Summary
Genomic imprinting is a parent-of-origin dependent monoallelic expression of genes. Previous studies showed that conversion of primed human pluripotent stem cells (hPSCs) into naive pluripotency is accompanied by genome-wide loss of methylation that includes imprinted loci. However, the extent of aberrant biallelic expression of imprinted genes is still unknown. Here, we analyze loss of imprinting (LOI) in a large cohort of both bulk and single-cell RNA sequencing samples of naive and primed hPSCs. We show that naive hPSCs exhibit high levels of non-random LOI, with bias toward paternally methylated imprinting control regions. Importantly, we show that different protocols used for the primed to naive conversion led to different extents of LOI, tightly correlated to FGF signaling. This analysis sheds light on the process of LOI occurring during the conversion to naive pluripotency and highlights the importance of these events when modeling disease and development or when utilizing the cells for therapy.
Keywords: human pluripotent stem cells, genomic imprinting, epigenetics, naive pluripotent stem cells
Highlights
-
•
Large-scale study of genomic imprinting in human naive pluripotent stem cells
-
•
Multiple imprinted genes are biallelically expressed following naive conversion
-
•
Imprinting aberrations occur in a locus-specific manner
-
•
FGF signaling is associated with the extent of loss of imprinting
Cleitman-Blackstien and Benvenisty present a comprehensive analysis of loss of imprinting in naive human pluripotent stem cells. They show that, upon conversion from primed to naive cells, multiple imprinted genes acquire biallelic expression in a locus-specific manner with higher loss of imprinting incidence of genes regulated by paternally methylated regions. Comparison between different protocols links FGF signaling levels with the loss of imprinting process.
Introduction
Genomic imprinting is presented in mammals as parent-of-origin dependent monoallelic expression of a subset of genes and is required for normal growth and development (Reik and Walter, 2001; Tucci et al., 2019). Imprinting is established mostly in the germline of the developing embryo and involves DNA methylation and acquisition of various histone marks along imprinted differentially methylated regions (iDMRs), which are termed imprinting control regions (ICRs), in a sex-specific manner. The imprinting signature is maintained during the massive demethylation process that occurs post fertilization and is only erased in the second wave of demethylation, which takes place in primordial germ cells (Tucci et al., 2019). Various genetic disorders result from imprinting aberrations, and the disrupted expression of imprinted genes has been shown to contribute to the progression of different common diseases, including cancer (Peters, 2014).
Human pluripotent stem cells (hPSCs) have the potential to differentiate into the three embryonic germ layers and are capable of indefinite self-renewal in culture. There are three major types of hPSCs, which are obtained by different methods. The first, termed human embryonic stem cells (hESCs), are obtained from the inner cell mass of preimplantation blastocysts that originate from in vitro fertilization (Thomson, 1998). The other two are obtained through a reprogramming process and include somatic cell nuclear transfer (SCNT)-ESCs, where a somatic cell nucleus is inserted into an enucleated oocyte, and induced PSCs (iPSCs), which are obtained through the expression of key pluripotency factors in a somatic cell (Takahashi and Yamanaka, 2006; Wilmut et al., 1997).
Previous studies examined the imprinting status of hPSCs and reported that hESCs exhibit some extent of imprinting aberrations but are relatively stable, while hSCNT-ESCs and hiPSCs are more prone to imprinting aberrations (Adewumi et al., 2007; Bar et al., 2017; Johannesson et al., 2014; Kim et al., 2007; Ma et al., 2014; Nazor et al., 2012; Pick et al., 2009; Rugg-Gunn et al., 2005, 2007).
hESCs grown in conventional culture conditions resemble in multiple aspects mouse post-implantation epiblast stem cells (mEpiSCs) and are thus referred to as being in a "primed" state. These similarities include the reliance on FGF/Erk and Activin/Nodal signaling for self-renewal, a global hyper-methylation of the genome and a tendency for the inactivation of one of the two X chromosomes in female cells (Dong et al., 2019; Yilmaz and Benvenisty, 2019). In contrast, mouse ESCs (mESCs) can be obtained under conditions that make them more similar to the preimplantation epiblast and are thus referred to as being in a "naive" state. These cells rely on leukemia inhibitor factor (LIF) and inhibition of FGF/Erk signaling, present dome-shaped colonies, exhibit genome-wide demethylation, harbor two active X chromosomes, and efficiently contribute to the formation of chimeric mice when injected into blastocysts (Dong et al., 2019; Yilmaz and Benvenisty, 2019).
Different strategies were developed to convert primed hPSCs into the naive state by using MEK and GSK3 inhibitors (2i), LIF, and either the overexpression of key pluripotent factors or the addition of different chemical inhibitor cocktails (Yilmaz and Benvenisty, 2019). Importantly, while both naive hPSCs and the preimplantation epiblast cells share a genome-wide hypomethylation status, in contrast to the preimplantation epiblast, naive hPSCs were shown to lose methylation at iDMRs and do not regain methylation at these loci upon re-priming (Pastor et al., 2016; Theunissen et al., 2016). However, while global LOI was demonstrated at the methylation level, no global analysis of LOI was performed regarding the biallelic expression of imprinted genes in naive hPSCs.
Here, we present the first large-scale analysis of LOI in naive hPSCs. By analyzing a large cohort of bulk and single-cell RNA sequencing (scRNA-seq) samples obtained from different studies and different conversion protocols, we show that multiple imprinted genes are biallelically expressed following the conversion from primed into naive pluripotency. Remarkably, the LOI events show gene- and parent-specific patterns and the extent of the imprint loss is well correlated with FGF signaling, which varies among different conversion protocols.
Results
Naive cells show higher levels of LOI than primed cells
To study LOI processes in naive hPSCs, we collected a total of 129 bulk RNA-seq samples from 16 different studies, consisting of 103 ESCs and 26 iPSCs. These totaled to 47 and 82 primed and naive samples, respectively (Table S1). Next, we analyzed LOI in these samples by identifying biallelic expression of imprinted genes through the calculation of their allelic ratios and biallelic scores (see experimental procedures, and supplemental experimental procedures and Figure 1A). If an imprinted gene is heterozygous to a specific single-nucleotide variant (SNV) in a certain position, monoallelic expression of that gene would result in reads harboring only one type of nucleotide within that position. However, LOI of that gene may lead to biallelic expression, which would result in reads with two alternate nucleotides in the heterozygote position.
