Summary
Pluripotent stem cell-derived organoids have transformed our ability to recreate complex three-dimensional models of human tissue. However, the directed differentiation methods used to create them do not afford the ability to introduce cross-germ-layer cell types. Here, we present a bottom-up engineering approach to building vascularized human tissue by combining genetic reprogramming with chemically directed organoid differentiation. As a proof of concept, we created neuro-vascular and myo-vascular organoids via transcription factor overexpression in vascular organoids. We comprehensively characterized neuro-vascular organoids in terms of marker gene expression and composition, and demonstrated that the organoids maintain neural and vascular function for at least 45 days in culture. Finally, we demonstrated chronic electrical stimulation of myo-vascular organoid aggregates as a potential path toward engineering mature and large-scale vascularized skeletal muscle tissue from organoids. Our approach offers a roadmap to build diverse vascularized tissues of any type derived entirely from pluripotent stem cells.
Keywords: reprogramming, differentiation, vascularized organoids, human pluripotent stem cells, neuro-vascular organoids, myo-vascular organoids
Highlights
-
•
Neuro- and myo-vascular tissue built via genetic reprogramming in vascular organoids
-
•
Neuro-vascular organoids were sustained in vitro for at least 45 days
-
•
Neuro-vascular organoids demonstrated functional neural and vascular cells
-
•
Myo-vascular organoid maturation could be driven by chronic electrical stimulation
Building vascularized tissue with cross-germ-layer cell types is a major challenge in organoid engineering. Here, the authors leverage genetic reprogramming in conjunction with standard chemically directed differentiation to generate lineage-specific vascularized organoids. By expressing lineage-specifying transcription factors during organoid differentiation, they demonstrate a robust protocol for generating myo-vascular and neuro-vascular tissues entirely from pluripotent stem cells.
Introduction
The ability to recapitulate organogenesis and create complex human tissue in vitro has been a long-standing goal for the stem cell and tissue engineering field. The advent of organoid technology has recently made it possible to create three-dimensional (3D), self-organized, pluripotent stem cell (PSC)-derived tissues for in vitro developmental and disease modeling that closely mimic the cellular, spatial, and molecular architecture of endogenous human tissue (Clevers, 2016; Lancaster and Knoblich, 2014). These advances have enabled substantial progress in building fully PSC-derived, functional human organs in vitro (Takebe and Wells, 2019). However, the absence of vasculature in most organoid techniques limits their utility, principally in two ways. Firstly, it is widely accepted that vasculature plays a crucial role in development and disease (Daniel and Cleaver, 2019; Petrova and Koh, 2018). Secondly, vasculature is necessary to prevent necrosis in tissues that grow beyond 1 mm in size (Grebenyuk and Ranga, 2019; Lancaster, 2018), which deems vasculature critical for building large-scale tissue models.
To address this, several groups have demonstrated progress on developing methods for vascularizing organoids (Cakir et al., 2019; Garreta et al., 2019; Guye et al., 2016; Homan et al., 2019; Low et al., 2019; Mansour et al., 2018; Pham et al., 2018). Some groups have succeeded in vascularizing organoids after transplanting them in vivo (Mansour et al., 2018; Pham et al., 2018), but requiring an in vivo host limits experimental control, increases cost, and diminishes its potential for clinical applications. Others transiently overexpressed GATA6 to introduce a nascent vascular network in liver-bud organoids (Guye et al., 2016); however, there is no current evidence this system can be translated to other organoids.
Organoid platforms leverage knowledge of development to provide temporally appropriate chemical cues to self-assembled PSC-derived embryoid bodies, modulating key organogenesis-specific signaling pathways to drive the directed differentiation of organ-specific cells in physiologically faithful architectures. Thus, cells that do not belong to those specific organ compartments or arise from different germ layers are absent from the final organoid. To introduce cell types outside of those available from directed differentiation, a promising strategy is to combine genetic overexpression with directed differentiation. Recently, vascularization of cerebral organoids has been reported, in which PSCs were engineered to ectopically express human ETS variant 2 (ETV2) (Cakir et al., 2019), a known driver of differentiation to endothelial cells from PSCs (Lindgren et al., 2015; Parekh et al., 2018). Although this method has exciting potential, it suffers from two limitations: one, it induces only a low degree of vascularization, and, two, it does not induce the full panoply of vascular lineages, such as smooth muscle cells (SMCs) and mesenchymal stem cells (MSCs), which are critical for blood vessel development and function (Ferland-McCollough et al., 2017). Thus, there is a need to explore alternative methods.
A recently described vascular organoid (VO) differentiation approach (Wimmer et al., 2019) yields complete blood vessel networks, including SMCs, MSCs, and endothelial cells, but these organoids lack organ-specific parenchymal cells, limiting their utility for broader disease modeling and regenerative medicine. Here, we overlay reprogramming with the directed differentiation of VOs to derive cross-germ-layer and cross-lineage organoids with complete vascular networks. We introduce a parenchymal cell component into VOs via transcription factor (TF) overexpression and demonstrate this approach by building neuro-vascular and myo-vascular organoids. This is done by the induced overexpression, in developing vascular organoids, of NEUROD1 (iN) to form neuro-vascular organoids (iN-VOs), and the induced overexpression of MYOD1 plus BAF60C (iMB) to form myo-vascular organoids (iMB-VOs).
