Abstract
Rationale & Objective
Biomarker studies are important for generating mechanistic insight and providing clinically useful predictors of chronic kidney disease (CKD) progression. However, variability across studies can often muddy the evidence waters. Here we evaluated real-world variability in biomarker studies using two published studies, independently conducted, of the novel plasma marker soluble urokinase-type plasminogen activator receptor (suPAR) for predicting CKD progression in children with CKD.
Study Design
A comparison of 2 prospective cohort studies.
Setting & Participants
541 children from the Chronic Kidney Disease in Children (CKiD) study, median age 12 years, median glomerular filtration rate (GFR) of 54 mL/min/1.73m2.
Outcome
The first occurrence of either a 50% decline in GFR from baseline or incident end-stage kidney disease.
Analytical Approach
The suPAR plasma marker was measured using the Quantikine ELISA immunoassay in the first study and Meso Scale Discovery (MSD) platform in the second. The analytical approaches varied. We used suPAR data from the 2 assays and mimicked each analytical approach in an overlapping subset.
Results
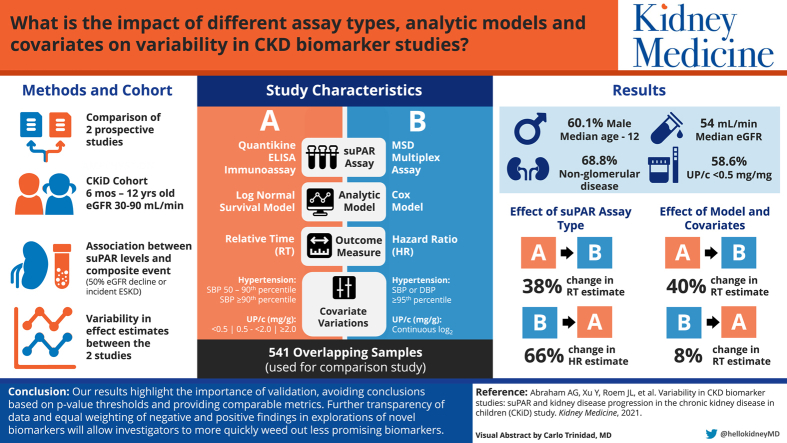
We found that switching assays had the greatest impact on inferences, resulting in a 38% to 66% change in the magnitude of the effect estimates. Covariate and modeling choices resulted in an additional 8% to 40% variability in the effect estimate. The cumulative variability led to different inferences despite using a similar sample of CKiD participants and addressing the same question.
Limitations
The estimated variability does not represent optimal repeatability but instead illustrates real-world variability that may be present in the CKD biomarker literature.
Conclusions
Our results highlight the importance of validation, avoiding conclusions based on P value thresholds, and providing comparable metrics. Further transparency of data and equal weighting of negative and positive findings in explorations of novel biomarkers will allow investigators to more quickly weed out less promising biomarkers.
Index Words: suPAR, kidney disease progression, chronic kidney disease, biomarker variability
Graphical abstract
Plain-Language Summary.
We used 2 published studies of a novel plasma marker soluble urokinase-type plasminogen activator receptor (suPAR) to look at noise in biomarker studies that can lead to different results. Both studies looked at the association between suPAR and kidney disease progression in children. We found that using different assays for suPAR changed the association by up to 66%. We also found that different ways of analyzing the data changed the association by up to 40%. Future biomarker studies should show raw data of biomarkers and associations, consider how different methods may change the associations using sensitivity analyses, and avoid using P values to draw conclusions.
The discovery of novel biomarkers associated with progression of chronic kidney disease (CKD) is an active area of research, both for mechanistic insights and for the development of prediction formulas that can aid in clinical care. However, the results from biomarker studies may be sensitive to a number of different factors that can have profound implications for the conclusions.
The use of different biomarker assays or nonstandardized assays across time or laboratories can introduce substantial variability in the biomarker levels, which could impact study results.1, 2, 3, 4, 5 Study design choices as well as varying analytical methods can also add variability,6 as can biological variation over time within individuals7 and across populations.8 The resulting variability in biomarker study results can obscure the value of some biomarkers while overstating the value of others. The degree to which these various factors impact a study’s inferences regarding the utility of novel biomarkers for mechanism and prediction is often left unexplored. Quantitative assessments of measurement imprecision require added expensive and precious repository samples, which are particularly limited in pediatric CKD research. Hence, replication studies to evaluate the reproducibility of results are rare in the biomarker literature.
However, without any data on the impact of different biomarker assays and different analytic methods across studies, biomarker performance cannot be adequately assessed. Even in studies with similar patient populations where biologic variability is expected to be lower, the influence of these factors on results may be enough to yield conflicting conclusions. This ambiguity may lead clinicians and researchers to ignore important biomarkers that truly have information about disease progression.
Here we quantify the variability across 2 studies of plasma soluble urokinase-type plasminogen activator receptor (suPAR) levels that came to different conclusions about the value of suPAR for predicting risk of CKD progression. We asked the question, What were the biggest contributors to different study results and conclusions? We took advantage of a chance replication where 2 published studies asking the same question about suPAR were independently conceived, conducted, and analyzed.9,10 We hypothesized that both assay differences and analytic choices substantially contributed to variability in estimated effect sizes and differing conclusions.
Methods
Sample Selection
The Chronic Kidney Disease in Children (CKiD) study is a prospective cohort of children, ages 6 months to 16 years old who have a glomerular filtration rate (GFR) of 30 to 90 mL/min/1.73 m2, from 55 participating sites in the United States and Canada. Plasma samples are collected at each study visit and stored at −80°C at a central biorepository (National Institute of Diabetes and Digestive and Kidney Diseases Biorepository at Precision for Medicine). As a result of 2 separate initiatives with different goals, 2 ancillary studies independently selected plasma samples from the biorepository collected at the 6-month CKiD visit to measure the same biomarker, suPAR, with different assays using slightly different inclusion criteria.
