Abstract
The presence of a 500-bp fragment which amplifies a region from the genome of Mycobacterium bovis (J. G. Rodriguez, G. A. Meija, P. Del Portillo, M. E. Patarroyo, and L. A. Murillo, Microbiology 141:2131–2138, 1995) was evaluated by carrying out PCR on 121 M. bovis isolates. The M. bovis strains, previously characterized by culture and biochemical tests, were isolated from cattle in different regions of Argentina, Mexico, and Colombia. Four additional strains isolated from sea lions that belong to the M. tuberculosis complex were also included in the study. All of the isolates tested were PCR positive, rendering the expected 500-bp band and giving a correlation of 100% with previous microbiological characterization. Southern blot analysis revealed a common band of 1,800 bp and a polymorphic high-molecular-mass hybridization pattern. The results show that this assay may be useful for diagnosis and identification of M. bovis in cattle.
Bovine tuberculosis remains an important zoonosis in many countries of the world. Cases of human tuberculosis of bovine origin have increased in recent years (3, 5, 8, 19), and this zoonosis has become a public health problem, as well as the cause of significant economic losses. Mycobacterium bovis, the causative agent of bovine tuberculosis, infects both animals of agricultural importance and wild mammals, which act as a reservoir for the organism, making it difficult to control the disease (12).
The bovine tuberculin test is easy to perform on a large scale on livestock, but it has the inconvenience of having a broad range of specificity and sensitivity (13). Confirmation of the diagnosis is achieved by culture and biochemical assays. Despite the fact that microbiological culture is highly specific, a positive result takes a long time to obtain and in most cases is achieved after the animal has been sacrificed.
It is necessary to develop new diagnostic methods for bovine tuberculosis which could identify M. bovis directly in biological samples, such as milk or blood, without having to culture them and which would also improve the predictive value of the tuberculin test. Although the PCR has been successfully applied for the diagnosis of tuberculosis, routine application of a PCR-based method requires that the target sequence be highly specific for the microorganism and that it be present in all of the strains isolated.
Rodríguez et al. (14) reported a PCR assay which amplifies a 500-bp fragment from the M. bovis genome by using the JB21-JB22 primer pair. However, only a small number of isolates were used in that study. The present work was performed to determine whether this 500-bp fragment could be amplified from the genome of different, previously characterized, M. bovis isolates.
Mycobacterial isolates and DNA extraction.
A total of 121 isolates identified as M. bovis on the basis of growth in the presence of pyruvate (scarce growth in glycerol), colony morphology, and biochemical and enzymatic tests (niacin negative, nitrate reduction negative, catalase negative, urease positive, pyrazinamidase negative) were used in this study (9). Susceptibility to thiophene-2-carboxylic acid hydrazide and p-aminosalicylic acid was determined in egg solid medium with glutamate added and without glycerol (9). In some cases, guinea pigs and rabbits were inoculated in order to confirm an M. bovis identification. M. tuberculosis H37Rv and M. bovis AN5 were used as reference strains. One hundred twelve of these were bovine strains from Argentina (taken from six different regions of the country), two were bovine strains from Mexico, and seven were from Colombia. Four isolates belonging to the M. tuberculosis complex were obtained from sea lions in Argentina and were also included in the study. Lymph nodes (40%), lung tissue (45%), liver (10%), and samples from other locations (5%) were collected during necropsy and cultured in Stonebrink broth (16). All of them showed macroscopic lesions typical of bovine tuberculosis. M. tuberculosis H37Rv, M. microti, M. africanum, and M. paratuberculosis were also analyzed in the study. Chromosomal DNAs were isolated as described by van Soolingen et al. (17), and 100 ng of each DNA was used for PCR amplification.
PCR assay.
