Summary
We here demonstrate that microsatellite (MS) alterations are elevated in both mouse and human induced pluripotent stem cells (iPSCs), but importantly we have now identified a type of human iPSC in which these alterations are considerably reduced. We aimed in our present analyses to profile the InDels in iPSC/ntESC genomes, especially in MS regions. To detect somatic de novo mutations in particular, we generated 13 independent reprogramed stem cell lines (11 iPSC and 2 ntESC lines) from an identical parent somatic cell fraction of a C57BL/6 mouse. By using this cell set with an identical genetic background, we could comprehensively detect clone-specific alterations and, importantly, experimentally validate them. The effectiveness of employing sister clones for detecting somatic de novo mutations was thereby demonstrated. We then successfully applied this approach to human iPSCs. Our results require further careful genomic analysis but make an important inroad into solving the issue of genome abnormalities in iPSCs.
Keywords: microsatellite alterations, InDels, mouse iPSCs, mouse ntESCs, human iPSCs, genome reprogramming, whole-genome sequencing, sister clones set, hotspots of microsatellite alteration, cord blood erythroblasts
Graphical abstract

Highlights
-
•
InDels and microsatellite alterations are elevated in iPSCs
-
•
These alterations are reduced in human iPSCs derived from cord blood erythroblasts
-
•
Employing sister clones is an effective way to detect somatic de novo mutations
In this article, Abe and colleagues conclusively demonstrate that InDels and microsatellite alterations are elevated in reprogrammed pluripotent stem cells, both mouse and human, by employing sister clones and conducting large-scale validation experiments. Furthermore, they show that these alterations are considerably reduced in human iPSCs derived from cord blood erythroblasts.
Introduction
Induced pluripotent stem cells (iPSCs) hold great promise for regenerative medicine (Takahashi and Yamanaka, 2006). However, the genetic aberrations observed in these cells remain a major impediment to their medical application due to possible immunogenic and/or tumorigenic effects (Araki et al., 2013; Gore et al., 2011; Liang and Zhang, 2013). Thus far, a significant number of point mutations have been revealed in iPSC genomes via genome-wide analyses of these stem cells (Bhutani et al., 2016; Cheng et al., 2012; Gore et al., 2011; Ji et al., 2012; Rouhani et al., 2016; Sugiura et al., 2014; Young et al., 2012). Notably, however, the overall picture of structural variants, such as insertions/deletions (InDels) and translocations in iPSC genomes still remains elusive, despite the magnitude of their biological impacts.
In previous studies of the coding regions, i.e., exons in human iPSC lines, several InDels at one to two locations in these regions were described (Cheng et al., 2012; Mandai et al., 2017). On the other hand, Young et al. (2012) noted the technical limitations of comprehensive InDel detection through their three genome-wide experiments. Because these authors observed too many false positives in two out of the three experiments, they did not conduct validation after the calling of several hundred InDel candidates through their informatics analyses. In the remaining one experiment, however, they reported that only 32 InDels among the several hundred candidates called by the informatics and expressed confidence that they were not false positives as they were also detectable in the sister iPSC clones. Those results indicated that almost all of the signals called by the InDel detection system were false positives and that the small population in the candidates that were considered to be true signals were pre-existing and not de novo. Notably, a more recent informatics study utilizing whole-genome sequencing (WGS) suggested that substantial numbers of InDel candidates were present in human iPSC genomes, 100–300 sites per genome, although validation tests were not performed (Bhutani et al., 2016).
In our current study, we attempted to clarify the somatic de novo InDel profile in reprogrammed PSCs. To obtain precise and comprehensive data in this regard, we employed an inbred mouse strain and, most importantly, created an “ideal” set of cells including more than 10 sister clones generated from the same MEFs. In addition, we refined our informatics approaches to this InDel analysis. We demonstrate that we can now detect InDels (from short to middle InDels) with few false positives not only in non-repetitive regions but also in the short tandem repeat (STR) regions known as microsatellites (MSs).
Results
A newly constructed system for detecting de novo InDels with few false positives
Because of the limited accuracy of current InDel detection methods, the status of these aberrations in reprogrammed stem cells has remained elusive to date. Indeed, in our previous study on human iPSCs, a substantial number of false-positive InDel signals were observed and only 6.6% of the candidates turned out to be true signals (Araki et al., 2020). We therefore here attempted to establish a reliable genome-wide somatic de novo InDel investigation system for analyzing reprogrammed PSCs, such as iPSCs and nuclear-transferred (nt) embryonic stem cells (ESCs). To this end, we focused on informatics and the materials to be analyzed, i.e., the target and control cells.
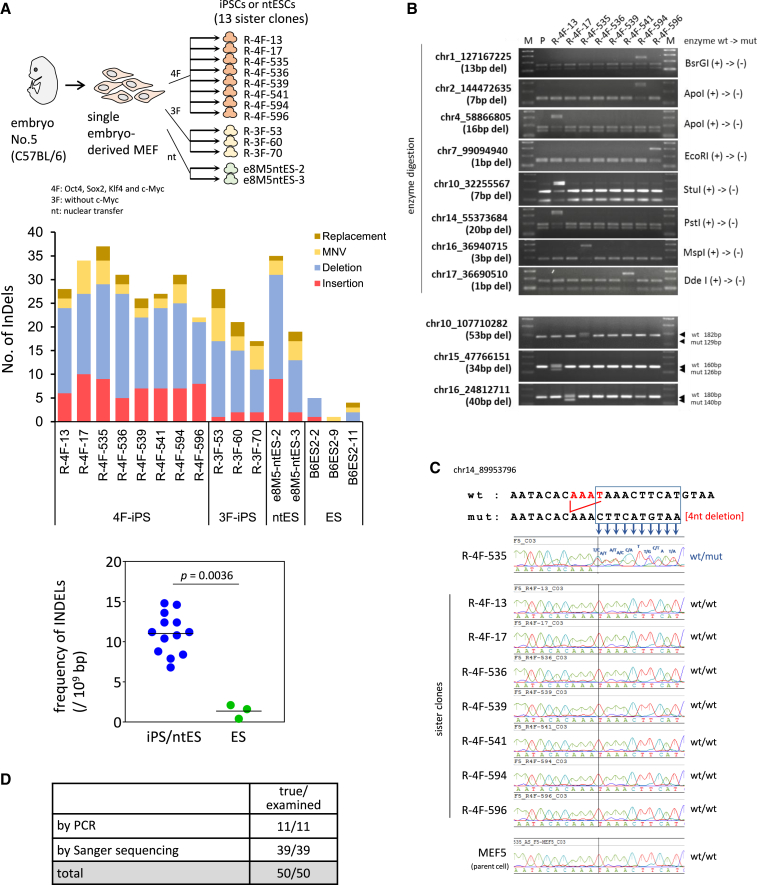
To first optimize the informatics processes for WGS data we designed 179 kinds of model InDels, i.e., from 1 to 70 bp insertions or deletions at 179 genome regions and obtained 4,468 model reads in total using BAMSurgeon software (Ewing et al., 2015) (Figure S1A; Data S1). In addition, we employed eight sister iPSC clones, R-4F-iPSCs, generated from a single embryo of an inbred mouse strain, C57BL/6, to remove pre-existing aberrations more efficiently (Figure 1A).
Figure 1.
InDels in iPSCs, ntESCs, and ESCs
(A) InDels arise frequently in reprogrammed PSCs. Upper: cells used for the mouse InDel analysis. A mouse inbred strain and sister iPSC and ntESC lines generated from single embryo-derived MEFs were used in this study. Middle: the definitive number of each type of InDels was determined after manual inspection. “Replacement” denotes an event where one or more bases have been replaced and where the identified allele has a length different from the reference, e.g., “TTT” to “GG”. MNV (multiple nucleotide variant) indicates that the reference and alternate sequences are of the same length and have to be greater than 1 and that all nucleotides in the sequences differ from one another, e.g., “GC” to “TA” (http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/700/Variant_types.html, https://m.ensembl.org/info/genome/variation/prediction/classification.html). Lower: statistically significant differences found in the number of InDels between reprogrammed and non-reprogrammed PSCs. The Mann-Whitney U test was employed for this statistical analysis.
(B) Validation assessments by restriction enzyme digestion or amplicon length. Validation of the deletions by digestion with a restriction enzyme or by length determination are shown. Eight sister clones were used. M, molecular weight markers; P, parental cells (MEF5).
(C) A representative case validated by Sanger sequencing. The sequences of the mutated clone, R-4F-535, and eight controls (remaining seven sister clones and parental MEFs) are shown.