This yields a facile method for co-differentiation of tissue-specific parenchymal cells and the entire blood vessel lineage from a single PSC line. We present this approach as a proof of concept for the introduction of other parenchymal cell types, via the overexpression of lineage-specifying TFs, in the context of a VO scaffold.
Results
Inducible cell line construction and validation
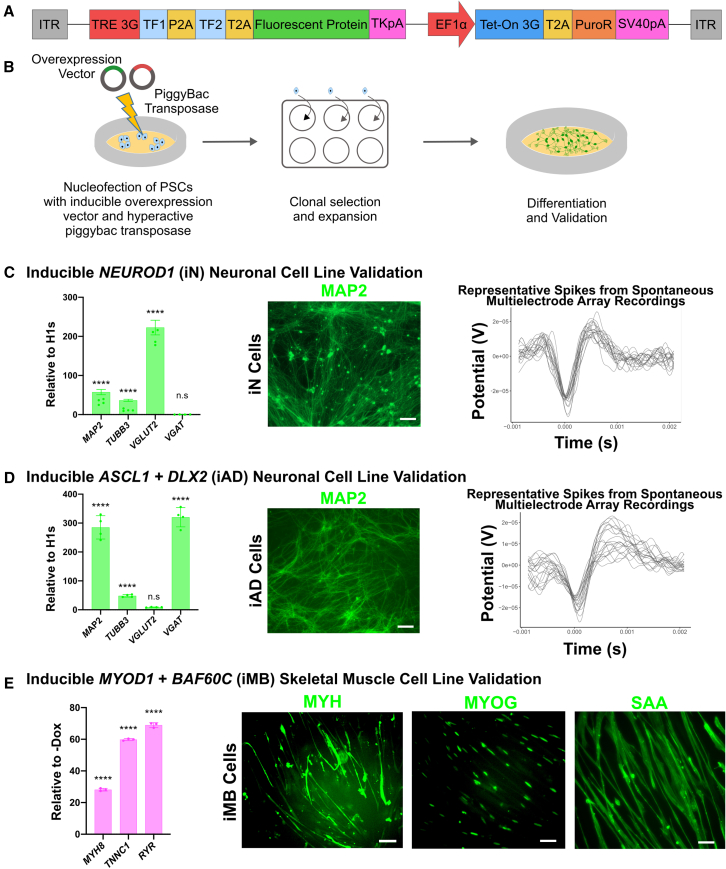
For the inducible expression of TFs, we designed a PiggyBac transposon-based overexpression vector that allowed a single vector to package the complete Tet-On system for doxycycline inducible expression, along with one or more TFs to be overexpressed in conjunction with a reporter fluorescent protein (Figure 1A). To establish the utility of our PiggyBac overexpression platform, we constructed an array of cell lines and validated their ability to differentiate into functional tissue upon adding doxycycline. For neural differentiation, we chose to overexpress NEUROD1 to generate glutamatergic excitatory neurons (Parekh et al., 2018; Zhang et al., 2013), and ASCL1+DLX2 for GABAergic inhibitory neuron differentiation (Yang et al., 2017). For mesodermal tissue differentiation, we chose to overexpress MYOD1+BAF60C (Albini et al., 2013) for skeletal muscle differentiation. Stable, dox-inducible NEUROD1 (iN), ASCL1+DLX2 (iAD), and MYOD1+BAF60C (iMB) human embryonic stem cell (hESC) lines were generated by nucleofecting hESCs with the respective overexpression vector, along with a hyperactive PiggyBac transposase (Yusa et al., 2011).
Figure 1.
Construction and characterization of inducible cell lines
(A) Schematic of PiggyBac transposon-based inducible overexpression vector.
(B) Schematic of cell line generation and validation process.
(C) Inducible NEUROD1 (iN) cell line validation at 3 weeks post induction via (1) qRT-PCR analysis of signature neuronal markers MAP2, TUBB3, VGLUT2, and VGAT; data represent the mean ± SD (n = 4 independent experiments); (2) immunofluorescence micrograph of MAP2+ cells (scale bars, 50 μm); and (3) representative spike plots from MEA measurements of spontaneously firing iN cells.
(D) Inducible ASCL1+DLX2 (iAD) cell line validation at 3 weeks post induction via (1) qRT-PCR analysis of signature neuronal markers MAP2, TUBB3, VGLUT2, and VGAT; data represent the mean ± SD (n = 4 independent experiments); (2) immunofluorescence micrograph of MAP2+ cells (scale bars, 50 μm); and (3) representative spike plots from MEA measurements of spontaneously firing iAD cells.