Primers JB21 and JB22 were synthesized on a Pharmacia synthesizer. Primer sequences and performance of the PCR were as reported previously by Rodriguez et al. (14). The reactions were performed in a final volume of 50 μl containing 1× reaction buffer (Promega), 2.5 U of Taq polymerase (Promega), 0.2 mM each deoxynucleoside triphosphate, 1.5 mM magnesium chloride, and 20 pmol of each primer. Target DNA was denatured by incubation for 5 min at 94°C before amplification for 30 cycles of 94°C for 1 min, annealing at 68°C for 1 min, and extension at 72°C for 1 min. All reactions were carried out in an automated thermal cycler (Biometra). After amplification, 1/10 of the PCR mixture was analyzed by gel electrophoresis in 1% agarose gels containing 0.5 μg of ethidium bromide per ml.
Hybridization analysis.
Genomic DNA was hydrolyzed with the PvuII restriction enzyme (Gibco BRL). After separation through agarose gel electrophoresis, the hydrolysate was transferred to a nylon membrane (Amersham) and hybridized with the 500-bp amplified fragment labeled with [α-32P]dCTP by random priming (Rediprime; Amersham).
Results.
PCR was carried out on 121 M. bovis isolates by using primers JB21 and JB22, which amplify a 500-bp fragment of M. bovis (14). The 500-bp genomic fragment was present in all of the M. bovis isolates used in this study, giving a 100% correlation with the microbiological characterization. The fragment was also amplified from the genome of the four M. tuberculosis complex strains isolated from sea lions. These isolates were characterized as M. tuberculosis complex strains because they shared molecular markers from both M. tuberculosis and M. bovis (2, 15). No amplification was observed for M. tuberculosis H37Rv, M. africanum, M. microti, and four M. paratuberculosis strains.
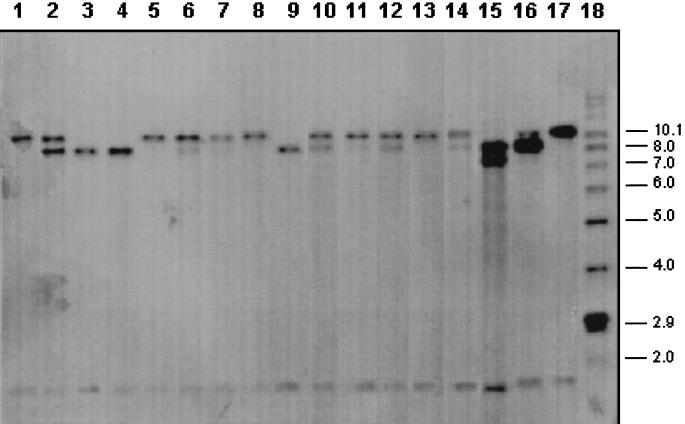
To determine whether this 500-bp sequence is present as a unique fragment in the M. bovis genome, Southern blot analysis was carried out by using 17 M. bovis isolates selected as representatives of the total strains and the 500-bp fragment was used as a hybridization probe. As shown in Fig. 1, the 500-bp fragment hybridized to a 1.8-kb band present in all of the samples tested, indicating a common location within the M. bovis genome. In addition, positive signals were obtained with one or two high-molecular-weight bands, between 7 and 10 kb. Hybridization with these high-molecular-weight bands was polymorphic among the different isolates tested and also present when M. tuberculosis genomic DNA was used (data not shown).
FIG. 1.
Southern blot analysis of different isolates of M. bovis. Genomic DNA was digested with restriction enzyme PvuII, and the 500-bp fragment of M. bovis was used as a probe. Lanes 1 to 17 contain the following isolates of M. bovis: 1, 520; 2, 476; 3, 468; 4, 555; 5, 548; 6, 478; 7, T-372; 8, T-482; 9, 559; 10, 565; 11, 482; 12, 521; 13, 531; 14, 545; 15, 540; 16, 543; 17, 558. Lane 18 contains molecular weight markers. The values on the right are molecular weights in thousands.
Discussion.
