D., Llorca A., Marin O. Development and functional diversification of cortical interneurons. Neuron. 2018;100:294–313. doi: 10.1016/j.neuron.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A.J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O. Human cortical interneurons take their time. Cell Stem Cell. 2013;12:497–499. doi: 10.1016/j.stem.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Marin O., Anderson S.A., Rubenstein J.L. Origin and molecular specification of striatal interneurons. J. Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof A.M., Keros S., Tyson J.A., Ying S.W., Ganat Y.M., Merkle F.T., Liu B., Goulburn A., Stanley E.G., Elefanty A.G. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Losa M., Tracy T.E., Ma K., Verret L., Clemente-Perez A., Khan A.S., Cobos I., Ho K., Gan L., Mucke L. Nav1.1-Overexpressing interneuron transplants restore brain rhythms and cognition in a mouse model of alzheimer's disease. Neuron. 2018;98:75–89 e75. doi: 10.1016/j.neuron.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T., Sutcliffe J.S., Fang P., Galjaard R.J., Jiang Y.H., Benton C.S., Rommens J.M., Beaudet A.L. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Mayer C., Hafemeister C., Bandler R.C., Machold R., Batista Brito R., Jaglin X., Allaway K., Butler A., Fishell G., Satija R. Developmental diversification of cortical inhibitory interneurons. Nature. 2018;555:457–462. doi: 10.1038/nature25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moarefi A.H., Chedin F. ICF syndrome mutations cause a broad spectrum of biochemical defects in DNMT3B-mediated de novo DNA methylation. J. Mol. Biol. 2011;409:758–772. doi: 10.1016/j.jmb.2011.04.050. [DOI] [PubMed] [Google Scholar]
- Nicholas C.R., Chen J., Tang Y., Southwell D.G., Chalmers N., Vogt D., Arnold C.M., Chen Y.J., Stanley E.G., Elefanty A.G. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M., Korsakova E., Allen D., Lee P., Fu K., Vargas B.S., Cinkornpumin J., Salas C., Park J.C., Germanguz I. Loss of MECP2 leads to activation of P53 and neuronal senescence. Stem Cell Reports. 2018;10:1453–1463. doi: 10.1016/j.stemcr.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra H.K., Turner A.P. The potential legacy of cancer nanotechnology: cellular selection. Trends Biotechnol. 2014;32:21–31. doi: 10.1016/j.tibtech.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Polioudakis D., de la Torre-Ubieta L., Langerman J., Elkins A.G., Shi X., Stein J.L., Vuong C.K., Nichterwitz S., Gevorgian M., Opland C.K. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron. 2019;103:785–801.e8. doi: 10.1016/j.neuron.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic S., Kanatani S., Hunt D., Faux C., Cariboni A., Chiara F., Khan S., Wansbury O., Howard B., Nakajima K. Cdk5 phosphorylation of ErbB4 is required for tangential migration of cortical interneurons. Cereb. Cortex. 2015;25:991–1003. doi: 10.1093/cercor/bht290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagie S., Ellran E., Katzir H., Shaked R., Yehezkel S., Laevsky I., Ghanayim A., Geiger D., Tzukerman M., Selig S. Induced pluripotent stem cells as a model for telomeric abnormalities in ICF type I syndrome. Hum. Mol. Genet. 2014;23:3629–3640. doi: 10.1093/hmg/ddu071. [DOI] [PubMed] [Google Scholar]
- Samarasinghe R.A., Miranda O.A., Buth J.E., Mitchell S., Ferando I., Watanabe M., Allison T.F., Kurdian A., Fotion N.N., Gandal M.J. Identification of neural oscillations and epileptiform changes in human brain organoids. Nat Neurosci. 2021 doi: 10.1038/s41593-021-00906-5. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Macosko E.Z., Wysoker A., Goldman M., Krienen F.M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174:1015–1030.e6. doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman B., Machold R.P., Hashikawa Y., Fuzik J., Fishell G.J., Rudy B. Four unique interneuron populations reside in neocortical layer 1. J. Neurosci. 2019;39:125–139. doi: 10.1523/JNEUROSCI.1613-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama K., Brunori A., Mayer M.L. Glial uptake of excitatory amino acids influences neuronal survival in cultures of mouse hippocampus. Neuroscience. 1989;32:779–791. doi: 10.1016/0306-4522(89)90298-4. [DOI] [PubMed] [Google Scholar]
- Turner C.P., Pulciani D., Rivkees S.A. Reduction in intracellular calcium levels induces injury in developing neurons. Exp. Neurol. 2002;178:21–32. doi: 10.1006/exnr.2002.8027. [DOI] [PubMed] [Google Scholar]
- Turner R.W., Lemon N., Doiron B., Rashid A.J., Morales E., Longtin A., Maler L., Dunn R.J. Oscillatory burst discharge generated through conditional backpropagation of dendritic spikes. J. Physiol. Paris. 2002;96:517–530. doi: 10.1016/S0928-4257(03)00007-X. [DOI] [PubMed] [Google Scholar]
- Ure K., Lu H., Wang W., Ito-Ishida A., Wu Z., He L.J., Sztainberg Y., Chen W., Tang J., Zoghbi H.Y. Restoration of Mecp2 expression in GABAergic neurons is sufficient to rescue multiple disease features in a mouse model of Rett syndrome. eLife. 2016;5:e14198. doi: 10.7554/eLife.14198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Cervino V., Kappeler C., Nobrega-Pereira S., Henkemeyer M., Rago L., Nieto M.A., Marin O. Molecular mechanisms controlling the migration of striatal interneurons. J. Neurosci. 2015;35:8718–8729. doi: 10.1523/JNEUROSCI.4317-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamsley B., Fishell G. Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat. Rev. Neurosci. 2017;18:299–309. doi: 10.1038/nrn.2017.30. [DOI] [PubMed] [Google Scholar]
- Wamsley B., Jaglin X.H., Favuzzi E., Quattrocolo G., Nigro M.J., Yusuf N., Khodadadi-Jamayran A., Rudy B., Fishell G. Rbfox1 mediates cell-type-specific splicing in cortical interneurons. Neuron. 2018;100:846–859.e7. doi: 10.1016/j.neuron.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders C.P., Anderson S.A. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Yang N., Chanda S., Marro S., Ng Y.H., Janas J.A., Haag D., Ang C.E., Tang Y., Flores Q., Mall M. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods. 2017;14:621–628. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Munoz-Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated or used in this study and are deposited in NIH GEO (GSE180132 and GSE181715) and are summarized in Table S1.