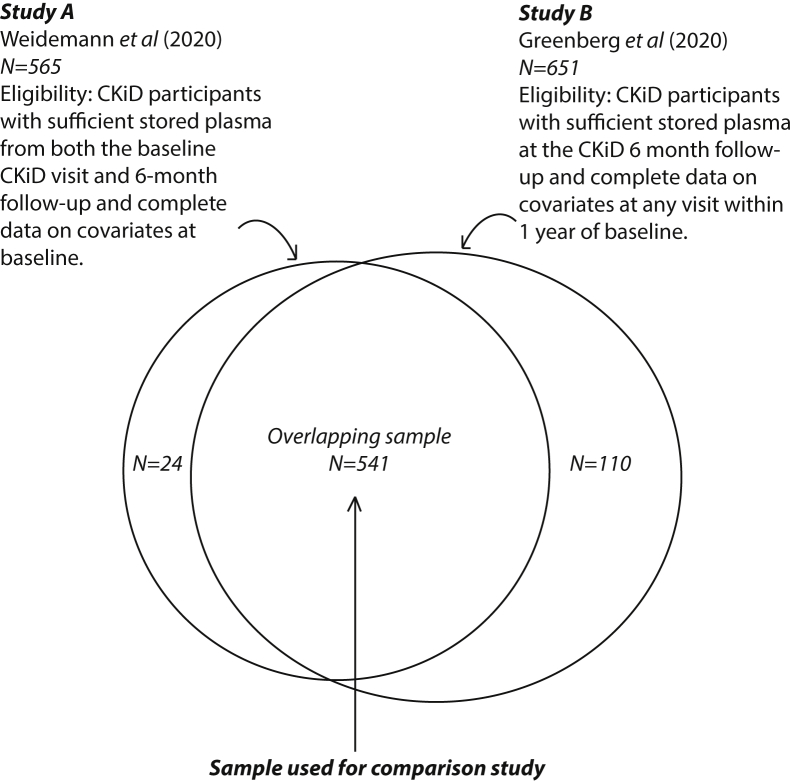
As shown in Figure 1, study A selected 565 plasma samples provided by CKiD participants who had sufficient biorepository sample volumes available from both the 6-month CKiD visit and then a follow-up visit 6-months later (1 year after enrollment in CKiD).10 Study B, which evaluated a larger panel of biomarkers, selected 651 plasma samples from those who had sufficient volume of stored plasma at the 6-month study and available data at study entry on blood pressure, estimated GFR (eGFR), urinary protein-creatinine ratio (UPCR), and body mass index (BMI).9 The overlapping sample used from the present comparison study included 541 individuals.
Figure 1.
Flow chart showing the sample of participants for the published studies A and B, as well as the overlapping sample used for the comparison study. Eligibility criteria are shown for both studies. CKiD, Chronic Kidney Disease in Children cohort.
Differences in the samples reflected the different budgetary constraints, sampling availability, eligibility criteria, and sample size targets of the 2 studies. Both studies investigated the association between suPAR levels and time from study baseline in CKiD until the composite event of a 50% decline in GFR or incident end-stage kidney disease (ESKD; dialysis or transplantation). The participants were censored in both studies at death, loss to follow-up, or the end of study. Further details on characteristics of the individual study samples and study designs can be found in the original publications.9,10 Written informed consent was obtained from all parents or legal guardians, along with assent, when appropriate, from the children. The CKiD study was approved by the institutional review board of each participating institution. The CKiD study is registered at ClinicalTrials.gov with the identifier NCT00327860.
SuPAR Assays
For study A, suPAR was measured from plasma collected at the 6-month CKiD visit in duplicate using Quantikine Enzyme-Linked Immunosorbent Assay (ELISA) immunoassay (catalog #DUP00; R&D Systems). The range was 33 to 4,415 pg/mL with a lower limit of detection of 33 pg/mL. The assay used 96-well polystyrene microplates (12 strips of 8 wells) coated with a monoclonal antibody specific for human uPAR (manufacturer #890714). The detection antibodies were proprietary polyclonal antibody specific for human uPAR conjugated to horseradish peroxidase with preservatives (manufacturer #890715). Plasma samples were diluted 1 in 5. From control samples run on each plate, the intra-assay coefficient of variation (CV) ranged from 2.7% to 3.9% and interassay CV ranged from 8.3% to 8.8%.10
For study B, suPAR was measured from plasma collected at the 6-month CKiD in duplicate using a multiplex assay on the Meso Scale Discovery (MSD) platform.9 The assay range was 53 to 64,000 pg/mL, and the lower limit of detection was 53 pg/mL. The capture antibodies were human uPAR monoclonal and clone 62022 (Bio-techne, catalog # MAB807), and the detection antibodies were human uPAR polyclonal (Bio-techne, catalog # AF807). Plasma samples were diluted (1 in 5 dilution) by mixing 25 μL of sample with 100 μL of Diluent 100 (R50AA-2; MSD). Measurements were repeated if 2 or more analytes had an intra-assay CV of >15%, and the final intra-assay CV was 8.3% after repeating measurements on 28 samples. The interassay CV was 8.4%, using an external sample of 35 children with glomerular disease not enrolled in the CKiD study to minimize the plasma volume required from the CKiD biorepository. All samples for both studies had one freeze-thaw cycle.
Covariates
Covariates were primarily obtained from the first annual CKiD visit for both studies, which occurred approximately 6 months before the suPAR measurements. The eGFR was estimated using the CKiD estimating equation that is based on serum creatinine, cystatin C, and blood urea nitrogen (BUN) concentrations, all captured from a standard renal laboratory panel assayed in a central laboratory for the CKiD study.11
The definition of hypertension varied between the studies. For study A, systolic blood pressure was categorized into 50th to 90th percentile and ≥90th percentile for age, sex, and height; for study B, hypertension was defined as either a systolic or diastolic blood pressure of ≥95th percentile for age, sex, and height.12 Urinary protein to creatinine ratio (UPCR) was categorized as < 0.5 mg/mg, 0.5 to <2.0 mg/mg, or ≥2.0 mg/mg for study A; it was used as the continuous log2 transformed UPCR in study B. BMI was age and gender standardized. Kidney disease subtypes were classified as either glomerular or nonglomerular etiology.13
Demographic data and use of hypertensive medication were collected during the study visit at clinical sites. In the case of missing data, study A replaced missing central laboratory values for CO2 with measures from the local site laboratory. Other missing data were excluded from the analysis. In study B, missing values in baseline data were drawn from data taken at the 12-month visit.