(E) Inducible MYOD1+BAF60C (iMB) cell line validation at 2 weeks post induction via (1) qRT-PCR analysis of signature skeletal muscle markers MYH8, TNNC1, and RYR; data represent the mean ± SD (n = 3 independent experiments); and (2) immunofluorescence micrograph of MYH+-, MYOG+-, and SAA+-labeled cells (scale bars, 50 μm). (C–E) ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001; ns, not significant.
For validation of neural differentiation of iN and iAD lines, qRT-PCR analysis demonstrated upregulation of neuronal markers MAP2 and TUBB3 for both iN and iAD cells (Figures 1C and 1D) compared with undifferentiated hESCs. iN neurons were confirmed to be glutamatergic via upregulation of excitatory marker VGLUT2 with no detectable expression of the inhibitory marker VGAT (Figure 1C), while iAD neurons were confirmed to be GABAergic, via upregulation of VGAT with minimal expression of VGLUT2 (Figure 1D). MAP2+ cells with classic neuronal morphology were confirmed by immunofluorescence for both iN and iAD cells (Figures 1C and 1D). Finally, iN and iAD neurons were confirmed to be functional by detection of spontaneous firing when co-cultured with glia and measured on a microelectrode array (MEA) at 3–5 weeks post induction (Figures 1C and 1D).
Skeletal muscle differentiation of iMB cells was validated by gene expression and immunofluorescence assays (Figure 1E). qRT-PCR analysis confirmed upregulation of skeletal muscle markers MYH8, TNNC1, and RYR. Immunofluorescence confirmed the presence of characteristic spindle-shaped MYH+ and SAA+ skeletal muscle morphology, along with MYOG+ nuclei.
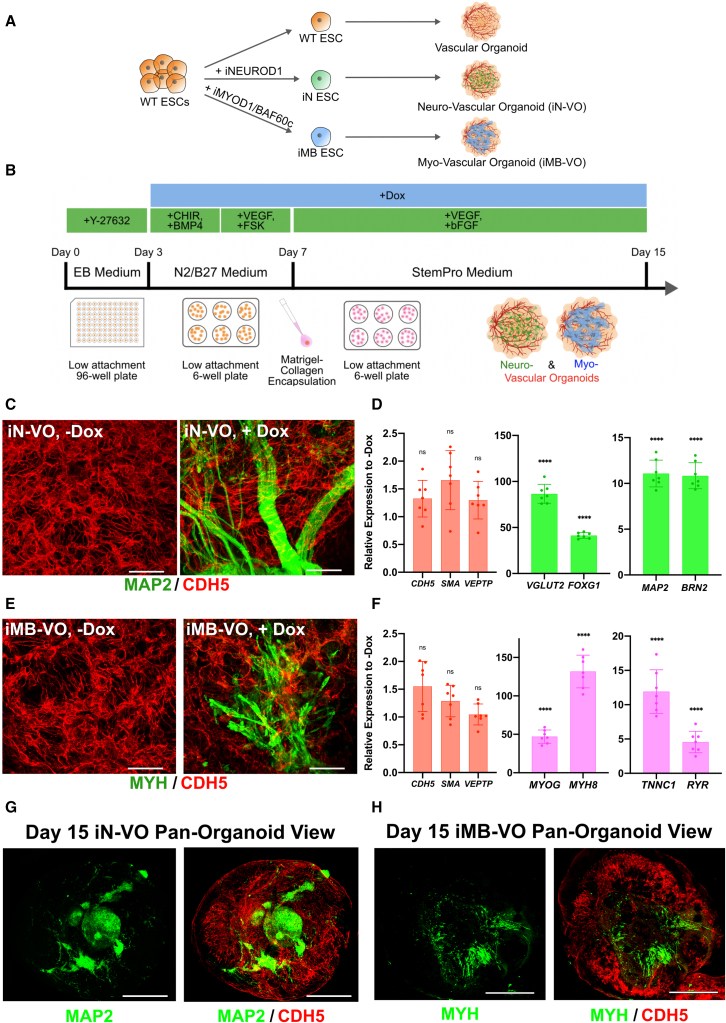
Constructing neuro-vascular and myo-vascular organoids
After clonal hESC lines were established, we developed a modular, generalizable, TF overexpression-based strategy to differentiate parenchymal cell types in VOs (Figure 2A). We conducted an optimization experiment (Note S1) to determine if wild-type ESCs are required for maintaining the vascular compartment in our organoids (Figure S1A). Given the higher expression of MAP2 and comparable expression of CDH5, subsequent experiments were conducted with iN-VOs formed from 100% iN cells (Figure S1B). Immunostaining was performed on both induced (+dox, iN-VOs) and uninduced (−dox, iN-VOs) organoids, confirming MAP2+ cells forming into bundle-like structures of neurons only in induced organoids, along with dense, interpenetrating CDH5+ vascular networks in both conditions (Figure 2C). Furthermore, pan-organoid confocal images show that endothelial networks span all layers of the organoid, as do neurons, although they are not uniformly distributed and form clusters and bundles of cells (Figures 2G and S1C; Videos S1 and S3). Further analysis via qRT-PCR showed an upregulation of the excitatory marker VGLUT2, along with cortical neuron markers BRN2 and FOXG1 (Zhang et al., 2013) in day 15 iN-VOs (Figure 2D). Moreover, we observed expression levels comparable with uninduced organoids of the endothelial markers CDH5 and VEPTP, along with the smooth muscle marker SMA (Figure 2D).
