Summary
Skeletal muscle satellite cells (SCs) are stem cells responsible for muscle development and regeneration. Although CRISPR/Cas9 has been widely used, its application in endogenous SCs remains elusive. Here, we generate mice expressing Cas9 in SCs and achieve robust editing in juvenile SCs at the postnatal stage through AAV9-mediated short guide RNA (sgRNA) delivery. Additionally, we reveal that quiescent SCs are resistant to CRISPR/Cas9-mediated editing. As a proof of concept, we demonstrate efficient editing of master transcription factor (TF) Myod1 locus using the CRISPR/Cas9/AAV9-sgRNA system in juvenile SCs. Application on two key TFs, MYC and BCL6, unveils distinct functions in SC activation and muscle regeneration. Particularly, we reveal that MYC orchestrates SC activation through regulating 3D genome architecture. Its depletion results in strengthening of the topologically associating domain boundaries thus may affect gene expression. Altogether, our study establishes a platform for editing endogenous SCs that can be harnessed to elucidate the functionality of key regulators governing SC activities.
Keywords: CRISPR/Cas9, muscle stem cell, 3D chromatin, MYC
Highlights
-
•
CRISPR/Cas9/AAV9-sgRNA system yields robust editing in juvenile SCs
-
•
Quiescent SCs are resistant to CRISPR/Cas9-mediated in vivo editing
-
•
Key TFs play diversified functions during SC early activation
-
•
MYC promotes SC activation through impinging on 3D chromatin architecture
In this article, Wang and colleagues generate a CRISPR/Cas9/AAV9-sgRNA in vivo genome editing system to achieve robust editing in juvenile satellite cells (SCs). Application of the system reveals diversified functions of key transcription factors (TFs) during SC activation and uncovers that MYC promotes SC activation through regulating 3D genome architecture. Quiescent SCs, however, are resistant to the in vivo editing.
Introduction
Skeletal muscle is built up by numerous multinucleated myofibers and presents excellent regeneration potential after injury, which is executed by adult muscle stem cells also called satellite cells (SCs) (Ancel et al., 2021). Normally, juvenile SCs characterized by paired-box gene 7 (PAX7) expression emerge about 2 days before birth in mice and continue to proliferate to form adult muscle through undergoing postnatal myogenesis. About 2–4 weeks after birth, quiescent SCs (QSCs) appear to build the SC pool (Tajbakhsh, 2009). In healthy adult skeletal muscle, PAX7+ QSCs are located in a specialized niche beneath the basal lamina. Upon traumas, these dormant SCs quickly activate and re-enter the cell cycle to give rise to myoblasts characterized by elevated expression of two basic helix-loop-helix (bHLH) transcription factors (TFs), myogenic factor 5 (MYF5) and MYOD1. Then a large portion of myoblasts downregulate PAX7 expression and undergo myogenic differentiation to form new muscle fibers by inducing MYOGENIN and myogenic regulatory factor 4 (MRF4) expression. Meanwhile, a sub-population of activated SCs retaining high PAX7 expression return to quiescence to restore the SC pool (Ancel et al., 2021).
Transcriptional regulation orchestrated by TFs represents the key intrinsic mechanism driving SC fate transition and lineage progression (Ancel et al., 2021). However, a systematic approach is needed to identify potential key TFs modulating SC quiescence and early activation. Moreover, an effective approach is needed to test the functionality of the above predicted key TFs. In particular, since quiescence cannot be maintained once the cells are isolated (Machado et al., 2017), TFs that function in QSCs thus need to be manipulated in vivo, which calls for an effective in vivo genome editing system.
Recently, the in vivo application of CRISPR/Cas9 is emerging to generate mouse models and to correct genetic diseases by viral or non-viral based Cas9/short guide RNA (sgRNA) delivery (Doetschman and Georgieva, 2017). In particular, several reports described its utility in skeletal muscle tissue (Doetschman and Georgieva, 2017; Tabebordbar et al., 2016) treating Duchenne muscular dystrophy (DMD). In these attempts, co-transduction of multiple adeno-associated virus (AAV) vectors is required to deliver the Cas9 and sgRNAs separately considering the restricted packaging capacity of AAV virus (~4.7 kb), which, however, limited the modification efficiency since successful editing only occurs in nuclei simultaneously receiving all the components. In addition, as most of the genomic modifications were conducted in post-mitotic myofibers, it remains to be determined whether such AAV/Cas9/sgRNA-based tools can be applied to edit SCs in vivo. Moreover, muscle-related studies would tremendously benefit from a facile in vivo platform which allows disruption of gene expression in endogenous SCs instead of isolating them or generating multiallelic transgenic mice (Goldstein et al., 2019). However, several issues need to be clarified before the generation of such a tool. The most conflicting one is whether AAV vectors are able to transduce SCs in vivo. While a previous study concluded that QSCs are resistant to AAV transduction (Arnett et al., 2014), two recent studies reported efficient transduction of multiple AAV serotypes to SCs in adult mice by a Cre/lox fluorescent reporter tracking system (Goldstein et al., 2019; Nance et al., 2019). Even with efficient transduction, the ability of CRISPR/Cas9 to modify genomes in endogenous SCs still needs to be tested. To this end, one study demonstrated successful editing in proliferating juvenile SCs with modest efficiency (2%–4% of endogenous SCs were edited) by injecting the AAV virus at postnatal stage (Tabebordbar et al., 2016), and another recent study found weak genome modification of CRISPR/Cas9 in adult mice by using a muscle graft model (Nance et al., 2019). Since both of the editing events occurred during active myogenesis, these findings hint that CRISPR/Cas9 may be able to modify SCs in certain settings. Nevertheless, whether QSCs in the adult stage are permissive to CRISPR/Cas9-mediated genome editing is still unknown.
Here, in this study, we generate a mouse line expressing Cas9 in SCs to evaluate the feasibility of CRISPR/Cas9-based genome modification in endogenous SCs. Cas9 expression in SCs does not have obvious impact on SC homeostasis and regenerative capacity. Robust editing by CRISPR/Cas9 was observed in juvenile SCs undergoing active myogenesis; up to 95% editing efficiency was achieved on the Myod1 locus, which resulted in a complete depletion of MYOD1 protein. Using this in vivo editing system, we tested the functionality of two potential key TFs, MYC and BCL6, and uncovered their distinctive importance in SC activation and muscle regeneration. Furthermore, we found that MYC promotes SC activation through remodeling 3D genome architecture. Interestingly, application of the above system in adult SCs at quiescent stage did not yield successful editing even with efficient AAV9 transduction. Altogether, our study thus establishes a CRISPR/Cas9/AAV9-sgRNA genome editing system in endogenous SCs, which can be harnessed to identify and elucidate the functions of key TFs or other regulators orchestrating SC activities.
