Abstract
Effective biomarkers for the diagnosis of colorectal cancer (CRC) are essential for improving prognosis. Imbalance in regulation of N6-methyladenosine (m6A) RNA has been associated with a variety of cancers. However, whether the m6A RNA levels of peripheral blood can serve as a diagnostic biomarker for CRC is still unclear. In this research, we found that the m6A RNA levels of peripheral blood immune cells were apparently elevated in the CRC group compared with those in the normal controls (NCs) group. Furthermore, the m6A levels arose as CRC progressed and metastasized, while these levels decreased after treatment. The area under the curve (AUC) of the m6A levels was 0.946, which was significantly higher than the AUCs for carcinoembryonic antigen (CEA; 0.817), carbohydrate antigen 125 (CA125; 0.732), and carbohydrate antigen 19-9 (CA19-9; 0.771). Moreover, the combination of CEA, CA125, and CA19-9 with m6A levels improved the AUC to 0.977. Bioinformatics and qRT-PCR analysis further confirmed that the expression of m6A modifying regulator IGF2BP2 was markedly elevated in peripheral blood of CRC patients. Gene set variation analysis (GSVA) implied that monocyte was the most abundant m6A-modified immune cell type in CRC patients’ peripheral blood. Additionally, m6A modifications were negatively related to the immune response of monocytes. In conclusion, our results revealed that m6A RNA of peripheral blood immune cells was a prospective non-invasive diagnostic biomarker for CRC patients and might provide a valuable therapeutic target.
Keywords: N6-methyladenosine, colorectal cancer, biomarker, therapeutic target, peripheral blood
Introduction
Colorectal cancer (CRC) is a common malignancy and the fourth leading cause of cancer-related deaths globally (1). If diagnosed in the early stage, the 5-year survival rate of CRC patients is as high as 70%–90% (2). Nevertheless, CRC patients with tumor metastases present a worse prognosis, with a 5-year survival rate of only approximately 20% (3). Furthermore, due to changes in people’s dietary and lifestyle habits, a growing number of patients with CRC are diagnosed at an advanced stage, which leads to challenging therapeutic resection of primary tumors and metastases (4).
Consequently, improving the prognosis of CRC patients largely depends on early and accurate diagnosis. At present, colonoscopy and tissue biopsy are the most efficient methods for CRC screening (5). Nonetheless, colonoscopy is an invasive procedure that can be traumatic for subjects, and the whole operation is occasionally hard to complete due to poor compliance of patients with CRC (2). Additionally, considering the invasiveness and cost of these operations, it is impractical to perform comprehensive screening as part of a general physical examination. Therefore, there is an urgent demand for more noninvasive and efficacious biomarkers for clinical diagnosis. Over recent years, the identification of blood biomarkers has become an important issue because of the pain-free operation of blood biomarkers testing (6). Blood biomarkers such as carbohydrate antigen 19-9 (CA19-9), carbohydrate antigen 125 (CA125), and carcinoembryonic antigen (CEA) are broadly applied for CRC detection (7, 8). Yet, these three biomarkers, alone or in combination, are not sufficient for diagnosing CRC due to their poor specificity and sensitivity (8, 9). Hence, there is an urgent need to optimize the diagnosis of CRC by other efficient blood biomarkers.
N6-methyladenosine (m6A) modification, which was encoded by the methyltransferase complex consisting of “writers”, “erasers”, and “readers”, has emerged as a critical regulator in a multitude of diseases (10, 11). The modification of m6A is enriched close to the 3′ untranslated terminal region (UTR) and the stop codon, thus influencing RNA transcription, processing, and translation (12, 13). Over recent years, activation of m6A modification has been reported in CRC tumor cells (10, 13). Upregulated m6A modification contributes to tumor progression by maintaining SOX2 expression in CRC cells through IGF2 mRNA binding proteins 2 (IGF2BP2)-dependent mechanisms (14, 15). Moreover, activating the glycolytic pathway by m6A methylation promotes CRC tumorigenesis, indicating that m6A modification of CRC tumor cells might become a therapeutic target (16, 17). Besides, the m6A-modified status of peripheral blood has been recently reported as a new promising hallmark in diabetes and gastric cancer (18, 19). Nevertheless, whether the m6A modification of peripheral blood RNA may act as a new diagnostic biomarker or therapeutic target for CRC remains unclear.
In this study, we examined the levels of m6A in peripheral blood RNA of CRC patients and NCs to assess its value as a diagnostic biomarker. We also used bioinformatics, which revealed that elevated m6A levels were mainly associated with monocytes and suppressed their immune response, indicating that m6A modifications of peripheral blood immune cells might become a therapeutic target for CRC.
Materials and Methods
Human Samples
The Institutional Review Board of Zhongshan People’s Hospital approved this retrospective study (IRB number: K2020-20) on March 20, 2020. Between March 2020 and June 2021, peripheral blood samples from 105 CRC patients and 64 NCs who had no history of basic or chronic diseases were collected from the Zhongshan People’s Hospital, using EDTA anticoagulation tubes. Whole blood (0.5 ml) and 1 ml of red blood cell lysate (TIANGEN, Beijing, China) were mixed and centrifuged. The precipitate was taken and dissolved with 1 ml TRIzol to stabilize RNA, after which the mixed samples were stored at −80°C for no longer than 6 months. All CRC patients were diagnosed on the basis of the histopathology by biopsy or endoscopic examination, and informed consent was obtained for all participants. A total of 105 CRC patients’ peripheral blood samples were collected at the time of diagnosis before surgery or radiochemotherapy. Of these, peripheral blood was collected for the first time on admission and for the second time 14 days after surgery in 33 CRC patients. Ethics approval was obtained from the Ethics Committee of the Zhongshan People’s Hospital. The clinical and biological characteristics of the patients are described in Table 1 .
Table 1.
