Abstract
Rationale & Objectives
Artificial intelligence driven by machine learning algorithms is being increasingly employed for early detection, disease diagnosis, and clinical management. We explored the use of machine learning–driven advancements in kidney research compared with other organ-specific fields.
Study Design
Cross-sectional bibliometric analysis.
Setting & Participants
ISI Web of Science database was queried using specific Medical Subject Headings (MeSH) terms about the organ system, journal International Standard Serial Number, and research methodology. In parallel, we screened the National Institutes of Health (NIH) RePORTER website to explore funded grants that proposed the use of machine learning as a methodology.
Predictors
Number of publications using machine learning as a research method.
Outcome
Articles were characterized by research methodology among 5 organ systems (brain, heart, kidney, liver, and lung). Grants funded by NIH for machine learning were characterized by study sections.
Analytical Approach
Percentages of articles using machine learning and other research methodologies were compared among 5 organ systems.
Results
Machine learning-based articles that are focused on the kidney accounted for 3.2% of the total relevant articles from the 5 organ systems. Specifically, brain research published over 19-fold higher number of articles than kidney research. As compared with machine learning, conventional statistical approaches such as the Cox proportional hazard model were used 9-fold higher in articles related to kidney research. In general, a lower utilization of machine learning–based approaches was observed in organ-specific specialty journals than the broad interdisciplinary journals. The digestive disease, kidney, and urology study sections funded 122 applications proposing machine learning–based approaches compared to 265 applications from the neurology, neuropsychology, and neuropathology study sections.
Limitations
Observational study.
Conclusions
Our analysis suggests lowest use of machine learning as a research tool among kidney researchers compared with other organ-specific researchers, underscoring a need to better inform the kidney research community about this emerging data analytic tool.
Index Words: Bibliometric analysis, kidney, machine learning, NIH funding, research methods
Plain-Language Summary.
Machine learning is an exciting research tool that increasingly is considered for early detection, disease diagnosis, and clinical management. We explored the use of machine learning–driven advancements in nephrology compared with other medical subspecialties. We did a bibliometric analysis employing a Web of Science database using specific search terms for organ systems and research methods. Our analysis suggested the lowest use of machine learning in nephrology compared with other medical subspecialties. Our study results highlight the importance of informing the kidney research community about this emerging data analytic tool.
Editorial, p. 693
Machine learning is rapidly emerging as an integral element in the repertoire of data analytic tools in a broad range of medical applications. With advances in hardware and software, advanced machine learning frameworks such as deep neural networks are increasingly being considered to process a range of biomedical datasets.1,2 In the context of kidney diseases and kidney health, a few examples include the application of machine learning to predict acute kidney injury using electronic health record data,3,4 use of digitized human kidney biopsies and deep learning to segment kidney structures5, 6, 7, 8 as well as predict clinical phenotypes,9 and analysis of radiological imaging data to measure total kidney volume.10 More examples can be found in a few recently published review articles,11, 12, 13, 14, 15 which are focused on educating the nephrology and the nephropathology communities on the merits and limitations of machine learning approaches.
Machine learning is a powerful data analytic tool that provides systems with the ability to automatically learn and improve from experience without being explicitly programmed. It is similar to several other tools that are available to the scientific community. When used appropriately, it has the potential to unravel interesting findings, such as how genome-wide association studies can identify new loci associated with kidney function and chronic kidney disease.16 Whether research in nephrology uses machine learning to the same extent as other fields is unknown. To better understand if kidney research has been keeping up with the pace of machine learning–driven advancements seen in other organ-specific fields, we conducted a bibliometric analysis to compare the number of manuscripts published using machine learning as a methodology among different organ systems and research areas. We also compared the funding sources of the machine learning manuscripts and the number of grants awarded that proposed machine learning as a research methodology.
Methods
Study Design and Data Collection
In this cross-sectional bibliometric analysis, we used the ISI Web of Science research database (WoS) to identify articles using machine learning methodologies.17 The WoS covers articles published since January 1, 1864. We performed our search on October 10, 2020. A detailed explanation of the Medical Subject Headings (MeSH) terms and Boolean commands used for the search strategy is available in Items S1 and S2. Briefly, we identified articles using machine learning and other methodologies in different journals using the International Standard Serial Number (ISSN) for journals and organ-specific MeSH words refined by specific research areas. We used the National Institutes of Health (NIH) RePORTER website18 to identify research grants given by NIH institutions for research projects using machine learning as a methodology. The database covers grant data between 1985 and 2020.
