Abstract
Environmental pressure to reduce our reliance on agrochemicals and the necessity to increase crop production in a sustainable way have made the rhizosphere microbiome an untapped resource for combating challenges to agricultural sustainability. In recent years, substantial efforts to characterize the structural and functional diversity of rhizosphere microbiomes of the model plant Arabidopsis thaliana and various crops have demonstrated their importance for plant fitness. However, the plant benefiting mechanisms of the rhizosphere microbiome as a whole community rather than as an individual rhizobacterium have only been revealed in recent years. The underlying principle dominating the assembly of the rhizosphere microbiome remains to be elucidated, and we are still struggling to harness the rhizosphere microbiome for agricultural sustainability. In this review, we summarize the recent progress of the driving factors shaping the rhizosphere microbiome and provide community-level mechanistic insights into the benefits that the rhizosphere microbiome has for plant fitness. We then propose the functional compensatory principle underlying rhizosphere microbiome assembly. Finally, we suggest future research efforts to explore the rhizosphere microbiome for agricultural sustainability.
Keywords: Rhizosphere, Driving factor, Plant beneficial function, Assembly pattern, Sustainable agriculture
1. Introduction
Plants harbor diverse microbiome inhabitants, including bacteria, fungi, viruses, protists, and nematodes. These microorganisms are key components of the host plant and can colonize outside and inside of the plant tissue, referring to the rhizosphere (a narrow zone influenced by plant roots), phyllosphere (aboveground plant parts, particularly the leaves), anthosphere (a zone around the flowers, a subdivision of phyllosphere), spermosphere (a habitat surrounding the seeds where the soil, germinating seeds, and the microbial communities interact) and endosphere microbiomes (inside plant parts) [1]. The plant and its associated microbiomes are proposed to function as a holobiont, which is a consequence of evolutionary selection between plants and microorganisms [2]. Compared to the other plant compartments and the bulk soil, the rhizosphere, in which the microbial abundance, density and activity are largely increased, is considered the second genome of plants [3]. Therefore, the rhizosphere is a hotspot for plant-microbiome-soil interactions and serves as the gateway for plants to uptake nutrients and the first line of defense against different biotic and abiotic stresses [4]. Consequently, the rhizosphere microbiome can potentially be manipulated to increase crop yields and to reduce chemical fertilizer and pesticide inputs [5].
The beneficial roles of some plant growth-promoting rhizobacteria (PGPR) have been extensively studied over the past decades [6]. For example, some Bacillus spp. and Pseudomonas spp. members can not only stimulate induced systemic resistance (ISR) of the host plant and produce antibiotics to suppress pathogens, but also secrete secondary metabolites to promote growth or to enhance abiotic stress tolerance of the host plant [7], [8]. However, these functions depend on the abiotic conditions and the biological interactions for these members to exert their plant beneficial properties in a rhizosphere community with high species diversity [9], [10]. To achieve a more comprehensive understanding of how to make full use of the plant beneficial functions of rhizosphere microorganisms, first, it is important to characterize the principles that govern the assembly process and drive the diversity and composition of the rhizosphere microbiome. Next, the complicated interactions within the rhizosphere microbiome and between the rhizosphere microbiome and its host plant and their consequences on promoting plant growth and development, improving plant nutrient acquisition, increasing abiotic stress tolerance and enhancing pathogen suppression of the plants need to be identified. Furthermore, integrated strategies exploiting microbial functions and plant traits need to be implemented in the sustainable development of agriculture.
In this review, we summarize the recent progress of the driving factors underlying the assembly of the rhizosphere microbiome. Next, we review mechanistic insights into the benefits of the rhizosphere microbiome on plant fitness. Finally, we discuss the new reductionist approaches for the development of synthetic microbial inoculants and the challenges for exploiting the beneficial outcomes of the rhizosphere microbiome.
2. Factors driving the assembly of the rhizosphere microbiome
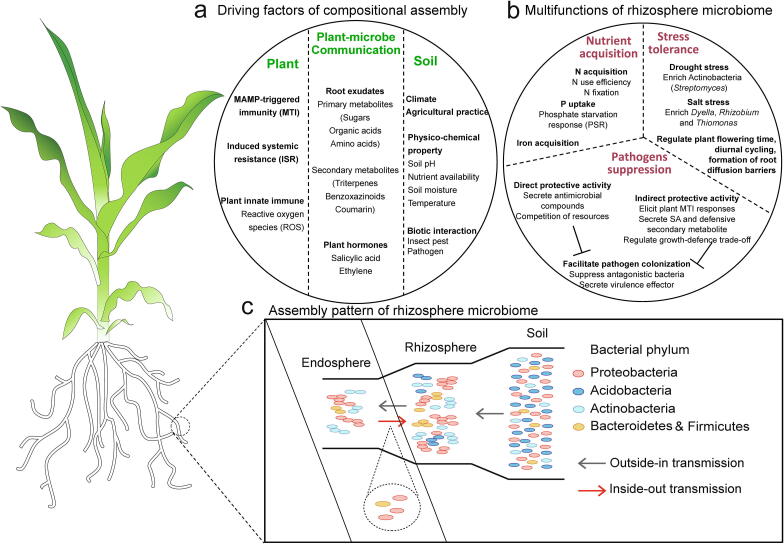
Most of the dominant members in the rhizosphere microbial community are generally fast-growing r-strategists that are attracted by and feed on the abundant carbon substrates released by the plant root [11]. Independent studies have depicted that the bacterial taxa detected in the rhizosphere mostly belong to the phylum Proteobacteria, followed by Actinobacteria and Firmicutes [12], [13], [14]. Numerous studies have shown that the composition and relative abundance of rhizosphere microbial populations are plant species-specific (plant dominated) or location-specific (soil dominated) [14], [15], [16]. The assembly of the rhizosphere microbiome is governed by the abiotic factors of soil properties and climate and the biotic factors of plant species, plant immunity and biotic interactions [15], [17], [18], [19] (Fig. 1a).