Correlation between the levels of m6A and clinicopathological characteristics in CRC.
| Characteristics | No. of patients | Peripheral blood m6A levels % (mean ± SD) | p-value |
|---|---|---|---|
| Age | |||
| ≤60 | 57 | 0.268 ± 0.057 | 0.649 |
| >60 | 48 | 0.273 ± 0.040 | |
| Gender | |||
| Female | 36 | 0.276 ± 0.064 | 0.386 |
| Male | 69 | 0.267 ± 0.043 | |
| Clinical stage | |||
| I | 6 | 0.243 ± 0.031 | 0.682 |
| II | 20 | 0.263 ± 0.031 | |
| III | 31 | 0.260 ± 0.048 | |
| IV | 26 | 0.302 ± 0.063 | |
| T classification | |||
| T1–T2 | 15 | 0.268 ± 0.040 | 0.739 |
| T3–T4 | 64 | 0.274 ± 0.056 | |
| N classification | |||
| N0 | 29 | 0.273 ± 0.066 | 0.933 |
| N1–N2 | 50 | 0.272 ± 0.046 | |
| N classification | |||
| N0–N1 | 57 | 0.269 ± 0.056 | 0.291 |
| N2 | 22 | 0.283 ± 0.047 | |
| M classification | |||
| M0 | 57 | 0.260 ± 0.041 | <0.001 |
| M1 | 26 | 0.302 ± 0.063 | |
| Differentiation | |||
| Poor | 14 | 0.273 ± 0.030 | 0.975 |
| Moderate/Well | 70 | 0.273 ± 0.056 | |
| Tumor budding | |||
| Bd1–Bd2 | 12 | 0.262 ± 0.043 | 0.861 |
| Bd3 | 16 | 0.259 ± 0.042 | |
| HER2 expression | |||
| Negative | 26 | 0.256 ± 0.040 | 0.368 |
| Positive | 26 | 0.267 ± 0.044 | |
| KRAS genotyping | |||
| Wild type | 10 | 0.277 ± 0.042 | 0.360 |
| Mutation type | 7 | 0.299 ± 0.053 | |
| BRAF genotyping | |||
| Wild type | 17 | 0.279 ± 0.049 | 0.600 |
| Mutation type | 3 | 0.295 ± 0.031 | |
| CEA (ng/ml) | |||
| <5 | 44 | 0.265 ± 0.040 | 0.202 |
| ≥5 | 54 | 0.278 ± 0.057 | |
| CA125 (ng/ml) | |||
| <35 | 68 | 0.269 ± 0.043 | 0.298 |
| ≥35 | 30 | 0.280 ± 0.063 | |
| CA19-9 (ng/ml) | |||
| <35 | 66 | 0.271 ± 0.054 | 0.742 |
| ≥35 | 32 | 0.275 ± 0.041 | |
RNA Isolation and qRT-PCR
Total RNA was extracted using TRIzol (Thermo Scientific, MA, USA) according to the manufacturer’s protocol. First-strand cDNA synthesis was performed using 500 ng of total RNA, and the qRT-PCR analysis system was performed using iQ SYBR Green Supermix (Accurate Biology, Changsha, China) and the iCycler Real-time PCR Detection System (Bio-Rad, California, USA). β-actin was used for normalization. Primers of targeted genes are listed in Supplementary Table S1 .
Monocyte Isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood samples from CRC patients and normal subjects via density gradient centrifugation. Whole blood was collected in EDTA tubes. The blood was diluted 1:1 with PBS free of calcium and magnesium. PBMCs were obtained by Ficoll density gradient isolation (Stemcell Technologies, Cologne, Germany). From the freshly isolated PBMCs, CD14+ monocytes were isolated using the EasySep Human Monocyte Isolation Kit (Stemcell Technologies, Cologne, Germany).
RNA m6A Quantification
The m6A levels in total RNA were measured using EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (Epigentek, New York, USA) according to the manufacturer’s protocol. RNA (200 ng) was added to assay wells covered with binding solution. Capture antibody solution, detection antibody solution, and enhancer solution were sequentially added to assay wells with diluted concentration, as specified in the manufacturer’s instructions. Developer solution and stop solution were added to the color reaction, after which the absorbance of each well at a wavelength of 450 nm was measured. The m6A levels were calculated based on the standard curve.
Bioinformatics Analysis
The RNA-seq data and clinical data of the peripheral blood of CRC and NCs were obtained from GEO (Gene Expression Omnibus) databases (GSE164191). Differential expression analysis was conducted by “limma” package of R studio (3.6.1) software. Gene set variation analysis (GSVA) was performed to estimate m6A modified pathways based on GO molecular function N6 methyladenosine containing RNA binding gene set and Figure 4B listed genes. Immune infiltrates of peripheral blood were estimated via MCP-counter method. Gene Set Enrichment Analysis (GSEA) was manipulated to predict the GO biological process gene sets of the Molecular Signature Database v7.4 (http://www.broadinstitute.org/gsea/msigdb) based on IGF2BP1/IGF2BP2/IGF2BP3 high and low expressed phenotype. A leading edge analysis was performed by GSEA 4.1.0 to elucidate key genes related to selected genes sets. EnrichmentMap plugin in Cytoscape 3.8.2 software was utilized with the following parameters: p-value cutoff = 0.05; similarity coefficient cutoff = 0.5. The protein–protein interaction (PPI) networks were constructed using The Search Tool for the Retrieval of Interacting Genes (STRING), which is a publicly available software for assessing the interaction between proteins and proteins (https://string-db.org/).
Figure 4.