(D) Summary of the validation tests. PCR tests indicate validation by digestion with a restriction enzyme or by length determination by electrophoresis, respectively. In the Sanger sequencing, InDels were verified basically in both the forward and reverse directions.
See also Figures S1 and S2, Tables S1 and S2, Data S2 and S3.
Using these experimental materials, which were ideally suited for the informatics analysis of WGS data due to their identical genetic backgrounds, we could optimize each informatics step, as denoted by the red characters in Figure S1B. Two types of algorithms were employed, one for short and one for medium InDel detection. We could thereby successfully identify InDels with high fidelity and high coverage (97.2%). The efficacy of each step is shown in Figure S1C.
In brief, we designed model InDel reads based on some of the WGS reads of the MEF5 (mouse embryonic fibroblast 5) cells that had been used for iPSC/ntESC generation and mixed them back into the remaining large pool of WGS reads for these cells in silico (referred to as “MEF5+mInDels”) for subsequent informatics analysis. We conducted a direct comparison between MEF5+mInDels and MEF5 itself also, and then detected only model InDels with high coverage, 97.2%, and extremely high fidelity (Figure S1C). On the other hand, five false negatives (179 [input] – 174 [called]) appeared, comprising one deletion and four insertions. The 60 bp deletion in this group can be explained by the fact that it was filtered out at step 5 because the read depth exceeded “100”. In the case of the four insertions, a 60 bp and a 70 bp insertion were also filtered out at step 5 because the number of variant reads were less than the cutoff value of 10, and the remaining two, both 70 bp, could not be called because of the absence of reads that could cover the breakpoints and the entire regions of the insertions. As the length of the inserted sequence becomes longer, the number of mappable reads gradually decreases, resulting in a decreased detection sensitivity (Figure S1D).
Accordingly, our pilot analysis using model InDels demonstrated that our mouse analyzing system enables us to identify these aberrations with high accuracy and coverage over the 1–70 bp range (Figure S1D).
Considerable numbers of InDels are present in mouse reprogrammed PSCs
Using our new detection approach, we analyzed 13 reprogrammed PSC lines, iPSCs/ntESCs, and an ESC control. We examined eight iPSCs generated by four factors, Oct4, Sox2, Klf4, and c-Myc (4F-iPSCs), which were utilized for the abovementioned informatics optimization, three iPSCs generated with only three factors without c-Myc, two ntESCs, and three ESC lines (Table S1). More importantly, to obtain conclusive results on the InDel profile in reprogrammed PSCs, we employed an inbred mouse strain instead of human cells and analyzed a set of sister clones, all of which were established from identical single embryo-derived MEFs (Figure 1A, upper) (Araki et al., 2020). As anticipated, our analysis showed a high fidelity, as 87.6% of our candidates were indicated to be true by manual inspection, and we detected a considerable number of InDels in reprogrammed PSC lines compared with ESCs (Figure 1A, middle and lower; Data S2).
Experimental validation of the InDel candidates called by the informatics
Although our pilot experiments employing model WGS reads demonstrated the high fidelity of our method, we also conducted a validation test of our InDel candidates because we still speculated whether the InDels detected by our actual analysis of iPSCs and ntESCs were true results. We randomly chose 56 of our InDel candidates for this evaluation for which conclusive results could be obtained for 50. We could not successfully design effective PCR primers for the remaining six candidates. Eight candidates harbored recognition sites for restriction enzymes within the corresponding regions of their parent wild alleles. A non-digested band will therefore be observed for the mutant allele, resulting in a heterozygous, non-digested, and digested pattern for each candidate (Figure 1B, upper). Furthermore, the length of the amplicons could be readily distinguished between wild and mutant by electrophoresis for three candidates (Figure 1B, lower). Sanger sequencing verified the 39 candidates (Figure 1C; Data S3). In all cases, no InDel was detectable in seven sister clones, in addition to the control parent somatic fibroblasts (MEF5). Thus, the InDels identified by our analysis were definitively clone specific (Figure 1D). Notably also, from ultra-deep sequencing of the iPSCs in which the candidates were identified, parent cells (MEF5) and ESCs (B6ES2-2), derived previously from another mouse individual (Sugiura et al., 2014) for the randomly chosen 23 candidates, strongly suggested that 22 out of the 23 were not pre-existing InDels (Figure S2A; Table S2).
Our analysis concluded that iPSC and ntESC genomes harbor a significant number of InDels at a much higher frequency than ESCs. Deletions thus occur often in reprogrammed PSCs. In addition, considerable numbers of “replacement” and/or “MNVs” were also observed in most lines (Figure 1A, middle). The distribution of the InDels is shown in Figure S2B. InDel length histograms are also shown in Figure S2C.
Intriguingly, several InDels were found within coding regions: five in 4F-iPSCs, one in 3F-iPSCs, and one in ntESCs (Figure S2D). Furthermore, three InDels were detected in UTRs. Four out of the seven InDels identified in coding regions caused a frameshift that resulted in a truncated product, and the remaining three InDels caused missense amino acid substitutions (Figure S2E). Notably, all of these seven InDels were experimentally verified by Sanger sequencings (Data S3). Our present results thus suggested that the frequency of InDels in coding regions including UTRs is less than one per line for mouse reprogrammed PSCs. In addition, it is noteworthy that possible tumorigenic implications have been discussed previously for the Znrf3 and Smug1 genes (Assie et al., 2014; Kemmerich et al., 2012; Wang et al., 2013). Smug1in particular encodes a DNA glycosylase and is a DNA repair gene. Its loss of expression has been reported to cause phenotypic abnormalities (Abdel-Fatah et al., 2013), suggesting that a heterozygous loss of this gene could affect genomic stability.
MS alterations caused by InDels in mouse iPSCs and ntESCs
There has been no effective way to comprehensively analyze InDels that have arisen in STR regions due to limitations in precise mapping/alignment of WGS reads over the reference genome sequence. However, with the improvements in InDel detection procedures we observed in reprogrammed PSCs that many occur in STR regions known as MSs, although these were beyond the scope of our first InDel detection analysis. We thus sought to develop a method of analyzing such MS alterations.
A representative InDel identified by our system to occur in STR sequences is a “TATA” sequence inserted into only one allele, resulting in a heterozygous “TA” × 7/”TA” × 8 (Figure 2A). Our informatics identified a considerable number of InDel candidates in STR regions in iPSC/ntESC genomes and we evaluated these via manual inspection, which has strong experimental reliability, as mentioned below. To achieve a robust validation, we randomly picked approximately 25% of such candidates from every chromosome for all cell lines. We finally evaluated 1,131 MS alteration candidates (26.9% of the total) and calculated the true rate for each cell clone (Figure S3A). We successfully estimated the frequency of MS alterations in the entire genome based on these true rate values. In conclusion, MS alterations are sharply elevated in reprogrammed PSCs, iPSCs, and ntESCs, compared with ESCs (Figure 2B).
Figure 2.
Microsatellite alterations in mouse iPSCs/ntESCs
(A) Identification of the InDels in tandem repeat regions. Candidates for microsatellite (MS) alterations were obtained from those removed at step 6 in the screening of InDels in non-repetitive regions shown in Figure S1B. A representative instance of MS alteration is also shown, which was detected in iPSC clone R-4F-17, chr6_ 26,444,460.
(B) Statistically significant differences in the number of MS alterations between reprogrammed and non-reprogrammed pluripotent mouse stem cells. The estimated numbers of MS alterations were based on the number of candidates detected by the newly developed investigation system in this study. The true rates are shown in Figure S3A. The Mann-Whitney U test was employed for these statistical analyses.
(C) Results of quantitative amplicon sequencing. Parent somatic MEF5 cells and B6ES2-2 cells were used as negative controls. In addition, seven sister clones of the iPSC line were also used as negative controls for each candidate. Red bars indicate altered MS regions and blue bars indicate wild MSs. The red bar indicates clear clone-specific phenomena, and it was notable also that the background signals (gray bar) appeared identical among the negative controls including the sister clones. The total number of the reads obtained by amplicon sequencing is indicated at the top left of each graph (k = 1,000 reads, e.g., 621k means 621,000 reads). Representative examples are shown. All of the remaining results are shown in Data S4.
(D) Summary of the validation testing. We designed PCR primers for 54 candidate InDels at MS regions for the validation, but only 29 were analyzable. The presence of all of these 29 candidates was clearly confirmed by amplicon sequencing.
See also Figures S3 and S5, Table S1 and Data S2, S3, and S4.