The accurate diagnosis of bovine tuberculosis remains, to this day, an elusive goal because no method has been developed which can precisely detect the presence of the microorganism in live animals. The tuberculin assay currently used around the world renders highly variable results due to problems with sensitivity and specificity. The tuberculin test depends on several factors, including high-quality reagents, as well as the immunological status of the animal. Furthermore, a negative tuberculin test does not means that the animal is not infected; on the other hand, a positive test can only mean a delayed hypersensitivity reaction due to previous exposure. A PCR-based assay, such as the one described here, could be used to detect the presence of M. bovis in biological samples (such as milk, blood, or nasal swabs) and thus become an important tool for the control and eventual eradication of the disease.
A reliable PCR-based diagnostic assay must have a target DNA sequence that is specific for the microorganism to be detected and that must also be present in most, if not all, isolates of the organism. The 500-bp fragment amplified by primers JB21 and JB22 fulfills the first requirement, since it is capable of discriminating M. bovis from related strains, such as M. avium, which is commonly isolated from cattle, and whose tuberculin is used in the comparative intradermal tuberculin test, and M. paratuberculosis, which is also pathogenic to cattle (1). This fragment also fulfills the second requirement: the results of this study confirm that the region amplified by primers JB21 and JB22 is conserved, since it was found in 121 isolates obtained from different geographic regions of Latin America. It is also important to note that the M. tuberculosis complex strains isolated from sea lions were PCR positive, indicating that this sequence is also present in isolates from other mammals, even thought they belong to a unique cluster clearly different from M. bovis strains, and closely related to M. tuberculosis (2, 4, 11, 15).
Initially, the fragment amplified by primers JB21 and JB22 was proposed by us to be exclusive to M. bovis and able to discriminate between M. bovis and M. tuberculosis. However, only 20 M. tuberculosis strains were included in that study (14). As we included larger numbers of M. tuberculosis isolates, we have observed that there are some strains which render a 500-bp amplification band with the JB21-JB22 set of primers. This, however, does not necessarily detract from the potential benefit of carrying out a PCR assay based on this sequence in biological samples extracted from cattle, inasmuch as a positive amplification is indicative of the presence of an infectious agent, be it human M. tuberculosis or M. bovis bovine tuberculosis. Studies are currently being carried out using a large panel of M. tuberculosis strains to determine the percentage of JB21-JB22-positive amplifications. We are also using different molecular markers, such as spoligotyping (10), oxyR (7), and mtp40 (6), with the aim of determining whether these strains might belong to a cluster of M. tuberculosis, similar to what is occurring with the sea lion isolates.
PCR-based assay has been successfully used for the detection of M. tuberculosis. Recently, however, it has been shown that some M. tuberculosis strains lack specific target sequences such as IS6110 or mtp40 (11, 18). This could be due to the presence of mutations or genomic rearrangements. So far, all of the M. bovis strains tested by the assay described here contain the 500-bp target sequence, indicating that this fragment is conserved among M. bovis strains. Field test evaluation of this PCR as a diagnostic tool for M. bovis detection is being carried out by using milk and blood as biological samples in order to demonstrate the validity of the test for the detection of bovine tuberculosis.