Results
Prolonged expression of Cas9 protein has no obvious impact on SC function
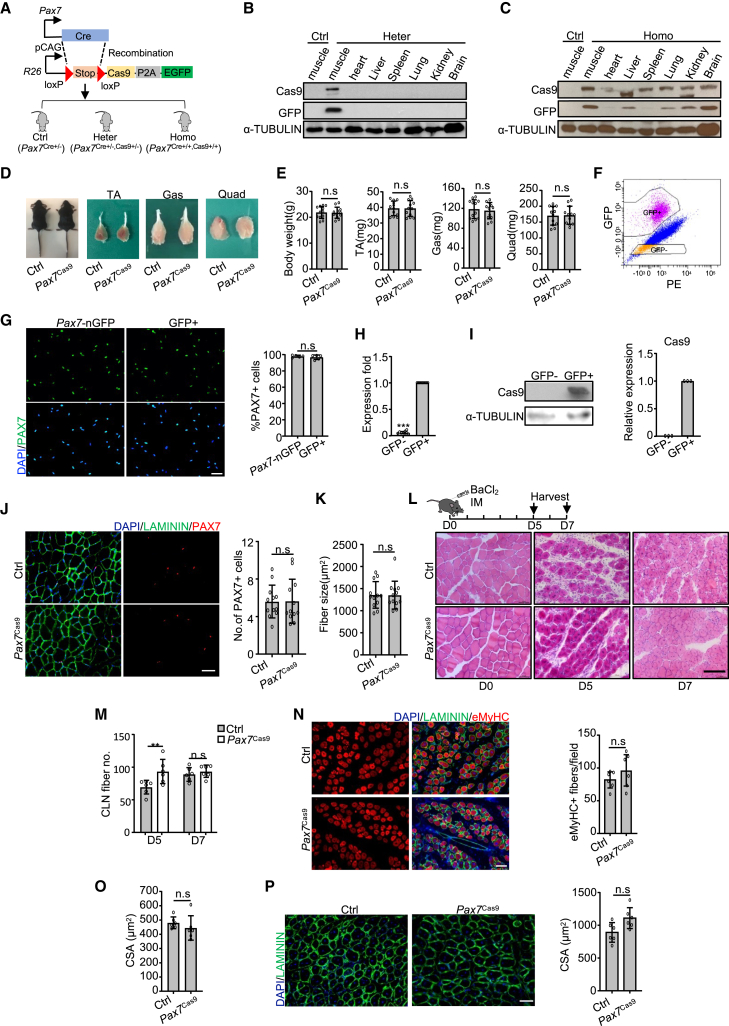
To demonstrate the feasibility of harnessing CRISPR/Cas9 to modify the genome in endogenous SCs, we first generated a mouse line that expressed Cas9 in SCs. We crossed Cre-dependent Rosa26Cas9−EGFP knockin mouse (Platt et al., 2014) with a Pax7Cre knockin mouse to generate heterozygous (Pax7Cre+/−, Cas9+/−) and homozygous (Pax7Cre+/+, Cas9+/+) mice along with the control (Ctrl) (Pax7Cre+/−) (Figure 1A). Co-expression of GFP facilitated us to monitor the Cas9-expressing cells. Cas9 and GFP proteins were restrictively expressed in muscle but not in other tissues of heterogeneous mice or muscle tissue of Ctrl mice (Figure 1B). PAX7 is known to be expressed in brain, especially in the hypothalamus (Hosoyama et al., 2010); the absence of detected expression in brain tissue is probably because only a small part, not the whole brain, was collected for detection. However, in some homozygous mice, Cas9 was expressed in multiple tissues (Figure 1C), possibly because PAX7 was expressed in a rare sub-population of spermatogonia of mice and thus turned on Cas9 expression as early as in the zygote (Aloisio et al., 2014). Therefore, the heterogeneous mice were used for the following experiments and referred to as Pax7Cas9 mice. The Pax7Cas9 progenies showed no overt morphological abnormalities (Figures 1D and 1E). To confirm the expression of Cas9 protein, SCs were isolated by fluorescence-activated cell sorting (FACS). Two distinguishable sub-populations appeared on the sorting plot based on GFP signals (Figure 1F) and up to 97% of GFP+ cells also expressed PAX7 (Figure 1G); the purity was similar to that of SCs isolated from Pax7-nGFP mice (96.74% ± 2.81% versus 97.85% ± 1.4%) (Figure 1G). High expression of Cas9 mRNA and protein were detected in GFP+ SCs but not in GFP− cells (Figures 1H and 1I). Furthermore, the number of PAX7+ SCs was not affected by prolonged Cas9 expression (Figure 1J) and the average fiber size showed no difference compared with the Ctrl mice (Figure 1K). To further investigate the effect of ectopic expression of Cas9 on SC function, acute muscle injury was induced by injection of barium chloride (BaCl2) into the tibialis anterior (TA) muscles (Figures 1L). Although more regenerating myofibers with centrally localized nuclei (CLN) were detected in Pax7Cas9 versus Ctrl at 5 days post injury, the difference disappeared at day 7 (Figures 1L and 1M). Immunostaining of eMyHC, a marker of newly formed fibers that may not have formed CLNs, on the other hand, revealed that the number of eMyHC+ fibers was comparable at 5 days after injury (Figure 1N). Moreover, the cross-sectional areas (CSA) of the newly formed fibers at days 5 and 7 revealed no obvious difference (Figures 1O and 1P). Altogether, the above results demonstrate that Cas9 expression in SCs does not cause obvious impact on SC homeostasis and regenerative capacity.
Figure 1.
Prolonged expression of Cas9 protein has no obvious impact on SC function
(A) Generation of mice expressing Cas9 in SCs. KI, knock in.
(B and C) Cas9 and GFP expressions were examined in heterozygous (B) and some homozygous mice (C).
(D and E) No obvious difference was detected between Ctrl and Pax7Cas9 littermates (TA, tibialis anterior; Gas, gastrocnemius; Quad, quadriceps). Representative images (D) and quantification (E) are shown. n = 12 mice.
(F) The gating strategy to isolate Cas9-GFP positive SCs. The phycoerythrin (PE) channel (biexponential scale) was used to separate GFP+ and GFP− cells.
(G) Left: IF staining of PAX7. n = 5 mice (an average of six fields/mouse). Scale bar, 50 μm.
(H) Cas9 mRNAs were detected (normalized to 18S mRNA). n = 9 independent experiments.
(I) Cas9 expression was examined and the band intensity was quantified. n = 3 independent experiments.
(J) Left: immunostaining of PAX7 and LAMININ on the TA muscle from 8-week-old mice. Right: the number of PAX7+ cells per 100 fibers. n = 12 mice (an average of six fields/mouse). Scale bar, 50 μm.
(K) Average fiber size. n = 12 mice.
(L and M) H&E staining on day 5 and 7 post injury (L). Myofibers with CLN per field (M). n = 6 mice (an average of five fields/mouse). Scale bar, 100 μm.
(N) Left: immunostaining of eMyHC and LAMININ at day 5 post injury. Right: the number of eMyHC+ myofibers. n = 6 mice (an average of six fields/mouse). Scale bar, 50 μm.
(O) CSA of the fibers with CLN at day 5 post injury. n = 6 mice.
(P) Left: immunostaining of LAMININ at day 7 post injury. Right: CSA of the fibers with CLN. n = 6 mice (an average of three fields/mouse). All the bar graphs are presented as mean ± SD. ∗∗p < 0.01. ns, no significance.
CRISPR/Cas9 in conjunction with AAV9-mediated sgRNAs delivery yields robust editing in juvenile SCs
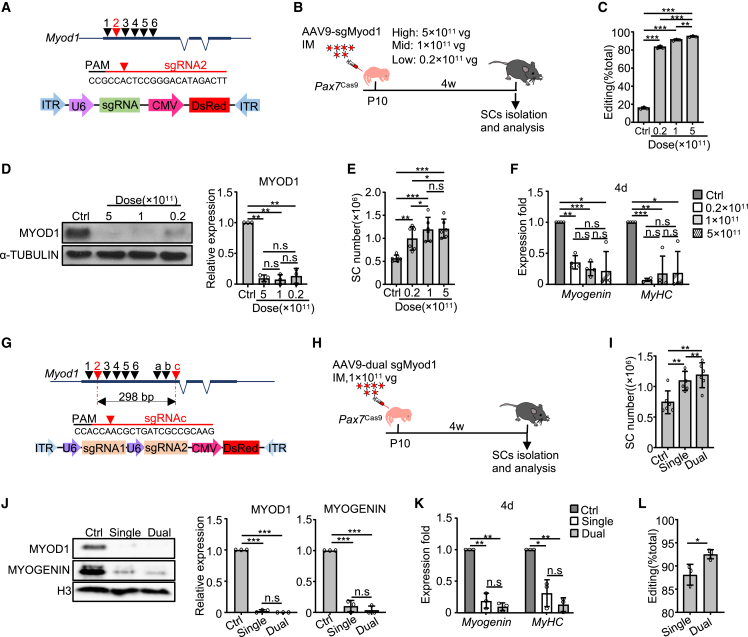
Next, we sought to test whether SCs can be efficiently edited in vivo using the above Pax7Cas9 mouse. To this end, we first tested whether this is feasible using juvenile SCs since a previous study reported mild editing efficiency in SCs at postnatal stage (Tabebordbar et al., 2016). As a proof of concept, the Myod1 gene was selected due to the wealth of knowledge of its function in SCs as a master TF orchestrating myogenic program (Peng et al., 2017; Yamamoto et al., 2018). A total of six sgRNAs against the first coding exon of Myod1 were designed (Figure 2A) and each was cloned into a Cas9-GFP-expressing vector and transfected into C2C12 cells followed by FACS to enrich transfected cells (Figure S1A); sgRNA2 showing the highest editing efficiency (19.5%) by Surveyor nuclease assay was chosen for further use. To deliver the sgRNA in vivo, sgRNA2 was placed into a U6-driven pAAV9-sgRNA vector (Figure 2A), which also carries a fluorescent dsRed gene; this AAV9-sgMyod1 resulted in an obvious cleavage at Myod1 locus (28.3%) in vitro (Figure S1B).
Figure 2.
CRISPR/Cas9 in conjunction with AAV9-mediated sgRNAs delivery yields robust editing in juvenile SCs
(A) Top: sgRNAs for targeting the Myod1 locus. Middle: selected sgRNA2 and its targeted sequence. Bottom: illustration of the vector. PAM, protospacer-adjacent motif.
(B) Illustration of the experimental design.
(C) Quantification of the editing efficiency (calculated as the ratio of the edited reads to the total reads). n = 3 mice.
(D) SCs were cultured for 2 days and the MYOD1 protein level was examined (left); the band intensity was quantified by ImageJ (right). n = 3 mice.
(E) The number of purified SCs. n = 6 mice.
(F) SCs were cultured for 4 days and the Myogenin or MyHC mRNAs were detected. n = 4 mice.
(G) Top: dual sgRNAs targeting Myod1. Middle: the targeted sequence for sgRNAc. Bottom: illustration of the vector.
(H) Illustration of the experimental design.
(I) The number of sorted FISCs. n = 6 mice.
(J) SCs were cultured for 2 days and the protein levels of MYOD1 and MYOGENIN were examined (left); the band intensity was quantified (middle and right). n = 3 mice.
(K) Myogenin and MyHC mRNAs in SCs cultured for 4 days. n = 3 mice.