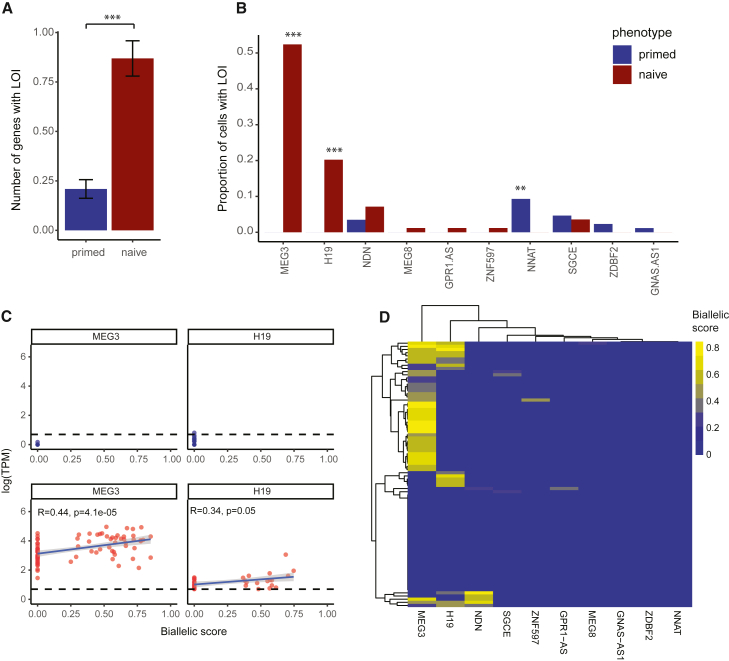
Figure 1.
Naive hPSCs present higher levels of LOI than their primed counterparts
(A) Schematic representation of the variant calling and biallelic score calculation pipeline.
(B and C) Comparison of the average number of genes that lost imprinting in each naive or primed sample per study-per cell line (B), or in all studies (C). ∗∗∗p = 3.0 × 10−5 by paired t test.
(D) Distribution of the number of genes with LOI per sample for primed and naive cells.
RNA-seq samples of hPSCs are often obtained from cells grown on mouse embryonic fibroblasts as feeders. This is especially true for naive cell cultures, which in most protocols rely on feeders (Szczerbinska et al., 2019). The presence of RNA fragments that originate from mouse cells were shown to be a source for the potential detection of false-positive SNVs if they are mistakenly aligned to the human genome (Avior et al., 2021; Stirparo et al., 2021). To remove such false-positive calls, prior to the variant calling step, we used the XenofilteR software, which removes reads that were aligned to both human and mouse genomes (Kluin et al., 2018). Because the XenofilteR might also filter human reads as well, we used this tool for all the samples, including cells which were grown without feeders to prevent any biases between studies. We also focused our analysis on a list of 34 single-isoform imprinted genes, to avoid the biallelic detection of specific isoforms that are not imprinted or are imprinted in a subset of tissues (Bar et al., 2017; Stelzer et al., 2015). Finally, we made sure that the total number of biallelically expressed imprinted genes per sample did not correlate with its retained read coverage after applying the XenofilteR (Figure S1A).
It should be noted that lack of a heterozygous SNV in an imprinted gene in a specific sample of RNA-seq can be attributed to either monoallelic expression of the gene, or simply lack of an SNV along this gene’s exons in this specific sample. Utilizing the dbSNP build 154 database (Sherry, 2001), we show that 32 out of the 34 analyzed imprinted genes contained SNP positions, strengthening the potential for these genes to harbor a heterozygous SNP in a given cell line (Figure S1B). To get a better approximation of the heterozygous SNP availability along imprinted genes in our samples, we searched for their existence in high-coverage whole-genome sequencing (WGS) samples of two hESC lines (WA01 and WA09), which constitute a large fraction of the samples included in this analysis (see Table S1, experimental procedures, and supplemental experimental procedures). Notably, 23 and 18 imprinted genes contained heterozygous SNPs in WA01 and WA09, respectively (Figure S1B), and together cover about 80% of the analyzed imprinted genes. In agreement with the loss of methylation at different imprinted DMRs (Theunissen et al., 2016), naive cells presented a significantly higher number of imprinted genes with biallelic expression compared with their primed counterparts (Figures 1B and 1C). This observation was consistent for almost every study included in the analysis (Figure 1B). A total of 68.1% of the primed samples did not show any genes with LOI, while 27.7% showed one LOI occurrence. The maximum number of genes to lose imprinting in the primed samples was two, harbored by only 4.3% of samples (Figure 1D). In contrast, only 11.0% of naive samples showed no LOI occurrence, while 31.7% presented one to two genes, 48.8% showed three to four genes and 8.5% showed more than four LOI instances (Figure 1D). Indeed, the 11% of the naive samples that showed no LOI were generated under the LIF-3i and 2iLI protocols which were reported to exhibit lower levels of genome-wide demethylation (discussed in greater detail below). Because conversion to naive pluripotency may cause chromosomal aberrations (Di Stefano et al., 2018), which could potentially alter the observed allelic proportions along aberrated loci, we searched for chromosomal aberrations in the primed and naive hPSCs samples by using eSNP karyotyping (Weissbein et al., 2016). This approach enables the identification of per-RNA-seq sample chromosomal aberrations through the detection of chromosomal regions with altered expression of allelic ratios. The analysis showed that most chromosomal regions in most of the samples exhibited an allelic ratio that corresponds to normal diploid karyotypes (Weissbein et al., 2016) (Figure S1C). This was true, except for a few samples in the naive samples from Hu et al. (2020), which showed noisy allelic ratios. Indeed, naive hPSCs exhibited higher levels of chromosomal aberrations than primed hPSCs. These aberrations included duplications in chromosomes 1 and 12, which are commonly detected in hPSCs (Halliwell et al., 2020), together with other abnormalities (Figure S1C). Nevertheless, these abnormalities could not explain the increased LOI observed in naive hPSCs. Specifically, regions that showed recurrent LOI exhibited relatively stable karyotypes (Figures S1C and S2C). Together, these results suggest that conversion of primed hPSCs into naive pluripotency is accompanied both by loss of methylation at imprinted loci and aberrant biallelic expression of imprinted genes.
It was previously shown that iPSCs acquire higher levels of imprinting aberrations than ESCs (Johannesson et al., 2014). Therefore, we set out to determine whether these differences will also be reflected in the sensitivity to LOI during the resetting to naive pluripotency. Interestingly, iPSCs did not show higher levels of LOI during the resetting (Figure S1D), suggesting that the epigenetic changes that occur during the reprogramming process do not confer extra epigenetic instability for the conversion into naive pluripotency, at least in the context of LOI. Finally, previous reports from mESCs suggested that the presence of two active X chromosomes in female cells can lead to increased erasure of methylation at the ICR (Yagi et al., 2017; Zvetkova et al., 2005). Nonetheless, we could not detect significant differences in the extent of LOI that occurs during the conversion to naive pluripotency between male and female hESCs, suggesting there is no sex-specific bias in LOI in naive hESCs (Figure S1E).