Experimental validation of MS alterations through the amplicon sequencing of sister clones enables clone-specific alterations to be distinguished from background signals
For MS alteration analysis in particular, validation of the candidates called by our informatics using additional wet experimental systems is essential because a significant number of false positives are frequently observed. Furthermore, it must be noted that similar alterations also readily and frequently occur in STR regions via in vitro DNA polymerization during the detection process. For this reason, various alterations are detected in negative control cells at various intensities, which can lead to many true signals being overlooked. To overcome this particular difficulty in analyzing repetitive sequence regions, we performed a quantitative comparison with background signals that were reproducibly observed in all control cells and sister clones with the same genetic background, in addition to using a PCR-free library preparation kit for WGS to minimize this problem associated with DNA polymerization.
In these evaluations, we designed PCR primers for 54 randomly chosen candidates in STR (repeat unit: ≥ 2 bp) regions. In general, not only the design of the unique primers but also the subsequent PCR amplification on STR regions are often quite difficult compared with non-repetitive sequence regions. Comprehensive validation testing of genetic aberrations in repetitive sequencing regions, such as satellites, minisatellites, and MSs, is thus problematic. Indeed, only 31 regions out of the 54 candidates could be amplified, and then only 29 of these 31 could be finally analyzed because the sequencing reaction did not work well for the other 2 candidates. To determine whether these candidates were clone-specific alterations, we employed a panel of 10 cell types: MEF5 (parent somatic cell fraction), B6ES2-2 (the gold standard of PSCs generated from unrelated mouse individuals), and 8 sister iPSC clones including one sample iPSC line in which alterations were suggested for each candidate. We thereby successfully observed abnormal MSs in a clone-specific manner (red) and, more importantly, also clearly observed identical background signal patterns (gray) due to in vitro DNA polymerization in all of the cell lines except for the mutated clone (Figure 2C; Data S4). Further, we observed that these background signals are dependent on the type of DNA polymerase used; the pattern of background signals drastically changes depending on the type of DNA polymerase, although it was identical among the control MEF5 and ESCs (Figure S3B). As a result, alterations called by informatics were confirmed to be true and clone specific by amplicon sequencing for all of these 29 candidates (Figure 2D). We also conducted Sanger sequencing to confirm the sequence structure of the InDels for five cases, which were randomly chosen (Data S3).
In addition, it was statistically shown that none of the 29 candidate MS alterations examined by amplicon sequencing, without exception, were present in their parental cells, or were pre-existing (Data S4). We have thus succeeded in comprehensively identifying somatic de novo MS alterations for the first time by employing a set of control cell lines derived from a single embryo of a mouse inbred strain, which thus had an identical genetic background, and an improved InDel analysis system.
MS alterations in various types of human iPSCs—A lower frequency is found in CB erythroblast-derived integration-free lines
Using HipSTR, we examined 34 human iPSC lines for MS alterations comprising 14 integration-free iPSC lines derived from cord blood erythroblasts (CB-epi iPSCs), 5 integration-free iPSCs from dermal fibroblasts (E-HDFa iPSCs), 3 retrovirus-mediated iPSC lines from dermal fibroblasts (R-HDFa iPSCs), 2 Sendai virus-mediated iPSCs from BJ cells (SeV-BJ iPSCs), and 10 Sendai virus-mediated iPSCs from skin tissue (HipSci Database iPSCs). Of these, the former four types of human iPSCs, CB-epi, E-HDFa (Figure S4), R-HDFa, and SeV-BJ iPSCs were established in our laboratory and were subjected to WGS (Figure 3A; Table S1). On the other hand, the remaining 10 cell lines (HipSci Database iPSCs, https://www.hipsci.org/) were previously published together with their sister clones on a public database (2 iPSC clones/individual × 5 individuals). These were examined to confirm the results we obtained for the iPSCs. We could, however, only conduct experimental validation on our own in-house iPSCs and not on the HipSci Database iPSCs. The frequencies of STR alterations (repeat unit: ≥ 2 bp) and homopolymer alterations in these iPSCs are shown (Figure 3B). There are statistically significant differences between CB-epi-iPSCs and E-HDFa iPSCs/R-HDFa iPSCs/HipSci Database iPSCs. Meanwhile, we could not draw any statistical conclusions in relation to SeV-BJ iPSCs due to only having two samples, although there was a trend toward a difference in this case.
Figure 3.
MS alterations in human iPSCs
(A) Schematic depiction of the sister clones used in the analysis. R, retrovirus system; HDF, human dermal fibroblasts; SeV, Sendai virus system; BJ, human BJ cells; CB, cord blood; epi, episomal vector system. ∗Although we conducted MS alteration analysis on three iPSC lines generated using the Sendai virus system, we could obtain InDel data from only two of them as the remaining line was strongly suggested to have arisen from a fusion of two iPSC clones. This third line was still useful as a control for any false signals called in the other two lines.
(B) Statistically significant differences in the MS alterations between CB erythroblast-derived and other human iPSCs. Frequency of STR alterations (repeat unit: ≥ 2 bp) and homopolymer alterations are shown. For STR alterations, we conducted manual inspections on all candidates and obtained final results (left). Meanwhile, on homopolymers we evaluated 179 alteration candidates, which were randomly picked, also by manual inspection, and revealed that 164 out of the 179 candidates were true (true rate: 91.6%). Therefore, here the number per clone corrected by the positive rate is shown (right). The Mann-Whitney U test was employed for the statistical analysis.
(C) Results of quantitative amplicon sequencing. Parent somatic cell line, CB-1, was used. In addition, four sister clones of the iPSC line were also used as negative controls for each candidate. The details of these graphs in terms of the bar colors are equivalent to Figure 2C. All of the remaining results are shown in Data S4.
(D) Summary of the validation testing. PCR primers could be designed for 72 candidate InDels at STR regions, but only 32 were analyzable. The presence of all of these 32 candidates was clearly confirmed by amplicon sequencing.
See also Figures S4–S6, Table S1, Data S3, S4, and S6.
Experimental validations were conducted on the candidates called by informatics and 32 sites (unit: ≥ 2 bp) could be amplified from a randomly chosen panel of 134 candidates. Subsequent next-generation sequencing (NGS) of these amplicons for the sample iPSC clone and negative controls, including sister clones and parent cells, clearly showed all of them to be true and non-preexisting in their parent cells (Figures 3C and 3D; Data S4).
Frequently altered MS regions containing more than one type of InDel
A number of MS alterations were observed in iPSCs and ntESCs compared with ESCs, but little or no genome-wide regional bias was observed (Figure 4A). We thus assessed the profile of the alterations, which were verified by manual inspection. A maximum of 9 di-nucleotide repeats were deleted (18 bp) and 4 di-nucleotide sequence insertions (8 bp) were observed in mouse iPSCs (Figure 4B). We detected 11 di-nucleotide repeat deletions (22 bp) and 6 di-nucleotide repeat insertions (12 bp) in human iPSCs (Figure 4C). In ntESCs, a maximum deletion of 2 di-nucleotide repeats and insertion of 3 di-nucleotide repeats were observed. Notably, both insertions and deletions frequently occurred in mouse iPSCs, but insertions predominantly occurred in ntESCs in which deletions were not so remarkable. In contrast, deletions were also not very remarkable in most of the human iPSC lines we tested.
Figure 4.
Length profile of MS alterations
(A) Distribution of MS alterations throughout the genome.
(B) Length profile of the MS alterations (mouse). The results of eight 4F-iPSC lines, two ntESC lines, and three ESC lines are summarized.
(C) Length profile of the MS alterations (human). The results of 14 CB-iPSCs, 8 HDF iPSCs (3 retro-mediated and 5 episomal-mediated), 2 BJ iPSCs, and 10 fibroblast (HipSci) iPSCs are summarized.
See also Data S2.
An important observation was that more than one InDel mutation pattern, i.e., frequent alteration, were detected for 18 MSs in MEF5-derived 10 iPSC/ntESCs (Table 1). Intriguingly, two different types of alterations were observed in an identical MS region among sister lines. These results suggested the existence of alteration-prone MS regions, i.e., hotspots. To further investigate this, we also analyzed ntESCs established from the tail tip fibroblasts (TTFs) of independent individual mice (Araki et al., 2017) and successfully identified different alterations in six STR regions in which MS alterations were identified in iPSCs (Data S5A). Thus, we also detected different alterations in an identical STR region among reprogrammed PSCs derived from a different type of parent somatic cells prepared from independent individual mice. Furthermore, also in human iPSCs, 13 sites of STR (Table 2) and 113 sites of homopolymer were identified as alteration-prone regions (Table 3; Data S5B).