Fig. 1.
The mechanisms underlying assembly of the plant rhizosphere microbiome. (a) The driving factors of rhizosphere microbiome assembly that are subjected to soil, plant and plant-microbe communications. (b) The beneficial effects of the plant rhizosphere microbiome on promoting nutrient acquisition, soil-borne pathogen suppression and stress tolerance of the host plant. (c) The assembly patterns of the rhizosphere microbiome involve both outside-in recruitment of the soil microbiome and inside-out release of the endosphere microbiome. The vector graph of the maize plant is derived from https://bookstore.ksre.ksu.edu.
First, the soil microbiome is considered the microbial seed bank of the rhizosphere microbiome [20]. Physico-chemical properties, such as soil pH, nutrient availability, soil moisture and temperature, among different soil types or along biogeographical distance patterns shape the structure of the soil microbial community [21], [22], [23], [24], which determines the initiation of rhizosphere community assembly. In particular, anthropogenic activities, generally agricultural management practices, have profound impacts on the physico-chemical properties of the soil, consequently influencing the soil and the rhizosphere microbial community [25], [26]. Amplicon sequencing-based studies focusing on the rhizosphere microbial communities of different A. thaliana populations have demonstrated that soil type is the primary driving factor affecting the rhizosphere bacterial community [15]; however, climate is more important for the rhizosphere fungal community than soil type [27].
Compared to the soil type, another strong determinant of the rhizosphere microbiome composition is the host plant. The differences in plant genotype-induced specific rhizodeposits, among both inter- and intraspecies of the plant, can strongly affect the colonization of rhizosphere microbes [28]. A recent genome-wide association study revealed that several annotated candidate genes within the chromosome 4 locus of the sorghum genome are likely to control the high heritability of Planctomycetes and Verrucomicrobia in the rhizosphere microbiome [29]. Similar results in the rhizosphere bacterial and archaeal communities of common bean emerged, in which a set of core bacterial taxa of Proteobacteria, Acidobacteria, Actinobacteria and Verrucomicrobia were consistently detected, regardless of the growing region, plant development, and divergent genotypes [30]. These genotypic differences of plants are likely to attract different microbes through their differences in the biosynthesis of specific metabolites, which may trigger multiple responses in different soil microorganisms [31]. Three divergent pathways for the biosynthesis of root triterpenes in the roots of A. thaliana were reconstituted, and their irreplaceable role as carbon sources in establishing an Arabidopsis-specific root microbial community was confirmed [32]. Moreover, the synthesis of the plant defense hormone salicylic acid can alter the composition of the rhizosphere microbial community in A. thaliana through the selection of tolerant microbes [33]. These heritable or specific metabolite-attractive/resistant members discovered in the rhizosphere microbiome are in line with host preference. This host preference could probably be linked to specific transcriptional reprogramming in roots to format host species-specific rhizosphere niches; hence, commensal bacteria confer a competitive advantage in their cognate host [34]. However, these host species-specific interactions are not invariable. For instance, the development of high-yielding varieties and the agronomic management of modern agriculture inadvertently modified the root-microbe interactions in maize, in which fewer microorganisms responsible for nitrogen (N) acquisition but more members contributing to N losses were recruited in the rhizosphere of recently developed cultivars [35]. In contrast, adaptive evolution by improving the competitiveness for root exudates or enhancing the tolerance to the plant-secreted antimicrobial metabolites of a nonplant-associated microbe may promote its established reciprocal symbiosis relationship with the host plant [36].
In addition to soil properties and specific metabolites, the plant immune system has become a priority in understanding the mechanisms that govern the distribution and abundance of plant rhizosphere microbes. Numerous commensal species can express the immunogenic microbe-associated molecular patterns (MAMPs) that are found in both plant pathogens and beneficial members, indicating the potential of these microbes to trigger immune responses in their host plants, known as MAMP-triggered immunity (MTI) [37]. In response, these rhizosphere microbes have evolved mechanisms to suppress host defense responses. For instance, the pathogenic fungus Magnaporthe oryzae can produce effectors that target the host jasmonic acid (JA) signaling pathway [38], while commensal and beneficial microbes may evade recognition by programming diverse peptide derivatives of flagellin (flg22) [39]. In addition to MAMP perception, the reactive oxygen species (ROS) burst is a critical step of innate immune activation in plants. The model pathogenic bacterium Pseudomonas syringae pv. tomato DC3000 is able to synthesize a newly discovered small molecule, phevamine A, to suppress a potentiation effects of spermidine and L-arginine on the ROS burst generated upon recognition of bacterial flagellin [40]. Moreover, FERONIA-mediated ROS production in A. thaliana, which is largely independent of its immune scaffold function, plays a role in development and jasmonic acid autoimmunity and has been proven to regulate beneficial pseudomonads specifically in the rhizosphere [41].
3. Plant-beneficial functions of the rhizosphere microbiome
Plants recruit a set of rhizosphere microbial populations to provide essential functions for plant growth and health. Currently, many studies have focused on the description of the taxonomic composition of the rhizosphere microbiome. Insights into the functions of the rhizosphere microbiome are pivotal to improving plant fitness. However, the dissection of plant-microbiome interactions is highly complicated regarding the diversity of the rhizosphere microbiome. In recent years, a reductionist approach to the synthetic community (SynCom) has been developed to study the specific mechanisms that drive community assembly and the interactions among different members in a gnotobiotic system [42], [43]. For instance, the consistent differences between the maize phenotypes of inbred lines and hybrids and the composition of rhizosphere bacterial and fungal communities [44] have been further confirmed by inoculation with a simple community of seven bacterial strains [45].This reductionist approach would allow global research to replicate experiments easily through two standardized pipelines: high-throughput bacterial cultivation and identification [46], [47] and the establishment of plant growth systems [48].