Because all the data used in this study is available to the public and does not contain any protected health information, we did not seek institutional review board approval. The study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guidelines for cross-sectional studies.
Outcomes and Measures
The WoS was searched to identify articles using methodological approach MeSH terms (machine learning, Cox-proportional hazard model, etc) and organ-specific MeSH terms (brain, heart, kidney, liver, and lung). We used search terms such as ELISA, PCR, Cox-proportional hazard model to represent traditional research tools and terms such as machine learning, CRISPR/Cas9, and GWAS to represent novel research tools in this analysis. We restricted our search to the areas of “neurosciences & neurology,” “cardiovascular system & cardiology,” “urology & nephrology,” “gastroenterology & hepatology,” and “respiratory system,” as defined by the WoS search glossary.
In the final analysis, we included only those articles that mentioned a specific methodology and the organ of interest and were tagged with a specific research area. Similarly, specific journals were searched using ISSN, organ name, and methodology in WoS. The journals were selected according to impact factor. We included journals focused on specific organ systems with high impact factors in the final analysis. We compared manuscripts restricted to our query across 5 different organs and different methodologies. Articles including original research, reviews, and meeting abstracts in all languages were included in our final analysis.
Using the NIH RePORTER website, we extracted data for research grants funded by various NIH institutions from 1985 to 2020, using the MeSH term “machine learning.” We extracted information on the awarding institute and type of grant—for instance, career development grants (K series), fellowship and training grants (F & T series), and R01 grants. We also searched journals to identify the number of machine learning papers that acknowledged specific NIH-level sponsors.
Statistical Analysis
We used descriptive statistics and compared proportions using the χ2 test. Statistical significance was set at 2-tailed P < 0.05. Statistical analysis was performed using GraphPad Prism (GraphPad Software).
Results
Articles Focused on Organ Systems
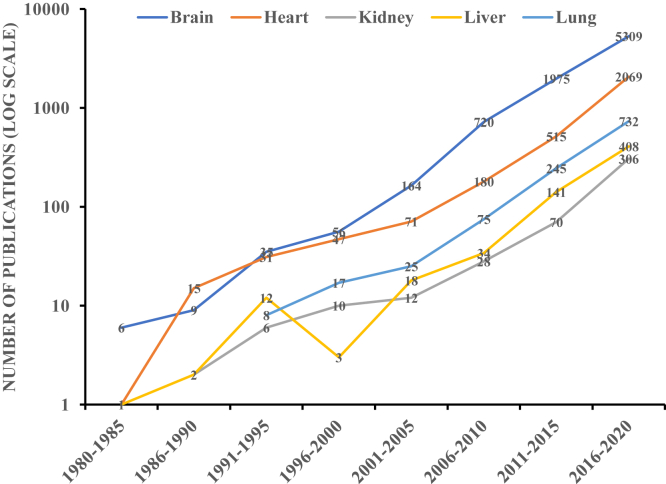
The WoS query identified a total of 388,169 articles across 5 research areas using organ-specific MeSH terms. Out of these articles, 13,373 (3.4%) belonged to the machine learning category. Among all the published machine learning articles, 434 (3.2%) articles were focused on kidney research (Table 1). Brain research had the highest number of research articles (61.9%, n = 8,278) published between 1952 and 2020 using machine learning, whereas kidney research had the lowest number of articles (3.2%, n = 434) published between 1989 and 2020. Also, the 5-year trends of published machine learning manuscripts across 5 organ-specific research areas indicated that brain research had the highest number of research articles whereas kidney research had the fewest research articles for a consistent duration (Fig 1).
Table 1.