(L) Quantification of the editing efficiency. n = 3 mice. All qRT-PCR data were normalized to 18S or Gapdh mRNA. All the bar graphs are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. ns, no significance.
See also Figures S1 and S2.
The above AAV9-sgMyod1 plasmid was then packed into virus particles and intramuscularly (IM) injected into the Pax7Cas9 mice with a single dose of 2×1011 viral genomes (vg)/mouse at postnatal day 10 (P10) (Figure S1C). At this stage of postnatal myogenesis, it is known that myogenic precursor cells actively proliferate to increase adult muscle; upon the termination of the postnatal myogenesis around P21 (Tajbakhsh, 2009), a portion of the edited myoblasts are expected to become quiescence SCs carrying the editing event (Figure S1D). For the Ctrl group, the same dose of AAV9 virus containing pAAV9-sgRNA backbone without any sgRNA insertion was injected. Four weeks after injection, we first evaluated the transduction efficiency of AAV9 virus into SCs. As expected, two distinguishable sub-populations were separated on the plot according to GFP expression (Figure S1E); intriguingly, the GFP+ SCs did not present differential dsRed expression. However, further examination of dsRed signal in the input cells (Figure S1E) by histogram showed there was indeed a separation of dsRed-high versus -low populations and GFP+ cells were located in the dsRed-high peak (Figure S1F), indicating that most GFP-expressing SCs were indeed infected by the virus. Moreover, immunofluorescence (IF) staining showed above 80% of the isolated SCs (GFP+) were dsRed positive (Figures S1G and S1H). These findings indicated that the above-administered AAV9 dosage was high enough to infect all the GFP+ SCs, thus the GFP+ population was isolated for following analyses. To estimate the editing efficiency, genomic DNAs from freshly isolated SCs (FISCs) were subject to Surveyor assay; around 30%–40% indel occurrence at the targeted Myod1 locus was detected in the AAV9-sgMyod1-injected mice but not in the Ctrl group (Figure S1I). Further Surveyor analysis of Myod1 transcripts from SCs cultured for 4 days also verified efficient mutagenesis (Figure S1J). Taken together, the above results demonstrate the juvenile SCs at postnatal stage can be efficiently edited in Pax7Cas9 mice.
A recent study implied that the administration of high-dose AAV9 virus particles to non-human primates and piglets caused severe toxicity (Hinderer et al., 2018), so we sought to determine the minimal dose of AAV9 needed to achieve effective editing. To this end, high (5×1011 vg/mouse), middle (1×1011 vg/mouse), or low (0.2×1011 vg/mouse) doses of AAV9-sgMyod1 virus particles were intramuscularly injected into Pax7Cas9 mice at P10 (Figure 2B) with no obvious abnormalities observed even in the high-dose group. Four weeks after injection, a slightly higher editing efficiency in the middle- versus low-dose group was observed, but no obvious difference between the middle- and high-dose groups (Figures S2A and S2B). To precisely determine the level of editing, sgMyod1 targeted locus was amplified from genomic DNAs in FISCs and subject to deep sequencing. The high-dose treatment led to a significantly higher level of mutations than the low- (95.08% ± 0.80% versus 83.16% ± 1.42%) or middle-dose groups (91.28% ± 0.92%) (Figure 2C); however, the percentage of frameshift mutations between the high- and middle-dose groups showed no obvious difference (77.43% ± 1.42% versus 75.58% ± 2.26%) (Figures S2C and S2D). A >80% indel formation frequency (Figure 2C) further confirmed that the AAV9 virus efficiently transduced SCs and the copy number of sgRNAs was high enough to achieve efficient editing even in the low-dose group.
At the protein level, we found that MYOD1 protein was completely eliminated in SCs from the high- or middle-dose groups, whereas a noticeable expression in the low-dose group was present (Figures 2D and S2E). However, we failed to observe a sharp decrease in Myod1 transcript levels (Figure S2F), suggesting that CRISPR/Cas9-mediated genome editing does not necessarily affect mRNA level which was recently described (Smits et al., 2019). To examine the impact of Myod1 editing on SCs, we noticed a robust increase of the number of FISCs (Figures 2E and S2G), in line with previous finding using Myod1 knockout mice (Yamamoto et al., 2018). When the SCs were cultured for differentiation for 2 or 4 days, Myogenin or MyHC was found to be significantly decreased (Figures 2F and S2H), confirming a defect in myoblast differentiation caused by MYOD1 loss; but the decreases were comparable among the groups with different doses. Taken together, these results suggest the middle dose (1×1011 vg/mouse) of AAV9-sgRNA administered at postnatal stage elicits almost complete depletion of MYOD1 expression in vivo. It is worth pointing out that a much higher dose (2×1014 genome copies per kilogram of body weight) of AAV9 virus particles was systemically administered by intravenous injection in the study by Hinderer et al. (2018); the dosages we used were much lower and locally administered IM. Nevertheless, the possible long-term effects remain to be tested.
Use of dual sgRNAs increases the genome editing efficiency in juvenile SCs
Several studies have demonstrated that simultaneous use of dual sgRNAs can induce indels as well as large deletions at the target loci to further enhance editing efficiency (Guo et al., 2017; Johansen et al., 2017). We thus sought to develop an AAV9-dual sgRNA system expressing two sgRNAs from a single vector (Figure 2G), which was shown to display higher cleavage potential compared with sgRNAs expressed from two separate vectors (Johansen et al., 2017). Two sgRNAs (sgRNA2 and sgRNAc) were selected to remove 298 bp in the exon 1 of Myod1 (Figures 2G and S2I). A middle dose of AAV9-dual sgMyod1 virus was injected (Figure 2H). For comparison, the same dose of single AAV9-Myod1-sgRNA2 virus was administered. Expectedly, the AAV9-dual sgMyod1 virus induced excision of the intervening sequence between the two sgRNA targeting sites (Figure S2J), which was not detected in the single sgRNA- or Ctrl virus-infected SCs. In addition, we noticed a further increase in FISC number in Pax7Cas9 mice injected with the dual versus single sgRNA (Figure 2I). However, the expression levels of MYOD1 protein (Figures 2J and S2K), Myod1 mRNA (Figure S2L), as well as its targets, Myogenin and MyHC (Figures 2J, 2K, and S2M), were comparable between the two groups. Subsequent deep sequencing (Figures 2L and S2N–S2P) confirmed that dual sgRNAs slightly outperformed single sgRNA in editing the Myod1 locus (92.55% ± 0.99% versus 88.13% ± 2.23%), favoring the application of dual sgRNA strategy in subsequent investigation.
CRISPR/Cas9 fails to efficiently modify SCs at quiescent stage
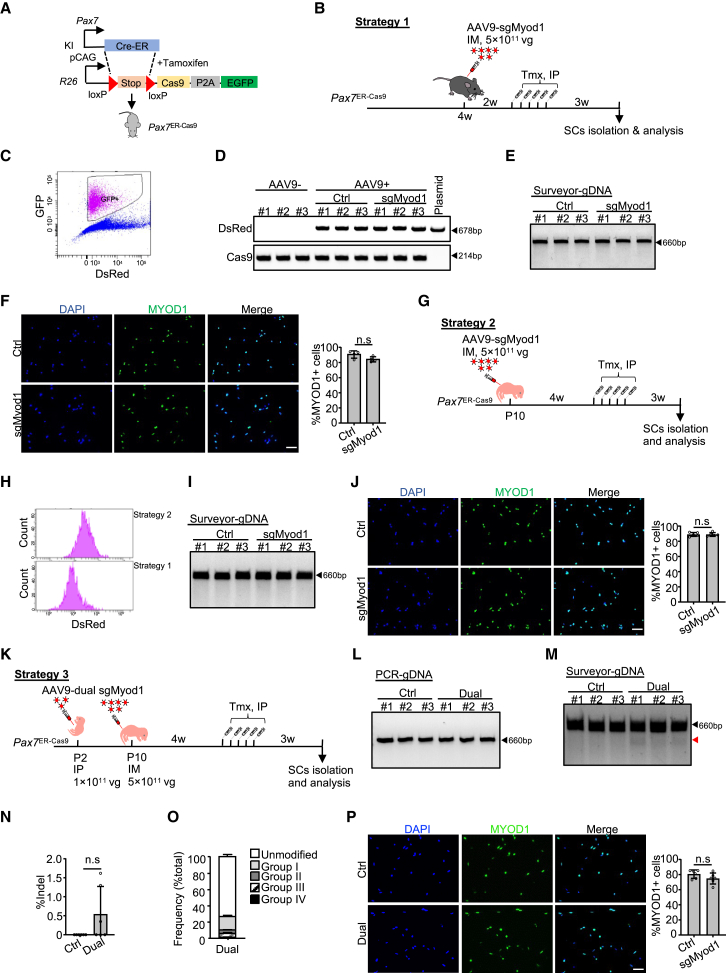
We next sought to test the performance of our CRISPR/Cas9/AAV9-sgRNA system in QSCs at adult stage. To this end, we generated an inducible Cas9-expressing mouse by crossing the homozygous Rosa26Cas9−EGFP mouse with the Pax7CreER knockin mouse, which contains an insertion of the CreER coding region downstream of the Pax7 gene; the resulting offspring were referred to as Pax7ER−Cas9 mice (Figure 3A). Unlike the Pax7Cas9 mice, in which Cas9 is induced in PAX7-descendant cells from the embryonic stage (Tajbakhsh, 2009), Cas9 induction in Pax7ER−Cas9 mice can be controlled by Tmx injection, thus restricting the editing to the quiescent stage. In the first strategy, a high-dose single AAV9-sgMyod1 virus was injected IM into 4-week-old Pax7ER−Cas9 mice followed by five consecutively intraperitoneal (IP) administrations of Tmx 2 weeks later. The mice were sacrificed for SC isolation after another 3 weeks (Figures 3B and S3A). Sufficient induction of Cas9 was confirmed (Figure 3C); successful AAV9 transduction in SCs was validated by PCR amplification of dsRed in FISC DNAs isolated from mice injected with AAV9 but not in those without AAV9 administration (Figure 3D). However, no editing event at the Myod1 locus was detected (Figure 3E) despite Myod1 transcript levels slightly decreased (Figure S3B); no changes in MYOD1 protein (Figure 3F), mRNA expression of Myogenin or MyHC (Figure S3B), and the SC number (Figure S3C) were observed.