The rhizosphere microbiome confers fitness advantages to the plant host, including growth promotion, nutrient acquisition, soil-borne pathogen suppression and stress tolerance (Fig. 1b). Many members of the rhizosphere microbiome have been shown to regulate plant growth [49]. Recent studies have demonstrated that a wide diversity of rhizosphere bacterial strains are able to inhibit root growth and can switch the plant response to low doses of glyphosate from growth promotion to growth inhibition [50]; however, the reversion of root growth inhibition could be assigned to a single bacterial genus of Variovorax, which contains a conserved auxin-degradation operon [51].
3.1. Nutrient acquisition
Plant-microbe interactions trigger the essential functions of the rhizosphere microbiome in improving plant nutrition. PGPRs can increase plant nutrient acquisition by either enhancing the availability of soil nutrients or improving the root system architecture of host plants. For instance, a better nitrogen use efficiency (NUE) of Oryza indica than Oryza japonica varieties is attribute to the recruitment of a higher proportion of N cycle-related bacteria in the rhizosphere of the O. indica varieties regulating by the host NRT1.1B gene [52]. Notably, inoculation of a 16-member indica-enriched SynCom confirmed its contribution to higher NUE in O. indica varieties. In maize varieties, a recent study established that root-derived flavones predominantly promote the enrichment of Oxalobacteraceae in the rhizosphere, which in turn improves LRT1-mediated lateral root development and promotes maize growth and N acquisition [53]. Sweetcorn displays a unique root exudate composition and recruits N-fixing bacterial taxa of Burkholderia, Rhizobium, and Sphingomonas in the rhizosphere among different maize germplasm groups [16]. To exploit the role of metabolites in activating plant-microbe interactions, Geddes et al. [54] expressed a synthetic pathway for the production of rhizopine scyllo-inosamine in both Medicago truncatula and barley, which was recognized by rhizobia associated with both the legume and cereal tissues.
The plant rhizosphere microbiome can also improve phosphorous (P) and iron nutrition under phosphate starvation and iron deprivation conditions, respectively. Under phosphate-limiting conditions, the master transcriptional regulator PHR1-mediated phosphate starvation response (PSR) in A. thaliana contributes to the assembly of the rhizosphere microbiome, and further inoculations of SynComs with different complexities demonstrate that the rhizosphere microbiome can enhance the transcription of plant PSR genes and promote the uptake of inorganic phosphate [55]. This PSR-induced rhizosphere microbiome assembly may encompass both plant-adaptive and opportunistic strategies, in which opportunistic colonizers are found to exacerbate phosphate starvation of the host plant [56]. In addition, the previously underestimated role of plant-fungus interactions in P cycling has been demonstrated to be enhanced in the rhizosphere of nonmycorrhizal plants to improve plant growth and P uptake in extremely P-limited soil [57]. It should be noted that the above study suggests a joint regulation of immune status associated with P acquisition regulated by the rhizosphere microbiome [55]. Similarly, the root-specific MYB72-regulated biosynthesis and BGLU42-dependent secretion of the coumarin scopoletin, an iron chelator with selective antimicrobial activity, altered the composition of the rhizosphere microbiome through its variable antimicrobial activity on specific bacterial taxa [58] and has important roles in both rhizobacteria-mediated iron acquisition and immune regulation under iron-limiting conditions [59].
3.2. Stress tolerance
The host plant is able to assemble a rhizosphere microbiome under abiotic or biotic stress conditions, which in turn modifies plant responses to environmental stresses. Drought is one of the most common environmental stresses. A coupled study of genome-resolved metagenomics and comparative genomics demonstrated that bacterial iron transport and metabolism functionality in the sorghum rhizosphere are highly correlated with their enrichment under drought stress [60]. Specifically, drought stress induced the loss of a phytosiderophore iron transporter and consequently led to the enrichment of Actinobacteria, while this enrichment could be disrupted by the exogenous application of iron. The prolonged drought-induced enrichment of endospheric Actinobacteria, especially Streptomyces, may have enduring impacts on rice and wheat [61], [62]. The regulation of the salt tolerance gene in rice under salt stress may involve the recruitment of some microbial species of Dyella, Rhizobium and Thiomonas [63]. Notably, Li et al. [64] suggest that it is a rhizosphere bacterial consortium, rather than individual members, that provides enduring resistance against salt stress. Furthermore, the plant flowering time has been proven to be altered by the rhizosphere microbiome [65]. The development of root diffusion barriers in the endodermis, especially suberin deposition, is critical for the mineral nutrient balance of plants and has been demonstrated to be mediated by the root microbial repression to the abscisic acid responses in the root [66]. These microbe-mediated regulations on plant flowering time and development root diffusion barriers have opened up multiple opportunities to develop crops that are more adapted to extreme environmental conditions.