Original Study Design and Analyses of Study A and Study B
As a result of differing priorities and study goals, different analytical strategies were chosen in the 2 studies, and because of differences in the study timing, administrative censoring occurred on August 1, 2017, for study A and March 1, 2018, for study B.
Study A used log-normal survival models to model relative time (RT) from the first CKiD study visit to the composite event by plasma suPAR quartiles. Fully adjusted log-normal survival models included age, male sex, Black race, Hispanic ethnicity, BMI z score, blood pressure categories, angiotensin-converting enzyme/angiotensin receptor blocker use, UPCR categories, glomerular diagnosis, and eGFR. A complete case analysis was used reducing the sample due to missing covariate values.
Study B used Cox models to assess hazard ratios (HR) of the association between the suPAR levels and time from the first CKiD study visit to the composite event by plasma suPAR quartiles. Fully adjusted Cox models included age, male sex BMI z score, hypertension, log2UPCR, glomerular diagnosis, and eGFR.
Statistical Analysis for Study Comparison
Using the 541 participants with suPAR measurements from both assays, we examined the agreement between suPAR assay measurements using Spearman correlation coefficient and Bland Altman analysis. We evaluated whether demographic and clinical characteristics contributed to assay differences by regressing the individual suPAR measurement difference (ELISA and MSD) on age, sex, eGFR, BMI, hypertension, glomerular diagnosis, log2 UPCR, and BUN. Hierarchical clustering analysis using complete linkage was also done comparing suPAR relationships with clinical variables (BMI z score, log2 UPCR, age, eGFR, BUN) across the 2 assays.
To assess the variability resulting from sample selection differences, assay differences, and analytic strategy we performed the following analyses. (1) We compared the results from the 541 participants from the unified sample to those from the original study B with 651 participants using the original methods. (2) In the overlapping sample of 541 participants, we repeated study A and study B using each set of suPAR assay results (ie, ELISA and MSD) holding the analytic strategy constant. (3) In the overlapping sample of 541 participants, we repeated study A and study B using each analytic strategy (ie, lognormal and Cox regression with respective covariate sets), holding the suPAR assay constant.
Because the RT and HR estimate magnitudes were difficult to compare, we used a parametric Weibull survival model, which is also a proportional hazards model, to provide a conversion between HR and RT. P values comparing effect estimates across quartiles of suPAR were estimated using both Wald and type III (effect across all levels of the categorical predictor) tests. Finally, to evaluate whether a categorical suPAR expression contributed to inferential differences, we estimated the best-fitting fully adjusted continuous Cox regression model based on the lowest akaike information criterion using data from each suPAR assay.
We used SAS 9.4 and RStudio for all analyses. Results with P < 0.05 were considered statistically significant.
Results
In this comparison study, we included the 541 participants with plasma suPAR measured on both ELISA and MSD platforms, representing 96% of the study A cohort and 83% of the study B cohort, respectively. The sample was 60.1% male with a median age of 12 years (interquartile range [IQR), 8-15). Most participants had a diagnosis of nonglomerular kidney disease (68.8%), with a median eGFR at baseline of 54 mL/min/1.73 m2 (IQR, 41-67), and 58.6% had a UPCR <0.5 mg/mg (Table 1). The median follow-up time was 4.7 years (IQR, 3.2-7.1, range: 0.6-11.7) for study A and 5.8 years (IQR, 4.0-7.8, range: 0.6-12.4) for study B. The difference in follow-up time was the result of differences in censoring time (August 2017 for study A and March 2018 for study B).
Table 1.
Baseline Demographic and Clinical Characteristics From the Original suPAR Studies and the Overlapping Sample of 541 Participants From the CKiD Study
| Baseline Characteristics | N = 541a | Study A (ELISA) N = 565 | Study B (MSD) N = 651 |
|---|---|---|---|
| Age (y) | 12 [8-15] | 12 [8-15] | 11 [8-15] |
| Male sex | 325 (60.1%) | 341 (60.4%) | 404 (62.1%) |
| Black race | 112 (20.7%) | 118 (20.9%) | 131 (20.1%) |
| Hispanic ethnicity | 65 (12.0%) | 67 (11.9%) | 86 (13.4%) |
| Glomerular diagnosisd | 169 (31.2%) | 173 (30.6%) | 195 (30.0%) |
| eGFR, mL/min per 1.73 m2 | 54 [41-67] | 53 [40.8-66.6] | 53 [40-67] |
| BMI (kg/m2) | 19 [16-22] | 19 [16-22] | 19 [16-22] |
| Height SDS < –2 | 57 (10.5%) | 60 (10.6%) | 63 (9.7%) |
| High blood pressureb | 116 (21.4%) | 118 (20.9%) | 117 (18.0%) |
| Antihypertensive use | 351 (64.9%) | 364 (64.4%) | 426 (65.4%) |
| UPCR categories (mg/mg Scr) | |||
| ≥2 | 61 (11.3%) | 61 (10.8%) | 76 (11.7%) |
| 0.5-2 | 163 (30.1%) | 164 (29.0%) | 188 (28.9%) |
| <0.5 | 317 (58.6%) | 320 (56.6%) | 387 (59.5%) |
| Anemia | 171 (31.6%) | 176 (31.2%) | 186 (29.1%) |
| Elevated CRP (>3 mg/L) | 93 (17.2%) | 96 (17.0%) | 111 (17.3%) |
| Acidosis (CO2 < 22 mmol/L)c | 279 (51.6%) | 291 (51.5%) | 197 (30.3%) |
| Hypoalbuminemic (<3.8 g/dL) | 46 (8.5%) | 47 (8.3%) | 54 (8.3%) |
Data are presented as median [interquartile range] or frequency (%).