Naive cells show gene-specific biases in LOI patterns
An important aspect of LOI during the naive resetting process relates not only to the number of genes that are biallelically expressed, but to whether these LOI events are random, or show biases toward specific genes. Interestingly, out of the 14 genes which lost imprinting in at least 1 sample, 12 had a higher proportion of LOI in naive samples compared with their primed counterparts. Out of these 12 genes, 6 genes (MEG3, H19, ZDBF2, NDN, RTL1, and GPR1-AS) showed significantly higher LOI occurrences in the naive cells (Figure 2A). Notably, MEG3 biallelic expression in naive cells was already detected in two of the studies included in this analysis (Giulitti et al., 2019; Theunissen et al., 2016). Interestingly, our analysis shows that MEG3 transitioned from not being expressed in most primed samples to being expressed in most naive samples (Figure S2A). Moreover, RTL1 also transitioned from being mostly turned off in the primed cells to being partly turned on in the naive cells, while GPR1-AS went from being completely silenced to being mostly turned on. H19 showed a heterogeneous expression pattern, being expressed by some primed cells and not by others, while being highly expressed by most naive samples. In contrast, NDN and ZDBF2 were expressed both in primed and naive cells (Figure S2A). These findings show that some genes that lose imprinting through the resetting process to naive state, transition from being completely or partly silenced in the primed state, to being both activated and biallelically expressed in the naive state. To find out whether the activation of some of the imprinted genes is an aberrant property of naive hPSCs or a property that reflects their similarity with the preimplantation epiblast, we analyzed the gene expression levels in published scRNA-seq data from preimplantation embryos (Petropoulos et al., 2016). This analysis showed that all the genes that were only activated in the naive state, except for RTL1, were expressed in most of the cells in the human preimplantation epiblast (Figure S2B). A subset of cells did show RTL1 expression but, similarly to naive hPSCs, it was not expressed in most cells. These results suggest that the biallelic expression, which likely accompanies the ICR methylation erasure, but not the activation per se of these imprinted genes, is the aberrant property of naive hPSCs.
Figure 2.
Naive hPSCs Show LOI patterns that differ from primed hPSCs
(A) Proportion of naive samples and their primed counterparts that presented LOI in each imprinted gene. Bars represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by two-sided paired t test.
(B) Venn diagram of genes that are highly prone to LOI only in primed cells, naive cells, or both.
(C) Proportion of naive samples and their primed counterparts that presented LOI in each imprinted gene ordered by locus.
(D) Mean delta naive-primed LOI per paternal/maternal methylated genes. Bars represent mean ± SEM. ∗∗∗p = 1.0 × 10−5 by two-sided unpaired t test.
(E) Spearman correlation between methylation of DMRs and the biallelic expression of their associated genes.
(F) Heatmap of each gene’s biallelic score in different time points during the naive conversion. Samples were analyzed from Collier et al. (2017).
Because LOI in primed samples was relatively scarce, searching for differences in LOI patterns between naive and primed cells would require a larger sample size of primed cells that would reflect their LOI signature more precisely. To this end, we pooled the primed samples that had naive counterparts along with 156 additional primed samples for a final pool of 203 samples originating from 52 different studies (Table S1; Figure S2C). Because of the LOI scale differences between primed and naive cells, we compared genes that showed biallelic expression in more than 5% of primed samples with genes that lost imprinting in at least 10% of naive samples. Indeed, the four genes (RTL1, GPR1-AS, H19, and MEG3) that are more active in naive cells (Figure S2A) show none-to-low levels of biallelic expression in primed cells (Figure S2C). One gene that is expressed both in primed and naive cells specifically lost imprinting during the conversion from primed to naive (NDN), three genes are prone to LOI in both primed and naive cells (ZDBF2, IGF2, and DLK1), and one gene (SGCE) already has very significant LOI in primed cells (Figures 2A, 2B, and S2C). When examining the regional distribution of genes prone to LOI in the two cell types, both genes that were prone to LOI in primed and naive and genes more unique to naive cells, were positioned on chr 2q33.3, chr 11p15.5, and chr 14q32.2, apart from NDN, which belongs to chr 15q11.2 (Figure 2C). Interestingly, SGCE, which was more unique to primed cells resides on the chr 7q21.3 imprinted region. These findings suggest that the imprinted regions on chr 2q33.3, chr 11p15.5, and chr 14q32.2 exhibit general sensitivity to LOI, while some genes in these regions are mostly silenced in primed cells and are turned on and lose imprinting during the naive conversion, which is accompanied by widespread demethylation. This is in parallel to regions that are more sensitive in primed cells (chr 7q21.3) and regions with stronger sensitivity only in naive cells (chr 15q11.2), which implies that the LOI process is not random and relates to the biological differences between the primed and naive state.
Another aspect of the sensitivity to LOI relates to the parent-specific regulation. A previous analysis of LOI in primed hPSCs showed that genes under the control of paternal germ line DMRs (gDMRs) are more prone to LOI than genes under the control of maternal gDMRs (Bar et al., 2017). Thus, we set out to examine whether any such bias occurs during the conversion to naive pluripotency. For each naive sample we calculated the mean LOI difference from the primed counterpart in genes under the control of paternal or maternal gDMRs. In accordance with the observation for primed hPSCs, during the conversion to naive pluripotency there was an increased sensitivity toward LOI for genes under the control of paternal gDMRs (Figures 2D and S2D). This result is strengthened considering that there are more genes under the control of maternal gDMRs, a trend that continues for the number of genes with SNPs in the dbSNP database and the number of genes with heterozygous SNPs in WA01/WA0 WGS samples that we analyzed (Figure S2E). This suggests that, despite having different patterns of LOI between primed and naive cells, genes under the control of paternal gDMRs are overall more prone to LOI than genes under the control of maternal gDMRs.
Next, as previous reports concerning imprinting aberrations focused mainly on the methylation of imprinted DMRs, we evaluated the connection between methylation and expression during primed to naive conversion. For this purpose, we analyzed whole-genome bisulfite sequencing samples from two studies (Guo et al., 2017; Takashima et al., 2014), reduced representation bisulfite sequencing samples from three studies (Di Stefano et al., 2018; Giulitti et al., 2019; Sahakyan et al., 2017), and MethylC-seq samples from one study (Theunissen et al., 2016), included in the original analyses. We focused on H19 gDMR, MEG3 somatic DMR (sDMR), and NDN sDMR as representatives of imprinted regions that showed sensitivity for LOI in naive cells. We divided the methylation levels at each DMR to either around 50% methylation level (30%–70%), which is expected in imprinted DMRs (which contain both hypermethylated and hypomethylated alleles), hypermethylated pattern (>70%), or hypomethylated pattern (<30%). As expected, naive cells mostly exhibited a hypomethylated state across the three DMRs (Figure 2E). All the DMRs showed a negative correlation between the methylation levels and the biallelic score of genes they regulate.