3.3. Pathogen suppression
To establish infection, plant pathogens secrete effectors, which are secreted molecules that support host colonization, to promote disease development during colonization of their hosts [67]. For instance, the fungal pathogen Verticillium dahlia can suppress antagonistic bacteria and manipulate the rhizosphere microbiome by secreting the virulence effector VdAve1, which may facilitate its colonization of tomato and cotton [68]. The dysbiosis of the protective gram-positive bacterial community promotes pathogen invasion and disease progress [69]. In response to pathogen invasion, rhizosphere microbes can modify plant evolutionary responses through multiple pathways [70]. First, the rhizosphere microbiome provides direct protection against soil-borne pathogens, including secretion of antimicrobial compounds to inhibit the growth and virulence of pathogens [71] and competition for resources such as carbon, nitrogen and iron [72], [73]. Then, plant immunity could be activated by rhizosphere microbes to enhance disease resistance [69]. Individual rhizosphere microbes can either elicit or dampen MTI responses to modulate host susceptibility to pathogens [74]. Interestingly, organic crop and soil management triggers enrichment of Micrococcales in the rhizosphere to induce high salicylic acid levels in plant and consequently improve plant resistance to herbivorous insects [75]. The PGPR strain Bacillus amyloliquefaciens GB03 elicits the release of β-caryophyllene, a plant volatile organic compound, by the host plant, which promotes downstream impacts on salicylic acid secretion in the roots of neighboring seedlings [76]. Finally, the rhizosphere microbiome can regulate the growth-defense trade-off of the host plant [77]. For instance, the transcriptional regulator MYC2 in A. thaliana regulates a microbiome-root-shoot circuit favoring growth over defense under suboptimal light [78]. Defensive secondary metabolites of the benzoxazinoids that are released by cereal roots increase jasmonate signaling for plant defense, however, plant growth can be decreased and the rhizosphere microbial composition is modified to improve insect herbivore resistance, even in the next plant generation [79]. The concept of a “soil-borne legacy” suggests a suppressive memory to benefit future plant generations growing in the same soil [80], [81], [82]. This suppressive memory to boost plant health is thought to be an beneficial microbial attractive result by the modification plant root exudates [83].
4. Assembly patterns of the rhizosphere microbiome
Despite the important roles of the rhizosphere microbiome in plant nutrient acquisition, stress tolerance and pathogen suppression, the current understanding of the assembly pattern in the rhizosphere is still in its infancy. Bulgarelli et al. [84] first proposed a two-step selection process for the root-associated microbiome, in which rhizodeposition fuels an initial substrate-driven community shift in the rhizosphere and the host genotype functions in further selection in the endosphere. On this basis, a three-step enrichment model for the assembly of the root-associated microbiome from bulk soil toward roots was presented, which introduced a microhabitat of the rhizoplane at the soil-root interface [85]. Besides, a multistep model suggests that the rhizoplane plays a selective gating role in root microbiome assembly from soil [14]. Recently, an “amplification-selection” model was proposed for the assembly of the rhizosphere microbiome [86]. However, these assembly models only considered the horizontal recruitment of root-associated microbiomes from the bulk soil. Generally, plants actively assemble their rhizosphere microbiomes from surrounding microbial reservoirs, including dominant horizontal transference from the bulk soil and a small portion of vertical transmission from the seed [87]. Recently, a new assembly pattern of “functional compensation”, i.e., the rhizosphere microbiome is assembled to compensate for the functional requirements of the host plant that cannot be completed by itself, has been proposed to dominate the assembly of the rhizosphere bacterial community [17]. This assembly pattern involves both outside-in recruitment of the soil microbiome and inside-out release of the endosphere microbiome (Fig. 1c). Shao et al. [88] detected a massive compensatory colonization of maize seed-borne phosphate-solubilizing bacteria, Burkholderia gladioli, in the rhizosphere when cultivated in P-limited soil. Therefore, plants recruit different rhizosphere bacterial communities to satisfy the functional requirements for their fitness under different soil conditions.
5. Future perspectives – exploiting the rhizosphere microbiome for sustainable agricultural production
The increasing world population for more food and feed production, the environmental concern for overuse of chemical fertilizers and pesticides, and the soil quality issues on plant debris turnover and organic carbon pool improvement are the major challenges for sustainable agriculture. Solutions to those challenges necessitate multiple approaches, and exploring the rhizosphere microbiome to support crop production is even more urgent. The rhizosphere microbiome can be manipulated or engineered to favor crop growth and health through agricultural management practices, and breeding for beneficial plant–microbe interactions will be an additional promising approach to make better use of the rhizosphere microbiome for sustainable agriculture.
Agricultural practices are usually recognized to have a negative effect on plant associations of the rhizosphere microbiome. Agrochemical applications, including fungicides and chemical soil disinfection used to suppress microbial pathogens, disrupt the dynamic balance of the microbial community and its functions [89]. Continued monoculture and soil tillage reduce soil and rhizosphere microbial diversity [90]. Precision agronomic practices integrating microbiome function will be a promising approach for the future. The agricultural practices that promote crop diversity (for example, rotations or intercropping), inoculation of beneficial microbial mixes (for example, N-fixing or P-solubilizing biological soil inoculant) and application of organic soil amendments (for example, manure or compost) or generally minimize environmental disturbance have been shown to promote rhizosphere microbiomes [91], [92]. Understanding how agricultural practices influence the rhizosphere microbiome may lead to strategies to modulate the rhizosphere microbiome in a desired direction.