Table 1.
Short tandem repeat regions, in which alterations occurred in multiple mouse iPSC/ntESC clones
| Chromosome_position | Region | Parenta wild/wild |
Mutant clonea | wild/mutantb | Δ | NGS data |
||
|---|---|---|---|---|---|---|---|---|
| Mutant reads |
Read depth |
Frequency (%) | ||||||
| chr1_31764385 | intergenic | (AG)28/28 | R-4F-535 | (AG)28/29 | +1 | 10 | 28 | 35.7 |
| R-3F-70 | (AG)28/27 | −1 | 11 | 40 | 27.5 | |||
| chr1_83786319 | intergenic | (AT)17/17 | R-3F-70 | (AT)17/18 | +1 | 14 | 43 | 32.6 |
| R-3F-53 | (AT)17/14 | −3 | 16 | 42 | 38.1 | |||
| chr1_102192243 | intron | (TC)26/26 | R-4F-536 | (TC)26/27 | +1 | 11 | 33 | 33.3 |
| R-4F-596 | (TC)26/25 | −1 | 10 | 35 | 28.6 | |||
| chr1_178540376 | intergenic | (TA)19/19 | R-4F-17 | (TA)19/20 | +1 | 10 | 29 | 34.5 |
| R-3F-60 | (TA)19/18 | −1 | 18 | 44 | 40.9 | |||
| chr3_145337695 | intergenic | (CA)18/18 | R-4F-17 | (CA)18/19 | +1 | 14 | 50 | 28.0 |
| R-4F-536 | (CA)18/17 | −1 | 15 | 37 | 40.5 | |||
| chr4_93696075 | intergenic | (AT)17/17 | R-4F-535 | (AT)17/18 | +1 | 26 | 52 | 50.0 |
| R-4F-594 | (AT)17/16 | −1 | 13 | 31 | 41.9 | |||
| chr4_111930759 | intron | (GT)21/21 | e8M5-ntES-2 | (GT)21/23 | +2 | 13 | 33 | 39.4 |
| R-4F-539 | (GT)21/20 | −1 | 12 | 27 | 44.4 | |||
| chr5_9557538 | intron | (TG)22/22 | e8M5-ntES-2 | (TG)22/21 | −1 | 12 | 34 | 35.3 |
| R-4F-536 | (TG)22/20 | −2 | 14 | 42 | 33.3 | |||
| chr5_111380751 | intron | (AT)20/20 | R-4F-596 | (AT)20/22 | +2 | 11 | 34 | 32.4 |
| R-4F-594 | (AT)20/19 | −1 | 11 | 39 | 28.2 | |||
| chr6_43702707 | intergenic | (TA)15/15 | R-4F-539 | (TA)15/16 | +1 | 12 | 37 | 32.4 |
| R-3F-53 | (TA)15/14 | −1 | 16 | 44 | 36.4 | |||
| chr8_130621418 | intergenic | (CA)20/20 | R-3F-60 | (CA)20/21 | +1 | 11 | 49 | 22.4 |
| R-4F-596 | (CA)20/17 | −3 | 16 | 43 | 37.2 | |||
| chr9_112229669 | intron | (TA)20/20 | R-4F-541 | (TA)20/21 | +1 | 19 | 32 | 59.4 |
| R-4F-13 | (TA)20/19 | −1 | 11 | 40 | 27.5 | |||
| chr11_82854381 | intergenic | (ATCT)12/12 | R-4F-536 | (ATCT)12/11 | −1 | 12 | 50 | 24.0 |
| R-4F-539 | (ATCT)12/10 | −2 | 15 | 65 | 23.1 | |||
| chr13_64860070 | intron | (AC)24/24 | R-4F-13 | (AC)24/25 | +1 | 16 | 44 | 36.4 |
| R-4F-596 | (AC)24/23 | −1 | 10 | 40 | 25.0 | |||
| chr14_6291411 | intron | (AC)24/24 | R-4F-13 | (AC)24/25 | +1 | 10 | 34 | 29.4 |
| R-4F-536 | (AC)24/21 | −3 | 24 | 37 | 64.9 | |||
| chr15_61099178 | intergenic | (GA)26/26 | R-4F-17 | (GA)26/24 | −2 | 10 | 26 | 38.5 |
| R-3F-53 | (GA)26/23 | −3 | 12 | 51 | 23.5 | |||
| chr16_88314677 | intergenic | (AC)13/13-(AG)19/19 | R-4F-539 | (AC)13/12-(AG)19/19 | −1 | 14 | 41 | 34.1 |
| R-4F-596 | (AC)13/13-(AG)19/14 | −5 | 11 | 24 | 45.8 | |||
| chr17_53143305 | intergenic | (AG)21/21 | R-4F-594 | (AG)21/22 | +1 | 21 | 52 | 40.4 |
| R-3F-70 | (AG)21/20 | −1 | 16 | 47 | 34.0 | |||
Table 2.
Short tandem repeat regions, in which alterations occurred in multiple human iPSC clones
| Chromosome_position | Region | Parenta wild/wild |
Mutant clonea | wild/mutantb | Δ | NGS data |
||
|---|---|---|---|---|---|---|---|---|
| Mutant reads |
Read depth |
Frequency (%) | ||||||
| chr9_29179157 | intron | (AT)15/15 | CB-epi-2-1-4 | (AT)15/16 | +1 | 12 | 27 | 44.4 |
| (AT)20/17 | CB-epi-2-3-4 | (AT)20/18 | +1 | 8 | 15 | 53.3 | ||
| chr12_23285465 | intergenic | (ATCT)12/14 | CB-epi-2-3-1 | (ATCT)12/13 | −1 | 6 | 17 | 35.3 |
| (ATCT)12/14 | CB-epi-2-3-4 | (ATCT)12/15 | +1 | 10 | 21 | 47.6 | ||
| chr3_143951213 | intergenic | (AT)22/24 | CB-epi-1-1 | (AT)22/26 | +2 | 4 | 13 | 30.8 |
| (AT)20/22 | E-HDFa-12-3 | (AT)20/23 | +1 | 8 | 15 | 53.3 | ||
| chr2_161668575 | intergenic | (AT)22/20 | CB-epi-2-2-4 | (AT)22/21 | +1 | 9 | 16 | 56.3 |
| (AT)23/23 | HPSI0614i-voce_1 | (AT)23/24 | +1 | 4 | 11 | 36.4 | ||
| chr2_162801693 | intron | (AT)15/22 | CB-epi-1-5 | (AT)15/23 | +1 | 8 | 18 | 44.4 |
| (AT)24/24 | HPSI0115i-iuad_2 | (AT)24/25 | +1 | 3 | 11 | 27.3 | ||
| chr3_191830148 | intergenic | (AT)17/21 | CB-epi-2-2-3 | (AT)17/20 | −1 | 4 | 11 | 36.4 |
| (AT)16/21 | HPSI0614i-uevq_4 | (AT)16/20 | −1 | 6 | 10 | 60.0 | ||
| chr5_21021412 | intergenic | (AT)16/17 | CB-epi-1-FF1 | (AT)16/18 | +1 | 13 | 20 | 65.0 |
| (AT)16/16 | HPSI0115i-iuad_2 | (AT)16/18 | +2 | 4 | 13 | 30.8 | ||
| chr3_126908708 | intergenic | (AT)13/16 | E-HDFa-4-2 | (AT)13/15 | −1 | 13 | 31 | 41.9 |
| (AT)23/24 | HPSI0614i-voce_1 | (AT)23/25 | +1 | 3 | 6 | 50.0 | ||
| chr6_63000210 | intergenic | (GT)17/17 | E-HDFa-4-2 | (GT)17/18 | +1 | 12 | 31 | 38.7 |
| (GT)17/17 | HPSI0614i-uevq_6 | (GT)17/15 | −2 | 12 | 23 | 52.2 | ||
| chr14_46721941 | intron | (GT)23/20 | E-HDFa-12-3 | (GT)23/21 | +1 | 13 | 24 | 54.2 |
| (GT)20/22 | HPSI0614i-voce_2 | (GT)20/23 | +1 | 11 | 28 | 39.3 | ||
| chr18_23370619 | intergenic | (AGAT)12/13 | E-HDFa-11-2 | (AGAT)12/14 | +1 | 12 | 24 | 50.0 |
| (AGAT)15/15 | HPSI0614i-voce_1 | (AGAT)15/16 | +1 | 6 | 19 | 31.6 | ||
| chr1_183339353 | intron | (AC)28/32 | R-HDFa-1-2 | (AC)28/33 | +1 | 13 | 22 | 59.1 |
| (AC)30/28 | HPSI0614i-uevq_6 | (AC)30/27 | −1 | 9 | 20 | 45.0 | ||
| chr5_166111593 | intergenic | (AT)21/22 | HPSI0115i-iuad_3 | (AT)21/23 | +1 | 4 | 14 | 28.6 |
| (AT)22/19 | HPSI0614i-voce_1 | (AT)22/20 | +1 | 11 | 17 | 64.7 | ||
(Repeat unit), number of unit/number of units.