Conversion of primed hPSCs to naive hPSCs occurs in multiple days. We thus set out to investigate the occurrence of LOI throughout the conversion in a time-dependent manner, by analyzing a subset of naive samples that were included in the original analysis and were taken from different time points along the conversion process (primed, naive day 10, and passages 5 and 10) (Collier et al., 2017). Specifically, the samples from day 10 of the conversion were sorted for naive markers and were shown to be early-stage naive cells, while samples from passage 5 onward were shown to be established mature naive cells (Collier et al., 2017). In line with other studies included in our analysis, primed samples did not harbor any LOI event. In contrast, day 10 samples showed LOI in H19, while passage 5 samples already showed LOI in H19, MEG3, NDN, and partially in ZNF597. Passage 10 samples showed a similar pattern of LOI with passage 5 samples, albeit with no apparent LOI in ZNF597 (Figure 2F). Notably, the same study from which these samples were obtained from, identified MEG3 as being one of the markers for the transition between early-to-late naive stages. Interestingly, the sharp increase of LOI events between the early and late naive cells coincides with re-activation of the inactive X chromosome and the activation of multiple naive-specific transposable elements (Collier et al., 2017), suggesting that, during the time period of the transition between early and late naive cells, the cells go through major epigenetic changes that might play a role in the LOI process. In this context, differentially expressed genes between early-to-late naive states are significantly enriched for regulation of transcription processes, with more than half of the genes in this category belonging to zinc-finger proteins, which are known to be involved in the regulation of imprinting (Collier et al., 2017; Langouët et al., 2018; Monteagudo-Sánchez et al., 2020; Takahashi et al., 2019).
It was previously shown that re-priming of naive cells results in the re-methylation of the genome, excluding imprinted DMRs, which maintain low methylation levels (Theunissen et al., 2016). Because the expression of imprinted genes might be influenced by other factors, such as secondary DMRs, we examined whether the biallelic expression of imprinted genes upon naive conversion are stable or can be reversible upon re-priming or differentiation. For this, we analyzed RNA-seq samples of naive re-primed cells and naive cells that were differentiated into either primitive endoderm or vascular progenitors from three of the studies included in the main analysis (Table S1). Indeed, re-primed cells exhibited an LOI pattern that matched many of the naive samples and even biallelically expressed another gene, suggesting the LOI was not reversed. Upon differentiation of naive cells into either primitive endoderm or vascular progenitors, most genes that lost imprinting in the naive cells maintained their biallelic expression, with the exception of a few cases (e.g., DLK1 during differentiation to primitive endoderm). However, a larger cohort of samples will be needed to determine whether these few cases truly reflect LOI reversal or simply expansion of specific clones. Notably, some differentiated cells showed LOI in new genes as well, possibly due to the extended culture periods (Figure S3). Taken together, these results suggest that LOI is generally not reversible upon re-priming or differentiation of naive cells, although a more comprehensive analysis of differentiated naive cells is needed to see if any genes might behave differently.
LOI analysis in scRNA-seq shows clone specificity of LOI
One of the noticeable disadvantages in performing the LOI analysis on bulk RNA-seq samples is the inability to evaluate whether LOI events in different genes are uniform throughout the culture or do different cells within a given sample harbor distinct LOI patterns. To overcome this limitation, we analyzed a cohort of 86 primed and 84 naive scRNA-seq samples (Messmer et al., 2019), using the same pipeline without the XenofilteR tool, because cells that passed the quality check are assumed to be human cells. Consistent with the elevated levels of LOI in the naive bulk RNA-seq, naive single cells showed higher levels of LOI compared with primed single cells (Figure 3A). In total, ten imprinted genes showed LOI in at least one cell. Out of those, five genes lost imprinting only in naive cells (MEG3, H19, MEG8, GPR1-AS, and ZNF597) (Figure 3B). In agreement with the bulk analysis, MEG3 and H19 exhibited a highly significant proportion of naive cells with LOI (52% and 20%, respectively) compared with primed cells (Figure 3B). Importantly, the heterogeneous pattern of LOI where only a subset of cells lose imprinting in each gene, suggests that LOI is clone specific and is not uniform throughout the culture. In contrast to the bulk analysis, which showed a heterogeneous expression pattern with some primed samples expressing MEG3/H19 and some not, the scRNA-seq data shows that the primed cells did not express MEG3/H19 at all (or at least not in high enough levels to be detected) while naive cells showed high expression of both genes (Figure 3C). This suggests that, as was shown for other genes (RTL1 and GPR1-AS) in the bulk analysis, conversion to naive pluripotency can lead to both activation of previously silenced imprinted genes and to their biallelic expression. Notably, both H19 and MEG3 showed a strong positive correlation between biallelic score and expression levels in naive cells (Figure 3C), suggesting that activation of two copies for these genes results in their upregulation. We also examined whether a cell loses imprinting in the same sequential pattern. Under this assumption, because the most common gene to lose imprinting in the naive population is MEG3 followed by H19, most of the cells with H19 LOI should exhibit LOI in MEG3. However, naive cells with H19 LOI did not have a significantly higher chance of also presenting MEG3 LOI (p = 0.19 by permutation test; 104 permutations; Figure 3D). Taken together, these results suggest that LOI is clone specific and that the imprint loss does not necessarily occur in the same sequential order.
Figure 3.
Single-cell RNA-seq analysis of naive hPSCs reveals culture heterogeneity in LOI patterns
(A) Comparison of the number of genes that lost imprinting in every naive or primed cell. Bars represent mean ± SEM. ∗∗∗p = 1.4 × 10−9 by unpaired two-sided t test.
(B) Proportion of naive and primed cells that presented LOI in each imprinted gene. ∗∗∗p < 0.001, ∗∗p = 6.6 × 10−3 by Fischer’s exact test.
(C) Spearman correlation test between MEG3 and H19 biallelic score and their log(TPM) expression values. Upper panel (blue) refers to primed cells and the lower panel (red) to naive cells. Only naive cells with TPM > 1 are shown.
(D) Heatmap grouped by hierarchical clustering representing the biallelic score of each cell in each imprinted gene.