The domestication and breeding of modern crop cultivars have affected the association of the rhizosphere microbiome. Modern crops have been shown to have a compromised ability to sustain relationships with mycorrhizal fungi and plant growth-promoting rhizobacteria [93]. Wild crops have evolved a specific rhizosphere microbiome, but this selection was disrupted with domestication, which has led not only to the loss of crop genetic diversity but also to a reduction in microbial diversity associated with crops and a loss of the capacity to interact with specific beneficial plant microorganisms [94]. It is possible to breed or select plants with the ability to attract a beneficial rhizosphere microbiome. Recent studies have demonstrated that the core rhizosphere functional microbiomes are determined by crop genetics [29], [52], which provides the possibility to include the traits of the rhizosphere microbiome as a breeding target. Yet, successful commercial modern crop cultivars targeting beneficial microbiomes are scarce, the limitations that impede these breeding efforts are the lack of understanding of plant mechanisms controlling the assembly of the rhizosphere microbiome and the infrequent of required microbes in highly diverse soil microbiomes. Future crop breeding should consider the associated rhizosphere microbiome within the holobiont to confer additional beneficial traits for sustainable agricultural production, which is highly promising in regard to delivering a new generation of microbe-improved plants.
The necessity to increase crop production in a sustainable way has become more urgent when considering climate change and world population growth; thus, harnessing the rhizosphere microbiome to fuel a future of sustainable agriculture is a promising strategy. However, a one-size-fits-all approach to better use rhizosphere microbiomes is unrealistic; there is a clear need for improved integration of microbial technology and host genetics to develop best management practices and breeding to sustain agricultural production.
Funding
This work was supported by funding from the National Natural Science Foundation of China (32072675) to W.X. and the Young Elite Scientists Sponsorship Program by CAST (2018QNRC001) to W.X., J.S., and R.Z.
CRediT authorship contribution statement
Weibing Xun: Conceptualization, Writing - review & editing, Funding acquisition. Jiahui Shao: Writing - original draft. Qirong Shen: Supervision. Ruifu Zhang: Conceptualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Berg G., Rybakova D., Grube M., Köberl M. The plant microbiome explored: implications for experimental botany. J Exp Bot. 2016;67(4):995–1002. doi: 10.1093/jxb/erv466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206(4):1196–1206. doi: 10.1111/nph.2015.206.issue-410.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- 3.Berendsen R.L., Pieterse C.M.J., Bakker P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17(8):478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R., Vivanco J.M., Shen Q. The unseen rhizosphere root–soil–microbe interactions for crop production. Curr Opin Microbiol. 2017;37:8–14. doi: 10.1016/j.mib.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Kavamura V.N., Mendes R., Bargaz A., Mauchline T.H. Defining the wheat microbiome: Towards microbiome-facilitated crop production. Comput Struct Biotechnol J. 2021;19:1200–1213. doi: 10.1016/j.csbj.2021.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63(1):541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 7.Aleti G., Sessitsch A., Brader G. Genome mining: Prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput Struct Biotechnol J. 2015;13:192–203. doi: 10.1016/j.csbj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zboralski A., Filion M. Genetic factors involved in rhizosphere colonization by phytobeneficial Pseudomonas spp. Comput Struct Biotechnol J. 2020;18:3539–3554. doi: 10.1016/j.csbj.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassan M.K., McInroy J.A., Kloepper J.W. The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: A review. Agriculture. 2019;9:142. doi: 10.3390/agriculture9070142. [DOI] [Google Scholar]
- 10.Sindhu S.S., Sehrawat A., Sharma R., Dahiya A., Khandelwal A. In: Plant-Microbe Interactions in Agro-Ecological Perspectives: Volume 2: Microbial Interactions and Agro-Ecological Impacts. Singh D.P., Singh H.B., Prabha R., editors. Springer; Singapore: 2017. Belowground microbial crosstalk and rhizosphere biology; pp. 695–752. [DOI] [Google Scholar]
- 11.Vandenkoornhuyse P., Mahe S., Ineson P., Staddon P., Ostle N., Cliquet J.-B. Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. Proc Natl Acad Sci USA. 2007;104(43):16970–16975. doi: 10.1073/pnas.0705902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeAngelis K.M., Brodie E.L., DeSantis T.Z., Andersen G.L., Lindow S.E., Firestone M.K. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3(2):168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 13.Na X., Xu T., Li M., Zhou Z., Ma S., Wang J. Variations of bacterial community diversity within the rhizosphere of three phylogenetically related perennial shrub plant species across environmental gradients. Front Microbiol. 2018;9:709. doi: 10.3389/fmicb.2018.0070910.3389/fmicb.2018.00709.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N.K., Bhatnagar S. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112(8):E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg D.S., Lebeis S.L., Paredes S.H., Yourstone S., Gehring J., Malfatti S. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters W.A., Jin Z., Youngblut N., Wallace J.G., Sutter J., Zhang W. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc Natl Acad Sci USA. 2018;115(28):7368–7373. doi: 10.1073/pnas.1800918115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren Y.i., Xun W., Yan H.e., Ma A., Xiong W.u., Shen Q. Functional compensation dominates the assembly of plant rhizospheric bacterial community. Soil Biol Biochem. 2020;150:107968. doi: 10.1016/j.soilbio.2020.107968. [DOI] [Google Scholar]