Figure 2.
Generation of iN-VOs and iMB-VOs
(A) General strategy for the generation of vascularized organ tissues via introduction of parenchymal cell types in VOs.
(B) Schematic of iN-VO and iMB-VO culture protocol.
(C) Immunofluorescence 100-μm z stack, maximum projection, confocal micrographs of MAP2- and CDH5-labeled uninduced (iN-VO, −Dox) and induced (iN-VO, +Dox) day 15 iN-VO organoids (scale bars, 50 μm).
(D) qRT-PCR analysis of signature endothelial genes CDH5 and VEPTP; signature smooth muscle gene SMA; and signature neuronal genes MAP2, VGLUT2, BRN2, and FOXG1 at day 15 of culture for iN-VO organoids. Data represent the mean ± SD (n = 7 organoids, from three independent experiments).
(E) Immunofluorescence 100-μm z stack, maximum projection, confocal micrographs of MYH- and CDH5-labeled uninduced (iMB-VO, −Dox) and induced (iMB-VO, +Dox) day 15 iMB-VO organoids (scale bars, 50 μm).
(F) qRT-PCR analysis of signature endothelial genes CDH5 and VEPTP; signature smooth muscle gene SMA; and signature skeletal muscle genes MYOG, MYH8, TNNC1, and RYR at day 15 of culture for iMB-VO organoids. Data represent the mean ± SD (n = 7 organoids, from three independent experiments).
(G) Pan-organoid tile-scan immunofluorescence confocal micrograph of a CDH5- and MAP2-labeled day 15 neuro-vascular organoid (scale bars, 500 μm). Image is a 200-μm z stack maximum intensity projection.
(H) Pan-organoid tile-scan immunofluorescence confocal micrograph of CDH5- and MYH-labeled day 15 myo-vascular organoid (scale bars, 500 μm). Image is a 200-μm z stack maximum intensity projection. (D and F) ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001; ns, not significant.
Once the formation of neuro-vascular organoids was established, we then sought to demonstrate if our system was compatible with other germ-layer or cross-lineage cell types. Using 100% iMB cells and the same protocol used to generate neuro-vascular organoids, we grew iMB-VOs (Figure 2B). iMB-VOs had MYH+ spindle-shaped cells (Figure 2E), along with expression of skeletal muscle marker genes MYOG, MYH8, TNNC1, and RYR (Figure 2F), confirming that our platform could also be used to grow vascularized tissue of mesodermal origin, specifically skeletal muscle. Pan-organoid tile-scan confocal images demonstrated MYH+ skeletal muscle cells present in many layers of the myo-vascular organoid, albeit not uniformly distributed throughout the organoid (Figures 2H and S1D; Videos S2 and S4).
Organoid is stained for CDH5 (red) and MAP2 (green).
Organoid is stained for CDH5 (red) and MYH (green).
Organoid is stained for CDH5 (red) and MAP2 (green). Scale bar, 100 μm.
Organoid is stained for CDH5 (red) and MYH (green). Scale bar, 100 μm.
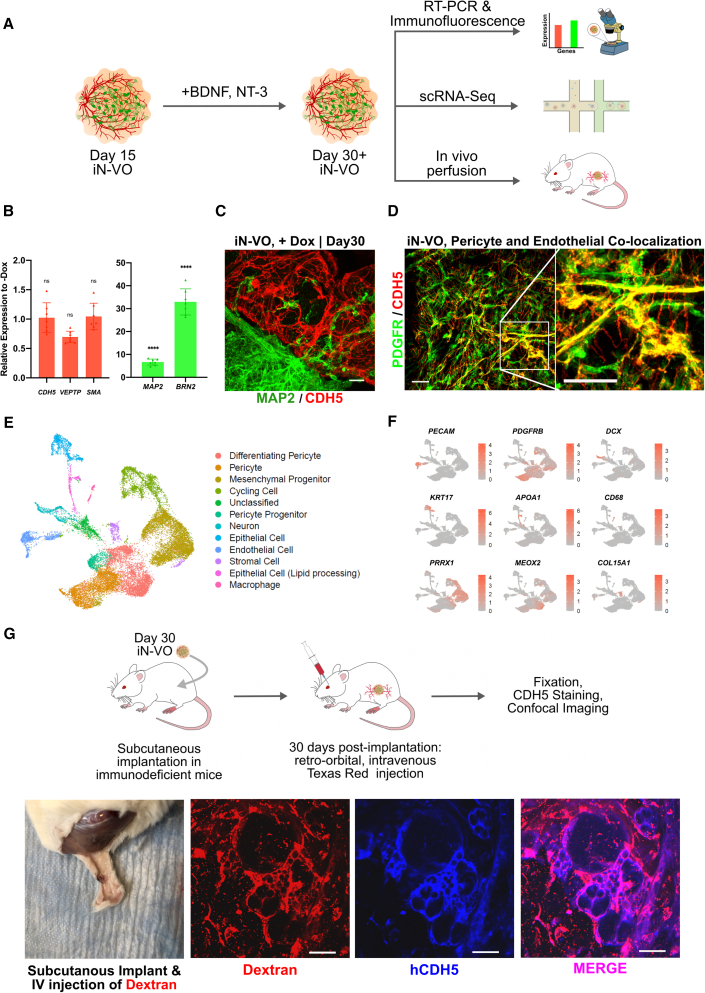
Comprehensive characterization of neuro-vascular organoids
We then sought to enable further long-term culture of the neuro-vascular organoids beyond the initial 15 days (Figure S1E). Media optimization experiments (Note S2) revealed that brain-derived neurotrophic factor (BDNF) and neurotrophin-3 (NT-3) supplementation from day 15 ensured long-term survival of neurons with reliable maintenance of vascular lineages (Figure S1F).