Culture-dependent LOI in naive cells
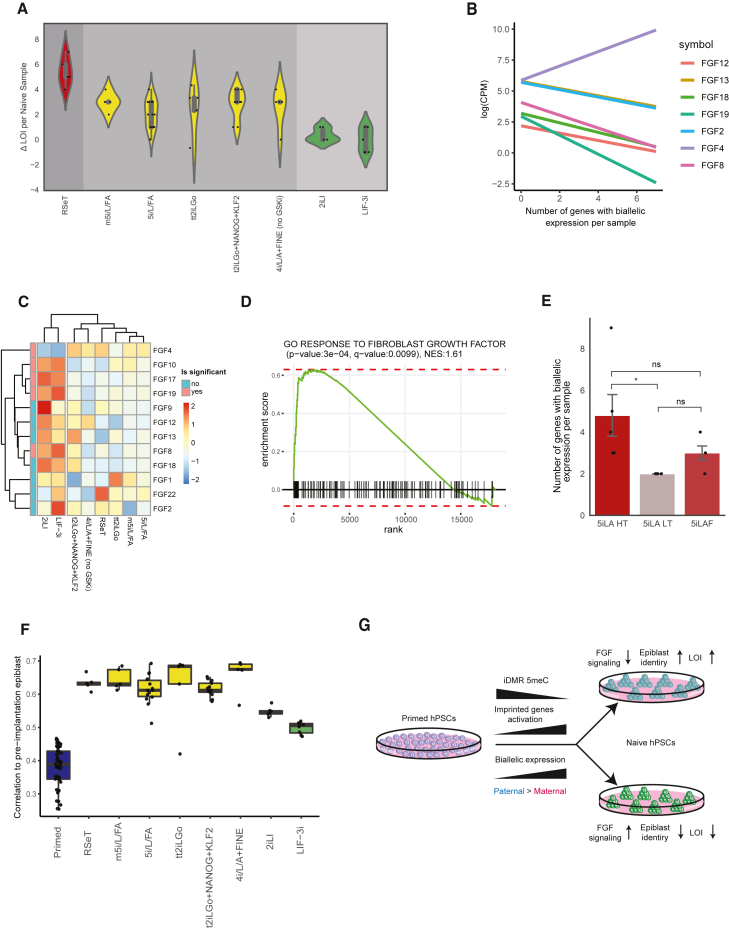
Because conversion into naive pluripotency can be obtained by various culture conditions, we set out to determine the influence of culture on the extent of LOI that occurs during the resetting process. To this end, we compared conversion protocols which had at least five naive samples as representatives. We also combined samples cultured in two different medium conditions (4i/L/A + FINE; Szczerbinska et al., 2019; Theunissen et al., 2016) that do not contain GSK3i, which was shown to reduce methylation in imprinted loci in mice (Popkie et al., 2010). This comparison showed a significant association between naive conversion protocol and LOI extents and divided the protocols into three distinct groups, which we denote as having high, medium, and low LOI levels (Figure 4A). Notably, the two culture conditions that showed low levels of LOI (2iLI and LIF-3i) were already shown to possess lower levels of imprinted DMR methylation erasure (Hu et al., 2020; Zimmerlin et al., 2016), highlighting the tight connection between DMR methylation and the biallelic expression of imprinted genes. Because we observed that the WA01 cell line had a larger number of genes with heterozygous SNPs in the genome compared with WA09 (Figure S1B; 23 as supposed to 18), we made sure that there was no significant difference in LOI extents between these two cell lines, which might imply that there is a technical heterogeneity of LOI extents between other cell lines in this analysis. However, no significant differences in LOI levels were seen between WA01 and WA09 cell lines, making the cell line-dependent LOI heterogeneity less likely (Figure S4A). Together, these results suggest that the extent of the imprint loss is correlated with the conversion medium, with different types of media showing different levels of LOI.
Figure 4.
Culture-dependent variability in LOI of naive hPSCs
(A) Comparison of the LOI extents between different conversion protocols. Each dot represents the delta number of genes with LOI of each naive sample compared with its primed counterpart. p = 1.0 × 10−5 by Kruskal Wallis test. Groups are separated by unique colors and were determined by post hoc Conover's test FDR (false discovery rate)-corrected p values.
(B) Linear regression lines of FGF members with significant correlation between expression levels and LOI extents (FDR-corrected p < 0.05 by Pearson test).
(C) Hierarchical clustering of the scaled mean (log(CPM)) values for different FGF members in different naive conversion protocols. FDR-corrected p < 0.05 for genes annotated as significant.
(D) GSEA results for "response to fibroblast growth factor" GO term in the comparison between low and medium/high LOI groups.
(E) Comparison of the LOI extents between the different naive samples from An et al. Bars represent mean ± SEM. ∗FDR-corrected p = 1.1 × 10−2 by Mann-Whitney U test.
(F) Per sample correlation to the preimplantation epiblast in different conversion protocols.
(G) Schematic model of the LOI process which occurs during the primed to naive conversion.
In mice, it was shown that inhibition of Mek1/2 downstream of FGF signaling in the 2i state results in a genome-wide demethylation, including imprinted ICRs (Choi et al., 2017; Ficz et al., 2013). Moreover, a recent study suggested that complete blockage of autocrine FGF signaling enables the generation and maintenance of a homogeneous population of human naive cells, which resemble the preimplantation epiblast (An et al., 2020). Because these cells also exhibited genome-wide hypomethylation compared with cells that still had active FGF signaling (specifically FGF2), we tested whether LOI levels in all the naive samples included in our analysis correlate with the expression of different FGF family members. Indeed, multiple FGF members, including FGF2, showed a significant negative correlation with LOI occurrences, apart from FGF4, which presented a highly positive correlation (Figure 4B; Table S2). The FGF-LOI correlation was consistent when removing samples belonging to low LOI protocols (Figure S4B; Table S2), suggesting that it can also explain the LOI variation of medium/high LOI protocols together with protocols that were not represented enough to enter the comparison of protocols in Figure 4A. Differential expression analysis between samples from protocols with low versus medium/high LOI levels showed the same pattern, with upregulation of multiple FGF members and a downregulation of FGF4 in the low LOI group (Figures 4C; Table S3). Accordingly, genes upregulated in the low LOI group were highly enriched in "response to fibroblast growth factor" GO term (Figure 4D; Table S4). Importantly, the upregulation of FGF signaling factors in the low LOI group included FGFR1 and the ERK response genes, which were shown to be upregulated when autocrine FGF2 is not fully blocked (Figures S4C and S4D) (An et al., 2020), implicating the FGF pathway ligand, receptor, and target upregulation in the low LOI group. To further establish the connection between FGF signaling and LOI, we obtained 5iLA naive RNA-seq samples from An et al. that were either (1) labeled as HT (highly expressing TFCP2L1 and tdTomato), which were shown to express higher levels of naive markers and a downregulation of autocrine FGF2 signaling, (2) LT (lowly expressing TFCP2L1 and tdTomato), which expressed lower levels of naive markers and higher autocrine FGF2 signaling, and (3) naive samples obtained with the 5iLA protocol supplemented with FGF2 (5iLAF) (Table S1). Interestingly, HT cells presented very high levels of LOI, while LT cells exhibited low levels of LOI, consistent with the FGF-LOI correlation observed in the other studies (Figure 4E). Notably, obtaining the naive cells under the same conditions with the addition of FGF2 resulted in intermediate levels of imprint loss, further supporting the connection between FGF signaling and LOI (Figure 4E). Together, these results imply that the LOI process is associated with FGF signaling, with cells having more active FGF signaling presenting low LOI levels and vice versa.