- 18.Chaparro J.M., Badri D.V., Vivanco J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8(4):790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiffer J.A., Spor A., Koren O., Jin Z., Tringe S.G., Dangl J.L. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA. 2013;110(16):6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lennon J.T., Jones S.E. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9(2):119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 21.Xun W., Huang T., Zhao J., Ran W., Wang B., Shen Q. Environmental conditions rather than microbial inoculum composition determine the bacterial composition, microbial biomass and enzymatic activity of reconstructed soil microbial communities. Soil Biol Biochem. 2015;90:10–18. doi: 10.1016/j.soilbio.2015.07.018. [DOI] [Google Scholar]
- 22.Xun W., Li W., Xiong W.u., Ren Y.i., Liu Y., Miao Y. Diversity-triggered deterministic bacterial assembly constrains community functions. Nat Commun. 2019;10:3833. doi: 10.1038/s41467-019-11787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahram M., Hildebrand F., Forslund S.K., Anderson J.L., Soudzilovskaia N.A., Bodegom P.M. Structure and function of the global topsoil microbiome. Nature. 2018;560(7717):233–237. doi: 10.1038/s41586-018-0386-6. [DOI] [PubMed] [Google Scholar]
- 24.van den Hoogen J., Geisen S., Routh D., Ferris H., Traunspurger W., Wardle D.A. Soil nematode abundance and functional group composition at a global scale. Nature. 2019;572(7768):194–198. doi: 10.1038/s41586-019-1418-6. [DOI] [PubMed] [Google Scholar]
- 25.Xun W., Zhao J., Xue C., Zhang G., Ran W., Wang B. Significant alteration of soil bacterial communities and organic carbon decomposition by different long-term fertilization management conditions of extremely low-productivity arable soil in South China. Environ Microbiol. 2016;18(6):1907–1917. doi: 10.1111/1462-2920.13098. [DOI] [PubMed] [Google Scholar]
- 26.Lekberg Y., Arnillas C.A., Borer E.T., Bullington L.S., Fierer N., Kennedy P.G. Nitrogen and phosphorus fertilization consistently favor pathogenic over mutualistic fungi in grassland soils. Nat Commun. 2021;12:3484. doi: 10.1038/s41467-021-23605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiergart T., Durán P., Ellis T., Vannier N., Garrido-Oter R., Kemen E. Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat Ecol Evol. 2020;4(1):122–131. doi: 10.1038/s41559-019-1063-3. [DOI] [PubMed] [Google Scholar]
- 28.Yeoh Y.K., Dennis P.G., Paungfoo-Lonhienne C., Weber L., Brackin R., Ragan M.A. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat Commun. 2017;8:215. doi: 10.1038/s41467-017-00262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng S, Caddell DF, Xu G, Dahlen L, Washington L, Yang J, et al. Genome wide association study reveals plant loci controlling heritability of the rhizosphere microbiome. ISME J 2021. In press. 10.1038/s41396-021-00993-z. [DOI] [PMC free article] [PubMed]
- 30.Stopnisek N, Shade A. Persistent microbiome members in the common bean rhizosphere: an integrated analysis of space, time, and plant genotype. ISME J 2021. in press. 10.1038/s41396-021-00955-5. [DOI] [PMC free article] [PubMed]
- 31.Zhalnina K., Louie K.B., Hao Z., Mansoori N., da Rocha U.N., Shi S. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3(4):470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 32.Huang A.C., Jiang T., Liu Y.-X., Bai Y.-C., Reed J., Qu B. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science. 2019;364(6440):eaau6389. doi: 10.1126/science:aau6389. [DOI] [PubMed] [Google Scholar]
- 33.Lebeis S.L., Paredes S.H., Lundberg D.S., Breakfield N., Gehring J., McDonald M. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349(6250):860–864. doi: 10.1126/science:aaa8764. [DOI] [PubMed] [Google Scholar]
- 34.Wippel K, Tao K, Niu Y, Zgadzaj R, Kiel N, Guan R, et al. Host preference and invasiveness of commensal bacteria in the Lotus and Arabidopsis root microbiota. Nat Microbiol 2021. In press. 10.1038/s41564-021-00941-9. [DOI] [PMC free article] [PubMed]
- 35.Favela A., Bohn M.O., Kent A.D. Maize germplasm chronosequence shows crop breeding history impacts recruitment of the rhizosphere microbiome. ISME J. 2021;15(8):2454–2464. doi: 10.1038/s41396-021-00923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li E., de Jonge R., Liu C., Jiang H., Friman V.-P., Pieterse C.M.J. Rapid evolution of bacterial mutualism in the plant rhizosphere. Nat Commun. 2021;12:3829. doi: 10.1038/s41467-021-24005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teixeira P.J.P., Colaianni N.R., Fitzpatrick C.R., Dangl J.L. Beyond pathogens: microbiota interactions with the plant immune system. Curr Opin Microbiol. 2019;49:7–17. doi: 10.1016/j.mib.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Plett J.M., Daguerre Y., Wittulsky S., Vayssieres A., Deveau A., Melton S.J. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc Natl Acad Sci USA. 2014;111(22):8299–8304. doi: 10.1073/pnas.1322671111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colaianni N.R., Parys K., Lee H.-S., Conway J.M., Kim N.H., Edelbacher N. A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe. 2021;29(4):635–649.e9. doi: 10.1016/j.chom.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 40.O’Neill E.M., Mucyn T.S., Patteson J.B., Finkel O.M., Chung E.-H., Baccile J.A. Phevamine A, a small molecule that suppresses plant immune responses. Proc Natl Acad Sci USA. 2018;115(41):E9514–E9522. doi: 10.1073/pnas.1803779115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Y.i., Wilson A.J., Zhang X.-C., Thoms D., Sohrabi R., Song S. FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat Plants. 2021;7(5):644–654. doi: 10.1038/s41477-021-00914-0. [DOI] [PubMed] [Google Scholar]