Figure 3.
CRISPR/Cas9 fails to efficiently modify SCs at quiescent stage
(A) Illustration to generate the Pax7ER−Cas9 mice.
(B) Illustration of the first strategy.
(C) FACS plot showing induction of Cas9-GFP expression.
(D) PCR to amplify the dsRed or Cas9 coding region. The AAV9-Myod1-sgRNA2 plasmid was used as control. n = 7 mice.
(E) Surveyor assay to detect the editing efficiency at Myod1 locus. n = 7 mice.
(F) SCs were cultured for 2 days and IF stained for Myod1. n = 4 mice (an average of six fields/mouse). Scale bar, 50μm.
(G) Illustration of the second strategy.
(H) FACS histograms to compare the dsRed signal of the Cas9-GFP positive SCs between strategies 1 and 2.
(I) Editing efficiency was examined as in (E). n = 5 mice.
(J) SCs were cultured for 2 days and IF stained for MYOD1. n = 5 mice (an average of six fields/mouse). Scale bar, 50μm.
(K) Illustration of the third strategy.
(L) PCR analysis targeting Myod1 locus. n = 6 mice.
(M and N) Editing efficiency was examined as in (E). The black and red arrowheads indicate wild and cleaved bands respectively (M). The frequency of indel formation was quantified (N). n = 6 mice.
(O) Distribution of the total sequencing reads in genomic DNA that showed faint editing in (N). n = 3 mice.
(P) SCs were cultured for 2 days and IF stained for MYOD1. n = 6 mice (an average of eight fields/mouse). Scale bar, 50 μm. All the bar graphs are presented as mean ± SD. ns, no significance.
See also Figure S3.
Next, we performed the injection of the AAV9-sgMyod1 virus at P10 hoping the increased exposure time to sgRNA would improve AAV transduction efficiency and Cas9 editing efficiency. In this strategy, juvenile SCs were infected by the virus and a portion of them were expected to become quiescence carrying the sgRNAs (Figures 3G and S3D). Compared with the first strategy, an evident increase of dsRed signal in GFP+ SCs was observed during FACS isolation (Figures 3H and S3E), and successful AAV9 transduction was confirmed (Figure S3F). Still, no editing was evident at Myod1 locus (Figures 3I, 3J, S3G, and S3H).
We next sought to determine whether the use of dual sgRNAs could possibly enhance the editing efficiency. To ensure successful AAV9 transduction into SCs, two injections were performed: a middle dose of AAV9 virus was injected via IP at P2 (Figures 3K and S3I) and reinforced by a high dose via IM at P10, which was then followed by Tmx administration 4 weeks later. The mice were sacrificed for SC isolation after another 3 weeks, and successful AAV9 transduction was confirmed (Figure S3J). Although the AAV9-dual sgMyod1 virus failed to excise the intervening sequence between the two sgRNA targeting sites (Figure 3L), Surveyor assay revealed an inconsistent editing at the Myod1 locus (Figures 3M and 3N). Nevertheless, by deep sequencing, we determined ~27% (26.87% ± 2.15%) of the total reads were actually edited and about 1% (1.18% ± 0.37%) elicited deletions (Figures 3O, S3K, and S3L). However, we found that the above limited level of editing failed to induce obvious change of SC number (Figure S3M) or expressions of Myod1, Myogenin, or MyHC (Figures 3P and S3N). To speculate the possible reasons for the unsuccessful editing in QSCs, we looked into the chromatin accessibility of the Myod1 locus as it is known that condensed heterochromatic regions are resistant to Cas9-mediated editing (Doetschman and Georgieva, 2017). However, evident H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) signals (Machado et al., 2017) were observed at the above sgRNA target sites (Figure S3O), indicating inaccessibility to Cas9 may not account for the failed editing in QSCs. Taken together, our results demonstrate that the in vivo CRISPR/Cas9/AAV9-sgRNA system fails to induce efficient genome editing at the Myod1 locus in QSCs.
Key TFs regulating SC quiescence and activation are predicted through SEs
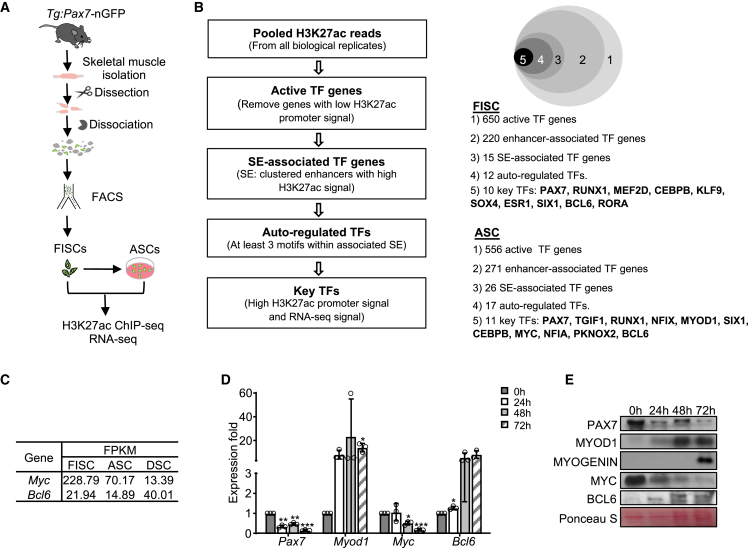
Next, we sought to extend our CRISPR/Cas9/AAV9-sgRNA system to dissect the functions of key TFs in myogenesis. Recent study has shown that key TFs bind clusters of enhancers termed super-enhancers (SEs) and that key TFs themselves are often driven by SEs, which can be used to predict key TFs (Saint-André et al., 2016). Therefore, ChIP-seq of H3K27ac was performed in FISCs from Pax7-nGFP mice and activated SCs (ASCs) cultured for 24 h (Figures 4A, S4A, and S4B). Enhancer constituents were then identified following the standard analysis pipeline (Peng et al., 2017) (Figure S4C; Table S1), showing dynamic remodeling of enhancer landscape during SC activation (Figure S4D; Table S1). A modified ROSE algorithm (Peng et al., 2017) was then applied to define 57 and 163 SEs in FISCs and ASCs, respectively (Figures S4E and S4F; Table S2). Based on the interconnected property between SEs and key TFs (Saint-André et al., 2016), key TFs were then predicted (Figures 4B and S4G). Ten in FISCs were defined (PAX7, RUNX1, MEF2D, CEBPB, KLF9, SOX4, ESR1, SIX1, BCL6, and RORA) and 11 in ASCs (PAX7, TGIF1, RUNX1, NFIX, MYOD1, SIX1, CEBPB, MYC, NFIA, PKNOX2, and BCL6; Figures 4B and S4E and Table S2). Of note, PAX7 was present in both FISC and ASC lists, while MYOD1 only appeared in the ASC list (Figures S4E and S4H).
Figure 4.
Key TFs regulating SC quiescence and activation are predicted through SEs
(A) Illustration of collection of FISCs and ASCs.
(B) Left: illustration of the pipeline for predicting key TFs. Right: Venn diagram showing the identified key TFs.
(C) Expression levels of two selected key TFs examined by RNA-seq.
(D) mRNA expression of the above TFs. n = 3 independent experiments. The data were normalized to 18S mRNA and presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(E) The protein levels of the above TFs.
Among these key TFs, BLC6 is a transcription repressor, while MYC normally functions to activate target gene expression. In muscle system, MYC is known to inhibit C2C12 differentiation through preventing myoblast fusion (Crescenzi et al., 1994), whereas BLC6 plays a pro-differentiating role by restraining apoptotic cell death (Kumagai et al., 1999). Moreover, our RNA sequencing (RNA-seq) showed these two TFs displayed distinct expression dynamics during SC lineage progression (Figure 4C): Myc exhibited the highest expression in FISCs and continued to decrease in ASCs and differentiated SCs (DSCs); however, Bcl6 expression decreased slightly in ASCs versus FISCs but increased dramatically in DSCs. It is interesting that, despite the high expression of Myc in FISCs, no associated SE was identified (Figure S4I) thus not predicted as a key TF in FISCs. The above RNA-seq results were validated by qRT-PCR in SCs cultured for 24, 48, and 72 h, during which period SCs were activated (24 h), proliferated (48 h), and differentiated (72 h) (Figure 4D). At the protein level, the two TFs exhibited definitely opposite trends consistent with their mRNA changes (Figure 4E). Based on their expression dynamics and known functions, MYC and BLC6 may play different roles during SC lineage progression and were selected for further functional investigation.