Abbreviations: BMI, body mass index; CKiD, Chronic Kidney Disease in Children; CRP, C-reactive protein; eGFR, estimated glomerular filtration rate; ELISA, enzyme-linked immunosorbent assay (Quantikine); MSD, Meso Scale Discovery; Scr, serum creatinine; SDS, standard deviation score; suPAR, soluble urokinase plasminogen activator receptor; UPCR, urinary protein-creatinine ratio.
For describing the combined sample, covariate definitions from study A were used.
For study A, hypertension was defined as systolic blood pressure greater than 90th percentile for age, sex, and height; For study B, hypertension was defined as either systolic or diastolic blood pressure >95 percentile for age, sex, and height.
For study A, acidosis was defined based on data from study baseline central laboratory CO2 measurements; when missing, the study used baseline local site laboratory CO2 measurements. For study B, acidosis was defined based on data from study baseline central laboratory CO2 measurements; when missing, the study used central laboratory CO2 measurements from the 12-month visit.
The most common glomerular disease diagnoses were focal segment glomerulosclerosis (29% in overlapping sample), hemolytic uremic syndrome (20% in overlapping sample), and systemic immunologic disease such as systemic lupus erythematous nephritis (14% in overlapping sample).
Differences in suPAR Distributions and Agreement Between the Assays
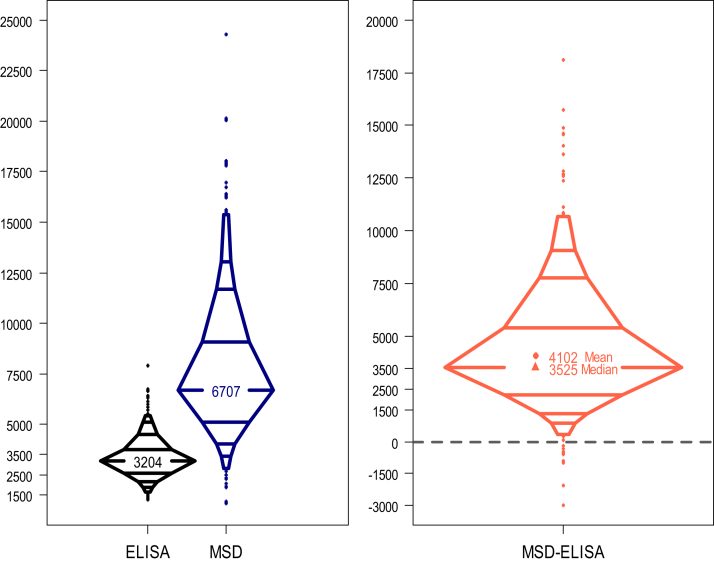
The distributions of suPAR from the 2 assays were characteristically different, as can be seen in Figure 2. The median suPAR level on the ELISA platform was 3,204 pg/mL (IQR, 2,605-3,761), and the median on the MSD platform was 6,707 pg/mL (IQR, 5,111-9,074). Looking at the discrepancies in suPAR levels from the 2 assays across the sample, the median difference was 3,526 pg/mL, and the mean difference on the natural scale was 4,102 pg/mL (95% CI, 3,989-4,216).
Figure 2.
Comparison of suPAR levels on ELISA vs MSD platforms. The left panel shows the distribution of suPAR measurements from the two suPAR assays. The right panel shows the distribution of the individual differences in measurements between the two assays. Abbreviations: ELISA, enzyme-linked immunosorbent assay (Quantikine); MSD, Meso Scale Discovery; suPAR, soluble urokinase plasminogen activator receptor.
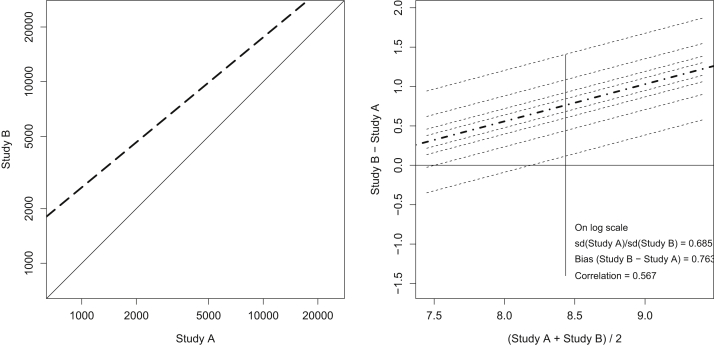
From the Bland Altman analysis,14 using the natural log of suPAR to normalize the distributions, the agreement between assay measurement was modest, with a bias of 0.76, a ratio of standard deviations of 0.69 and a Pearson correlation of 0.57 (Fig 3). As suPAR was treated as a categorical variable in both studies, we examined the concordance between quartiles. Only 47% of participants (253 of 541) maintained their placement in the same suPAR quartile by ELISA and MSD; hence the concordance was poor (κ = 0.44 [IQR, 0.38-0.49]).
Figure 3.
Agreement of suPAR measurements from 2 assays. The left panel shows the deviation of agreement from the line of identity on the natural scale showing both a shift of the central tendency and a slope change. The right panel shows the results from Bland Altman analysis after natural log transformation to normalize distributions showing a bias, difference in the spread of the data and modest linear correlation. Study A measurements were performed with ELISA, and study B measurements were performed with MSD. The bias was estimated as the mean of the differences in measurements. The Pearson correlation and ratio of standard deviations are also shown. Abbreviations: ELISA, enzyme-linked immunosorbent assay (Quantikine); MSD, Meso Scale Discovery; suPAR, soluble urokinase plasminogen activator receptor.