- 42.Vorholt J.A., Vogel C., Carlström C.I., Müller D.B. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe. 2017;22(2):142–155. doi: 10.1016/j.chom.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Bengtsson-Palme J. Microbial model communities: To understand complexity, harness the power of simplicity. Comput Struct Biotechnol J. 2020;18:3987–4001. doi: 10.1016/j.csbj.2020.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner M.R., Roberts J.H., Balint‐Kurti P., Holland J.B. Heterosis of leaf and rhizosphere microbiomes in field-grown maize. New Phytol. 2020;228(3):1055–1069. doi: 10.1111/nph.v228.310.1111/nph.16730. [DOI] [PubMed] [Google Scholar]
- 45.Wagner MR, Tang C, Salvato F, Clouse KM, Bartlett A, Vintila S, et al. Microbe-dependent heterosis in maize. Proc Natl Acad Sci USA 2021;118:e2021965118. 10.1073/pnas.2021965118. [DOI] [PMC free article] [PubMed]
- 46.Zhang J., Liu Y.-X., Guo X., Qin Y., Garrido-Oter R., Schulze-Lefert P. High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat Protoc. 2021;16(2):988–1012. doi: 10.1038/s41596-020-00444-7. [DOI] [PubMed] [Google Scholar]
- 47.Bai Y., Müller D.B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528(7582):364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 48.Kremer J.M., Sohrabi R., Paasch B.C., Rhodes D., Thireault C., Schulze-Lefert P. Peat-based gnotobiotic plant growth systems for Arabidopsis microbiome research. Nat Protoc. 2021;16(5):2450–2470. doi: 10.1038/s41596-021-00504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Backer R., Rokem J.S., Ilangumaran G., Lamont J., Praslickova D., Ricci E. Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci. 2018;9:1473. doi: 10.3389/fpls.2018.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramirez-Villacis D.X., Finkel O.M., Salas-González I., Fitzpatrick C.R., Dangl J.L., Jones C.D. Root microbiome modulates plant growth promotion induced by low doses of glyphosate. MSphere. 2020;5(4):00484-20. doi: 10.1128/mSphere.00484-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finkel O.M., Salas-González I., Castrillo G., Conway J.M., Law T.F., Teixeira P.J.P.L. A single bacterial genus maintains root growth in a complex microbiome. Nature. 2020;587(7832):103–108. doi: 10.1038/s41586-020-2778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J., Liu Y.-X., Zhang N.a., Hu B., Jin T., Xu H. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat Biotechnol. 2019;37(6):676–684. doi: 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- 53.Yu P., He X., Baer M., Beirinckx S., Tian T., Moya Y.A.T. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat Plants. 2021;7(4):481–499. doi: 10.1038/s41477-021-00897-y. [DOI] [PubMed] [Google Scholar]
- 54.Geddes B.A., Paramasivan P., Joffrin A., Thompson A.L., Christensen K., Jorrin B. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat Commun. 2019;10:3430. doi: 10.1038/s41467-019-10882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castrillo G., Teixeira P.J.P.L., Paredes S.H., Law T.F., de Lorenzo L., Feltcher M.E. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543(7646):513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finkel O.M., Salas-González I., Castrillo G., Spaepen S., Law T.F., Teixeira P.J.P.L. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 2019;17(11):e3000534. doi: 10.1371/journal.pbio.3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almario J., Jeena G., Wunder J., Langen G., Zuccaro A., Coupland G. Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc Natl Acad Sci USA. 2017;114(44):E9403–E9412. doi: 10.1073/pnas.1710455114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harbort C.J., Hashimoto M., Inoue H., Niu Y., Guan R., Rombolà A.D. Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe. 2020;28(6):825–837.e6. doi: 10.1016/j.chom.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stringlis I.A., Yu K.e., Feussner K., de Jonge R., Van Bentum S., Van Verk M.C. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci USA. 2018;115(22):E5213–E5222. doi: 10.1073/pnas.1722335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L., Dong Z., Chiniquy D., Pierroz G., Deng S., Gao C. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat Commun. 2021;12:3209. doi: 10.1038/s41467-021-23553-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santos-Medellín C., Liechty Z., Edwards J., Nguyen B., Huang B., Weimer B.C. Prolonged drought imparts lasting compositional changes to the rice root microbiome. Nat Plants. 2021;7(8):1065–1077. doi: 10.1038/s41477-021-00967-1. [DOI] [PubMed] [Google Scholar]
- 62.Si J., Froussart E., Viaene T., Vázquez-Castellanos J.F., Hamonts K., Tang L. Interactions between soil compositions and the wheat root microbiome under drought stress: From an in silico to in planta perspective. Comput Struct Biotechnol J. 2021;19:4235–4247. doi: 10.1016/j.csbj.2021.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lian T., Huang Y., Xie X., Huo X., Shahid M.Q., Tian L. Rice SST variation shapes the rhizosphere bacterial community, conferring tolerance to salt stress through regulating soil metabolites. MSystems. 2020;5(6):e00721-20. doi: 10.1128/mSystems.00721-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, La S, Zhang X, Gao L, Tian Y. Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J 2021. in press. 10.1038/s41396-021-00974-2. [DOI] [PMC free article] [PubMed]
- 65.Lu T., Ke M., Lavoie M., Jin Y., Fan X., Zhang Z. Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome. 2018;6:231. doi: 10.1186/s40168-018-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salas-González I., Reyt G., Flis P., Custódio V., Gopaulchan D., Bakhoum N. Coordination between microbiota and root endodermis supports plant mineral nutrient homeostasis. Science. 2021;371(6525):eabd0695. doi: 10.1126/science:abd0695. [DOI] [PubMed] [Google Scholar]