CRISPR/Cas9/AAV9-sgRNA-mediated genome editing of Myc hinders SC activation and muscle regeneration
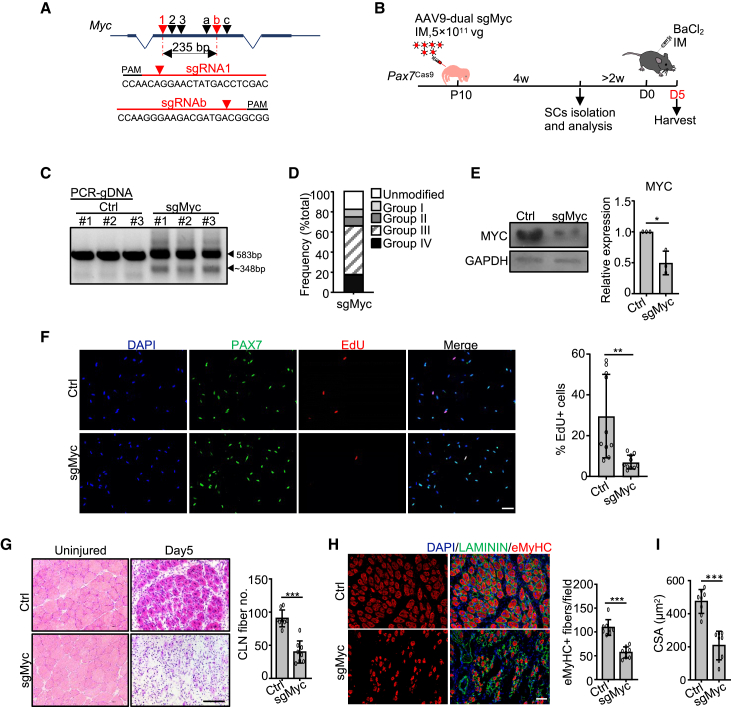
We next sought to validate the functionality of MYC and BLC6 in early stages of SC fate transition through leveraging our established CRISPR/Cas9/AAV9-sgRNA system. To edit Myc, two sgRNAs (sgRNA1 and sgRNAb) were selected (Figures 5A and S5A); the AAV9-dual sgMyc vector succeeded in excising the Myc locus in vitro (Figure S5B). Following the above established administration strategy (Figure S5C), a middle dose of the dual sgMyc virus was injected to the Pax7Cas9 muscle at P10, which, however, led to inefficient editing (Figures S5D and S5E). Increased dose of 5×1011 vg/mouse (Figure 5B) yielded successful deletion of a 235 bp of the Myc exon (Figures 5C and S5F). Deep sequencing revealed ~83% editing at the Myc locus (Figures 5D and S5G) and ~18% of the total reads were detected as deletions (Figure 5D, group IV). The level of Myc mRNA was not significantly changed (Figure S5H), but MYC protein level was remarkably decreased (Figure 5E). We further attempted the editing in QSCs; as expected, no indel occurrence was detected (Figures S5I and S5J) despite the sgRNA target sites appeared to be at open chromatin regions marked by H3K27ac (Figures S5K).
Figure 5.
CRISPR/Cas9/AAV9-sgRNA-mediated genome editing of Myc hinders SC activation and muscle regeneration
(A) Dual sgRNAs targeting Myc locus.
(B) Illustration of the experimental design.
(C) PCR to test the cleavage efficiency. Wild-type (583 bp) and cleaved (~348 bp) fragments are indicated by arrowheads.
(D) Distribution of the total sequencing reads.
(E) Left: MYC protein level. Right: the band intensity. n = 3 mice.
(F) FISCs were labeled with EdU for 24 h and the percentage of EdU+ cells was quantified. n = 10 mice (an average of 25 fields/mouse). Scale bar, 50 μm.
(G) Left: H&E staining at 5 days post injury. Right: the myofibers with CLN per field. n = 8 mice (an average of six fields/mouse). Scale bar, 100 μm.
(H) Left: immunostaining of eMyHC and LAMININ. Right: the number of eMyHC+ myofibers. n = 8 mice (an average of seven fields/mouse). Scale bar, 50 μm.
(I) The CSA of the newly formed fibers with CLN. n = 8 mice. All the bar graphs are presented as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Figures S5 and S6.
Next, when analyzing the impact of Myc editing on SCs, no obvious difference in the number of sorted SCs was detected in AAV9-dual sgMyc versus Ctrl group (Figure S5L), suggesting the loss of MYC may not influence SC homeostasis. To characterize MYC function on SC activation, the edited SCs were cultured in vitro for 24 h in the presence of 5-ethynyl-2'-deoxyuridine (EdU) as it is commonly believed that it takes about 24–36 h for SCs to enter the first cell cycle and become fully activated (Rodgers et al., 2014). MYC loss led to a significant delay of SC activation (Figure 5F), suggesting that MYC protein functions to promote SC activation. The promoting effect appeared to be in discrepancy with its downregulation in ASCs versus FISCs (Figures 4C–4E); however, further examination of Myc expression in QSCs (obtained by prior in situ fixation with paraformaldehyde [PFA] before FACS, which is believed to avoid the isolation induced early activation of SCs; Machado et al., 2017) revealed it was indeed induced in FISCs versus QSCs (Figure S5M), which was also confirmed by IF staining of MYC protein (Figure S5N). To further examine whether loss of MYC impairs damage induced muscle regeneration, Pax7Cas9 mice were administered with high doses of AAV9-dual sgMyc virus at P10 followed by BaCl2 injection in the TA muscle 6 weeks later (Figure 5B). Without injury, the body weight and limb muscle weight of the mice injected with sgMyc virus did not display evident difference (Figure S5O). Moreover, the PAX7+ SC number was not affected (Figure S5P), again indicating that loss of MYC may not affect SC homeostasis in adult mice. The average fiber size in the uninjured muscles showed no difference either (Figure S5Q). However, 5 days post injury, we observed a significantly reduced number of regenerating myofibers with CLN in sgMyc versus Ctrl group (Figure 5G). Consistently, IF staining of eMyHC revealed a marked decrease in the number of newly formed fibers (Figure 5H) as well as fiber size (Figure 5I). Together, the above findings demonstrate MYC is important for SC activation and acute-injury-induced muscle regeneration.
To further demonstrate the usage of the in vivo genome editing system, we expanded to include BLC6. The designed dual sgRNAs yielded obvious cleavage in vivo (Figures S6A–S6D) but also failed to edit the Bcl6 locus in QSCs (Figures S6E and S6F); Interestingly, different from Myod1 and Myc cases, we found the sgRNA target sites were located in regions void of H3K27ac signal (Figures S6G). About 38% of indel formation was detected (Figure S6H). Similar to the Myc case, editing of Bcl6 induced obvious decrease of its protein expression without altering the mRNA level (Figures S6I and S6J). Although the number of sorted FISCs was not affected (Figure S6K), editing of Bcl6 led to accelerated SC activation (Figure S6L). However, the muscle regeneration process after acute injury appeared to be delayed (Figures S6M–S6P). We think this discrepancy may be caused by the pleiotropic roles BLC6 plays in different stages of SC lineage progression. Although it inhibits SC activation, it is known to promote myoblast differentiation (Kumagai et al., 1999); therefore, the net effect on regeneration appeared to be negative.