Bold shows mutant MS.
Table 3.
Homopolymer regions, in which alterations occurred in multiple human iPSC clones
| Chromosome_position | Region | Parenta wild/wild |
Mutant clonea | wild/mutantb | Δ | NGS data |
||
|---|---|---|---|---|---|---|---|---|
| Mutant reads |
Read depth |
Frequency (%) | ||||||
| chr1_29769347 | intron | (A)19/19 | CB-epi-1-FF1 | (A)19/18 | −1 | 10 | 15 | 66.7 |
| (A)19/19 | CB-epi-1-8 | (A)19/20 | +1 | 7 | 21 | 33.3 | ||
| chr1_89867798 | intron | (T)17/17 | SeV-BJ-2 | (T)17/16 | −1 | 15 | 30 | 50.0 |
| (T)15/17 | HPSI0714i-kute_4 | (T)15/18 | +1 | 10 | 30 | 33.3 | ||
| chr1_191672724 | intron | (T)23/21 | SeV-BJ-2 | (T)23/22 | +1 | 14 | 24 | 58.3 |
| (T)21/21 | R-HDFa-1-2 | (T)21/22 | +1 | 8 | 14 | 57.1 | ||
| chr1_201082903 | intron | (A)25/25 | HPSI0614i-uevq_6 | (A)25/26 | +1 | 5 | 20 | 25.0 |
| (A)25/28 | CB-epi-1-FF1 | (A)25/27 | −1 | 6 | 15 | 40.0 | ||
| chr2_29939863 | intron | (A)23/23 | E-HDFa-1-3 | (A)23/22 | −1 | 4 | 8 | 50.0 |
| (A)23/23 | CB-epi-2-3-7 | (A)23/24 | +1 | 7 | 17 | 41.2 | ||
| chr2_34021849 | intron | (T)17/16 | HPSI0714i-kute_5 | (T)17/15 | −1 | 20 | 32 | 62.5 |
| (T)16/16 | HPSI0115i-iuad_2 | (T)16/15 | −1 | 11 | 23 | 47.8 | ||
| chr2_64657332 | intergenic | (T)18/21 | SeV-BJ-3 | (T)18/20 | −1 | 9 | 22 | 40.9 |
| (T)21/18 | HPSI0714i-kute_4 | (T)21/17 | −1 | 13 | 33 | 39.4 | ||
| chr2_80591682 | intron | (T)25/20 | R-HDFa-1-12 | (T)25/19 | −1 | 15 | 21 | 71.4 |
| (T)20/20 | HPSI0614i-uevq_6 | (T)20/19 | −1 | 13 | 29 | 44.8 | ||
| chr2_101110042 | intron | (T)18/18 | HPSI0614i-uevq_4 | (T)18/17 | −1 | 13 | 25 | 52.0 |
| (T)16/18 | CB-epi-2-2-1 | (T)16/17 | −1 | 19 | 30 | 63.3 | ||
| chr2_162369788 | intergenic | (T)24/24 | R-HDFa-1-12 | (T)24/23 | −1 | 8 | 14 | 57.1 |
| (T)24/24 | CB-epi-2-2-1 | (T)24/23 | −1 | 9 | 18 | 50.0 | ||
| chr3_768553 | intron | (A)16/16 | HPSI0614i-voce_1 | (A)16/15 | −1 | 18 | 39 | 46.2 |
| (A)16/16 | HPSI0514i-letw_5 | (A)16/15 | −1 | 24 | 48 | 50.0 | ||
| chr3_3924878 | intergenic | (T)20/20 | HPSI0514i-letw_5 | (T)20/19 | −1 | 13 | 32 | 40.6 |
| (T)20/20 | HPSI0115i-iuad_3 | (T)20/19 | −1 | 24 | 35 | 68.6 | ||
| chr3_12746169 | intergenic | (T)24/24 | CB-epi-2-1-5 | (T)24/23 | −1 | 6 | 15 | 40.0 |
| (T)24/24 | CB-epi-1-5 | (T)24/23 | −1 | 7 | 16 | 43.8 | ||
| chr3_66963504 | intergenic | (A)17/17 | SeV-BJ-3 | (A)17/16 | −1 | 9 | 25 | 36.0 |
| (A)17/17 | E-HDFa-12-3 | (A)17/16 | −1 | 16 | 30 | 53.3 | ||
| chr3_109103351 | intergenic | (T)24/20 | HPSI0514i-letw_5 | (T)24/19 | −1 | 7 | 11 | 63.6 |
| (T)20/24 | E-HDFa-1-3 | (T)20/23 | −1 | 13 | 23 | 56.5 | ||
| chr3_121475187 | intergenic | (T)25/25 | R-HDFa-1-19 | (T)25/24 | −1 | 7 | 25 | 28.0 |
| (T)25/25 | HPSI0614i-voce_2 | (T)25/26 | +1 | 7 | 25 | 28.0 | ||
| chr3_127226236 | intron | (A)13/13 | SeV-BJ-2 | (A)13/12 | −1 | 12 | 32 | 37.5 |
| (A)13/12 | E-HDFa-4-2 | (A)13/11 | −1 | 18 | 42 | 42.9 | ||
| chr3_129394880 | intron | (A)25/25 | CB-epi-2-2-1 | (A)25/24 | −1 | 15 | 27 | 55.6 |
| (A)25/25 | CB-epi-2-1-4 | (A)25/24 | −1 | 11 | 26 | 42.3 | ||
| chr3_137373146 | intergenic | (T)16/16 | R-HDFa-1-19 | (T)16/15 | −1 | 6 | 15 | 40.0 |
| (T)16/16 | E-HDFa-4-2 | (T)16/15 | −1 | 9 | 36 | 25.0 | ||
| chr3_138059967 | intergenic | (T)19/19 | R-HDFa-1-2 | (T)19/18 | −1 | 9 | 26 | 34.6 |
| (T)19/21 | HPSI0714i-kute_5 | (T)19/20 | −1 | 18 | 26 | 69.2 | ||
The hotspots detected in chr1-3. See Data S5B for other chromosomes.
(Repeat unit), number of unit/number of units.
Bold shows mutant MS.
Discussion
MS alteration analyses can be broadly divided into two types of approaches, one which focuses on the differences among individuals, and the other that relates specifically to somatic de novo alterations, such as those associated with tumorigenesis. Identifying the latter type was the aim of this study, i.e., genome reprogramming-associated de novo InDels. Such InDels contained in iPSCs/ntESCs are clone specific, even if they are generated from cells derived from an identical individual. Although MS alterations in human iPSCs have been investigated previously, the focus has been on the differences among individuals (Jakubosky et al., 2020). The MS alterations observed in every cell line derived from the same individual in that study were defined as positive, even though they were pre-existing. Conversely, we here focused on iPSC clone-specific, somatic de novo alterations.
InDel detection software is currently available for most of the systems developed for NGS, but typically produce a substantial number of false-positive signals. Conventional informatics, especially for human genomes, frequently overlook InDels due to limitations in the accuracy of aligning read sequences on the reference genome. Kloosterman et al. (2015) convincingly demonstrated a 60% false-positive rate in their extensive validation test sequencing of over 1,000 InDel candidates. In addition, the detection of somatic de novo InDels, which is required for the InDel analysis of iPSCs/ntESCs, is more complex. Hence, not only subsequent manual inspection but also experimental validation assessing tests are urgently required to determine the true positives. However, InDels in iPSCs have been yet discussed without these extensive investigations, such as comprehensive and experimental validation tests. To overcome these difficulties and accurately depict the InDels in iPSCs/ntESCs, we employed an inbred mouse strain and examined substantial numbers of sister cell lines of a sample cell line, all of which were generated from an identical MEF fraction prepared from a single embryo, enabling us to achieve a highly accurate alignment of WGS read sequences with the reference genome, and efficiently exclude false-positive signals. Namely, we conducted InDel analysis and subsequent validation tests using an ideal combination of mouse C57BL/6 cell lines and various negative control cells with an identical genetic background. As a result, we obtained robust and conclusive results for the occurrence of InDel aberrations in non-repetitive regions of iPSC/ntESC genomes.