Aside from distributional differences, the relationships between suPAR measurements and several other key variables differed. The MSD suPAR levels were more strongly associated with eGFR than ELISA suPAR levels, with Spearman correlation coefficients of −0.62 versus −0.46, respectively. When the difference in assay values was regressed on a number of clinical and demographic factors, BMI z score, log2 UPCR, eGFR, and BUN were all significant predictors of the difference, explaining 29% of the variability. The difference was primarily explained by an inverse association with eGFR (R2 = 0.242).
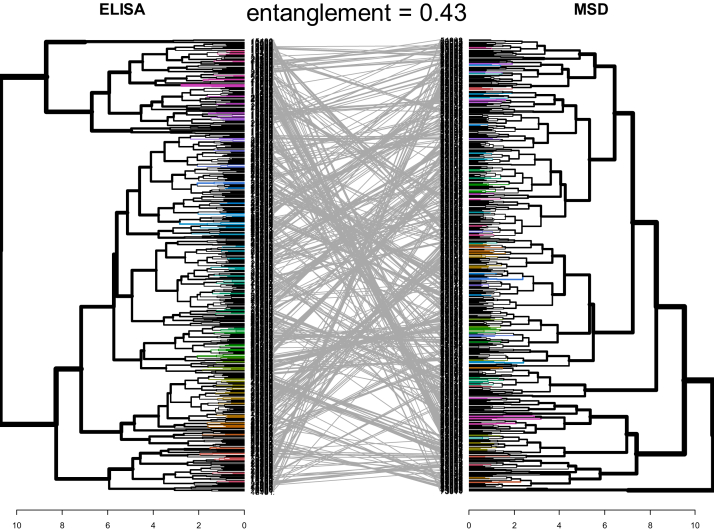
Using clustering analysis, we examined the way individuals grouped together based on values of covariates and suPAR, comparing ELISA and MSD suPAR measurements. Figure 4 shows the resulting dendrograms. The crossing of the grey lines indicates the shifting of clustering patterns depending upon the assay used for suPAR. The entanglement value of 0.43 suggests only moderate alignment of the dendrograms. This indicates relationships between suPAR and clinical factor values are not maintained when the assay changes from ELISA to MSD.
Figure 4.
Comparison of dendrograms from hierarchical clustering analysis using complete linkage. Each leaf or line corresponds to 1 observation. Observations that are similar to each other are combined (fused) as the dendrogram flows away from the center. The height of the fusion along the horizontal axis indicates the (dis)similarity between 2 observations. The farther away from the center the fusion occurs, the less similar the observations are. The dendrogram on the left shows relationships between participants’ given values of ELISA suPAR, BMI z score, log2(UPCR), age, eGFR, and BUN. The dendrogram on the right shows relationships between participants’ given values of MSD suPAR, BMI z score, log2(UPCR), age, eGFR, and BUN. Grey lines illustrate how individuals re-sort depending on whether ELISA or MSD is used for suPAR measurement. The quality of the alignment of the 2 trees is indicated by the entanglement. Entanglement is a measure between 1 (full entanglement) and 0 (no entanglement). A lower entanglement coefficient corresponds to a good alignment. Abbreviations: BMI, body mass index; eGFR, estimated glomerular filtration rate; ELISA, enzyme-linked immunosorbent assay (Quantikine); MSD, Meso Scale Discovery; BUN, blood urea nitrogen; suPAR, soluble urokinase plasminogen activator receptor; UPCR, urinary protein-creatinine ratio.
Differences in Associations Between suPAR and the Composite Event
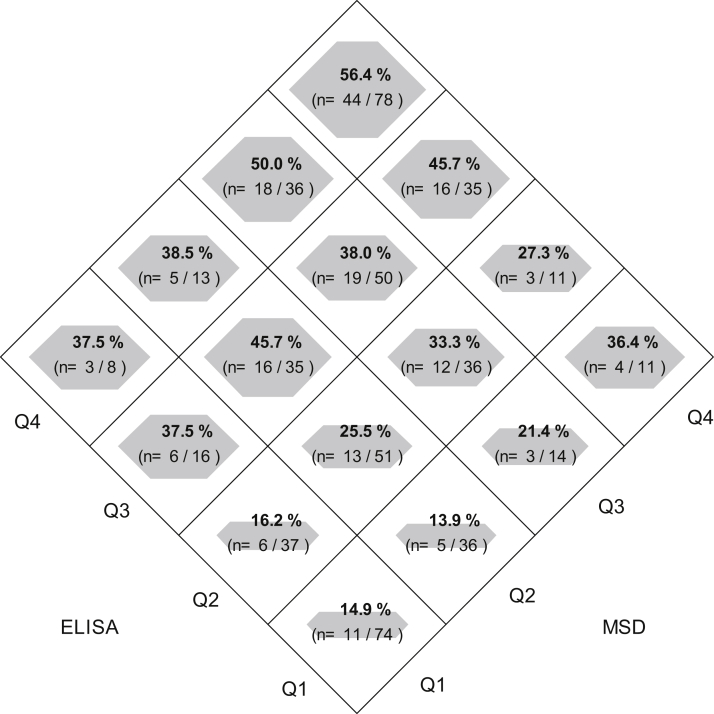
The diamond plot in Figure 5 shows the risk of the composite event across quartiles of ELISA- and MSD-based suPAR. The figure illustrates how participants change suPAR categories depending on the assay. However, in general, trends in risk appear qualitatively similar across the 2 assays. Table 2 shows the results of exchanging analytic strategies and assays.
Figure 5.
Probability of composite event of end-stage kidney disease or >50% decline in glomerular filtration rate based on quartile (Q) categories. Area of the diamond within each square represents magnitude of the risk of the composite event. Numerators are the number of events and denominators are the number of individuals in each cross category of ELISA and MSD quartile. Abbreviations: ELISA, enzyme-linked immunosorbent assay (Quantikine); MSD, Meso Scale Discovery.
Table 2.