The need to block FGF signaling to obtain the bona fide naive state both in humans and mice, together with the report showing that full blockage of autocrine FGF signaling enables the induction of a homogeneous population of naive hPSCs (An et al., 2020), implies that the connection between LOI and FGF signaling might coincide with distinct levels of "naivety." This possibility is also supported by the upregulation of the preimplantation epiblast marker FGF4 in the medium/high LOI groups (Figures 4B and 4C). We thus examined the similarity between naive hPSCs belonging to different LOI groups, and a pseudo-bulk RNA-seq sample obtained from scRNA-seq samples of preimplantation human epiblasts (Petropoulos et al., 2016). To this end, we calculated the correlation of each naive sample with the pseudo-bulk epiblast sample based on the expression of an unbiased list of 47 preimplantation epiblast markers, obtained from an integrated analysis of human preimplantation scRNA-seq datasets (Stirparo et al., 2018). As expected, all naive samples showed stronger correlation to the preimplantation epiblast compared with primed samples (Figure 4F). Nonetheless, samples belonging to the low LOI group showed consistent weaker correlation to the epiblast compared with samples belonging to the medium/high LOI groups. In summary, naive samples belonging to low LOI groups exhibit both elevated FGF signaling and a lower transcriptional similarity to the preimplantation epiblast based on the expression of epiblast markers.
Discussion
Naive hPSCs present a valuable resource for both the study of human preimplantation development and the potential use in regenerative medicine. However, reports regarding the erasure of methylation from imprinted DMRs observed in these cells (Pastor et al., 2016; Theunissen et al., 2016) called for a better understanding of the mechanisms and the outcomes of this methylation erasure, especially considering the tight involvement of imprinting aberrations in different pathologies (Peters, 2014). This study presents the first comprehensive analysis of LOI in naive hPSCs, focusing on the biallelic expression of different imprinted genes by integrating a large cohort of both bulk and scRNA-seq samples, which were obtained from different studies, conversion protocols, and cell lines.
We show that naive cells exhibit a much higher number of biallelically expressed imprinted genes compared with their primed counterparts. Our analysis suggests that the LOI process is not random, but rather that some imprinted loci show increased sensitivity compared with other loci. Specifically, genes under the control of paternally methylated iDMR seem to bear higher sensitivity to imprint loss compared with genes under the control of maternal iDMRs. This could be attributed to the underlying differences between paternal iDMRs, which usually map to intergenic regions and maternally methylated iDMRs, which usually map to promoters and intragenic regions (Edwards and Ferguson-Smith, 2007). Bar et al. (2017) suggested that the differences between paternal and maternal DMRs may in part be related to specific factors that are required for the methylation acquisition specifically in oocytes and the differences of the demethylation processes that occur in the paternal versus maternal pronuclei after fertilization. Another factor that should be taken into consideration is that the methylation-acquisition process in the oocyte is mainly transcriptionally coupled, both generally and specifically in imprinted loci, which, as mentioned above, are mainly positioned in promoters and intragenic regions (Chotalia et al., 2009; Smallwood et al., 2011). This attribute may give an easier access to repressive machinery that will maintain imprinting during the methylation erasure that occurs in the primed to naive conversion. However, this hypothesis still needs to be tested.
We also show that the changes in gene expression of naive cells include the activation of previously silenced or partly silenced imprinted genes (RTL1, GPR1-AS, H19, and MEG3), which, upon activation, show biallelic expression in many samples. Importantly, the activation of these genes by itself does not seem aberrant but rather a manifestation of preimplantation epiblast identity in the naive cells, suggesting that only the biallelic expression of these genes is aberrant. For genes that were generally expressed both in naive and primed samples we observed that NDN seemed to lose imprinting solely in naive cells. This could be a direct result of the methylation erasure seen in the NDN DMR (Figure 2E). However, the presence of samples, which on one hand showed low methylation levels of NDN DMR and on the other presented monoallelic expression of NDN suggests that the demethylation by itself might not suffice for the activation of the imprinted copy, and that other interventions, such as the replacement of repressive histone marks with active ones, are necessary. Notably, while SGCE showed very high sensitivity to imprint loss in the primed cells (Figure S2C), its imprinting was mostly stable during the conversion to naive pluripotency. Because of the massive demethylation that occurs globally in the naive cells, it seems more likely that, in the case of SGCE, other molecular processes are involved during the LOI, which are methylation independent and do not specifically occur during the conversion. The final group of genes that lost imprinting both in primed and, to a greater extent in naive cells (DLK1, IGF2, and ZDBF2), likely reside in regions that are generally sensitive to LOI, particularly as they are controlled by paternally methylated DMRs, which are more sensitive to LOI. We were also able to follow the kinetics of the LOI events and to position them along the conversion trajectory. This demonstrated that the major events of LOI occur during the transition from early-to-late naive cells, similarly to other major epigenetic processes, such as X chromosome re-activation and the activation of multiple naive-specific transposable elements (Collier et al., 2017).
Through analyzing LOI in scRNA-seq of naive cells we show that LOI is heterogeneous around the culture with different cells harboring from none to varying occurrences of LOI. This finding comes in agreement with the heterogeneity of the naive cells, which was attributed to the incomplete blockage of autocrine FGF2 signaling (An et al., 2020).
Finally, we were able to discriminate between conversion protocols that were associated with low versus higher levels of LOI. We show that LOI magnitudes in the naive cells correlate with the expression of multiple FGF members and overall weaker FGF signaling in cells with higher LOI levels. In this context, we show that, while HT cells present extremely high levels of LOI and LT present very low levels, the addition of FGF2 to the 5iLA medium lowers the LOI levels toward the numbers seen in the LT cells, which adds a causative factor to the observed FGF correlations. Importantly, naive samples from all the conversion protocols showed a stronger correlation to the human preimplantation epiblast based on the expression of preimplantation epiblast markers. However, this correlation was consistently weaker for samples taken from the low LOI protocols compared with the medium/high LOI protocols. These results are logical considering the need to block FGF signaling to maintain naivety. Indeed, other pathways besides "response to fibroblast growth factor" were enriched in the differential expression analysis between low and medium/high LOI groups (Table S4), suggesting other differences between these cells. Nevertheless, due to the importance of FGF blockage for naivety maintenance, the strong FGF signaling in the low LOI protocols (Figures 4C and 4D) and lower LOI occurrences when naive cells are cultured with FGF2 (Figure 4E), we believe that FGF signaling might play a central role in the extent of imprint erasure during conversion to naive pluripotency. Thus, we propose a model that includes the LOI patterns, epiblast identity, and the connection to FGF signaling (Figure 4G). Because the two protocols that showed lower LOI levels use the same concentration of PD03 as in the higher LOI groups (aside from the m5i/L/FA protocol), it is more plausible that the other inhibitors used in the higher LOI groups are the ones that push the naive cells toward a more aggressive imprint erasure. This makes sense as the kinases that are blocked aside from GSK3 and MEK/ERK in the higher LOI protocols also affect the FGF signaling pathway (Taei et al., 2020). This is also supported by the samples from the m5i/L/FA protocol, which includes SRCi + RAFi that also affect FGF signaling (Taei et al., 2020). These samples showed high LOI levels despite having lower PD03 concentrations (0.5 μM), suggesting that the additive inhibition of different protein kinases might lead to reduced FGF signaling and end in a more aggressive imprinting erasure. The connection between preimplantation epiblast identity and LOI implies that a trade-off might exist between obtaining what was previously termed as "bona fide naive pluripotent cells" and the retention of imprinting. We suggest that this trade-off should be taken into consideration when deciding which protocols to use.