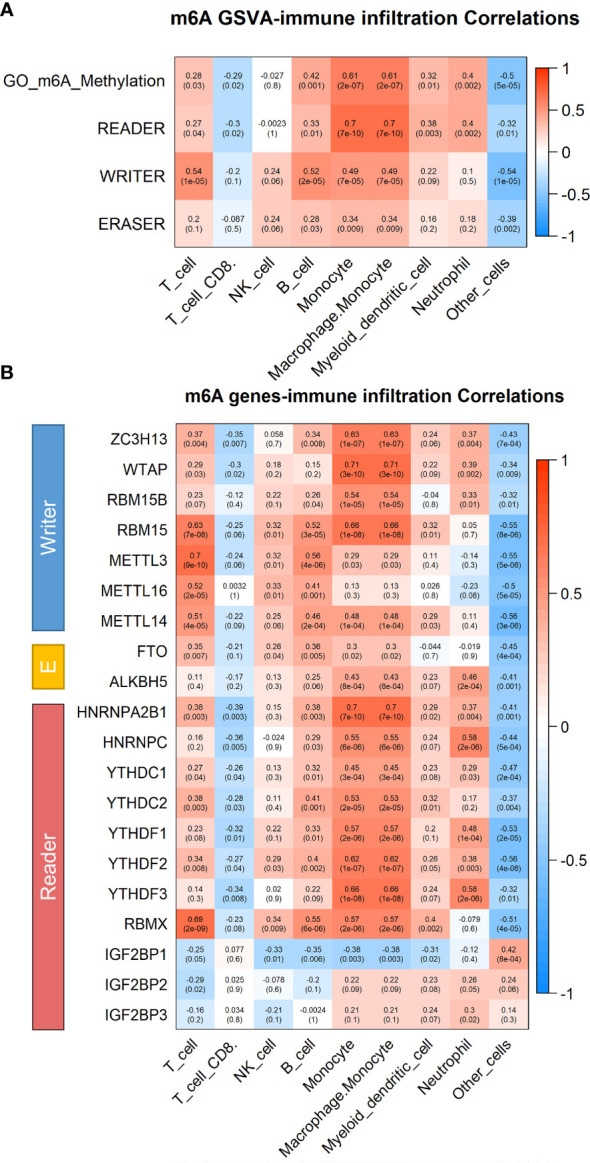
Correlation between immune infiltrating cell types and m6A modification in peripheral blood immune cells of CRC patients. (A) Heatmap of correlation between immune infiltrating cell types and m6A modification pathways in peripheral blood of CRC patients by Gene set variation analysis by GSVA (n = 59). (B) Heatmap of correlation between immune infiltrating cell types and m6A modification related gene in peripheral blood of CRC patients by GSVA.
Statistical Analysis
The variability of the data, which was presented as the SD (mean ± SD), was assessed with unpaired Student’s t test between two groups for normally distributed data. Otherwise, the data were analyzed by nonparametric Mann–Whitney test. Paired t-tests were used to analyze the effects of treatment on m6A levels. For multiple groups, significant differences were determined using one-way ANOVA. Pearson correlation analysis was conducted to determine the correlation between GSVA scores and immune infiltrates. Forest plot of multivariate logistic regression analysis was performed to access risk indicators associated with CRC diagnosis. Statistical significance was defined at p < 0.05.
Results
The m6A RNA Levels of Peripheral Blood Immune Cells in CRC Patients and NCs
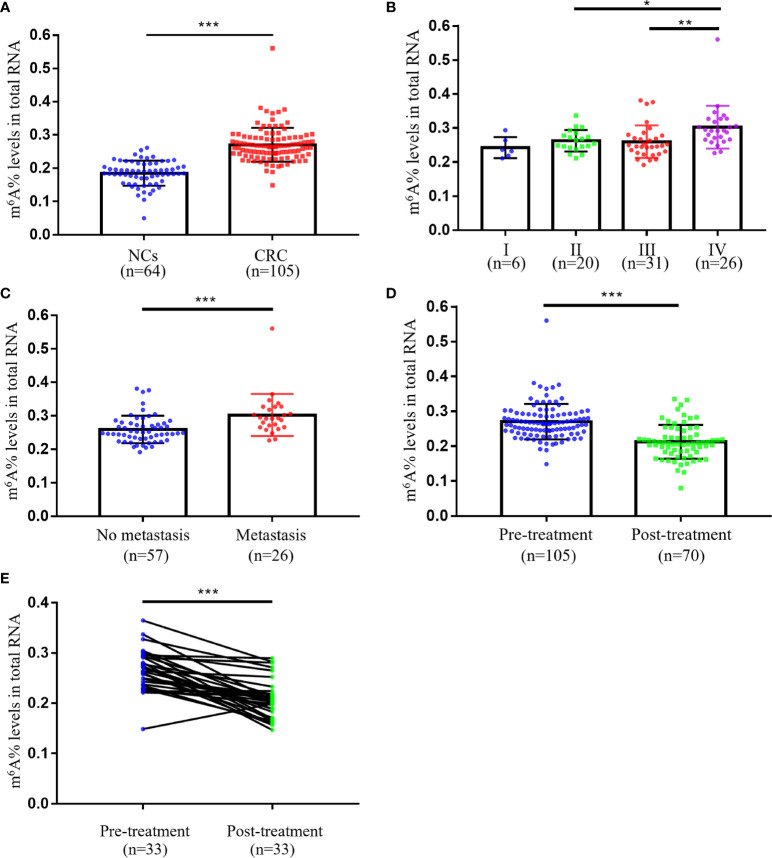
First, we analyzed the m6A levels of total RNA in NCs (n = 64) and CRC patients (n = 105) so as to evaluate the status of m6A modification in peripheral blood immune cells. The m6A levels in peripheral blood immune cells were remarkably increased in patients with CRC (0.271 ± 0.051) than in NCs (0.185 ± 0.038; Figure 1A ). Furthermore, statistical analyses of the relationship between the m6A levels and clinicopathological features of CRC are performed in Table 1 . Our data indicated that the m6A levels correlated with M classification (p < 0.001), but not with clinical stage, T classification, N classification, differentiation, tumor budding, as well as other common CRC tumor markers, including CEA, CA125, and CA19-9 ( Table 1 ). As shown in Figure 1B , the levels of m6A were dramatically elevated in the stage IV group (n = 26, 0.302 ± 0.063) than in stage I (n = 6, 0.243 ± 0.031), II (n = 20, 0.263 ± 0.031), or III groups (n = 31, 0.260 ± 0.048). In addition, CRC patients with distant tumor metastasis (n = 26, 0.302 ± 0.063) had apparently increased m6A levels compared to those without distant metastasis (n = 57, 0.259 ± 0.041; Figure 1C ). These results suggested that peripheral blood m6A RNA levels could partially distinguish the various pathological stages in patients with CRC.
Figure 1.