MYC orchestrates SC activation through impinging on 3D chromatin architecture
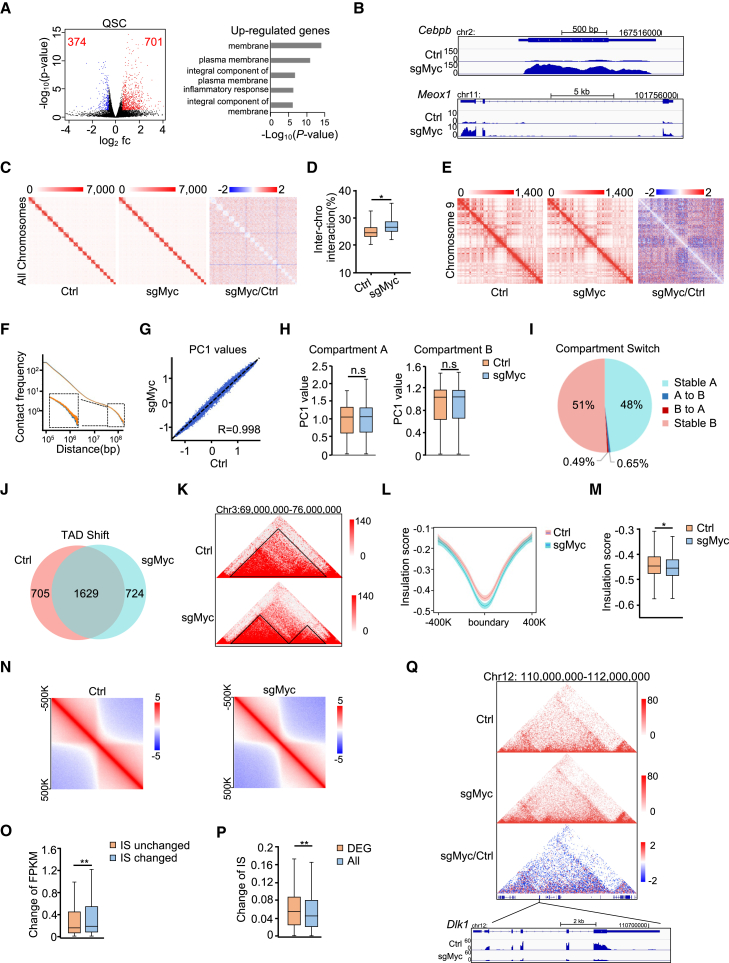
Next, we attempted to elucidate the mechanism underlying the above-described MYC function in SC activation. RNA-seq analysis was conducted to dissect the transcriptomic changes induced by MYC depletion in QSCs isolated from Pax7Cas9 mice infected with sgMyc virus (Figure S7A). A total of 701 and 374 genes were up- or downregulated, respectively (Figure 6A and Table S3). The upregulated genes were highly enriched for GO terms such as “membrane” and “plasma membrane.” It is known that QSCs express various glycoproteins distributed on the cell membrane as extracellular sensors to integrate signals from the niche to actively maintain SC in quiescent stage (Ancel et al., 2021). It is thus likely that loss of MYC promotes SC quiescence and delays SC early activation through increasing the expression of these glycoproteins. Additionally, among the upregulated genes, we also found Cebpb (Lala-Tabbert et al., 2020) and Mexo1 (Nguyen et al., 2017) (Figure 6B), which are known to maintain SC quiescence and inhibit SC activation. The downregulated genes were not enriched for any significant GO terms (Table S3); however, some genes, for example Dlk1, are known to promote SC activation (Zhang et al., 2021). Moreover, the expression of some well-known MYC targets, including Kat2a and Itgb4 (Kress et al., 2015) (Figure S7B), were repressed and activated respectively. Since the gene expression program is tightly orchestrated by the 3D genome conformation at multiple levels, including compartments, topologically associate domains (TADs), and chromatin loops (Ke et al., 2017), and MYC has a potential role in remodeling global genome structure (Kieffer-Kwon et al., 2017), in situ high-throughput chromosome conformation capture (Hi-C) was then performed in the above QSCs (Figures S7A, S7C, and S7D) to identify MYC-depletion-induced 3D genome alterations. Globally, MYC depletion resulted in a higher ratio of inter-chromosomal interactions in QSCs (Figures 6C and 6D), indicating a potential role of MYC in chromosome segregation. At the compartment level, the compartmentalization matrices displayed a similar pattern (Figure 6E), despite a mild increase being observed in long-range chromatin interactions upon MYC loss (Figures 6F and S7E), indicating MYC may promote chromatin decompaction, as suggested by a recent study (Kieffer-Kwon et al., 2017). Overall, the A and B compartments remained constant, as compartmentalization strength (Figure S7F), compartment profiles by principal component 1 (PC1) values (Figures 6G and S7G and Table S4), as well as compartment score (Figure 6H) did not display significant alterations. Only about 0.65% of the genome switched from A compartment to B and 0.49% from B to A (Figure 6I and Table S4).
Figure 6.
MYC orchestrates SC activation through impinging on 3D chromatin architecture
(A) RNA-seq for Ctrl and sgMyc QSCs. Left: DEGs were identified. Right: GO analysis of the upregulated genes. Fc, fold change.
(B) Two examples of upregulated genes.
(C) Hi-C interaction matrices in Ctrl (left) and sgMyc (middle) QSCs. Heatmap of differential inter-chromosomal interactions (right). Resolution: 5 Mb.
(D) The ratio of inter-chromosomal in total interactions for each chromosome.
(E) Normalized interaction matrices for chromosome 9 in Ctrl (left) and sgMyc (middle) QSCs. Heatmap of differential interactions (right). Resolution: 250 kb.
(F) Overall scaling of normalized interaction frequency as a function of genomic distance in Ctrl (blue) and sgMyc (orange) QSCs.
(G) PC1 values of each compartment.
(H) Boxplots showing the compartment score of A (left) and B compartments (right).
(I) Pie chart showing compartment switching.
(J) Venn diagram showing the number of shifted TADs.
(K) Snapshot of Hi-C matrices to show TAD shift. The black triangles indicate the identified TADs.
(L) Average IS distribution around TAD boundaries (±400 kb).
(M) Boxplots showing TAD IS.
(N) Aggregate Hi-C maps of TAD borders.
(O) Boxplot showing the absolute fragments per kilobase per million (FPKM) changes for genes located within boundaries with at least 50% IS value change upon MYC depletion compared with those residing in IS unchanged boundaries.
(P) Boxplots showing the absolute IS changes for DEG-containing boundaries compared with all boundaries.
(Q) Top: snapshot of Hi-C data showing differential contact map at genomic regions encompassing Dlk1 locus. Bottom: differential expression of Dlk1 in QSCs determined by RNA-seq. Statistical analyses in H, M, O, and P were done by Wilcoxon rank-sum test; ∗p < 0.05, ∗∗p < 0.01. ns, no significance.
See also Figure S7 and Table S3. DEGs in QSCs infected with AAV9-dual sgMyc versus control viruses, Table S4. Genomic distribution and PC1 value of each compartment in QSCs infected with AAV9-dual, Table S5. Genomic distribution of each TAD in QSCs infected with AAV9-dual sgMyc versus control viruses, Table S6. GO analysis of DEGs residing in TAD boundaries, related to Figure 6.
Next, a total of 2,334 and 2,353 TADs were identified in Ctrl and sgMyc groups, respectively (Figure 6J and Table S5); TAD size was largely unaltered with the median length of 720 kb (Figure S7H); and TAD numbers per chromosome were also comparable (Figure S7H). Although the majority of TADs (1,629) were stable, MYC depletion resulted in shifting of 30.2% (705) of the TADs (Figures 6J and 6K), in which cases the TADs split, merged, disappeared, built, or rearranged (Ke et al., 2017) (Figure S7I and Table S5). Interestingly, such TAD shifting was also uncovered in B cells when we examined the published Hi-C data (Kieffer-Kwon et al., 2017) (Figure S7J); a total of 2,625 TADs were identified in B cells and 38.4% (1,007) were found to be shifted upon MYC deletion. Of note, although 1,976 TAD boundaries were conservatively identified in both B cells and QSCs (Figure S7K), only 70 boundaries were changed in both cells upon MYC deletion (data not shown), indicating MYC impact on TAD boundaries may be cell type specific. Further analysis revealed that loss of MYC in QSCs caused a decrease of TAD insulation score (IS) (Figures 6L-6N), indicating strengthened boundary formation and decreased inter-TAD interactions (Figures S7L and S7M). Interestingly, intra-TAD interactions also decreased (Figure S7L). To further determine whether MYC-dependent change of TAD boundary strength correlated with disrupted expression of genes located at the boundary regions, we found that genes located within the boundaries with at least 50% IS value change (ΔIS) underwent higher expression changes upon MYC depletion compared with those residing within IS unchanged boundaries (1.44 versus 1.19) (Figure 6O); with a different cutoff of ΔIS ≥20%, the expression changes remained significant (1.31 versus 1.21) (Figure S7N). Furthermore, differentially expressed gene (DEG)-containing boundaries displayed higher change of IS compared with average IS change of all boundaries (Figures 6P and S7O), indicating a correlation between TAD alteration and gene expression change orchestrated by MYC in QSCs (Figure 6Q). Further GO analysis of these boundary-located DEGs revealed that they were enriched for membrane-related terms including “membrane,” “potassium channel activity,” and “integral component of plasma membrane” (Figure S7P and Table S6), consistent with the findings from Figure 6A and reinforcing the notion that MYC may control SC quiescence and early activation through regulating membrane-related glycoproteins. Lastly, we found 80.2% (1,005) of the H3K27ac positive peaks within the TAD boundary regions contained MYC binding motifs (Figure S7Q). Since MYC has been reported to recruit histone-modifying enzymes, including the histone acetyltransferases TIP60 and KAT2A and the serine/threonine kinase PIM1 (Kress et al., 2015), it is possible that MYC may interact with these chromatin-modifying enzymes on the open boundary regions, which subsequently affect boundary organization. In sum, our data suggest that MYC may potentially function to modulate 3D genome architecture to regulate gene expression program in SCs.