Experimental procedures
Data collection
Most of the RNA-seq samples were downloaded from the SRA database (http://www.ncbi.nlm.nih.gov/sra) (Wheeler et al., 2007). Samples from An et al. (2020) were downloaded from the GSA database (http://bigd.big.ac.cn/gsa) (Wang et al., 2017) (Table S1). WGS samples were obtained from the SRA database (http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR6377128 and SRR2070629. Fastq files were generated from the sra files using the sra-toolkit (http://ncbi.github.io/sra-tools/). Analysis of the samples for variant calling is described in supplemental experimental procedures.
eSNP-karyotyping analysis
eSNP-karyotyping was performed basically as described by Weissbein et al. (2016) (see supplemental experimental procedures).
Statistical analysis
Hierarchical clustering was performed using the pheatmap (Kolde and Vilo, 2015). pheatmap: Pretty Heatmaps. R Package version 1.0.8. https://CRAN.R-project.org/package=pheatmap). Scatterplots, boxplots, violin plots, and barplots were generated either in Python using the Seaborn library or in R using the ggplot2 package (Wickham, 2016).
Author contributions
G.K. and N.B. designed the research. G.K. established the bioinformatical pipeline and performed the analysis. G.K. and N.B. interpreted the results and prepared the manuscript. N.B. supervised the study and secured funding.
Conflict of interests
N.B. is CSO of NewStem Ltd.
Acknowledgments
We thank Dr. Ido Sagi and all members of The Azrieli Center for Stem Cells and Genetic Research for their input and critical reading of the manuscript. This work was partially supported by the Israel Science Foundation (494/17), the Rosetrees Trust, and by Azrieli Foundation. N.B. is the Herbert Cohn Chair in Cancer Research.
Published: September 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.09.002.
Supplemental information
References
- Adewumi O., Aflatoonian B., Ahrlund-Richter L., Amit M., Andrews P.W., Beighton G., Bello P.A., Benvenisty N., Berry L.S., Bevan S. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- An C., Feng G., Zhang J., Cao S., Wang Y., Wang N., Lu F., Zhou Q., Wang H. Overcoming autocrine FGF signaling-induced heterogeneity in naive human ESCs enables modeling of random X chromosome inactivation. Cell Stem Cell. 2020;27:482–497. doi: 10.1016/j.stem.2020.06.002. [DOI] [PubMed] [Google Scholar]
- Avior Y., Lezmi E., Eggan K., Benvenisty N. Cancer-related mutations identified in primed human pluripotent stem cells. Cell Stem Cell. 2021;28:10–11. doi: 10.1016/j.stem.2020.11.013. [DOI] [PubMed] [Google Scholar]
- Bar S., Schachter M., Eldar-Geva T., Benvenisty N. Large-scale analysis of loss of imprinting in human pluripotent stem cells. Cell Rep. 2017;19:957–968. doi: 10.1016/j.celrep.2017.04.020. [DOI] [PubMed] [Google Scholar]
- Choi J., Huebner A.J., Clement K., Walsh R.M., Savol A., Lin K., Gu H., Di Stefano B., Brumbaugh J., Kim S.-Y. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotalia M., Smallwood S.A., Ruf N., Dawson C., Lucifero D., Frontera M., James K., Dean W., Kelsey G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier A.J., Panula S.P., Schell J.P., Chovanec P., Plaza Reyes A., Petropoulos S., Corcoran A.E., Walker R., Douagi I., Lanner F. Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell. 2017;20:874–890. doi: 10.1016/j.stem.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Fischer L.A., Theunissen T.W. Recent insights into the naïve state of human pluripotency and its applications. Exp. Cell Res. 2019;385:111645. doi: 10.1016/j.yexcr.2019.111645. [DOI] [PubMed] [Google Scholar]
- Edwards C.A., Ferguson-Smith A.C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.-L., Walter J. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulitti S., Pellegrini M., Zorzan I., Martini P., Gagliano O., Mutarelli M., Ziller M.J., Cacchiarelli D., Romualdi C., Elvassore N. Direct generation of human naive induced pluripotent stem cells from somatic cells in microfluidics. Nat. Cell Biol. 2019;21:275–286. doi: 10.1038/s41556-018-0254-5. [DOI] [PubMed] [Google Scholar]
- Guo G., von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W. Epigenetic resetting of human pluripotency. Development. 2017;144:2748–2763. doi: 10.1242/dev.146811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell J., Barbaric I., Andrews P.W. Acquired genetic changes in human pluripotent stem cells: origins and consequences. Nat. Rev. Mol. Cell Biol. 2020;21:715–728. doi: 10.1038/s41580-020-00292-z. [DOI] [PubMed] [Google Scholar]
- Hu Z., Li H., Jiang H., Ren Y., Yu X., Qiu J., Stablewski A.B., Zhang B., Buck M.J., Feng J. Transient inhibition of mTOR in human pluripotent stem cells enables robust formation of mouse-human chimeric embryos. Sci. Adv. 2020;6:eaaz0298. doi: 10.1126/sciadv.aaz0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson B., Sagi I., Gore A., Paull D., Yamada M., Golan-Lev T., Li Z., LeDuc C., Shen Y., Stern S. Comparable frequencies of coding mutations and loss of imprinting in human pluripotent cells derived by nuclear transfer and defined factors. Cell Stem Cell. 2014;15:634–642. doi: 10.1016/j.stem.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Kim K.-P., Thurston A., Mummery C., Ward-van Oostwaard D., Priddle H., Allegrucci C., Denning C., Young L. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007;17:1731–1742. doi: 10.1101/gr.6609207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin R.J.C., Kemper K., Kuilman T., de Ruiter J.R., Iyer V., Forment J.V., Cornelissen-Steijger P., de Rink I., ter Brugge P., Song J.-Y. XenofilteR: computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinform. 2018;19:366. doi: 10.1186/s12859-018-2353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R., Vilo J. GOsummaries: an R Package for visual functional annotation of experimental data. F1000Res. 2015;4:574. doi: 10.12688/f1000research.6925.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langouët M., Glatt-Deeley H.R., Chung M.S., Dupont-Thibert C.M., Mathieux E., Banda E.C., Stoddard C.E., Crandall L., Lalande M. Zinc finger protein 274 regulates imprinted expression of transcripts in Prader-Willi syndrome neurons. Hum. Mol. Genet. 2018;27:505–515. doi: 10.1093/hmg/ddx420. [DOI] [PubMed] [Google Scholar]