The m6A RNA levels of peripheral blood immune cells in CRC patients and NCs. (A) The m6A levels of peripheral blood RNA in NCs (n = 64) and CRC patients (n = 105). (B) The m6A levels of peripheral blood RNA at different clinical stages of CRC patients (stage I, n = 6; stage II, n = 20; stage III, n = 31; stage IV, n = 26). (C) Comparison of m6A levels of peripheral blood RNA between CRC patients with (n = 26) and without (n = 57) metastasis. (D) Comparison of m6A levels of peripheral blood RNA between CRC patients with (n = 70) and without (n = 105) treatment. (E) The m6A levels of peripheral blood RNA in CRC patients (n = 33) before and after 14 days of treatment. Bars represent the mean ± SD of the results from replicate measurements; *p < 0.05, **p < 0.01 and ***p < 0.001.
To elucidate whether m6A could be used to assess treatment status in CRC patients, we compared the m6A levels of peripheral blood between the pre-treatment group and post-treatment group. The obtained results demonstrated that m6A levels were markedly reduced in the post-treatment group ( Figure 1D ). We also observed significant changes in m6A levels before and after surgery (14 days) in 33 CRC patients, indicating that m6A RNA levels of peripheral blood immune cells could be used as a promising indicator for post-treatment follow-up ( Figure 1E ).
Clinical Utility for CEA, CA125, CA19-9, and the m6A RNA Levels of Peripheral Blood Immune Cells to Diagnose CRC Patients
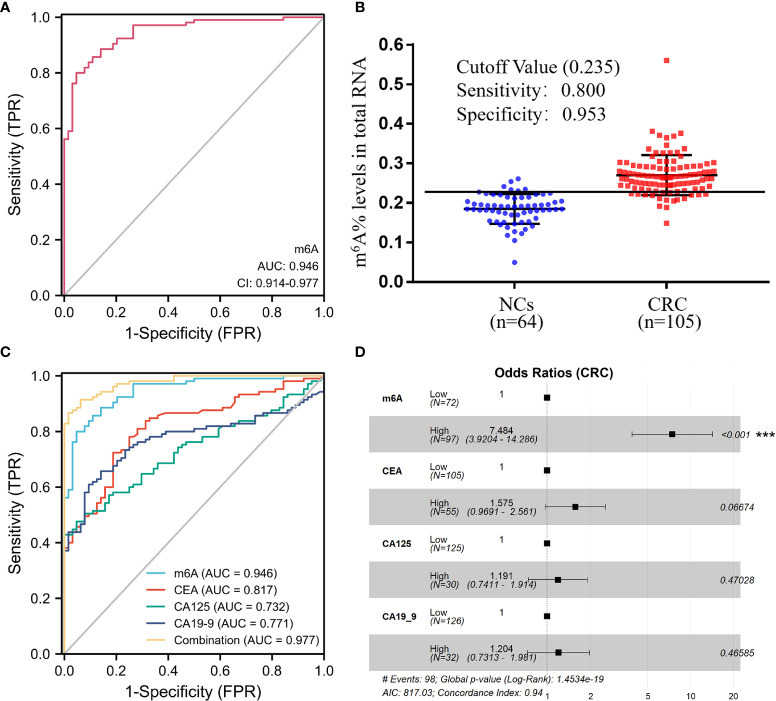
We plotted ROC curves to further assess the diagnostic capability of m6A RNA levels of peripheral blood immune cells for CRC. The area under the curve (AUC) of m6A was up to 0.946 (95% CI, 0.914–0.977), indicating that m6A levels could differentiate CRC patients from NCs ( Figure 2A ). Also, the optimum m6A cutoff value was 0.235 (specificity, 0.953; sensitivity, 0.800; Figure 2B ). Impressively, the diagnostic ability of m6A was superior to the usual CRC blood biomarkers, such as CEA, CA125, and CA19-9, with AUCs of 0.817, 0.732, and 0.771, respectively ( Figure 2C and Table 2 ). Moreover, the ROC curve for the multivariate combination of m6A, CEA, CA125, and CA19-9 increased the AUC to 0.977 (95% CI, 0.961–0.994; Figure 2C ). Furthermore, the forest plot of multivariate logistic regression analysis demonstrated that the m6A levels were an independent factor associated with CRC diagnosis ( Figure 2D ). Taken together, these results clarified that the m6A RNA levels of peripheral blood immune cells presented satisfactory diagnostic utility for CRC patients.
Figure 2.
Clinical utility for CEA, CA125, CA19-9, and the m6A RNA levels of peripheral blood immune cells to diagnose CRC patients. (A, B) ROC curve (A) and cutoff value (B) of the m6A levels of peripheral blood RNA in NCs (n = 64) and CRC patients (n = 105). (C) ROC curve of the m6A levels of peripheral blood RNA compared and combined diagnosis with CEA, CA125, and CA19-9. (D) Forest plot of multivariate logistic regression analysis demonstrated that the m6A levels were an independent factor associated with CRC diagnosis; ***p < 0.001.
Table 2.
Sensitivity and specificity of the diagnostic value of various markers alone and in combination.