Discussion
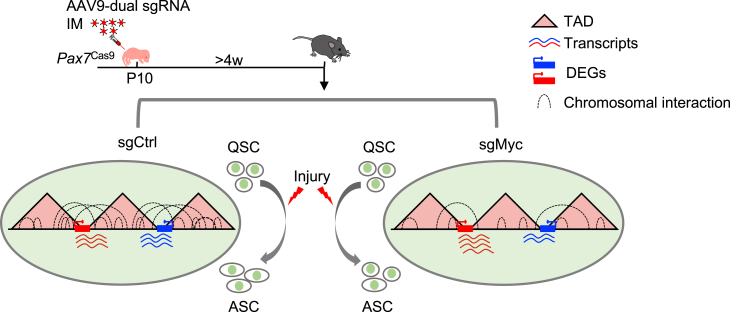
In this study, leveraging the Cas9 knockin mouse and AAV9-mediated sgRNA delivery, we documented in vivo genome editing by CRISPR/Cas9 in active juvenile SCs but not in quiescent SCs at adult stage. Using Myod1 locus as a proof of concept, this CRISPR/Cas9/AAV9-sgRNA system efficiently introduced mutagenesis at the target locus and disrupted its expression in SCs. Application of this system on key TFs predicted by SEs, MYC, and BLC6 revealed their distinct functions in the early stage of SC activation and acute-damage-induced muscle regeneration. Further mechanistic investigation uncovered a possible role for MYC in regulating SC activation through modulating 3D genome organization (Figure 7).
Figure 7.
CRISPR/Cas9/AAV-sgRNA-mediated in vivo genome editing uncovers the possible role of MYC in 3D genome organization in SCs
Pax7Cas9 mice were administered with AAV9-dual sgRNA virus at postnatal stage to delete MYC expression in QSCs. MYC depletion results in a global strengthening of the TAD boundaries and decrease of both of the intra- and inter-TAD interactions. The change of TAD boundary strength is associated with disrupted gene expression, which may lead to delayed SC activation.
One of our key findings is that the CRISPR/Cas9/AAV9-sgRNA system achieved a remarkable editing efficiency in endogenous juvenile SCs. Overall, our system manifests several advantages. The most prominent one is that it allows for manipulation of SC genomes in situ without requiring cell isolation or culture, thereby making it possible to study the impact on SC quiescence, homeostasis in their native niche, and early activation upon damage. On the paradigm of Myod1 locus, it achieved up to 95% editing efficiency and nearly complete depletion of MYOD1 protein even at the low dose of AAV9-sgRNA (0.2×1011 vg/mouse) (Figure 2). The attempt on Myc locus was also successful with around 83% genome editing and a significant depletion of MYC protein. However, a high dose of AAV (5×1011 vg/mouse) was necessitated for Myc locus. The editing efficiency on Bcl6 (38%) was not high even with the use of the high dose of virus and dual sgRNAs, suggesting that the editing efficiency is locus dependent, which may be due to the inherent quality of different protospacer sequences or the chromatin state of each locus (Doetschman and Georgieva, 2017).
It is foreseeable that our system will accelerate the pace at which gene function and interaction can be interrogated in endogenous SCs compared with commonly employed transgenic and gene knockout-based models. Traditionally, it is time consuming and expensive to generate knockout mice, and this shortcoming would be exacerbated when multiple genes need to be disrupted simultaneously. However, in our system, the entire procedure from sgRNA selection to phenotypic dissection only takes several weeks. In addition, our system can be adapted to enable rapid and direct in vivo screening of candidate and novel targets suspected to influence SC phenotypes. The ability to achieve multiplex genetic perturbations using the Cas9 mouse also enables the possibility of interrogating multigenic effects. In addition to coding genes, this system can also be used to interrogate non-coding genome such as enhancers and long non-coding RNAs (lncRNAs). For example, in a recent study (Zhao et al., 2019), we applied it to edit an enhancer RNA (eRNA) in skeletal muscle, which facilitated the investigation of the eRNA function in regulating myoblast differentiation.
Of note, this system has several limitations. One caveat is that the AAV9 administration can also edit muscle fibers. Since PAX7 expression emerges as early as embryonic day 10.5 (E10.5) in muscle progenitor cells to drive the formation of muscle compartments (Tajbakhsh, 2009), Cas9 is expressed not only in SCs but also in the fibers of Pax7Cas9 mice. We attempted to solve this problem by using inducible Pax7ER−Cas9 mice that will allow SC restricted expression of Cas9, but the results are not compelling at the current stage. Second, this system does not generate a uniform knockout mouse. Although the transduction efficiency appeared to be close to 100% (Figure S1H), the editing efficiency per SC and the editing events largely varied (Figures S2D and S5G), leading to mosaic mutagenesis. Mosaic editing for some genes may not be sufficient to elicit any phenotypical alterations, in which case the traditional knockout models are needed to confirm the findings.
Another unexpected observation in our study is the incompetence of CRISPR/Cas9 to edit QSCs. The inefficient transduction of AAV virus into the adult SCs was thought to be the reason (Arnett et al., 2014). However, findings from detecting an AAV9-specific element using a sensitive PCR-based method (Figure 3D) demonstrated successful transduction of AAV9 into QSCs, which was substantiated by two recent studies showing high transduction efficiency of multiple AAV serotypes to adult SCs by a Cre/lox fluorescent reporter tracking system (Goldstein et al., 2019; Nance et al., 2019). Still, no obvious editing was achieved after Tmx induction of Cas9 expression (Figure 3E). In a second strategy, we also attempted to inject the AAV9 virus at juvenile stage followed by Cas9 induction after the infected SCs became quiescence (Figure 3G). Augmented transduction efficiency was achieved (Figures 3H and S3E), but the editing remained unsuccessful at all three loci, Myod1 (Figure 3I), Myc (Figures S5I and S5J), and Bcl6 (Figures S6E and S6F). On the Myod1 locus, however, we noticed a weak editing identified by in-depth sequencing when dual sgRNAs were applied (Figure 3O). Since active myogenesis may occur in adult mice to repair subtle injuries routinely happening during normal muscle activity (Ancel et al., 2021), we cannot rule out the possibility that the observed weak editing at the Myod1 locus may happen in activated SCs but not in QSCs. Altogether, our findings suggest insufficient copy of sgRNAs was not the reason for the failed editing. Cas9 inaccessibility to condensed heterochromatin regions may cause failed editing (Doetschman and Georgieva, 2017); however, the sgRNA target sites for both Myod1 (Figure S3O) and Myc (Figure S5K) are located at accessible DNA regions. Another possibility is that the amount of Cas9 in the heterogeneous Pax7ER−Cas9 mice used in our study may not be sufficient to induce the editing, as it has been shown that the concentration of Cas9 is critical for efficient editing (Doetschman and Georgieva, 2017). This puzzle thus remains to be answered in the future.
Lastly, leveraging the system, our study unraveled previously uncharacterized functions of two key TFs, MYC and BLC6, in early stages of SC activation. Loss of MYC led to evident impairment of SC activation, thus delaying acute-injury-induced regenerating process. Taking advantage of the recent developed in situ fixation method (Machado et al., 2017), we conducted RNA-seq and Hi-C analyses in QSCs. Our data for the first time presented a global change of 3D chromatin structure induced by MYC depletion in QSCs. Specifically, at the TAD level, we found the insulation strength of TADs, especially those containing DEGs, was dramatically altered upon MYC depletion. Our findings thus provide new insights into MYC regulation of the 3D genome, which may render its ability to modulate transcriptional program during SC activation (Figure 7). Future efforts will be needed to tease out how MYC orchestrates TAD organization through direct binding to TAD boundaries. Compared with MYC, loss of BLC6, on the other hand, slightly promoted SC activation.
In summary, our study revealed robust genome editing in active juvenile SCs but not QSCs using the Cas9 knockin mouse line in conjunction with AAV9-mediated sgRNA delivery. Importantly, we generated a versatile platform that allows efficient genome modification in endogenous SCs. Further application of this system enabled dissecting the functionality of key TFs in SC activation and muscle regeneration.
Experimental procedures
For further details, see supplemental experimental procedures.
Mice
All animal experiments were performed according to guidelines for experimentation with laboratory animals set in institutions and approved by the Chinese University of Hong Kong (CUHK) Animal Ethics Committee.
SC isolation, culture, and EdU incorporation assay
SCs were isolated from Pax7-nGFP, Pax7Cas9, or Pax7ER−Cas9 mice by the FACS method based on GFP signal as reported previously (Machado et al., 2017).
sgRNA design, selection, and Surveyor nuclease assay
Site-specific sgRNAs were predicted using the Web tool Crispor (http://crispor.tefor.net/). Surveyor nuclease assay was conducted according to the manufacturer's protocol.
AAV9 virus production, purification, and injection
AAV9 virus particles were produced in HEK293FT cells by the triple transfection method.
qRT-PCR
Total RNAs from cells were extracted using TRIzol reagent (Life Technologies) according to the manufacturer's instructions.
RNA-seq and data analysis
RNA-seq was performed as described previously (Zhao et al., 2019).
ChIP-seq and data analysis
ChIP-seq and the data analysis were performed following the procedures described previously (Peng et al., 2017).
Hi-C and data analysis
The in situ Hi-C libraries were prepared as previously reported (Ke et al., 2017).
Deep sequencing
Deep sequencing was performed as previously described (Johansen et al., 2017).