Here, we found seven InDels within coding regions, three of which were in cancer-related genes, including the Smug1 gene. The deletion in Smug1 is worth highlighting, because this gene encodes a DNA repair enzyme, the inactivation of which can cause genome instability, and the low expression of which has been reported to cause cancer (Abdel-Fatah et al., 2013). Hence, losses causing a phenotypic deficiency must be carefully investigated, because heterozygous effects have been recently reported for some genes. On the other hand, however, our current results suggest an InDel frequency of less than one per cell line, indicating that it will likely be feasible to isolate coding region InDel-free iPSC/ntESC lines.
The analysis of STR regions is very tricky despite their biological significance. It is practically unfeasible to perform comprehensive analysis of this nature in human genomes because STR regions often have considerable sequence variations between alleles and between individuals, so as to be used as polymorphic markers. In human cells therefore, the reference genome and germline sequences do not match and are often heterozygous. Hence, it is thought that misalignment is likely to occur very frequently. In addition, the error rate of the DNA polymerase used in the sequencing process when analyzing MSs is markedly higher than in other non-repetitive regions. Both false positives and false negatives also occur frequently during the identification of InDels within MSs, because identical alterations often appear in negative control genomes also. Tandem repeat regions have thus been typically excluded when conducting conventional InDel analysis. In our current study, however, by employing an inbred strain mice (C57BL/6) and a number of sister iPSC lines, and through our refinement of the informatics processes with model InDel reads, we have largely overcome these difficulties. A considerable level of false positives are still obtained, and this necessitates a subsequent extensive manual inspection step. In laboratory mice, however, the genomic regions that match the reference genome are significantly larger than those in humans and, in principle, few heterozygous alleles would be expected in an inbred strain.
After the success of our current MS alteration analysis using an inbred mouse system, we conducted similar analysis also on human iPSCs by combining our newly developed informatics approach and utilizing sister iPSC clones. Unfortunately, however, this analysis was unsuccessful. We randomly chose 24 InDel candidates that were called from within STR regions and subsequently performed manual inspections. All turned out to be false positives, i.e., no true signals; 2 out of them were the result of mis-mapping due to long repetitive sequences (more than 80 bp), and the remaining 22 were the result of a misalignment due to polymorphisms among parent cells and sister iPSC clones. From these results, we considered that the heterogeneity of most genetic loci and the presence of vast amounts of SNPs in humans would impede the genome-wide analysis of MS abnormalities. Hence, instead of simply narrowing down the alterations in STR regions in the genome by informatics using WGS data, we adopted a large-scale analysis approach focusing on only the STR regions, and not on the entire genome. Information on these regions, including their flanking regions, has already been obtained, enabling us to achieve an accurate alignment of our WGS data with reference sequences. In the analysis, we utilized an algorithm called “HipSTR” that covers 1,620,030 human STR regions. Although HipSTR has been mainly used for investigating MS differences between individuals (Willems et al., 2017), we clearly demonstrate in our present analysis that this program can also analyze somatic de novo MS alterations in human iPSCs by combination with the use of sister clones.
Importantly, profiling of the MS alterations in iPSCs/ntESCs revealed preferential STR regions, suggesting the presence of hotspots. We thus subsequently searched for hotspots of MS alterations, and successfully identified 18 in the mouse genome and 126 (13 of STR and 113 of homopolymer regions) in the human genome (Tables 1, 2, and 3; Data S5). It was further notable that the variations observed in six hotspot regions were derived from different parent cells: one came from MEF5 cells and the other from TTFs, which had been prepared from independent mouse individuals (Data S5A). Of particular note, drastic deletions occurred independently in iPSCs and ntESCs at chr9_77599501. Although the wild-type allele shows 23 repeats, only a 17 repeat unit was detected in an iPSC clone from MEF5, whereas an 18 repeat unit were observed in the other line, an ntESC clone generated from TTFs (Data S5A). In human cells, alteration-prone STR regions were often detected in (AT)-STR regions (8/13) (Table 2). This was consistent with our finding that the alterations were detected in only one cell line that exhibited AT preferentiality (Figure S5A). Thus, our study findings indicate the presence of hotspots of MS alteration, but more careful exploration will be needed to identify their entire profile.
Biological functions of MS regions have been discussed in a number of publications to date. Trinucleotide repeat disorders have been well characterized (Di Prospero and Fischbeck, 2005) and recently an association of MS regions with the expression regulation of non-coding transcripts has also been revealed (Ninomiya and Hirose, 2020). Although we investigated the loci of trinucleotide repeat disorders, no alterations were evident (Data S6). In addition, MS alteration regions utilized for tumor diagnoses were not observed among those observed in our present analyses (Data S6). This may indicate that the alterations observed in reprogrammed PSCs are not attributable to a defect of mismatch repair activities. Finally, because of a limited sequence read length (150 bp), the detection of alteration peaks occurred at around 20 repeats of a 2 bp STR unit. The longer the STR, the more difficult it becomes to accurately determine both ends of its regions, which will cause alterations to be overlooked in (AT)-STR regions of more than 20 repeats (Figure S5C). Hence, deep analysis of MS regions should be mandatory for iPSCs prior to their use in any medical applications. Information on hotspots and preferential sequences would also shed light on MS alterations in human cells, in which genome-wide investigations are currently still not feasible.
One of the most important findings of our present study is that certain human iPSCs, which were created using erythroblasts expanded from human CB by means of an episomal vector system, have fewer MS alterations. We recently reported that this type of iPSC shows a lower frequency of point mutations and InDels in non-repetitive regions (Araki et al., 2020). In this report, a decrease in the MS alteration frequency was observed in all 14 independent lines examined that were generated from the 4 individuals, without exception. This included 5 lines established from one individual and 3 lines from each of 3 individuals. In contrast, the other human iPSCs, which were viral-transduction based, showed more MS alterations, including 5 using episomal vector system, 3 generated with a retrovirus system from dermal fibroblasts, 2 using a Sendai virus system from BJ cells, and 10 also by Sendai virus from skin fibroblasts.
Thus, CB erythroblast-derived iPSCs, which are integration free, have a distinct advantage for possible future use in regenerative medicine. However, the mechanisms underlying this intriguing phenomenon remain elusive. Two possible factors were considered to be behind the lower number of alterations in these cells, i.e., the cell of origin and the reprogramming procedure. To assess this further, we attempted to generate integration-free iPSCs from the human dermal fibroblast (HDF) fraction using the episomal system identical to that employed for CB-iPSCs. However, no colonies appeared despite three attempts. We subsequently conducted a similar experiment using another episomal system that is currently widely used (Okita et al., 2011) (https://skip.stemcellinformatics.org/), and succeeded in generating five integration-free iPSC lines from the identical HDF fraction to that used for retroviral-mediated iPSCs (Figures 3A and S4). Our results showed a clear difference in the alteration frequency between the two types of integration-free iPSCs generated using episomal vectors, i.e., HDF- and CB-derived vectors (Figure 3B). Thus, although our results seem to suggest that the alteration frequency largely depends on the parental somatic cell, this conclusion would still be controversial due to the fact that the episomal system we used this time contained p53-dominant negatives in addition to the defined factors. To date, p53 suppression during genome reprogramming has been found to have little impact on the genome stability of iPSCs (Rasmussen et al., 2014). However, further analysis and appropriate comparisons with an identical episomal system will be needed to make a definitive conclusion on this issue.
It has not yet proved to be possible to directly distinguish genome reprogramming-induced from culture-induced genetic aberrations in iPSCs, and our present analyses could not do so either. On the other hand, we made some observations related to this issue in our experiments. In mice, the number of InDels/MS alterations in ESCs is lower than that in iPSCs or ntESCs (Figures 1A and 2B). Furthermore, it was also shown in our present experiments that error-prone STR sequences are clearly different between ESCs and reprogrammed PSCs (Figure S5B). Our findings of similarities between iPSCs and ntESCs, but not between reprogrammed cells and ESCs in this regard, seem to indicate that the InDels/MS alterations we detected are associated with reprogramming.