Articles Focused on Specific Organs in the Web of Science Bibliometric Database Using Combination of Organ and Methodology Specific MeSH Term and Boolean Operators
| Methodologies | Brain (n = 96,225) | Heart (n = 89,846) | Kidney (n = 48,207) | Liver (n = 87,191) | Lung (66,700) | Total No. of Manuscripts per Row |
|---|---|---|---|---|---|---|
| Cox proportional hazard model | 2,118 (8.09%) | 13,286 (50.7%) | 4,087 (15.6%) | 2,971 (11.3%) | 3,715 (14.2%) | 26,177 |
| CRISPR/Cas9 | 662 (29.3%) | 560 (24.8%) | 251 (11.1%) | 427 (18.8%) | 361 (15.9%) | 2,261 |
| ELISA | 10,263 (17.8%) | 16,417 (28.5%) | 7,599 (13.2%) | 12,578 (21.9%) | 10,670 (18.5%) | 57,527 |
| GWAS | 991 (29.1%) | 1,436 (42.2%) | 219 (6.4%) | 340 (10%) | 413 (12.1%) | 3,399 |
| Machine learning | 8,278 (61.9%) | 2,931 (21.9%) | 434 (3.2%) | 619 (4.6%) | 1,111 (8.3%) | 13,373 |
| PCR | 36,786 (22.8%) | 31,119 (19.3%) | 20,627 (12.8%) | 42,095 (26%) | 30,754 (19%) | 161,381 |
| Western blot | 37,127 (29.9%) | 24,097 (19.4%) | 14,990 (12.08%) | 28,161 (22.7%) | 19,676 (15.9%) | 124,051 |
Percentages are calculated per total number of manuscripts in a specific row. Example of search terms: SU = (Neurosciences & Neurology) AND (AB = Methodology term AND Brain OR AK = Methodology term AND Brain)
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; ELISA, enzyme-linked immunosorbent assay; GWAS, genome-wide association study; MeSH, Medical Subject Headings; PCR, polymerase chain reaction.
Figure 1.
Publication trends of manuscripts using machine learning as a research tool for 5-year intervals across 5 organ systems from 1980 to 2020.
Articles in Different Journals
Subject-specific and clinical journals had fewer publications using machine learning compared with science and multidisciplinary journals (Table 2). The highest number of machine learning manuscripts were published in Nature Communications followed by Circulation, whereas the kidney-based research published the lowest number of articles. These journals include the Journal of American Society of Nephrology (JASN) followed by Kidney International (KI). In a subgroup analysis, we compared machine learning articles related to kidney research versus those that were nonkidney related. We found that a smaller proportion of kidney research articles used machine learning methods versus other analysis methods such as the Cox-proportional hazard models, whereas this proportion was higher in the case of nonkidney related articles (χ2 (1, N = 39,550) = 1337.2, P < 0.001).
Table 2.
Articles Published in Specialty Journals Listed on the Web of Science Bibliometric Database Using ISSN Number of the Specific Journal and MeSH Term for the Methodology
| Kidney Int (n = 1,813) | J Am Soc Nephol (n = 680) | Brain (n = 402) | Hepatology (n = 4681) | Chest (n = 599) | Am J Respir Crit Care Med. (n = 1,210) | Circulation (n = 10,123) | New Engl J Med (n = 964) | Nat Med (n = 472) | Nat Commun (n = 1,766) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cox proportional hazard model | 139 (7.6%) | 94 (13.8%) | 20 (4.9%) | 192 (4.1%) | 119 (19.9%) | 86 (7.1%) | 2,311 (22.8%) | 82 (8.5%) | 3 (0.6%) | 2 (0.11%) |
| CRISPR/Cas9 | 9 (0.5%) | 13 (1.9%) | 6 (1.5%) | 48 (1.02%) | 0 | 5 (0.41%) | 121 (1.2%) | 5 (0.51%) | 27 (5.7%) | 341 (19.3%) |
| ELISA | 312 (17.2%) | 80 (11.8%) | 28 (6.9%) | 949 (20.3%) | 122 (20.4%) | 221 (18.3%) | 1,601 (15.8%) | 102 (10.6%) | 13 (2.7%) | 21 (1.2%) |
| GWAS | 6 (0.33%) | 16 (2.3%) | 9 (2.2%) | 16 (0.34%) | 2 (0.33%) | 16 (1.32%) | 299 (2.9%) | 0 | 11 (2.3%) | 212 (12%) |
| Machine learning | 9 (0.49%) | 7 (1.02%) | 30 (7.5%) | 33 (0.7%) | 13 (2.2%) | 22 (1.8%) | 206 (2.03%) | 14 (1.45%) | 25 (5.3%) | 292 (16.5%) |
| PCR | 767 (42.3%) | 251 (36.9%) | 183 (45.5%) | 2,018 (43.1%) | 282 (47.07%) | 597 (49.3%) | 2,443 (24.1%) | 661 (68.5%) | 255 (54%) | 518 (29.3%) |
| Western blot | 573 (31.6%) | 219 (32.2%) | 126 (31.3%) | 1,425 (30.4%) | 61 (10.2%) | 263 (21.7%) | 3,142 (31%) | 100 (10.3%) | 138 (29.2%) | 380 (21.5%) |
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; ELISA, enzyme-linked immunosorbent assay; GWAS, genome-wide association study; ISSN, International Standard Serial Number; MeSH, Medical Subject Headings; PCR, polymerase chain reaction.