| Marker | Sensitivity | Specificity | AUC | 95% CI |
|---|---|---|---|---|
| m6A | 0.800 | 0.953 | 0.946 | 0.914–0.977 |
| CEA | 0.724 | 0.812 | 0.817 | 0.754–0.881 |
| CA125 | 0.476 | 0.953 | 0.732 | 0.659–0.806 |
| CA19-9 | 0.657 | 0.859 | 0.771 | 0.700–0.842 |
| m6A+CEA+CA125+CA19-9 | 0.914 | 0.938 | 0.977 | 0.961–0.994 |
Expressions and Diagnostic Values of IGF2BP1, IGF2BP2, and IGF2BP3 in Peripheral Blood Immune Cells of CRC Patients
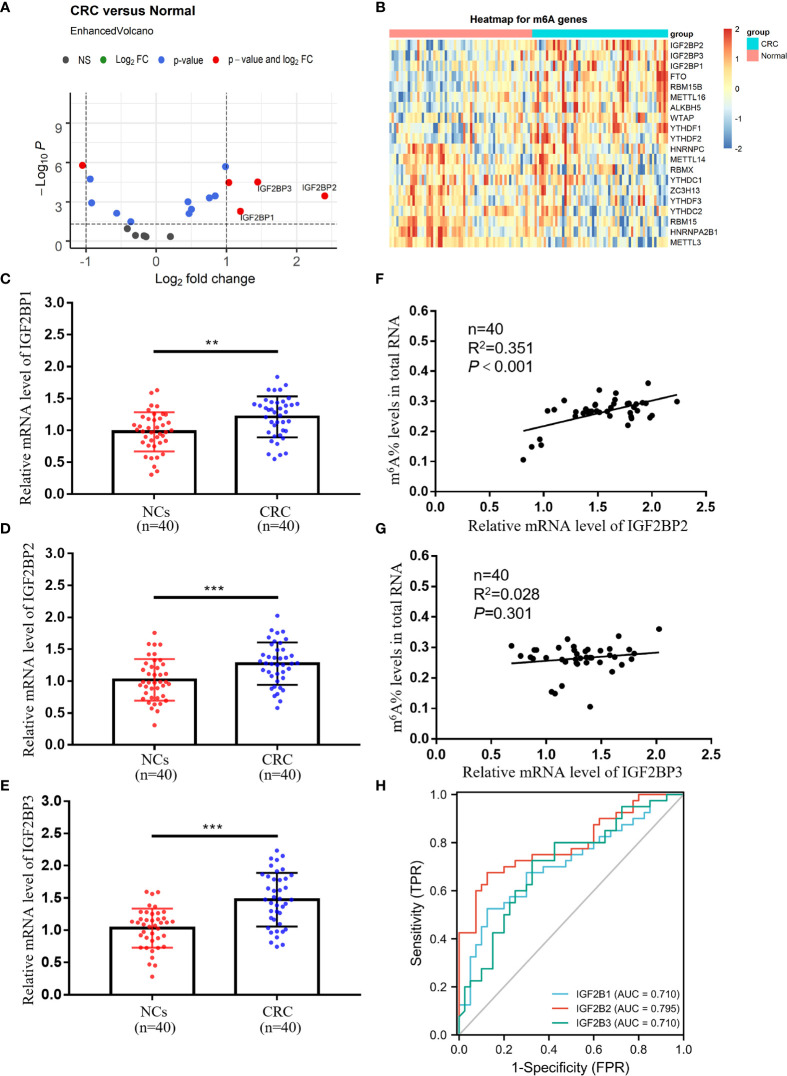
To screen for core molecules that regulate m6A modifications in peripheral blood immune cells RNA, we analyzed the GSE164191 dataset, containing RNA-seq data on peripheral blood leukocytes of CRC patients and normal subjects. Surprisingly, members of the IGF2BP family (IGF2BP1, IGF2BP2, and IGF2BP3) were the most dramatically altered molecules in the methyltransferase complex consisting of “writers”, “erasers”, and “readers” ( Figures 3A, B ). Meanwhile, the strongest increase in IGF2BP2 was observed in CRC patients, suggesting a potentially vital role in m6A modification of peripheral blood immune cells ( Figures 3A, B ). qRT-PCR analysis also proved significantly higher expression of IGF2BP1, IGF2BP2, and IGF2BP3 in CRC patients compared to normal subjects ( Figures 3C–E ). We further discovered a relationship between the levels of m6A and the expressions of IGF2BP2, but no correlation with the expressions of IGF2BP1 and IGF2BP3 ( Figures 3F, G and Supplementary Figure 1 ). The AUCs of IGF2BP1, IGF2BP2, and IGF2BP3 were 0.710, 0.795, and 0.710, respectively ( Figure 3H ). Their AUCs were similar to common CRC blood biomarkers CEA, CA125, and CA19-9 but still smaller than the AUC of m6A. Collectively, IGF2BP2 in peripheral blood immune cells was a potentially valuable diagnostic biomarker for CRC associated with m6A modification.
Figure 3.
Expressions and diagnostic values of IGF2BP1, IGF2BP2, and IGF2BP3 in peripheral blood immune cells of CRC patients. (A) Screening key molecules related to m6A modification in peripheral blood of CRC patients (n = 59) compared to normal subjects (n = 62) by limma differential analysis. (B) Heatmap of key molecules related to m6A modification in peripheral blood of CRC patients. (C–E) qRT-PCR analysis of IGF2BP1 (C), IGF2BP2 (D), and IGF2BP3 (E) mRNA expression levels in peripheral blood of NCs and CRC patients. (F, G) Correlation between the levels of IGF2BP2/IGF2BP3 and m6A in peripheral blood of CRC patients. (H) ROC curves of the IGF2BP1, IGF2BP2, and IGF2BP3 mRNA expression levels in peripheral blood of CRC patients. Bars represent the mean ± SD of the results from replicate measurements; **p < 0.01, ***p < 0.001.