The aforementioned analyses in the mouse cannot be conducted in humans as the preparation of human ESCs that are isogenic with iPSCs is not possible. Moreover, the generation of human ntESCs is completely impossible. Because of this, we attempted to investigate the age dependency of the alterations instead, i.e., if the number of alterations reflects the number of DNA replications, then an age-dependent increase may be seen. We compared iPSCs derived from individuals with ages ranging from 25 to 74 years but found no increase in the STR or homopolymer frequency with increasing age (Figure S6). This suggested that the detected alterations cannot be simply attributable to a higher number of DNA replications in the parental somatic cells.
Taken together, our current observations indicate that, although most of the InDels/STR alterations that we detected seemed to be induced by reprogramming, this will need to be validated in future studies. For that purpose, further analysis that can distinguish between culture-induced and genome reprogramming-induced variations will be required (Nguyen et al., 2014).
Experimental procedures
Cell lines
Mouse cell lines used in this study were established from C57BL/6J mice (Japan SLC, Hamamatsu) for which WGS data, except those from ESCs, and SNV analyses have been reported previously by our laboratory (Araki et al., 2017, 2020; Sugiura et al., 2014). Our mouse iPSC lines were established using single embryonic fibroblasts (MEF5) with four (Oct4, Sox2, Klf4, and c-Myc) or three (Oct4, Sox2, and Klf4) factors delivered via a retroviral transduction system (Araki et al., 2020). Nuclear transfer ESC lines were established using also the same parent MEF5 or tail tip fibroblast 2 (TTF2) nuclei (Araki et al., 2017). ESC lines were established from fertilization (natural mating) blastocysts (Sugiura et al., 2014). For this InDel study, we additionally conducted WGS of ESCs (Table S1).
With regard to the human cell lines used in the experiments, we previously used an episomal vector to establish 14 human iPSC lines from erythroblasts expanded from CB mononuclear cells, and a retroviral vector to generate 3 human iPSC lines from dermal fibroblasts from a single individual and performed WGS in all cases (Figure 3A) (Araki et al., 2020). We here also established five human iPSC lines from the dermal fibroblast fraction" with "dermal fibroblast fraction (FC-0024, Lifeline Cell Technology, Frederick, MD) that had been used for the generation of the 3 lines of retrovirus-mediated iPSCs by means of a Human iPS Cell Generation Episomal Vector Mix (Takara Bio, Otsu, Japan) (Okita et al., 2011) and three human iPSC lines from BJ fibroblasts (ATCC, CRL-2522, newborn foreskin) using CytoTune-iPS2.0 Sendai viral vectors (ID Pharma, Tokyo, Japan). PCR was carried out to assess the genome integration-free status of iPSCs that had been newly established with episomal vectors. The primer sequences were as follows: 5′-TTC CAC GAG GGT AGT GAA CC-3ʹ and 5ʹ-TCG GGG GTG TTA GAG ACA AC-3ʹ for oriP, 5ʹ-ATC GTC AAA GCT GCA CAC AG-3ʹ and 5ʹ-CCC AGG AGT CCC AGT CA-3ʹ for EBNA1, and 5ʹ-GGT TGG CCA ATC TAC TCC CAG G-3ʹ and 5ʹ-CAA CTT CAT CCA CGT TCA CC-3ʹ for β-globin. We downloaded WGS data from the HipSci collection for the 10 human iPSC lines in this study that had been previously established from skin fibroblasts using Sendai viral vectors (http://www.hipsci.org/).
WGS
Genomic DNA was prepared from cells with DNeasy (QIAGEN, Hilden, Germany) and then used for library construction with an Illumina TruSeq DNA PCR-free library prep kit. Sequencing was performed using HiSeq X sequencer (Illumina, San Diego, CA) with 151-base, paired-end reads as described in our recent study (Araki et al., 2020).
InDel analysis in mouse iPSCs/ntESCs
We conducted InDel analysis using the WGS reads for which reliability was shown using SNV analyses in our previous studies (Araki et al., 2017, 2020). The conditions used for mapping and calling (CLC genomics workbench, CLC Bio, Aarhus, Denmark) are shown in Figure S1B. To reduce the incidence of false positives, any variants that were shared among other the PSC lines analyzed in this study were excluded. Visual (manual) inspection of the read sequence alignments was performed for all InDel candidates. For MS alteration analysis, we analyzed the candidates that were removed at step 6 from among the InDel candidates identified in non-repetitive sequences (Figure 2A). In the case of ESC analysis, we first analyzed both the male and female parental genomes because no reference genome with an identical genetic background to the ESCs was available. With such analysis, however, there is a concern that the false-negative rate will increase, so we therefore conducted additional analysis using only the paternal genome as the reference and additionally removed any polymorphisms (step 4). Both sets of analyses elicited similar results.
Validation of InDel candidates by restriction enzyme digestion and Sanger sequencing
The regions incorporating the InDel candidate were amplified by PCR from genomic DNA; 5 ng of the amplified DNA was used for restriction enzyme digestion and 10 ng for Sanger sequencing with EX taq or Titanium Taq DNA polymerase (TAKARA BIO, Kusatsu, Japan). The PCR products were purified with MinElute (QIAGEN). Primer sequences and PCR conditions are available upon request. The iPSC lines in which the InDel was called were investigated along with many negative controls (seven sister iPSC lines, MEF5 parental somatic cells and ESCs). For Sanger sequencing, ExoSAP-IT and the BigDye Terminator v.3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA) were employed.
MS alteration analysis in human iPSCs
We used the program HipSTR v.0.6.2 (Willems et al., 2017) to identify the alteration of MSs with the following filters: --max-str-len 500; --min-reads 80; --def-stutter-model (default). If two or more mutant sequence reads were detected in any one of the negative control cells, parent cells, or sister clones, the mutation was excluded from our list of candidates. Additional conditions for these analyses were as follows: depth ≥ 6; variant read count ≥ 3; VAF ≥ 20%; stutter noise < 15%.
Validation of MS alteration candidates using amplicon sequencing
The regions incorporating the MS instability candidate were amplified by PCR from 10 ng (mouse) or 20 ng (human) of genomic DNA using PrimeSTAR MAX DNA polymerase (TAKARA BIO). In addition to each iPSC line in which MS alteration was called by our detection system, seven sister iPSC lines, their parental somatic cells and ESCs for mouse, and four sister iPSC lines and their parental somatic cells for human were examined as controls.
PCR products were mixed, followed by the purification via the MinElute PCR Purification Kit (QAIGEN), and sequenced using a NovaSeq 6000 or HiSeq X (Illumina). Primer sequences and PCR conditions are available upon request.
Data and code availability
The accession numbers for the sequences reported in this paper are DDBJ Sequence Read Archive (DRA): DRA009756, DRA011197 and DRA012278. The previously generated Raw Illumina sequencing reads analyzed during this study have also been deposited in the DRA under accession codes: DRA006232, DRA007325, DRA002956, DRA005423, DRA007336, DRA006622 and DRA008459.
Author contributions
S.K. analyzed the data and wrote the manuscript. T.S. performed the bioinformatics analyses and wrote the manuscript. M.S. performed the DNA preparations for NGS and the validation experiments. Y.H., K.I., M.F., and M.N. performed the iPSC generation and cell culture. R.A. and M.A. designed the experiments and wrote the manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We are grateful to S. Yamanaka (University of Kyoto) for providing the OSKM genes expressed in plasmid and retroviral vectors through Addgene and to T. Kitamura (University of Tokyo) for providing the PlatE cells. We also thank Y. Kasama and H. Yoshida for technical assistance. This study makes use of data generated by the HipSci Consortium, funded by The Wellcome Trust and the MRC. This work was partially funded by the Japan Society for the Promotion of Science (grant no. KAKENHI 17H03615, 17K20053, and 20K15707) and by the Takeda Science Foundation, Uehara Memorial Foundation, and the Mitsubishi Foundation.
Published: September 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.08.017.
Contributor Information
Ryoko Araki, Email: araki.ryoko@qst.go.jp.
Masumi Abe, Email: abe.masumi@qst.go.jp.
Supplemental information
(A) Site, mutation type, and read depth.
(B) Sequence of each model InDel read.
(A) List of the InDels detected in non-repetitive regions of mouse iPSCs/ntESCs.
(B) List of InDels detected in microsatellite regions of mouse iPSCs/ntESCs.
(C) List of InDels detected in microsatellite regions of human iPSCs
The results of forward sequencing validation of the InDels are shown. Most of these InDels were also confirmed by reverse sequencing.