REFERENCES
- 1.Bauerfeind R, Benazzi S, Weiss R, Schliesser T, Willems H, Baljer G. Molecular characterization of Mycobacterium paratuberculosis isolates from sheep, goats, and cattle by hybridization with a DNA probe to insertion element IS900. J Clin Microbiol. 1996;34:1617–1621. doi: 10.1128/jcm.34.7.1617-1621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernardelli A, Bastida R, Loureiro P, Michelis C, Romano M I, Cataldi A, Costa E. Tuberculosis in sea lions and fur seals from the southwestern Atlantic Ocean. Rev Sci Tech O I E (Off Int Epizoot) 1996;15:985–1005. doi: 10.20506/rst.15.3.963. [DOI] [PubMed] [Google Scholar]
- 3.Brett J L, Humble M W. Incidence of human tuberculosis caused by Mycobacterium bovis. N Z Med J. 1991;104:13–14. [PubMed] [Google Scholar]
- 4.Cousins D V, Francis B R, Gow B L, Collins D M, McGlashan C H, Gregory A, Mackenzie R M. Tuberculosis in captive seals: bacteriological studies on an isolate belonging to the Mycobacterium tuberculosis complex. Res Vet Sci. 1990;48:196–200. [PubMed] [Google Scholar]
- 5.de Kantor I N, Ritacco V. Bovine tuberculosis in Latin America and the Caribbean: current status, control and eradication programs. Vet Microbiol. 1994;40(1–2):5–14. doi: 10.1016/0378-1135(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 6.Del Portillo P, Murillo L A, Patarroyo M E. Amplification of a species-specific DNA fragment of Mycobacterium tuberculosis and its possible use in diagnosis. J Clin Microbiol. 1991;29:2163–2168. doi: 10.1128/jcm.29.10.2163-2168.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinosa de los Minteros L E, Galan J C, Gutierrez M, Samper S, Garcia Marin J F, Martin C, Dominguez L, de Rafael L, Baquero F, Gomez-Mampaso E, Blazquez J. Allele-specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: intraspecific M. bovis pncA sequence polymorphism. J Clin Microbiol. 1998;36:239–242. doi: 10.1128/jcm.36.1.239-242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanning A, Edwards S. Mycobacterium bovis infection in human beings in contact with elk (Cervus elaphus) in Alberta, Canada. Lancet. 1991;338:1253–1255. doi: 10.1016/0140-6736(91)92113-g. [DOI] [PubMed] [Google Scholar]
- 9.Grange J M, Yates M D. Guidelines for speciation within the Mycobacterium tuberculosis complex. WHO/Zoom./94.174. Geneva, Switzerland: World Health Organization Veterinary Public Health Unit; 1994. [Google Scholar]
- 10.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liebana E, Aranaz A, Francis B, Cousins D. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J Clin Microbiol. 1996;34:933–938. doi: 10.1128/jcm.34.4.933-938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Reilly L M, Daborn C J. The epidemiology of Mycobacterium bovis infection in animals and man: a review. Tubercle Lung Dis. 1995;76:1–46. doi: 10.1016/0962-8479(95)90591-x. [DOI] [PubMed] [Google Scholar]
- 13.O’Reilly L M. Specificity and sensitivity of tuberculin test. In: Moussa A A M, Lotfi O, Mahair S, et al., editors. Proceedings of the International Conference on Animal Tuberculosis in Africa and the Middle East. Cairo, Egypt: General Organization for Veterinary Services; 1992. pp. 83–139. [Google Scholar]
- 14.Rodriguez J G, Mejia G A, Del Portillo P, Patarroyo M E, Murillo L A. Species-specific identification of Mycobacterium bovis by PCR. Microbiology. 1995;141:2131–2138. doi: 10.1099/13500872-141-9-2131. [DOI] [PubMed] [Google Scholar]
- 15.Romano M I, Alito A, Bigi F, Fisanotti J C, Cataldi A. Genetic characterization of mycobacteria from South American wild seals. Vet Microbiol. 1995;47:89–98. doi: 10.1016/0378-1135(95)00103-h. [DOI] [PubMed] [Google Scholar]
- 16.Stonebrink B. The use of a pyruvate containing egg medium in the culture of isoniazid resistant strains of Mycobacterium tuberculosis var. Hominis. Acta Tuberc Scand. 1958;35:67–80. [Google Scholar]
- 17.Van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. The occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of IS-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weil A, Plikaytis B B, Butler W R, Woodley C L, Shinnick T M. The mtp40 gene is not present in all strains of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:2309–2311. doi: 10.1128/jcm.34.9.2309-2311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. Zoonotic tuberculosis (Mycobacterium bovis): memorandum from a WHO meeting (with the participation of FAO) Bull W H O. 1994;72:851–857. [PMC free article] [PubMed] [Google Scholar]