Upon determining an optimized protocol for long-term growth of iN-VOs, we comprehensively characterized these long-term cultured neuro-vascular organoids (Figure 3A). qRT-PCR analysis showed an upregulation of the neuronal markers MAP2, VGLUT2, BRN2, and FOXG1 in day 30 iN-VOs (Figure 3B), while maintaining the expression level of endothelial markers CDH5 and VEPTP, and smooth muscle marker SMA (Figure 3B). Immunofluorescence imaging confirmed the presence of MAP2+ neurons displaying robust neurite growth, along with interpenetrating CDH5+ vascular networks through 30 days in culture (Figure 3C). These organoids formed capillary networks that consisted of lumen-forming endothelial cells (Figure S3A) that were tightly associated with pericytes (Figure 3D).
Figure 3.
Molecular and functional characterization of iN-VOs
(A) Outline of long-term cultured iN-VO characterization.
(B) qRT-PCR analysis of signature endothelial genes CDH5 and VEPTP, signature smooth muscle gene SMA, and signature neuronal genes MAP2 and BRN2 at day 30 of iN-VO culture. Data represent the mean ± SD (n = 7 organoids, from three independent experiments).
(C) Immunofluorescence 100-μm z stack, maximum projection, confocal micrographs of MAP2+- and CDH5+-induced day 30 iN-VOs (scale bars, 100 μm).
(D) Immunofluorescence100-μm z stack, maximum projection, confocal micrographs of PDGFR+-and CDH5+-induced day 30 iN-VOs (scale bars, 50 μm).
(E) Uniform manifold approximation and projection (UMAP) visualization of cell types from day 45 iN-VOs. Two independent induction conditions, along with one non-induction condition.
(F) Cluster-specific expression of marker genes in day 45 iN-VOs.
(G) Experimental validation of iN-VO perfusibility in vivo by subcutaneous implantation of iN-VO, showing immunofluorescence micrographs of intravital Dextran, CDH5, and overlay (scale bar, 25 μm). Representative image from two independent experiments.
To characterize the cell-type composition of iN-VOs cultured for up to 45 days, we assayed them using single-cell RNA-sequencing (scRNA-seq). We assayed 26,959 cells across day 45 organoids grown under three conditions: 15 days of NEUROD1 overexpression (iN-VO, 15 days induction), 45 days of NEUROD1 overexpression (iN-VO, 45 days induction), and without overexpression (iN-VO, no induction). Using the Seurat pipeline (Butler et al., 2018), we identified that PRRX1+ mesenchymal progenitors, MEOX2+ differentiating pericytes, and cycling cells were present in all three types of organoids and constituted a majority of each of the organoids (Figures 3E, 3F, S2A, and S2B). Importantly, functional vascular cell types PDGFRβ+ pericytes and PECAM1+ endothelial cells were present in all types of organoids and constituted 16%–22% and 2.5%–6% of cells respectively (Figures 3E, 3F, S2A, and S2B). Endothelial and neural cells mapped with high fidelity to the endothelium and neurons from the Mouse Cell Atlas, respectively (Figure S2C). To further validate their identity, neurons from the neuro-vascular organoids were mapped to DCX+ neuronal clusters from a reference dataset profiling two-dimensional (2D) differentiation of neurons from hPSCs by NGN2 overexpression (Schörnig et al., 2021) (Figures S2D and S2E). After molecular characteristics of the iN-VOs were assayed, we assessed the functionality of both neural and vascular cell types in the organoid. First we assessed endothelial function in vitro by confirming uptake of acetylated low-density lipoprotein by endothelial cells in the organoids (Figure S3B). Further, we assayed the ability of the vascular networks to form perfusable blood vessels when implanted into mice. Specifically, iN-VOs were grown till day 30 in vitro, and then subcutaneously implanted in Rag2−/−;γc−/− immunodeficient mice. Thirty and 90 days post implantation, intravenous (IV) injection of a Texas red dye followed by organoid extraction, fixation, and immunofluorescence staining showed co-localization of the dye with human CDH5+ vascular cells and PDGFR+ pericytes in the iN-VO (Figures 3G and S3C). Endothelium function in vivo was sustained for at least 90 days post implantation, as demonstrated by confocal imaging of lumenized and perfused CDH5+ vessels (Figure S3Cii,iii). The absence of CDH5 signal in wild-type mouse kidneys stained with human-specific anti-CDH5 demonstrates the human specificity of the antibody (Figure S3D), confirming that the perfused CDH5+ vessels in the organoids are of human origin.