Comparison of Hazard Ratio (95% CI) and Relative Time (95% CI) to Composite Event of End-Stage Kidney Disease or >50% Decline in GFR Across Two Different Analytic/Design Strategies and Two Different suPAR Assays
| Lognormal Survival Analysisa | Study A (ELISA) |
Study B (MSD) |
||||||
|---|---|---|---|---|---|---|---|---|
| Model 1: Fully Adjusted Without eGFRb |
Model 2: Fully Adjusted With eGFRc |
Model 1: Fully Adjusted Without eGFRb |
Model 2: Fully Adjusted With eGFRc |
|||||
| RT (95% CI) | P | RT (95% CI) | P | RT (95% CI) | P | RT (95% CI) | P | |
| SuPAR quartiles | ||||||||
| 1 | 1 | Ref | 1 | Ref | 1 | Ref | 1 | Ref |
| 2 (vs 1) | 0.67 (0.50-0.89) | 0.006 | 0.76 (0.57-1.02) | 0.07 | 0.99 (0.75-1.31) | 0.94 | 1.18 (0.89-1.58) | 0.25 |
| 3 (vs 1) | 0.56 (0.42-0.74) | <.001 | 0.72 (0.54-0.96) | 0.02 | 0.57 (0.44-0.75) | <.001 | 0.82 (0.62-1.11) | 0.19 |
| 4 (vs 1) | 0.44 (0.33-0.58) | <.001 | 0.65 (0.48-0.87) | 0.004 | 0.54 (0.41-0.70) | <.001 | 0.90 (0.66-1.22) | 0.49 |
| Type III testd | <.001 | 0.04 | <.001 | 0.089 | ||||
| Cox Regression Analysise | Model 1: Fully Adjusted Without eGFRf |
Model 2: Fully Adjusted With eGFRg |
Model 1: Fully Adjusted Without eGFRf |
Model 2: Fully Adjusted With eGFRg |
||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| SuPAR quartiles | ||||||||
| 1 | 1 | Ref | 1 | Ref | 1 | Ref | 1 | Ref |
| 2 (vs 1) | 1.87 (1.13-3.09) | 0.02 | 1.44 (0.87-2.39) | 0.16 | 1.23 (0.72-2.10) | 0.46 | 0.85 (0.50-1.47) | 0.56 |
| 3 (vs 1) | 2.69 (1.64-4.41) | <.001 | 1.71 (1.03-2.84) | 0.04 | 2.33 (1.42-3.84) | <.001 | 1.04 (0.61-1.77) | 0.90 |
| 4 (vs 1) | 3.36 (2.07-5.46) | <.001 | 1.74 (1.03-2.95) | 0.04 | 3.00 (1.83-4.91) | <.001 | 1.05 (0.60-1.84) | 0.87 |
| Type III testd | <.001 | 0.17 | <.001 | 0.82 | ||||
Differences in the number of composite events arise from different administrative censoring times in the 2 studies.
Abbreviations: BMI, body mass index; BP, blood pressure; eGFR, estimated glomerular filtration rate; ELISA, enzyme-linked immunosorbent assay (Quantikine); HR, hazard ratio; MSD, Meso Scale Discovery; Ref, reference; RT, relative time; suPAR, soluble urokinase plasminogen activator receptor; UPCR, urinary protein-creatinine ratio.
For lognormal models, n = 518, events = 170.
Model 1: Adjusted for age, gender, race, ethnicity, hypertension (systolic BP percentiles), antihypertensive use, BMI, UPCR category, glomerular diagnosis; n = 23 were omitted due to missing.
Model 2: Model 1 plus eGFR; n = 23 were omitted due to missing.
Type III tests the overall effect of suPAR across all levels of suPAR.
For Cox models, n = 541, events = 184.
Model 1: Adjusted for age, gender, hypertension (systolic/diastolic BP percentiles), BMI z score, glomerular diagnosis, log base 2 UPCR.
Model 2: Model 1 plus eGFR.
For study A with ELISA-based suPAR, the original analysis using log normal survival models and limited to the joint sample of 541 participants indicated that children in the highest suPAR quartile level experienced a significantly shorter time to the composite event than those with lower levels in final models (quartile 4 vs 1: RT, 0.65; 95% CI, 0.48-0.87). Switching to the other analytic strategy using Cox models and a slightly different covariate set from study B also showed a significant effect of the highest suPAR quartile on time to the composite event (quartile 4 vs 1: HR, 1.74; 95% CI, 1.03-2.95). This magnitude of HR was approximately similar to an RT of 0.39 (95% CI, 0.16-0.94), suggesting that moving from the original analysis to the Cox model analysis resulted in a 40% change in the RT estimate. Comparisons from the Weibull can be seen in Table S1.
For study B with MSD-based suPAR, the original analysis using Cox models and limited to the joint sample of 541 participants showed that higher suPAR levels were not associated with the composite event after adjustment for baseline eGFR (quartile 4 vs quartile 1: HR, 1.05; 95% CI, 0.60-1.84). For study B, switching to lognormal models and the corresponding covariate set from study A, the estimates were similarly nonsignificant (RT, 0.90; 95% CI, 0.66-1.22). The HR from the Cox analysis was approximately equivalent to an RT of 0.98 (95% CI, 0.39-2.46), suggesting a resulting 8% change in the RT estimate by moving from the original analysis to the lognormal model analysis.
Although there were changes in the magnitude of the effect estimates as a result of the 2 different analytic strategies within a study, the effect estimates were qualitatively similar in that the effects were consistently strong in study A and consistently weak in study B. However, differences resulting from the 2 assay methods with the same analytical strategy were striking. There was a 38% change in the RT (0.65 vs 0.90, respectively) moving from ELISA to MSD for study A (lognormal model analysis) and a 66% change in HR (1.05 vs 1.74, respectively) moving from MSD to ELISA for study B (Cox model analysis), resulting in a change in inferences using a standard P value threshold to determine statistical significance (Table 2).
Because the use of quantile categorization has been criticized in the literature,15 potentially leading to a loss of power and thus potentially more variability, we also fit the optimal continuous model to each set of data. A linear model provided the best fit for the ELISA suPAR data while a nonlinear fit with a square term was the best fit for the MDR suPAR data. Effect estimate magnitudes with the nonlinear model are difficult to compare. However, effect estimates were significant in the MDR model and nonsignificant in the ELISA model using a P value of 0.05, yielding the opposite inference to that obtained from the categorical model (Table 3).