Funding Sources of Articles
The highest number of machine learning manuscripts, 573 (14%), acknowledged the National Institute of Neurological Disorders and Stroke (NINDS) as their funding source. The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) was only acknowledged by 70 (1.7%) total manuscripts based on machine learning approaches (Table 3).
Table 3.
Machine Learning–Based Articles Supported From Grants From Various NIH Institutions
| NIH Institutions | Brain | Heart | Kidney | Liver | Lung | N (%) |
|---|---|---|---|---|---|---|
| NIDDK | 0 | 21 | 29 | 16 | 4 | 70 (1.7%) |
| NIGMS | 127 | 70 | 22 | 13 | 21 | 253 (6.1%) |
| NCI | 124 | 36 | 19 | 38 | 111 | 328 (8.03%) |
| NCATS | 103 | 67 | 18 | 10 | 20 | 218 (5.3%) |
| NHLBI | 37 | 189 | 10 | 6 | 57 | 299 (7.3%) |
| NIBIB | 454 | 39 | 6 | 16 | 28 | 543 (13.3%) |
| NHGRI | 0 | 15 | 6 | 0 | 9 | 30 (0.7%) |
| NIAID | 0 | 0 | 4 | 7 | 15 | 26 (0.6%) |
| NIEHS | 0 | 12 | 4 | 8 | 8 | 32 (0.7%) |
| NINDS | 521 | 43 | 3 | 2 | 4 | 573 (14.0%) |
| NIA | 433 | 24 | 3 | 2 | 4 | 466 (11.4%) |
| NCRR | 131 | 30 | 2 | 4 | 16 | 183 (4.4%) |
| NIAAA | 31 | 0 | 0 | 4 | 0 | 35 (0.8%) |
| NICHD | 138 | 11 | 0 | 6 | 5 | 160 (3.9%) |
| NIAMS | 0 | 0 | 0 | 4 | 5 | 9 (0.2%) |
| NIMH | 522 | 13 | 0 | 0 | 7 | 542 (13.2%) |
| NIDA | 146 | 9 | 2 | 0 | 4 | 161 (3.9%) |
| NIDCD | 51 | 0 | 0 | 0 | 0 | 51 (1.2%) |
| NEI | 40 | 0 | 0 | 0 | 0 | 40 (0.9%) |
| NIMHD | 0 | 8 | 0 | 0 | 0 | 8 (0.19%) |
| NLM | 58 | 50 | 12 | 9 | 25 | 154 (3.7%) |
| 4,081 |
The data were extracted from the Web of Science bibliometric database from 1864 to 2020.
Abbreviations: NCATS, National Center for Advancing Translational Sciences; NCI, National Cancer Institute; NCRR, National Center For Research Resources; NEI, National Eye Institute; NHGRI, National Human Genome Research Institute; NHLBI, National Heart, Lung, and Blood Institute; NIA, National Institute on Aging; NIAAA, National Institute on Alcohol Abuse and Alcoholism; NIAID, National Institute of Allergy and Infectious Diseases; NIBIB, National Institute of Biomedical Imaging and Bioengineering; NICHD, National Institute of Child Health and Human Development; NIDA, National Institute on Drug Abuse; NIDDK, National Institute of Diabetes and Digestive and Kidney Diseases; NIGMS, National Institute of General Medical Sciences; NIMH, National Institute of Mental Health; NINDS, National Institute of Neurological Disorders and Stroke; NLM, National Library of Medicine.
Grants Funded by NIH
Out of all grants funded by 7 NIH institutions, the National Cancer Institute (NCI) funded the maximum number of grants, 428 (25.1%), whereas the NIDDK funded 122 (7.2%) grants (Table 4). In terms of the career development grants and fellowship grants, the National Heart, Lung, and Blood Institute (NHLBI) funded the most grants: 56 (31.5%) and 29 (25%), respectively.
Table 4.