To discriminate the background signals and abnormality-specific signals, multiple negative controls were employed, including parent somatic MEFs, ESCs generated from C57BL/6 mouse independently, and seven sister iPSC clones for each mutated line (mouse), and parent somatic CB-1 or HDFa cells and four sister iPSC clones for each mutated line (human). Amplicon sequencing clearly revealed the clone-specific nature of the alterations and background signals for the repetitive sequences.
(A) Short tandem repeat regions, in which alterations occurred in multiple mouse iPSC/ntESC clones established from different mouse individuals.
(B) Homopolymer regions, in which alterations occurred in multiple human iPSC clones (chromosome 4–22).
Trinucleotide repeat disorder-related MS regions in human iPSCs (Chintalaphani et al., 2021) and MSI marker regions in human iPSCs (Fujimoto et al., 2020; Wong et al., 2006) were analyzed. The repeat counts in human iPSCs, analyzed by HipSTR, are shown. nd, not detected; MSI, microsatellite instability.
References
- Abdel-Fatah T.M., Albarakati N., Bowell L., Agarwal D., Moseley P., Hawkes C., Ball G., Chan S., Ellis I.O., Madhusudan S. Single-strand selective monofunctional uracil-DNA glycosylase (SMUG1) deficiency is linked to aggressive breast cancer and predicts response to adjuvant therapy. Breast Cancer Res. Treat. 2013;142:515–527. doi: 10.1007/s10549-013-2769-6. [DOI] [PubMed] [Google Scholar]
- Araki R., Hoki Y., Suga T., Obara C., Sunayama M., Imadome K., Fujita M., Kamimura S., Nakamura M., Wakayama S. Genetic aberrations in iPSCs are introduced by a transient G1/S cell cycle checkpoint deficiency. Nat. Commun. 2020;11:197. doi: 10.1038/s41467-019-13830-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R., Mizutani E., Hoki Y., Sunayama M., Wakayama S., Nagatomo H., Kasama Y., Nakamura M., Wakayama T., Abe M. The number of point mutations in induced pluripotent stem cells and nuclear transfer embryonic stem cells depends on the method and somatic cell type used for their generation. Stem Cells. 2017;35:1189–1196. doi: 10.1002/stem.2601. [DOI] [PubMed] [Google Scholar]
- Araki R., Uda M., Hoki Y., Sunayama M., Nakamura M., Ando S., Sugiura M., Ideno H., Shimada A., Nifuji A. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- Assie G., Letouze E., Fassnacht M., Jouinot A., Luscap W., Barreau O., Omeiri H., Rodriguez S., Perlemoine K., Rene-Corail F. Integrated genomic characterization of adrenocortical carcinoma. Nat. Genet. 2014;46:607–612. doi: 10.1038/ng.2953. [DOI] [PubMed] [Google Scholar]
- Bhutani K., Nazor K.L., Williams R., Tran H., Dai H., Dzakula Z., Cho E.H., Pang A.W., Rao M., Cao H. Whole-genome mutational burden analysis of three pluripotency induction methods. Nat. Commun. 2016;7:10536. doi: 10.1038/ncomms10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Hansen N.F., Zhao L., Du Y., Zou C., Donovan F.X., Chou B.K., Zhou G., Li S., Dowey S.N. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Prospero N.A., Fischbeck K.H. Therapeutics development for triplet repeat expansion diseases. Nat. Rev. Genet. 2005;6:756–765. doi: 10.1038/nrg1690. [DOI] [PubMed] [Google Scholar]
- Ewing A.D., Houlahan K.E., Hu Y., Ellrott K., Caloian C., Yamaguchi T.N., Bare J.C., P'ng C., Waggott D., Sabelnykova V.Y. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat. Methods. 2015;12:623–630. doi: 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubosky D., Smith E.N., D'Antonio M., Jan Bonder M., Young Greenwald W.W., D'Antonio-Chronowska A., Matsui H., Stegle O., Montgomery S.B., DeBoever C. Discovery and quality analysis of a comprehensive set of structural variants and short tandem repeats. Nat. Commun. 2020;11:2928. doi: 10.1038/s41467-020-16481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Ng S.H., Sharma V., Neculai D., Hussein S., Sam M., Trinh Q., Church G.M., McPherson J.D., Nagy A. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- Kemmerich K., Dingler F.A., Rada C., Neuberger M.S. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung–/–Msh2–/– mice. Nucleic Acids Res. 2012;40:6016–6025. doi: 10.1093/nar/gks259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Francioli L.C., Hormozdiari F., Marschall T., Hehir-Kwa J.Y., Abdellaoui A., Lameijer E.W., Moed M.H., Koval V., Renkens I. Characteristics of de novo structural changes in the human genome. Genome Res. 2015;25:792–801. doi: 10.1101/gr.185041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai M., Watanabe A., Kurimoto Y., Hirami Y., Morinaga C., Daimon T., Fujihara M., Akimaru H., Sakai N., Shibata Y. Autologous induced stem-cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- Nguyen H.T., Markouli C., Geens M., Barbé L., Sermon K., Spits C. Human embryonic stem cells show low-grade microsatellite instability. Mol. Hum. Reprod. 2014;20:981–989. doi: 10.1093/molehr/gau059. [DOI] [PubMed] [Google Scholar]
- Ninomiya K., Hirose T. Short tandem repeat-enriched architectural RNAs in nuclear bodies: functions and associated diseases. Noncoding RNA. 2020;6:6. doi: 10.3390/ncrna6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Rasmussen M.A., Holst B., Tümer Z., Johnsen M.G., Zhou S., Stummann T.C., Hyttel P., Clausen C. Transient p53 suppression increases reprogramming of human fibroblasts without affecting apoptosis and DNA damage. Stem Cell Reports. 2014;3:404–413. doi: 10.1016/j.stemcr.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani F.J., Nik-Zainal S., Wuster A., Li Y., Conte N., Koike-Yusa H., Kumasaka N., Vallier L., Yusa K., Bradley A. Mutational history of a human cell lineage from somatic to induced pluripotent stem cells. PLoS Genet. 2016;12:e1005932. doi: 10.1371/journal.pgen.1005932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M., Kasama Y., Araki R., Hoki Y., Sunayama M., Uda M., Nakamura M., Ando S., Abe M. Induced pluripotent stem cell generation-associated point mutations arise during the initial stages of the conversion of these cells. Stem Cell Reports. 2014;2:52–63. doi: 10.1016/j.stemcr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wang X., Baek S.J., Eling T.E. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem. Pharmacol. 2013;85:597–606. doi: 10.1016/j.bcp.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems T., Zielinski D., Yuan J., Gordon A., Gymrek M., Erlich Y. Genome-wide profiling of heritable and de novo STR variations. Nat. Methods. 2017;14:590–592. doi: 10.1038/nmeth.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.A., Larson D.E., Sun C.W., George D.R., Ding L., Miller C.A., Lin L., Pawlik K.M., Chen K., Fan X. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Site, mutation type, and read depth.
(B) Sequence of each model InDel read.
(A) List of the InDels detected in non-repetitive regions of mouse iPSCs/ntESCs.
(B) List of InDels detected in microsatellite regions of mouse iPSCs/ntESCs.
(C) List of InDels detected in microsatellite regions of human iPSCs
The results of forward sequencing validation of the InDels are shown. Most of these InDels were also confirmed by reverse sequencing.
To discriminate the background signals and abnormality-specific signals, multiple negative controls were employed, including parent somatic MEFs, ESCs generated from C57BL/6 mouse independently, and seven sister iPSC clones for each mutated line (mouse), and parent somatic CB-1 or HDFa cells and four sister iPSC clones for each mutated line (human). Amplicon sequencing clearly revealed the clone-specific nature of the alterations and background signals for the repetitive sequences.
(A) Short tandem repeat regions, in which alterations occurred in multiple mouse iPSC/ntESC clones established from different mouse individuals.
(B) Homopolymer regions, in which alterations occurred in multiple human iPSC clones (chromosome 4–22).
Trinucleotide repeat disorder-related MS regions in human iPSCs (Chintalaphani et al., 2021) and MSI marker regions in human iPSCs (Fujimoto et al., 2020; Wong et al., 2006) were analyzed. The repeat counts in human iPSCs, analyzed by HipSTR, are shown. nd, not detected; MSI, microsatellite instability.
Data Availability Statement
The accession numbers for the sequences reported in this paper are DDBJ Sequence Read Archive (DRA): DRA009756, DRA011197 and DRA012278. The previously generated Raw Illumina sequencing reads analyzed during this study have also been deposited in the DRA under accession codes: DRA006232, DRA007325, DRA002956, DRA005423, DRA007336, DRA006622 and DRA008459.