Correlation Between Immune Infiltrating Cell Types and m6A Modification in Peripheral Blood Immune Cells of CRC Patients
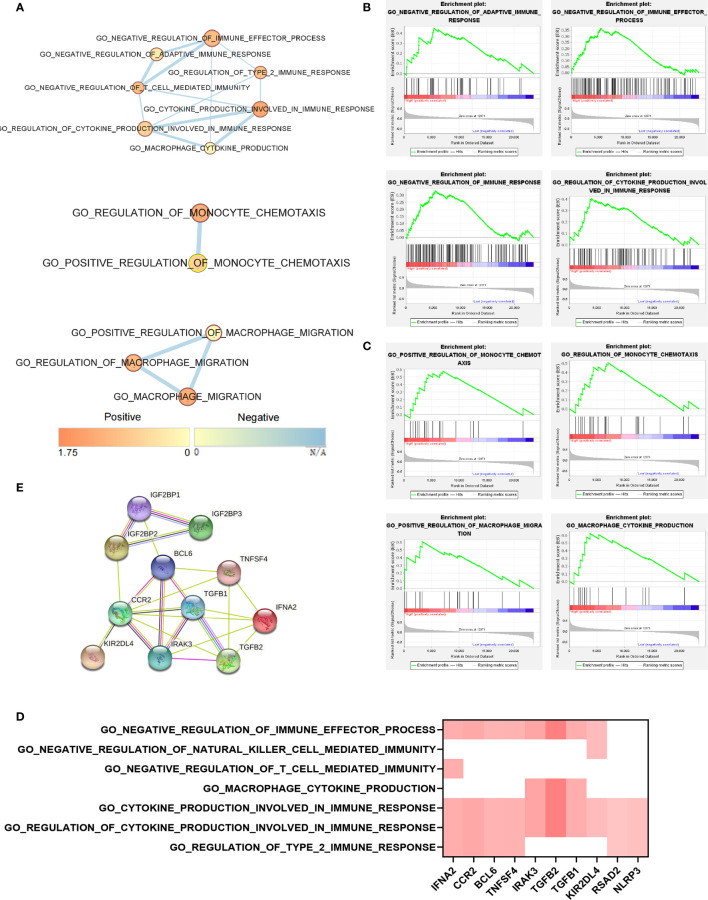
To further elucidate the specific immune cells associated with elevated m6A levels of peripheral blood in CRC patients, we analyzed the GSE164191 database by GSVA. The obtained results suggested that the methyltransferase complexes, consisting of “writer”, “eraser”, and “reader”, all exhibited the strongest positive correlation with monocytes infiltrating ( Figure 4A ). Detection of monocytes isolated from peripheral blood of CRC patients and normal subjects also revealed that monocytes from CRC patients possessed higher levels of m6A ( Supplementary Figure 2 ). Meanwhile, infiltration of monocytes was also markedly correlated with IGF2BP2 expression, consistent with the results in Figure 3 regarding the importance of IGF2BP2 in m6A modifications ( Figure 4B ). In conclusion, monocytes resulted as the specific immune cells most strongly associated with upregulated m6A levels of peripheral blood immune cells in CRC patients.
IGF2BP2 Involved in the Immune Response of Monocytes in Peripheral Blood of CRC Patients
The function of IGF2BP2 in the monocytes of the peripheral blood of CRC patients was investigated using the EnrichmentMap plugin in Cytoscape 3.8.2 software. The corresponding association network showed that the IGF2BP2 high-expression phenotype presented a robust positive association between several monocyte immune response pathways ( Figure 5A ). GSEA was applied to predict the biological processes of monocytes in peripheral blood based on IGF2BP2 expression. Likewise, high IGF2BP2 expression was mainly enriched in the immune response pathways, such as “Negative regulation of immune effector process”, “Regulation of monocyte chemotaxis”, and “Cytokine production” ( Figures 5B, C ). Additionally, the results of leading edge analysis identified the intersection of important genes associated with the immune response pathways ( Figure 5D ). Meanwhile, the PPI networks structured by the STRING database suggested that IGF2BP2 may interact with the above vital genes ( Figure 5E ). IGF2BP1 and IGF2BP3 also performed approximately the same immune functions as IGF2BP2 in monocytes ( Supplementary Figure 3 ). Taken together, IGF2BP2 exerted an essential role in the immune response of peripheral blood monocytes of CRC patients.
Figure 5.
IGF2BP2 involved in the immune response of monocytes in peripheral blood of CRC patients. (A) EnrichmentMap pathways network revealed overlaps among IGF2BP2 high-expressed phenotype enriched pathways relating to immunity in peripheral blood of CRC patients. Nodes are colored by Enrichment Score, and edges are sized on the basis of the number of genes shared by the connected pathways. (B) GSEA indicated that IGF2BP2 was negatively correlated with the immune response of monocytes. (C) GSEA indicated that IGF2BP2 was positively correlated with monocyte chemotaxis and cytokine production. (D) Leading edge analysis of their intersection genes indicates the vital genes shared by the IGF2BP2 high-expressed phenotype associated with the immune response of monocytes. (E) STRING database analysis revealed that IGF2BP2 interacted with the above vital genes related to the immune response of monocytes.
Discussion
Most patients are already at an advanced stage by the time they are diagnosed with CRC, which substantially contributes to the poor prognosis (4). Hence, improving the prognosis of CRC patients depends on an early and accurate diagnosis. However, the currently used clinical tumor biomarkers for CRC such as CEA, CA125, and CA19-9 are not specific or sensitive enough to detect CRC patients (7, 9). Therefore, optimizing the diagnosis of CRC with other validated biomarkers is of urgent importance. The present study identified the m6A status of peripheral blood immune cells as a novel marker for CRC screening. In addition, it might also serve as a new target for CRC treatment.
Despite a growing body of reports that have linked m6A dysregulation to various cancers, the role of m6A modifications in CRC tumor tissues remained controversial (10, 20). Stimulating m6A modification promotes β-catenin translation to drive the epithelial–mesenchymal transition of CRC cells, while some studies found that m6A regulation suppresses proliferation and metastasis (15, 21, 22). Our research revealed for the first time that the m6A RNA levels of peripheral blood immune cells were dramatically higher in patients with CRC than in healthy subjects ( Figure 1A ). Our results demonstrated that m6A RNA was more strongly modified in peripheral blood immune cells of CRC, yet m6A modification in CRC tumor tissue needs to be further explored. Additionally, the m6A status of peripheral blood immune cells was substantially elevated in CRC patients with distant metastases compared to those without metastases, implying that it could also discriminate if the tumor had metastasized ( Figures 1B, C ). Although the m6A levels were reduced in treated CRC patients, more clinical samples were requested to determine whether they could be used as an indicator of oncologic efficacy, such as relapse and drug resistance ( Figures 1D, E ). It has been discussed that the m6A levels might be applied as a biomarker for gastric cancer, but the regulation of m6A modification in different tumors varied significantly (18, 23). Therefore, it is worthwhile to investigate further whether the m6A levels had diagnostic value in other tumors.