Comparison of Grants Awarded by Various NIH Institutions for Research Projects That Proposed the Use of Machine Learning as a Research Tool From 1985 to 2020
| NIH Institution | Total No. of Grants (N = 1,704) | Career Development Grants (n = 178) | Fellowship and Training Grants (n = 116) |
|---|---|---|---|
| NCI | 428 (25.1%) | 24 (13.4%) | 24 (20.7%) |
| NHLBI | 285 (16.7%) | 56 (31.5%) | 29 (25%) |
| NINDS | 265 (15.5%) | 25 (14.04%) | 23 (19.8%) |
| NLM | 251 (14.7%) | 22 (12.3%) | 16 (13.8%) |
| NIBIB | 185 (10.8%) | 12 (6.7%) | 3 (2.6%) |
| NHGRI | 133 (7.8%) | 17 (9.5%) | 12 (10.3%) |
| NIDDK | 122 (7.2%) | 21 (11.8%) | 8 (6.9%) |
| NCATS | 35 (2.05%) | 1 (0.56%) | 1 (0.9%) |
Abbreviations: NCATS, National Center for Advancing Translational Sciences; NCI, National Cancer Institute; NHGRI, National Human Genome Research Institute; NHLBI, National Heart, Lung, and Blood Institute; NIBIB, National Institute of Biomedical Imaging and Bioengineering; NIDDK, National Institute of Diabetes and Digestive and Kidney Diseases; NINDS, National Institute of Neurological Disorders and Stroke; NLM, National Library of Medicine.
Discussion
Historically, the field of nephrology has lagged in using analytical approaches. For example, large observational studies on cardiovascular disease risk were published in the late 1950s and early 1960s,19,20 but similar studies were published only decades later in nephrology.21 In line with the historical perspective, we have shown that kidney disease research underutilizes machine learning as a research tool compared with other organs and organ systems. In terms of the 5-year trends related to the publication of machine learning-based articles, kidney-focused articles lag behind those for other organ systems. We also found that organ-specific journals have been publishing a smaller number of machine learning–based articles compared with multidisciplinary journals. Even within these journals, the kidney-specific journals are lagging behind in terms of publishing machine learning–based manuscripts. The lowest number of articles using machine learning approaches acknowledged NIDDK as a funding source.
These findings suggest underutilization of machine learning as a research tool in kidney research compared with other specialties. The question then arises as to the reason for such a discrepancy in kidney literature compared with other specialties. An approach or a technology employed in any scientific research is based on its appropriateness to address a question, the availability of the research tool, and the expertise and knowledge of the investigative team. These parameters likely dictate the publications and inclusion of such a technology in research proposals. Our results, which demonstrate the least number of machine learning research papers acknowledging NIDDK as a supporting agency and the least number of kidney research articles published in prime kidney journals (JASN and KI), are symptomatic of one or a combination of the aforementioned factors.
These results also raise the possibility of whether there is lukewarm enthusiasm among kidney researchers to embrace machine learning as an analysis tool or if researchers with machine learning expertise are not necessarily focused on kidney diseases per se. Interestingly, our analysis also suggested that kidney disease researchers have adopted other novel methods and techniques like CRISPR/Cas9 and GWAS (genome-wide association study) at higher rates than machine learning tools.
To address the issue of underutilization of machine learning as a research tool among trainees, clinicians, and kidney researchers, the following strategies can be considered. First, trainees in medical schools can be introduced to machine learning through courses focused on population health in general22 and by showcasing examples related to kidney diseases in particular to illustrate how this tool can impact disease prediction, risk stratification, and management. It is also possible to improve community-wide awareness about the advantages and limitations of machine learning by developing continuing medical education content and disseminating the material during conferences and workshops. During these events, dedicated research sessions and seminars on the applications of machine learning in nephrology and nephropathology could be organized. The presence of educators who are well versed in machine learning will be helpful so that they can illustrate its advantages and limitations to the nephrology and the nephropathology communities.
This gradual transformation would also be reflected in the constitution of peer review processes and NIH study sections. Special issues within kidney journals focusing on machine learning applications would also augment awareness of this technology. It is noteworthy that there are ongoing efforts to integrate machine learning in the Kidney Precision Medicine Project,23 the Chronic Renal Insufficiency Cohort (CRIC) study,24, 25, 26 the Cure Glomerulonephropathy (CureGN) study,27 and the Nephrotic Syndrome Study Network (NEPTUNE).28,29 Making the greater scientific community aware of these initiatives will likely increase data science research in kidney diseases.