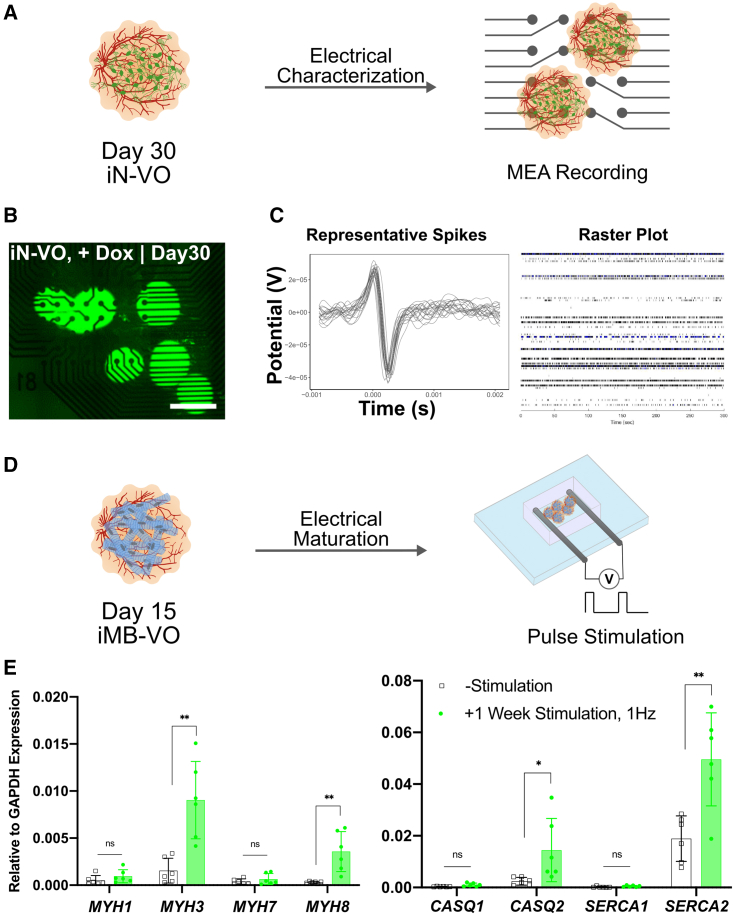
To assess functionality of neurons in the iN-VOs, we assayed the organoids for spontaneous electrical activity using MEAs (Figures 4A and 4B). Strikingly, while uninduced organoids displayed no spontaneous activity, spontaneous firing was repeatedly observed in iN-VOs (Figure 4C), confirming the presence of functional neurons. Taken together, we demonstrate the formation of functional neuro-vascular tissue with long-term culture capability.
Figure 4.
Electrical characterization and stimulation of organoids
(A) Schematic of iN-VO electrical characterization via MEA recordings.
(B) Image of day 30 iN-VOs plated on microelectrode array (scale bars, 500 μm).
(C) Representative spike plots from MEA measurements of spontaneously firing iN-VOs and corresponding raster plot. Representative image and plot from two independent experiments.
(D) Schematic of iMB-VO in vitro maturation by electrical stimulation.
(E) qRT-PCR analysis of skeletal muscle myosins: MYH2, MYH3, MYH7, and MYH8, and genes involved in calcium handling, CASQ1, CASQ2, SERCA1, SERCA2, and RYR for stimulated versus unstimulated iMB-VOs. Data represent the mean ± SD (n = 6 organoids, from two independent experiments). ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001; ns, not significant.
In vitro maturation of myo-vascular organoids
Finally, while our iMB-VO platform was able to differentiate vascularized skeletal muscle, maturation of this lineage in vitro is a long-standing challenge. To further mature the differentiated skeletal muscle, we applied chronic electrical stimulation (Khodabukus et al., 2019; Rao et al., 2018).
To subject organoids to stimulation, we encapsulated them in a fibrin and Matrigel blend, and placed them in a custom chip between two graphite rods. A pulsed constant-voltage stimulation was then applied for a week after encapsulation to drive maturation (Figure 4D). Organoids were then assayed for gene expression of muscle and calcium-handling genes (Figure 4E). In stimulated organoids, embryonic skeletal muscle myosin (MYH3) was upregulated, along with a small increase in expression of adult fast skeletal muscle myosin (MYH2). Additionally, the calcium-handling genes CASQ2 and SERCA2 were also highly upregulated. In summary, our iMB-VO approach is a promising method to generate mature, vascularized skeletal muscle tissue in vitro.