Table 3.
Hazard ratio (95% CI) for Composite Event of End-Stage Kidney Disease or >50% Decline in GFR Based on the Best Fit Model (Based on Akaike Information Criterion) With Log Base 2 suPAR as a Continuous Exposure
| Every 2-Fold Increase in suPAR | Model 1: Fully Adjusted Without eGFR (n = 541, events = 184) |
Model 2: Fully Adjusted With eGFR (n = 541, events = 184) |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| ELISAa | 2.61 (1.76-3.88) | <.001 | 1.36 (0.88-2.11) | 0.17 |
| MSDb | <.001 | 0.039 | ||
| Main effect | 1.6 × 10−4 (1 × 10−6, 0.026) | 3.7 × 10−3 (3.8 × 10−5, 0.370) | ||
| Square termc | 1.45 (1.19-1.76) | 1.25 (1.05-1.51) | ||
Abbreviations: BMI, body mass index; BP, blood pressure; eGFR, estimated glomerular filtration rate; ELISA, enzyme-linked immunosorbent assay (Quantikine); MSD, Meso Scale Discovery; suPAR, soluble urokinase plasminogen activator receptor; UPCR, urinary protein-creatinine ratio.
For ELISA, model 1 adjusted for age, gender, race, ethnicity, hypertension (systolic BP percentiles), antihypertensive use, BMI, UPCR, and glomerular diagnosis; model 2 additionally adjusted for eGFR.
For MSD, model 1 adjusted for age, gender plus hypertension (systolic/diastolic BP percentiles), BMI z score, glomerular diagnosis, and UPCR; model 2 additionally adjusted for eGFR.
Squared terms are interpreted as an interaction of suPAR with itself: the change in the main effect of a 2-fold increase in suPAR on the outcome with each 2-fold increase in suPAR.
Discussion
This real-world comparison of 2 published suPAR biomarker studies demonstrated that substantial differences in biomarker levels can arise when measuring the same biomarker in the same samples using different assays. The somewhat disheartening but eye-opening conclusion is that these differences are large enough that they can lead to divergent study conclusions.
Laboratory biomarker assay comparisons of SuPAR are scant in the literature but 1 prior investigation in the sepsis literature comparing a Luminex (8-plex) assay versus ELISA showed a much higher correlation coefficient of 0.95 than found in our study, as would be expected from a well-designed laboratory comparison study.16 However, a laboratory comparison study represents a highly controlled comparison, where factors like sample processing are standardized, and thus demonstrates a minimum amount of variability that can be expected. By contrast, the present study represents the effective differences in real-world biomarker studies, which arguably is more useful information for assessing the true variability in the biomarker literature.
Using the standard practices for judging significant results based on a 0.05 P value threshold, the associations between the ELISA assay suPAR levels in study A and progression were significant while associations between MSD assay suPAR levels in study B and progression were not after adjustment for known CKD progression risk factors, including baseline eGFR. The magnitude of the effect estimates changed by 38% to 66% as a result of switching assay protocols.
We also showed how different analytic choices can exacerbate differences in results, increasing the likelihood that dissimilar conclusions will be reached. Though results within a study were mostly consistent in inferences between analytic strategies, differences in magnitude and interpretation of effect estimates (RTs vs HRs) made the results hard to compare. Additional variability likely arose from analytic choices such as the handling of missing data, covariate inclusion, and administrative censoring times. Although the covariate choices and definitions were mostly consistent between the studies, there were differences that impacted the suPAR effect estimate and its interpretation. We estimated that the analytical strategy altered effect estimates by 8% to 40%, with larger magnitude effect estimates more affected. Our results are not necessarily generalizable to other studies, but they serve to illustrate that these differences can and do have an impact on results and conclusions.
Both the original published studies used quartiles to categorize the suPAR levels to explore associations with progression. The use of empirical thresholds is often a starting point for analyses of novel biomarkers for which clinically meaningful thresholds are unknown; however, valid criticisms of the approach have been detailed in the literature.15 Categories lead investigators to multiple testing of pairwise comparisons. In fact, we found that while using a global hypothesis test to calculate one P value across all categories, only study A (ELISA) using the Cox modeling strategy maintained a significant relationship between suPAR and progression. Further, data-driven cut points defining categories can make results difficult to compare across studies. However, nonlinear continuous models that may provide the best fit to the data can be challenging to interpret. In our reanalysis using continuous models, we found that the discrepancy in inferences between MSD and ELISA suPAR measures persisted using a standard P value threshold.
In conclusion, this biomarker study comparison highlights the variability that may exist in the current CKD biomarker literature and the need for care in the interpretation of results from novel CKD biomarker studies. Ideally, all studies would include a validation component that would provide some replication of associations. To improve efforts to rapidly evaluate novel biomarkers, new studies should consider providing results in metrics that allow for cross-comparison to other studies so the degree of uncertainty regarding the value of a new biomarker is more transparent. Figures that show raw associations can also highlight the strength or tenuous nature of biomarker relationships. Relying solely on P value thresholds to draw conclusions about meaningful relationships, particularly in early investigations of novel biomarkers, can lead to apparent conflict in results, as has been previously discussed in the epidemiologic literature.17 Sensitivity analyses can be used to provide realistic boundaries on the effect sizes. Finally, equal weight should be given to publication of both positive and negative biomarker studies to ensure the full weight of the evidence is accessible.
Article Information
Authors’ Full Names and Academic Degrees
Alison G. Abraham, PhD, Yunwen Xu, MHS, Jennifer L. Roem, MS, Jason H. Greenberg, MD, MHS, Darcy K. Weidemann, MD, MHS, Venkata S. Sabbisetti, PhD, Joseph V. Bonventre, MD, PhD, Michelle Denburg, MD, MSCE, Bradley A. Warady, MD, and Susan L. Furth, MD, PhD.