CEA, CA125, and CA19-9 are widely used in physical screening for CRC (9). Nevertheless, due to their poor specificity and sensitivity, these three biomarkers alone or in combination are not sufficient to diagnose CRC (7). As shown in Figure 2 , the AUC for m6A to differentiate CRC patients from healthy subjects was 0.946 (95% CI, 0.914–0.977), which was significantly higher than the AUC for CEA (0.817; 95% CI, 0.754–0.881), CA125 (0.732; 95% CI, 0.659–0.806), and CA19-9 (0.771; 95% CI, 0.700–0.842). The combination of CEA, CA125, and CA19-9 with m6A further increased the AUC to 0.977 (95% CI, 0.961–0.994). Besides, forest plots from multiple logistic regression analysis showed that the m6A levels were an independent risk factor associated with the diagnosis of CRC compared to these common tumor biomarkers ( Figure 2D ). Our study presented a considerable challenge to the value of these tumor biomarkers.
“Writers”, “erasers”, and “readers” together formed the methyltransferase complex responsible for m6A modification. Wilms tumor 1-associated protein (WTAP), Methyltransferase-like 3 (METTL3), and METTL14 were classified as “writers” catalyzing the formation of m6A (24–26). AlkB homolog 5 (ALKBH5) and Fat mass and obesity-associated protein (FTO) represented “erasers”, meaning they could induce selective removal of methylation code from the target mRNA (27, 28). “Readers” were able to decode m6A modification, comprising YT521-B homology domain-containing protein (YTHDF) as well as IGF2BP families (16, 29). m6A modifications altered the expression of target genes and changed the consequent biological features (30). To further understand the role of the elevated m6A levels in CRC tumor progression, we screened for the most variable “writers”, “erasers”, and “readers” in CRC peripheral blood immune cells by limma differential analysis. Members of the IGF2BP family (IGF2BP1, IGF2BP2, and IGF2BP3), which belonged to “readers”, were the most markedly changed molecules in the methyltransferase complex ( Figure 3 ). Simultaneously, IGF2BP2 revealed the greatest increase, thus suggesting a potentially crucial role in peripheral blood immune cell m6A modification ( Figure 3 ). Unlike other readers, IGF2BPs acted as a unique family of m6A readers that target a multitude of mRNA transcripts and enhance the conservation and stability of their candidate mRNAs in an m6A-dependent way (14, 15, 31). Our study further demonstrated that elevated IGF2BP2 might interact with several essential genes to negatively regulate immunity, such as cytokine production and chemotaxis ( Figure 5 and Supplementary Figure 3 ). Although we found that increased IGF2BPs expression combined with elevated m6A levels affected cancer immunity in CRC, we have not yet clarified the mechanism of increased IGF2BPs, which is also the biggest limitation of the current study. Taken together, m6A modification and IGF2BPs expression were likely to be novel targets for CRC treatment, but further in vivo experimental studies are required.
Previous studies reported that elevated m6A levels of peripheral blood in patients with gastric cancer might be due to downregulation of FTO and ALKBH5, which belonged to “erasers” (18). Our qRT-PCR results also revealed a slight downregulation of FTO and ALKBH5 in peripheral blood cells of CRC patients, partially explaining the increased m6A levels ( Supplementary Figure 4 ). Other unknown methylases and demethylases may also be involved in the changes of m6A levels that deserved further exploration (32). Additionally, monocytes were identified as the immune cells most strongly associated with the increased regulation of upregulated m6A levels in peripheral blood of CRC patients ( Figure 4 ). It has been noted that the presence of a large number of m6A-modified infiltrating immune cells in the tumor tissue microenvironment promotes tumor progression (33, 34). Furthermore, imbalanced m6A regulation strongly conferred immune disruption and tumor evasion, primarily by affecting immune cell migration, rather than apoptosis or survival (35). These observations were generally consistent with our findings in peripheral blood immune cells. Moreover, the number of monocytes in the CD14+CD16+HLA-DRhi subpopulation of patient’s peripheral blood was found to be the most accurate predictor of progression-free survival and overall survival after receiving PD-1 inhibitor therapy (36). Whether the subset of monocytes with elevated m6A levels had a similar role in tumor immunotherapy to the CD14+CD16+HLA-DRhi subset deserves further investigation.
In conclusion, the highlights of our research were the first identification of m6A RNA levels in peripheral blood immune cells as a novel biomarker for the diagnosis of CRC and the provision of a new strategy for the treatment of CRC by targeting m6A levels or IGF2BPs expression in peripheral blood immune cells.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Zhongshan People’s Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HY, HH, and CL conceived and designed this study. JX, ZH, and PJ performed the experiments and analyzed the data. RW and HJ contributed to the data analysis and discussion. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the fund from the National Nature Science Foundation of China (81900775; 81902693); Educational Commission of Guangdong Province (2017KTSCX155); Guangdong Basic and Applied Basic Research Foundation (2019A1515011318); Natural Science Foundation of Guangdong Province (2018A030310298); the Science Foundation of Guangzhou First People’s Hospital (Q2019004; KYQD0046); China Postdoctoral Science Foundation (2019M662991); Key Medical and Health Projects of Zhongshan City (2020K0012); Guangzhou Science and Technology Planning Project (202102020142).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.760747/full#supplementary-material
Correlation between the levels of IGF2BP1 and m6A in peripheral blood of CRC patients. Absence of correlation between the m6A levels and IGF2BP1 expression.
The m6A levels of monocytes isolated from peripheral blood of CRC patients and normal subjects. The m6A levels of monocytes isolated from CRC patients was higher than those in monocytes from normal subjects.
IGF2BP1 and IGF2BP3 expression are negatively associated with several immune response pathways. (A, B) EnrichmentMap pathways network exhibited connectivity among IGF2BP1 (A) and IGF2BP3 (B) high-expressed phenotype enriched pathways relating to immunity response in peripheral blood of CRC patients. (C, D) GSEA indicated that IGF2BP1 (C) and IGF2BP3 (D) were negatively correlated with the immune response of monocytes.
Expressions of FTO and ALKBH5 in peripheral blood RNA of CRC patients. (A, B) Q-PCR analysis of FTO (A) and ALKBH5 (B) mRNA expression levels in peripheral blood of NCs and CRC patients.