- Ma H., Morey R., O’Neil R.C., He Y., Daughtry B., Schultz M.D., Hariharan M., Nery J.R., Castanon R., Sabatini K. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer T., von Meyenn F., Savino A., Santos F., Mohammed H., Lun A.T.L., Marioni J.C., Reik W. Transcriptional heterogeneity in naive and primed human pluripotent stem cells at single-cell resolution. Cell Rep. 2019;26:815–824. doi: 10.1016/j.celrep.2018.12.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteagudo-Sánchez A., Hernandez Mora J.R., Simon C., Burton A., Tenorio J., Lapunzina P., Clark S., Esteller M., Kelsey G., López-Siguero J.P. The role of ZFP57 and additional KRAB-zinc finger proteins in the maintenance of human imprinted methylation and multi-locus imprinting disturbances. Nucleic Acids Res. 2020;48:11394–11407. doi: 10.1093/nar/gkaa837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazor K.L., Altun G., Lynch C., Tran H., Harness J.V., Slavin I., Garitaonandia I., Müller F.-J., Wang Y.-C., Boscolo F.S. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W.A., Chen D., Liu W., Kim R., Sahakyan A., Lukianchikov A., Plath K., Jacobsen S.E., Clark A.T. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell. 2016;18:323–329. doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 2014;15:517–530. doi: 10.1038/nrg3766. [DOI] [PubMed] [Google Scholar]
- Petropoulos S., Edsgärd D., Reinius B., Deng Q., Panula S.P., Codeluppi S., Plaza Reyes A., Linnarsson S., Sandberg R., Lanner F. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick M., Stelzer Y., Bar-Nur O., Mayshar Y., Eden A., Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- Popkie A.P., Zeidner L.C., Albrecht A.M., D’Ippolito A., Eckardt S., Newsom D.E., Groden J., Doble B.W., Aronow B., McLaughlin K.J. Phosphatidylinositol 3-kinase (PI3K) signaling via glycogen synthase kinase-3 (Gsk-3) regulates DNA methylation of imprinted loci. J. Biol. Chem. 2010;285:41337–41347. doi: 10.1074/jbc.M110.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W., Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Ferguson-Smith A.C., Pedersen R.A. Epigenetic status of human embryonic stem cells. Nat. Genet. 2005;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Ferguson-Smith A.C., Pedersen R.A. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum. Mol. Genet. 2007;16:R243–R251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S.A., Tomizawa S., Krueger F., Ruf N., Carli N., Segonds-Pichon A., Sato S., Hata K., Andrews S.R., Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B., Ueda M., Sabri S., Brumbaugh J., Huebner A.J., Sahakyan A., Clement K., Clowers K.J., Erickson A.R., Shioda K. Reduced MEK inhibition preserves genomic stability in naive human embryonic stem cells. Nat. Methods. 2018;15:732–740. doi: 10.1038/s41592-018-0104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer Y., Bar S., Bartok O., Afik S., Ronen D., Kadener S., Benvenisty N. Differentiation of human parthenogenetic pluripotent stem cells reveals multiple tissue- and isoform-specific imprinted transcripts. Cell Rep. 2015;11:308–320. doi: 10.1016/j.celrep.2015.03.023. [DOI] [PubMed] [Google Scholar]
- Stirparo G.G., Boroviak T., Guo G., Nichols J., Smith A., Bertone P. Correction: integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast (doi: 10.1242/dev.158501) Development. 2018;145:dev169672. doi: 10.1242/dev.158501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirparo G.G., Smith A., Guo G. Cancer-related mutations are not enriched in naive human pluripotent stem cells. Cell Stem Cell. 2021;28:164–169.e2. doi: 10.1016/j.stem.2020.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczerbinska I., Gonzales K.A.U., Cukuroglu E., Ramli M.N. Bin, Lee B.P.G., Tan C.P., Wong C.K., Rancati G.I., Liang H., Göke J. A chemically defined feeder-free system for the establishment and maintenance of the human naive pluripotent state. Stem Cell Reports. 2019;13:612–626. doi: 10.1016/j.stemcr.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taei A., Rasooli P., Braun T., Hassani S.-N., Baharvand H. Signal regulators of human naïve pluripotency. Exp. Cell Res. 2020;389:111924. doi: 10.1016/j.yexcr.2020.111924. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Coluccio A., Thorball C.W., Planet E., Shi H., Offner S., Turelli P., Imbeault M., Ferguson-Smith A.C., Trono D. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 2019;33:49–54. doi: 10.1101/gad.320069.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Friedli M., He Y., Planet E., O’Neil R.C., Markoulaki S., Pontis J., Wang H., Iouranova A., Imbeault M. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tucci V., Isles A.R., Kelsey G., Ferguson-Smith A.C., Bartolomei M.S., Benvenisty N., Bourc’his D., Charalambous M., Dulac C., Feil R. Genomic imprinting and physiological processes in mammals. Cell. 2019;176:952–965. doi: 10.1016/j.cell.2019.01.043. [DOI] [PubMed] [Google Scholar]
- Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B., Dong L., Ding N., Zhang Q. GSA: Genome Sequence Archive. Genom. Proteom. Bioinform. 2017;15:14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbein U., Schachter M., Egli Di., Benvenisty N. Analysis of chromosomal aberrations and recombination by allelic bias in RNA-seq. Nat. Commun. 2016;7:12144. doi: 10.1038/ncomms12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wilmut I., Schnieke A.E., McWhir J., Kind A.J., Campbell K.H.S. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Yagi M., Kishigami S., Tanaka A., Semi K., Mizutani E., Wakayama S., Wakayama T., Yamamoto T., Yamada Y. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548:224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- Yilmaz A., Benvenisty N. Defining human pluripotency. Cell Stem Cell. 2019;25:9–22. doi: 10.1016/j.stem.2019.06.010. [DOI] [PubMed] [Google Scholar]
- Zimmerlin L., Park T.S., Huo J.S., Verma K., Pather S.R., Talbot C.C., Agarwal J., Steppan D., Zhang Y.W., Considine M. Tankyrase inhibition promotes a stable human naïve pluripotent state with improved functionality. Development. 2016;143:4368–4380. doi: 10.1242/dev.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvetkova I., Apedaile A., Ramsahoye B., Mermoud J.E., Crompton L.A., John R., Feil R., Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.