Discussion
In this work, we have demonstrated that a combination of directed differentiation and the overexpression of a lineage-specific reprogramming factor can build cross-lineage and cross-germ-layer organoid systems. We used this approach to introduce neurons and skeletal muscle into VOs, as a proof of principle of such an approach. We then assayed these mixed-lineage organoids to demonstrate that this combined approach yielded neurons only upon induction of NEUROD1 overexpression, and skeletal muscle only upon induction of MYOD1+BAF60C overexpression, while retaining the architecture of the VO. The neuro-vascular organoids thus generated were further optimized for long-term culture, and comprehensively characterized for composition and to confirm function of both lineage branches. Finally, for the myo-vascular organoids, we demonstrated the maturation of these organoids via chronic electrical stimulation, which enhanced the expression of skeletal muscle myosins and calcium-handling genes.
While providing a modular and powerful approach to engineer vascularized multi-lineage organoids, several challenges still remain to be solved. While we have taken advantage of known reprogramming factors to differentiate neurons and skeletal muscle, such recipes are not known for all cell types and will have to be discovered. Known overexpression recipes may also need to be modified for organoid culture conditions, since recipes are typically optimized and validated only for specific 2D culture and media conditions. While we have restricted the current study to overexpression, knockdown approaches (Qian et al., 2020) may also be important tools in these engineering efforts. Additionally, improved control of this spatial organization will be critical for accurate tissue engineering, especially as reprogrammed cells transition directly from initial to final lineage states, and one may need to incorporate novel strategies like optogenetic control of gene expression (Nihongaki et al, 2015, 2017; Polstein and Gersbach, 2015) or synthetic biology approaches (Toda et al., 2018).
In summary, our demonstrated approach provides a blueprint for complete bottom-up engineering of vascularized organoids, where genetic perturbation methods for cell reprogramming can be overlaid onto our core protocol to introduce additional cell types. This framework could be a powerful platform to generate diverse vascularized organoids, thus laying the foundation for completely in vitro-derived, large-scale tissue systems for regenerative medicine purposes.
Experimental procedures
Detailed methods are provided in the supplemental experimental procedures.
iMB-VO and iN-VO generation
To generate iMB-VOs or iN-VOs, 9,000 iMB or iN cells, respectively, were seeded in ULA 96 well plates for embryoid body (EB) formation. EBs were grown in EB media (DMEM/F12 + 20% KOSR + 50 μM Y-27632) for 2 days and then transferred into an ULA 6-well plate, with about 8–12 EBs/well. This media was then replaced with 2 mL of Mesoderm Induction Media (N2/B27 + BMP4 + CHIR), with the addition of doxycycline to introduce either the muscle or neural compartment for iMB-VOs or iN-VOs, respectively. Doxycycline was spiked into the media for every media change onwards. Organoids were then cultured at 37°C, 5% CO2 for 3 days. Three days later, the medium was replaced with 2 mL of Vascular Induction Media (N2/B27 + VEGF + FSK) per well and cultured at 37°C, 5% CO2. This media was replaced 24 hours later and cultured at 37°C, 5% CO2 for 24 h. On day 7 of organoid formation, organoids were encapsulated in a blend of matrigel and collagen and then cultured in Vascular Maturation Media (StemPro + 15% FBS + VEGF + bFGF). Organoids were cultured at 37°C, 5% CO2 and Vascular Maturation Media was replaced every 3 days. For long-term growth of iN-VOs, Vascular Maturation Media was supplemented with BDNF and NT-3 from day 15 and onwards.
Data and code availability
All scRNA-seq data can be accessed via GEO (GEO: GSE164268).
Author contributions
Conceptualization and design, A.D., U.P., T.N.N., and P.M.; experiments, A.D., U.P., D.K., A.K., D.M., A.M., and P.B.; computational analyses, U.P.; writing, A.D., U.P., and P.M., with input from all authors.
Conflict of interests
A.D., U.P., and P.M. have filed patents based on this work. P.M. is a scientific co-founder of Shape Therapeutics, Boundless Biosciences, Navega Therapeutics, and Engine Biosciences, which have no commercial interests related to this study. The terms of these arrangements have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies.
Acknowledgments
The authors would like to thank James Yen, Yan Wu, Kian Kalhor, Reinner Wimmer, Cleber Trujillo, Hammza Khaliq, and Ramin Dailamy for useful discussions and support throughout all stages of the study; and Jennifer Santini from UCSD School of Medicine Microscopy Core (grant P30 NS047101). This work was generously supported by UCSD Institutional Funds and NIH grants (R01HG009285, RO1CA222826, RO1GM123313). This publication includes data generated at the UCSD IGM Genomics Center utilizing an Illumina NovaSeq 6000 that was purchased with funding from an NIH SIG grant (S10 OD026929).
Published: September 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.08.014.