Authors’ Contributions
Research idea and study design: AA, YX, JR, JG, DW; data acquisition: SF, JB, VS, BW, MD, JG, DW; data analysis/interpretation: AA, YX, JR, JG, DW, SF; statistical analysis: AA, YX, JR. Each author contributed important intellectual content during manuscript drafting or revision and accepts accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
Support
Data collection was supported by the Marion Merrell Dow Scholarship Award of the Children's Mercy Hospital and an NIH career development grant K08DK110536. Further support was provided by NIH (NIDDK K24DK078737 and U01DK66174) and specifically the CKD Biomarkers Consortium (NIDDK grants U01DK085689, U01DK102730, U01DK103225, U01DK085660). Data in this manuscript were collected by the Chronic Kidney Disease in Children prospective cohort study (CKiD) with clinical coordinating centers at Children’s Mercy Hospital and the University of Missouri–Kansas City and Children’s Hospital of Philadelphia, Central Biochemistry Laboratory at the University of Rochester Medical Center, and data coordinating center at the Johns Hopkins Bloomberg School of Public Health. The CKiD study is funded by the National Institute of Diabetes and Digestive and Kidney Diseases, with additional funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the National Heart, Lung, and Blood Institute (U01DK66143, U01DK66174, U01DK082194, U01DK66116). The CKiD website is located at http://www.statepi.jhsph.edu/ckid.
Financial Disclosure
The authors declare that they have no relevant financial interests.
Peer Review
Received October 28, 2020. Evaluated by 1 external peer reviewer, with direct editorial input by the Statistical Editor, an Associate Editor, and the Editor-in-Chief. Accepted in revised form April 4, 2021.
Footnotes
Complete author and article information provided before references
Table S1: Comparison of hazard ratio (95% CI) and relative time (95% CI) to composite event of end-stage kidney disease or >50% decline in GFR. Based on Weibull model.
Supplementary Material
Table S1.
References
- 1.Selvin E., Manzi J., Stevens L.A. Calibration of serum creatinine in the National Health and Nutrition Examination Surveys (NHANES) 1988-1994, 1999-2004. Am J Kidney Dis. 2007;50(6):918–926. doi: 10.1053/j.ajkd.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 2.Inker L.A., Eckfeldt J., Levey A.S. Expressing the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) cystatin C equations for estimating GFR with standardized serum cystatin C values. Am J Kidney Dis. 2011;58(4):682–684. doi: 10.1053/j.ajkd.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsson A., Hansson L.-O., Flodin M. Calibration of the Siemens cystatin C immunoassay has changed over time. Clin Chem. 2011;57(5):777–778. doi: 10.1373/clinchem.2010.159848. [DOI] [PubMed] [Google Scholar]
- 4.Voskoboev N.V., Larson T.S., Rule A.D. Importance of cystatin C assay standardization. Clin Chem. 2011;57(8):1209–1211. doi: 10.1373/clinchem.2011.164798. [DOI] [PubMed] [Google Scholar]
- 5.Maahs D.M., Jalal D., McFann K. Systematic shifts in cystatin C between 2006 and 2010. Clin J Am Soc Nephrol. 2011;6(8):1952–1955. doi: 10.2215/CJN.11271210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbe E., Levesque L., Suissa S. Variability of breast cancer risk in observational studies of hormone replacement therapy: a meta-regression analysis. Maturitas. 2004;47(3):175–183. doi: 10.1016/j.maturitas.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 7.Brambilla D., Reichelderfer P.S., Bremer J.W. The contribution of assay variation and biological variation to the total variability of plasma HIV-1 RNA measurements. The Women Infant Transmission Study Clinics. Virology Quality Assurance Program. AIDS. 1999;13(16):2269–2279. doi: 10.1097/00002030-199911120-00009. [DOI] [PubMed] [Google Scholar]
- 8.Waters K.M., Le Marchand L., Kolonel L.N. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1285–1289. doi: 10.1158/1055-9965.EPI-08-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg J.H., Abraham A.G., Xu Y. Plasma biomarkers of tubular injury and inflammation are associated with CKD progression in children. J Am Soc Nephrol. 2020;31(5):1067–1077. doi: 10.1681/ASN.2019070723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidemann D.K., Abraham A.G., Roem J.L. Plasma soluble urokinase plasminogen activator receptor (suPAR) and CKD progression in children. Am J Kidney Dis. 2020;76(2):194–202. doi: 10.1053/j.ajkd.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz G.J., Munoz A., Schneider M.F. New equations to estimate GFR in children with CKD. J Am Soc Nephrol. 2009;20(3):629–637. doi: 10.1681/ASN.2008030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynn J.T., Kaelber D.C., Baker-Smith C.M. Subcommittee on Screening and Management of High Blood Pressure in Children. Clinical practice guideline for screening and management of high blood pressure in children and adolescents. Pediatrics. 2017;140(3) doi: 10.1542/peds.2017-1904. Published correction appears in Pediatrics. 2018;142(3):e20181739. [DOI] [PubMed] [Google Scholar]
- 13.Warady B.A., Abraham A.G., Schwartz G.J. Predictors of rapid progression of glomerular and nonglomerular kidney disease in children and adolescents: The Chronic Kidney Disease in Children (CKiD) Cohort. Am J Kidney Dis. 2015;65(6):878–888. doi: 10.1053/j.ajkd.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- 15.Bennette C., Vickers A. Against quantiles: categorization of continuous variables in epidemiologic research, and its discontents. BMC Med Res Methodol. 2012;12:21. doi: 10.1186/1471-2288-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kofoed K., Schneider U.V., Scheel T. Development and validation of a multiplex add-on assay for sepsis biomarkers using xMAP technology. Clin Chem. 2006;52(7):1284–1293. doi: 10.1373/clinchem.2006.067595. [DOI] [PubMed] [Google Scholar]
- 17.Greenland S., Senn S.J., Rothman K.J. Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur J Epidemiol. 2016;31(4):337–350. doi: 10.1007/s10654-016-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.