Last but not least, national- and local-level research sponsors should consider increasing the funding priority for machine learning–based applications focused on kidney health and disease. Creative approaches such as dedicated fellowships and funding opportunities that are focused on data science would be educational and attract the interest of the broader community in pursuing kidney research.
Our study has a few strengths and limitations. To our knowledge, this bibliometric study is the first of its kind to identify the differences in the number of articles published, trends of publications, and research funding across different organ systems that have used machine learning as a methodology. The study’s limitation is that we worked only with Web of Science and NIH RePORTER. There are other public and commercially available bibliometric databases such as Scopus and Google Scholar, and no bibliometric database is superior to the others; the differences in the way data are organized in each database may lead to subtle differences in the search outputs.30 Therefore, we acknowledge that there might be possible discrepancies in identifying the exact set of manuscripts relevant to the scope of this study. Nevertheless, we maintained consistency across our search keywords and our approach, which provided us with the data needed to evaluate the extent to which machine learning is used as a methodology within the kidney research community. Also, our work mainly focused on US-specific databases such as NIH RePORTER, but this approach can be easily extended to European, Asian, and other funding agencies.
In conclusion, our bibliometric study based on querying public databases provided the direct insight that there are significant differences in the use of machine learning as a research tool among kidney researchers compared with those who are focused on other organ systems. The reasons for this critical gap should be explored, and the kidney research community should become better informed via various educational platforms and training programs about this exciting research tool.
Article Information
Authors’ Full Names and Academic Degrees
Ashish Verma, MBBS, Vipul C. Chitalia, MD, PhD, Sushrut S. Waikar, MD, MPH, and Vijaya B. Kolachalama, PhD.
Authors’ Contributions
Research idea and study design: AV, VK; data acquisition: AV, VK; data analysis/interpretation: AV, VK; statistical analysis: AV, VK; supervision: VC, SW, VK. Each author contributed important intellectual content during manuscript drafting or revision and accepts accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
Support
This work was supported by the following grants: Scientist Development Grant (17SDG33670323) and a Strategically Focused Research Network (SFRN) Center Grant (20SFRN35460031) from the American Heart Association; and NIH grants R01-AG062109, R01-AR070139, R21-CA253498, R01-CA175382, R01-HL132325, and R21-DK119740.
Financial Disclosure
The authors declare that they have no relevant financial interests.
Peer Review
Received January 3, 2021. Evaluated by 2 external peer reviewers, with direct editorial input by the Editor-in-Chief. Accepted in revised form April 4, 2021.
Footnotes
Complete author and article information provided before references.
Item S1: Search strategy for bibliometric analysis.
Item S2: Search strategy for NIH grants for research projects utilizing machine learning as research tool.
Supplementary Material
Items S1-S2.
References
- 1.Esteva A., Robicquet A., Ramsundar B. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 2.Wang F., Casalino L.P., Khullar D. Deep learning in medicine-promise, progress, and challenges. JAMA Intern Med. 2019;179(3):293–294. doi: 10.1001/jamainternmed.2018.7117. [DOI] [PubMed] [Google Scholar]
- 3.Tomašev N., Glorot X., Rae J.W. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572(7767):116–119. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyner J.L., Carey K.A., Edelson D.P., Churpek M.M. The development of a machine learning inpatient acute kidney injury prediction model. Crit Care Med. 2018;46(7):1070–1077. doi: 10.1097/CCM.0000000000003123. [DOI] [PubMed] [Google Scholar]