Statistical analysis
All the data are presented as the mean ± standard error of the mean (SD). All tests were two sided, and p < 0.05 was considered statistically significant.
Data and code availability
The accession code of the sequencing data used in this study is Gene Expression Omnibus (GEO): GSE134529.
Author contributions
H.W., H.S., L.H., Y.D., and Y.Z. conceived and designed the experiments. L.H., Y.Z., K.K.S., Y.L., Z.H., and X.C. performed the experiments. Y.D., X.L.P., and J.Y. analyzed the data. H.W. and L.H. wrote the paper. H.W., H.S., L.H., Y.D., and Y.Z. reviewed and edited the manuscript.
conflict of interests
The authors declare no competing interests.
Acknowledgments
This work was supported by General Research Funds (GRF) from the Research Grants Council (RGC) of the Hong Kong Special Administrative Region (14116918, 14120420, and 14120619 to H.S.; 14115319, 14100018, 14100620, 14106117 and 14106521 to H.W.); the National Natural Science Foundation of China (NSFC) to H.W. (Project code: 31871304); Collaborative Research Fund (CRF) from RGC to H.W. (C6018-19GF); NSFC/RGC Joint Research Scheme to H.S. (Project code: N_CUHK 413/18); Hong Kong Epigenomics Project (EpiHK) Fund to H.W. and H.S.; Area of Excellence Scheme (AoE) from RGC (Project number: AoE/M-402/20).
Published: September 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.08.011.
Contributor Information
Hao Sun, Email: haosun@cuhk.edu.hk.
Huating Wang, Email: huating.wang@cuhk.edu.hk.
Supplemental information
References
- Aloisio G.M., Nakada Y., Saatcioglu H.D., Peña C.G., Baker M.D., Tarnawa E.D., Mukherjee J., Manjunath H., Bugde A., Sengupta A.L. PAX7 expression defines germline stem cells in the adult testis. J. Clin. Invest. 2014;124:3929–3944. doi: 10.1172/JCI75943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancel S., Stuelsatz P., Feige J.N. Muscle stem cell quiescence: controlling stemness by staying asleep. Trends Cell Biol. 2021;31:556–568. doi: 10.1016/j.tcb.2021.02.006. [DOI] [PubMed] [Google Scholar]
- Arnett A.L., Konieczny P., Ramos J.N., Hall J., Odom G., Yablonka-Reuveni Z., Chamberlain J.R., Chamberlain J.S. Adeno-associated viral vectors do not efficiently target muscle satellite cells. Mol. Ther. Methods Clin. Dev. 2014;1:14038. doi: 10.1038/mtm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzi M., Crouch D.H., Tatò F. Transformation by myc prevents fusion but not biochemical differentiation of C2C12 myoblasts: mechanisms of phenotypic correction in mixed culture with normal cells. J. Cell Biol. 1994;125:1137–1145. doi: 10.1083/jcb.125.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T., Georgieva T. Gene editing with CRISPR/Cas9 RNA-directed nuclease. Circ. Res. 2017;120:876–894. doi: 10.1161/CIRCRESAHA.116.309727. [DOI] [PubMed] [Google Scholar]
- Goldstein J.M., Tabebordbar M., Zhu K., Wang L.D., Messemer K.A., Peacker B., Kakhki S.A., Gonzalez-Celeiro M., Shwartz Y., Cheng J.K. In situ modification of tissue stem and progenitor cell genomes. Cell Rep. 2019;27:1254–1264.e7. doi: 10.1016/j.celrep.2019.03.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., VanDusen N.J., Zhang L., Gu W., Sethi I., Guatimosim S., Ma Q., Jardin B.D., Ai Y., Zhang D. Analysis of cardiac myocyte maturation using CASAAV, a platform for rapid dissection of cardiac myocyte gene function in vivo. Circ. Res. 2017;120:1874–1888. doi: 10.1161/CIRCRESAHA.116.310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoyama T., Nishijo K., Garcia M.M., Schaffer B.S., Ohshima-Hosoyama S., Prajapati S.I., Davis M.D., Grant W.F., Scheithauer B.W., Marks D.L. A postnatal Pax7+ progenitor gives rise to pituitary adenomas. Genes Cancer. 2010;1:388–402. doi: 10.1177/1947601910370979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen A.K., Molenaar B., Versteeg D., Leitoguinho A.R., Demkes C., Spanjaard B., de Ruiter H., Akbari Moqadam F., Kooijman L., Zentilin L. Postnatal cardiac gene editing using CRISPR/Cas9 with AAV9-mediated delivery of short guide RNAs results in mosaic gene disruption. Circ. Res. 2017;121:1168–1181. doi: 10.1161/CIRCRESAHA.116.310370. [DOI] [PubMed] [Google Scholar]
- Ke Y., Xu Y., Chen X., Feng S., Liu Z., Sun Y., Yao X., Li F., Zhu W., Gao L. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell. 2017;170:367–381.e20. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- Kieffer-Kwon K.-R., Nimura K., Rao S.S.P., Xu J., Jung S., Pekowska A., Dose M., Stevens E., Mathe E., Dong P. Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol. Cell. 2017;67:566–578.e10. doi: 10.1016/j.molcel.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress T.R., Sabò A., Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- Kumagai T., Miki T., Kikuchi M., Fukuda T., Miyasaka N., Kamiyama R., Hirosawa S. The proto-oncogene Bcl6 inhibits apoptotic cell death in differentiation-induced mouse myogenic cells. Oncogene. 1999;18:467–475. doi: 10.1038/sj.onc.1202306. [DOI] [PubMed] [Google Scholar]
- Lala-Tabbert N., AlSudais H., Marchildon F., Fu D., Wiper-Bergeron N. CCAAT/enhancer binding protein β promotes muscle stem cell quiescence through regulation of quiescence-associated genes. Stem Cells. 2020;39:345–357. doi: 10.1002/stem.3319. [DOI] [PubMed] [Google Scholar]
- Machado L., de Lima J.E., Fabre O., Proux C., Legendre R., Szegedi A., Varet H., Ingerslev L.R., Barrès R., Relaix F. In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 2017;21:1982–1993. doi: 10.1016/j.celrep.2017.10.080. [DOI] [PubMed] [Google Scholar]
- Nance M.E., Shi R., Hakim C.H., Wasala N.B., Yue Y., Pan X., Zhang T., Robinson C.A., Duan S.X., Yao G. AAV9 edits muscle stem cells in normal and dystrophic adult mice. Mol. Ther. 2019;27:1568–1585. doi: 10.1016/j.ymthe.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P.D., Gurevich D.B., Sonntag C., Hersey L., Alaei S., Nim H.T., Siegel A., Hall T.E., Rossello F.J., Boyd S.E. Muscle stem cells undergo extensive clonal drift during tissue growth via Meox1-mediated induction of G2 cell-cycle arrest. Cell Stem Cell. 2017;21:107–119.e6. doi: 10.1016/j.stem.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Peng X.L., So K.K., He L., Zhao Y., Zhou J., Li Y., Yao M., Xu B., Zhang S., Yao H. MyoD-and FoxO3-mediated hotspot interaction orchestrates super-enhancer activity during myogenic differentiation. Nucleic Acids Res. 2017;45:8785–8805. doi: 10.1093/nar/gkx488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J.T., King K.Y., Brett J.O., Cromie M.J., Charville G.W., Maguire K.K., Brunson C., Mastey N., Liu L., Tsai C.-R. mTORC1 controls the adaptive transition of quiescent stem cells from G 0 to G Alert. Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-André V., Federation A.J., Lin C.Y., Abraham B.J., Reddy J., Lee T.I., Bradner J.E., Young R.A. Models of human core transcriptional regulatory circuitries. Genome Res. 2016;26:385–396. doi: 10.1101/gr.197590.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits A.H., Ziebell F., Joberty G., Zinn N., Mueller W.F., Clauder-Münster S., Eberhard D., Savitski M.F., Grandi P., Jakob P. Biological plasticity rescues target activity in CRISPR knock outs. Nat. Methods. 2019;16:1087–1093. doi: 10.1038/s41592-019-0614-5. [DOI] [PubMed] [Google Scholar]
- Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. J. Intern. Med. 2009;266:372–389. doi: 10.1111/j.1365-2796.2009.02158.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Legendre N.P., Biswas A.A., Lawton A., Yamamoto S., Tajbakhsh S., Kardon G., Goldhamer D.J. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Reports. 2018;10:956–969. doi: 10.1016/j.stemcr.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kubota M., Nakamura A., Kaji T., Seno S., Uezumi A., Andersen D.C., Jensen C.H., Fukada S. Dlk1 regulates quiescence in calcitonin receptor-mutant muscle stem cells. Stem Cells. 2021;39:306–317. doi: 10.1002/stem.3312. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhou J., He L., Li Y., Yuan J., Sun K., Chen X., Bao X., Esteban M.A., Sun H. MyoD induced enhancer RNA interacts with hnRNPL to activate target gene transcription during myogenic differentiation. Nat. Commun. 2019;10:5787. doi: 10.1038/s41467-019-13598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession code of the sequencing data used in this study is Gene Expression Omnibus (GEO): GSE134529.