Supplemental information
References
- Albini S., Coutinho P., Malecova B., Giordani L., Savchenko A., Forcales S.V., Puri P.L. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir B., Xiang Y., Tanaka Y., Kural M.H., Parent M., Kang Y.-J., Chapeton K., Patterson B., Yuan Y., He C.-S. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Daniel E., Cleaver O. Vascularizing organogenesis: lessons from developmental biology and implications for regenerative medicine. Curr. Top. Dev. Biol. 2019;132:177–220. doi: 10.1016/bs.ctdb.2018.12.012. [DOI] [PubMed] [Google Scholar]
- Ferland-McCollough D., Slater S., Richard J., Reni C., Mangialardi G. Pericytes, an overlooked player in vascular pathobiology. Pharmacol. Ther. 2017;171:30–42. doi: 10.1016/j.pharmthera.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreta E., Prado P., Tarantino C., Oria R., Fanlo L., Martí E., Zalvidea D., Trepat X., Roca-Cusachs P., Gavaldà-Navarro A. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 2019;18:397–405. doi: 10.1038/s41563-019-0287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenyuk S., Ranga A. Engineering organoid vascularization. Front. Bioeng. Biotechnol. 2019;7:39. doi: 10.3389/fbioe.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guye P., Ebrahimkhani M.R., Kipniss N., Velazquez J.J., Schoenfeld E., Kiani S., Griffith L.G., Weiss R. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan K.A., Gupta N., Kroll K.T., Kolesky D.B., Skylar-Scott M., Miyoshi T., Mau D., Valerius M.T., Ferrante T., Bonventre J.V. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodabukus A., Madden L., Prabhu N.K., Koves T.R., Jackman C.P., Muoio D.M., Bursac N. Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle. Biomaterials. 2019;198:259–269. doi: 10.1016/j.biomaterials.2018.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A. Brain organoids get vascularized. Nat. Biotechnol. 2018;36:407–408. doi: 10.1038/nbt.4133. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Lindgren A.G., Veldman M.B., Lin S. ETV2 expression increases the efficiency of primitive endothelial cell derivation from human embryonic stem cells. Cell Regen. 2015;4:1. doi: 10.1186/s13619-014-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J.H., Li P., Chew E.G.Y., Zhou B., Suzuki K., Zhang T., Lian M.M., Liu M., Aizawa E., Rodriguez Esteban C. Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell. 2019;25:373–387.e9. doi: 10.1016/j.stem.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A.A., Gonçalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y., Yamamoto S., Kawano F., Suzuki H., Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 2015;22:169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Nihongaki Y., Furuhata Y., Otabe T., Hasegawa S., Yoshimoto K., Sato M. CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods. 2017;14:963–966. doi: 10.1038/nmeth.4430. [DOI] [PubMed] [Google Scholar]
- Parekh U., Wu Y., Zhao D., Worlikar A., Shah N., Zhang K., Mali P. Mapping cellular reprogramming via pooled overexpression screens with paired fitness and single-cell RNA-sequencing readout. Cell Syst. 2018;7:548–555.e8. doi: 10.1016/j.cels.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova T.V., Koh G.Y. Organ-specific lymphatic vasculature: from development to pathophysiology. J. Exp. Med. 2018;215:35–49. doi: 10.1084/jem.20171868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M.T., Pollock K.M., Rose M.D., Cary W.A., Stewart H.R., Zhou P., Nolta J.A., Waldau B. Generation of human vascularized brain organoids. Neuroreport. 2018;29:588–593. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein L.R., Gersbach C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Kang X., Hu J., Zhang D., Liang Z., Meng F., Zhang X., Xue Y., Maimon R., Dowdy S.F. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature. 2020;582:550–556. doi: 10.1038/s41586-020-2388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L., Qian Y., Khodabukus A., Ribar T., Bursac N. Engineering human pluripotent stem cells into a functional skeletal muscle tissue. Nat. Commun. 2018;9:126. doi: 10.1038/s41467-017-02636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schörnig M., Ju X., Fast L., Ebert S., Weigert A., Kanton S., Schaffer T., Nadif Kasri N., Treutlein B., Peter B.M. Comparison of induced neurons reveals slower structural and functional maturation in humans than in apes. eLife. 2021;10:e59323. doi: 10.7554/eLife.59323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T., Wells J.M. Organoids by design. Science. 2019;364:956–959. doi: 10.1126/science.aaw7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S., Blauch L.R., Tang S.K.Y., Morsut L., Lim W.A. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer R.A., Leopoldi A., Aichinger M., Wick N., Hantusch B., Novatchkova M., Taubenschmid J., Hämmerle M., Esk C., Bagley J.A. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Chanda S., Marro S., Ng Y.-H., Janas J.A., Haag D., Ang C.E., Tang Y., Flores Q., Mall M. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods. 2017;14:621–628. doi: 10.1038/nmeth.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Zhou L., Li M.A., Bradley A., Craig N.L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U S A. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Organoid is stained for CDH5 (red) and MAP2 (green).
Organoid is stained for CDH5 (red) and MYH (green).
Organoid is stained for CDH5 (red) and MAP2 (green). Scale bar, 100 μm.
Organoid is stained for CDH5 (red) and MYH (green). Scale bar, 100 μm.
Data Availability Statement
All scRNA-seq data can be accessed via GEO (GEO: GSE164268).