- 5.Ginley B., Lutnick B., Jen K.Y. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol. 2019;30(10):1953–1967. doi: 10.1681/ASN.2018121259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermsen M., de Bel T., den Boer M. Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol. 2019;30(10):1968–1979. doi: 10.1681/ASN.2019020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh J.N., Matlock M.K., Kudose S. Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging. 2018;37(12):2718–2728. doi: 10.1109/TMI.2018.2851150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kannan S., Morgan L.A., Liang B. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep. 2019;4(7):955–962. doi: 10.1016/j.ekir.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolachalama V.B., Singh P., Lin C.Q. Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep. 2018;3(2):464–475. doi: 10.1016/j.ekir.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma K., Rupprecht C., Caroli A. Automatic segmentation of kidneys using deep learning for total kidney volume quantification in autosomal dominant polycystic kidney disease. Sci Rep. 2017;7(1):2049. doi: 10.1038/s41598-017-01779-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker J.U., Mayerich D., Padmanabhan M. Artificial intelligence and machine learning in nephropathology. Kidney Int. 2020;98(1):65–75. doi: 10.1016/j.kint.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barisoni L., Lafata K.J., Hewitt S.M., Madabhushi A., Balis U.G.J. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. 2020;16(11):669–685. doi: 10.1038/s41581-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sealfon R.S.G., Mariani L.H., Kretzler M., Troyanskaya O.G. Machine learning, the kidney, and genotype-phenotype analysis. Kidney Int. 2020;97(6):1141–1149. doi: 10.1016/j.kint.2020.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saez-Rodriguez J., Rinschen M.M., Floege J., Kramann R. Big science and big data in nephrology. Kidney Int. 2019;95(6):1326–1337. doi: 10.1016/j.kint.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 15.Niel O., Bastard P. Artificial intelligence in nephrology: core concepts, clinical applications, and perspectives. Am J Kidney Dis. 2019;74(6):803–810. doi: 10.1053/j.ajkd.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Köttgen A., Pattaro C., Böger C.A. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarivate Analytics Web of Science Core Collection. 2020. https://clarivate.com/webofsciencegroup/solutions/web-of-science-core-collection/
- 18.National Institutes of Health Research Portfolio Online Reporting Tools (RePORT). 2020. https://report.nih.gov/
- 19.Dawber T.R., Moore F.E., Mann G.V. Coronary heart disease in the Framingham study. Am J Public Health Nations Health. 1957;47(4):4–24. doi: 10.2105/ajph.47.4_pt_2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kannel W.B., Dawber T.R., Kagan A., Revotskie N., Stokes J., 3rd Factors of risk in the development of coronary heart disease—six year follow-up experience. The Framingham Study. Ann Intern Med. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 21.Lowrie E.G., Lew N.L. Death risk in hemodialysis patients: the predictive value of commonly measured variables and an evaluation of death rate differences between facilities. Am J Kidney Dis. 1990;15(5):458–482. doi: 10.1016/s0272-6386(12)70364-5. [DOI] [PubMed] [Google Scholar]
- 22.Kolachalama V.B., Garg P.S. Machine learning and medical education. NPJ Digit Med. 2018;1:54. doi: 10.1038/s41746-018-0061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong E., Wang L.L., Schaub J. Modelling kidney disease using ontology: insights from the Kidney Precision Medicine Project. Nat Rev Nephrol. 2020;16(11):686–696. doi: 10.1038/s41581-020-00335-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lash J.P., Go A.S., Appel L.J. Chronic Renal Insufficiency Cohort (CRIC) Study: baseline characteristics and associations with kidney function. Clin J Am Soc Nephrol. 2009;4(8):1302–1311. doi: 10.2215/CJN.00070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldman H.I., Appel L.J., Chertow G.M. The Chronic Renal Insufficiency Cohort (CRIC) Study: design and methods. J Am Soc Nephrol. 2003;14(7 Suppl 2):S148–S153. doi: 10.1097/01.asn.0000070149.78399.ce. [DOI] [PubMed] [Google Scholar]
- 26.Denker M., Boyle S., Anderson A.H. Chronic Renal Insufficiency Cohort Study (CRIC): overview and summary of selected findings. Clin J Am Soc Nephrol. 2015;10(11):2073–2083. doi: 10.2215/CJN.04260415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariani L.H., Bomback A.S., Canetta P.A. CureGN Study rationale, design, and methods: establishing a large prospective observational study of glomerular disease. Am J Kidney Dis. 2019;73(2):218–229. doi: 10.1053/j.ajkd.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gadegbeku C.A., Gipson D.S., Holzman L.B. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013;83(4):749–756. doi: 10.1038/ki.2012.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barisoni L., Nast C.C., Jennette J.C. Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE) Clin J Am Soc Nephrol. 2013;8(8):1449–1459. doi: 10.2215/CJN.08370812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plana N.M., Massie J.P., Bekisz J.M., Spore S., Diaz-Siso J.R., Flores R.L. Variations in databases used to assess academic output and citation impact. N Engl J Med. 2017;376(25):2489–2491. doi: 10.1056/NEJMc1616626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Items S1-S2.