- 67.Rovenich H., Boshoven J.C., Thomma B.P. Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr Opin Plant Biol. 2014;20:96–103. doi: 10.1016/j.pbi.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Snelders N.C., Rovenich H., Petti G.C., Rocafort M., van den Berg G.C.M., Vorholt J.A. Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat Plants. 2020;6(11):1365–1374. doi: 10.1038/s41477-020-00799-5. [DOI] [PubMed] [Google Scholar]
- 69.Lee S.-M., Kong H.G., Song G.C., Ryu C.-M. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2021;15(1):330–347. doi: 10.1038/s41396-020-00785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vannier N., Agler M., Hacquard S., Zipfel C. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019;15(6):e1007740. doi: 10.1371/journal.ppat.1007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y., Wang J., Yang N., Wen Z., Sun X., Chai Y. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat Commun. 2018;9:3429. doi: 10.1038/s41467-018-05683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei Z., Yang T., Friman V.P., Xu Y., Shen Q., Jousset A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun. 2015;6:8413. doi: 10.1038/ncomms9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu S., Wei Z., Shao Z., Friman V.-P., Cao K., Yang T. Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat Microbiol. 2020;5(8):1002–1010. doi: 10.1038/s41564-020-0719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma K.-W., Niu Y., Jia Y., Ordon J., Copeland C., Emonet A. Coordination of microbe–host homeostasis by crosstalk with plant innate immunity. Nat Plants. 2021;7(6):814–825. doi: 10.1038/s41477-021-00920-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blundell R., Schmidt J.E., Igwe A., Cheung A.L., Vannette R.L., Gaudin A.C.M. Organic management promotes natural pest control through altered plant resistance to insects. Nat Plants. 2020;6(5):483–491. doi: 10.1038/s41477-020-0656-9. [DOI] [PubMed] [Google Scholar]
- 76.Kong H.G., Song G.C., Sim H.-J., Ryu C.-M. Achieving similar root microbiota composition in neighbouring plants through airborne signalling. ISME J. 2021;15(2):397–408. doi: 10.1038/s41396-020-00759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heil M., Baldwin I.T. Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 2002;7:61–67. doi: 10.1016/S1360-1385(01)02186-0. [DOI] [PubMed] [Google Scholar]
- 78.Hou S., Thiergart T., Vannier N., Mesny F., Ziegler J., Pickel B. A microbiota–root–shoot circuit favours Arabidopsis growth over defence under suboptimal light. Nat Plants. 2021;7(8):1078–1092. doi: 10.1038/s41477-021-00956-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu L., Robert C.A.M., Cadot S., Zhang X.i., Ye M., Li B. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun. 2018;9:2739. doi: 10.1038/s41467-018-05122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bakker P.A.H.M., Pieterse C.M.J., de Jonge R., Berendsen R.L. The soil-borne legacy. Cell. 2018;172(6):1178–1180. doi: 10.1016/j.cell.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 81.Berendsen R.L., Vismans G., Yu K.e., Song Y., de Jonge R., Burgman W.P. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12(6):1496–1507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hannula S.E., Heinen R., Huberty M., Steinauer K., De Long J.R., Jongen R. Persistence of plant-mediated microbial soil legacy effects in soil and inside roots. Nat Commun. 2021;12:5686. doi: 10.1038/s41467-021-25971-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan J., Zhao J., Wen T., Zhao M., Li R., Goossens P. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome. 2018;6:156. doi: 10.1186/s40168-018-0537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bulgarelli D., Schlaeppi K., Spaepen S., van Themaat E.V.L., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64(1):807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 85.Reinhold-Hurek B., Bünger W., Burbano C.S., Sabale M., Hurek T. Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol. 2015;53(1):403–424. doi: 10.1146/annurev-phyto-082712-102342. [DOI] [PubMed] [Google Scholar]
- 86.Wang X., Wang M., Xie X., Guo S., Zhou Y., Zhang X. An amplification-selection model for quantified rhizosphere microbiota assembly. Sci Bull. 2020;65(12):983–986. doi: 10.1016/j.scib.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 87.Berg G., Raaijmakers J.M. Saving seed microbiomes. ISME J. 2018;12(5):1167–1170. doi: 10.1038/s41396-017-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shao J., Miao Y., Liu K., Ren Y.i., Xu Z., Zhang N. Rhizosphere microbiome assembly involves seed-borne bacteria in compensatory phosphate solubilization. Soil Biol Biochem. 2021;159:108273. doi: 10.1016/j.soilbio.2021.108273. [DOI] [Google Scholar]
- 89.Hussain S, Siddique T, Saleem M, Arshad M, Khalid A. Chapter 5 Impact of pesticides on soil microbial diversity, enzymes, and biochemical reactions. Advances in Agronomy, vol. 102, Academic Press; 2009, p. 159–200. 10.1016/S0065-2113(09)01005-0. [DOI]
- 90.Kim N., Zabaloy M.C., Guan K., Villamil M.B. Do cover crops benefit soil microbiome? A meta-analysis of current research. Soil Biol Biochem. 2020;142:107701. doi: 10.1016/j.soilbio.2019.107701. [DOI] [Google Scholar]
- 91.Ling N., Zhu C., Xue C., Chen H., Duan Y., Peng C. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol Biochem. 2016;99:137–149. doi: 10.1016/j.soilbio.2016.05.005. [DOI] [Google Scholar]
- 92.Hartman K., van der Heijden M.G.A., Wittwer R.A., Banerjee S., Walser J.-C., Schlaeppi K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome. 2018;6:14. doi: 10.1186/s40168-017-0389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Valente J., Gerin F., Le Gouis J., Moënne‐Loccoz Y., Prigent–Combaret C. Ancient wheat varieties have a higher ability to interact with plant growth-promoting rhizobacteria. Plant Cell Environ. 2020;43(1):246–260. doi: 10.1111/pce:v43.110.1111/pce:13652. [DOI] [PubMed] [Google Scholar]
- 94.Porter S.S., Sachs J.L. Agriculture and the disruption of plant–microbial symbiosis. Trends Ecol Evol. 2020;35(5):426–439. doi: 10.1016/j.tree.2020.01.006. [DOI] [PubMed] [Google Scholar]