References
- 1. Jacobs D, Zhu R, Luo J, Grisotti G, Heller DR, Kurbatov V, et al. Defining Early-Onset Colon and Rectal Cancers. Front Oncol (2018) 8:504. doi: 10.3389/fonc.2018.00504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tong GJ, Zhang GY, Liu J, Zheng ZZ, Chen Y, Niu PP, et al. Comparison of the Eighth Version of the American Joint Committee on Cancer Manual to the Seventh Version for Colorectal Cancer: A Retrospective Review of Our Data. World J Clin Oncol (2018) 9(7):148–61. doi: 10.5306/wjco.v9.i7.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cespedes Feliciano EM, Kroenke CH, Meyerhardt JA, Prado CM, Bradshaw PT, Dannenberg AJ, et al. Metabolic Dysfunction, Obesity, and Survival Among Patients With Early-Stage Colorectal Cancer. J Clin Oncol (2016) 34(30):3664–71. doi: 10.1200/JCO.2016.67.4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baars JE, Kuipers EJ, van Haastert M, Nicolai JJ, Poen AC, van der Woude CJ. Age at Diagnosis of Inflammatory Bowel Disease Influences Early Development of Colorectal Cancer in Inflammatory Bowel Disease Patients: A Nationwide, Long-Term Survey. J Gastroenterol (2012) 47(12):1308–22. doi: 10.1007/s00535-012-0603-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tfaily MA, Naamani D, Kassir A, Sleiman S, Ouattara M, Moacdieh MP, et al. Awareness of Colorectal Cancer and Attitudes Towards Its Screening Guidelines in Lebanon. Ann Glob Health (2019) 85(1):75. doi: 10.5334/aogh.2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner IC, Guimaraes MJ, da Silva LK, de Melo FM, Muniz MT. Evaluation of Serum and Pleural Levels of the Tumor Markers CEA, CYFRA21-1 and CA 15-3 in Patients With Pleural Effusion. J Bras Pneumol (2007) 33(2):185–91. doi: 10.1590/s1806-37132007000200013 [DOI] [PubMed] [Google Scholar]
- 7. Li L, Zhang L, Tian Y, Zhang T, Duan G, Liu Y, et al. Serum Chemokine CXCL7 as a Diagnostic Biomarker for Colorectal Cancer. Front Oncol (2019) 9:921. doi: 10.3389/fonc.2019.00921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bagaria B, Sood S, Sharma R, Lalwani S. Comparative Study of CEA and CA19-9 in Esophageal, Gastric and Colon Cancers Individually and in Combination (ROC Curve Analysis). Cancer Biol Med (2013) 10(3):148–57. doi: 10.7497/j.issn.2095-3941.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomas DS, Fourkala EO, Apostolidou S, Gunu R, Ryan A, Jacobs I, et al. Evaluation of Serum CEA, CYFRA21-1 and CA125 for the Early Detection of Colorectal Cancer Using Longitudinal Preclinical Samples. Br J Cancer (2015) 113(2):268–74. doi: 10.1038/bjc.2015.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen XY, Zhang J, Zhu JS. The Role of M6a RNA Methylation in Human Cancer. Mol Cancer (2019) 18(1):103. doi: 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao BS, He C. Fate by RNA Methylation: M6a Steers Stem Cell Pluripotency. Genome Biol (2015) 16:43. doi: 10.1186/s13059-015-0609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A Majority of M6a Residues are in the Last Exons, Allowing the Potential for 3’ UTR Regulation. Genes Dev (2015) 29(19):2037–53. doi: 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 Facilitates Tumor Progression via an M(6)a-IGF2BP2-Dependent Mechanism in Colorectal Carcinoma. Mol Cancer (2019) 18(1):112. doi: 10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 Regulates DANCR by Serving as an N6-Methyladenosine Reader. Cell Death Differ (2020) 27(6):1782–94. doi: 10.1038/s41418-019-0461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, et al. N(6)-Methyladenosine (M(6)a) RNA Modification in Gastrointestinal Tract Cancers: Roles, Mechanisms, and Applications. Mol Cancer (2019) 18(1):178. doi: 10.1186/s12943-019-1099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, et al. Lncrna LINRIS Stabilizes IGF2BP2 and Promotes the Aerobic Glycolysis in Colorectal Cancer. Mol Cancer (2019) 18(1):174. doi: 10.1186/s12943-019-1105-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, et al. M(6)a-Dependent Glycolysis Enhances Colorectal Cancer Progression. Mol Cancer (2020) 19(1):72. doi: 10.1186/s12943-020-01190-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ge L, Zhang N, Chen Z, Song J, Wu Y, Li Z, et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin Chem (2020) 66(2):342–51. doi: 10.1093/clinchem/hvz004 [DOI] [PubMed] [Google Scholar]
- 19. Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, et al. Decreased N(6)-Methyladenosine in Peripheral Blood RNA From Diabetic Patients is Associated With FTO Expression Rather Than ALKBH5. J Clin Endocrinol Metab (2015) 100(1):E148–54. doi: 10.1210/jc.2014-1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-Nucleotide-Resolution Mapping of M6a and m6Am Throughout the Transcriptome. Nat Methods (2015) 12(8):767–72. doi: 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song P, Feng L, Li J, Dai D, Zhu L, Wang C, et al. Beta-Catenin Represses Mir455-3p to Stimulate M6a Modification of HSF1 mRNA and Promote its Translation in Colorectal Cancer. Mol Cancer (2020) 19(1):129. doi: 10.1186/s12943-020-01244-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-Mediated N6-Methyladenosine Modification of SOX4 mRNA Inhibits Tumor Metastasis in Colorectal Cancer. Mol Cancer (2020) 19(1):106. doi: 10.1186/s12943-020-01220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li F, Kennedy S, Hajian T, Gibson E, Seitova A, Xu C, et al. A Radioactivity-Based Assay for Screening Human M6a-RNA Methyltransferase, METTL3-METTL14 Complex, and Demethylase ALKBH5. J Biomol Screen (2016) 21(3):290–7. doi: 10.1177/1087057115623264 [DOI] [PubMed] [Google Scholar]
- 24. Schumann U, Shafik A, Preiss T. METTL3 Gains R/W Access to the Epitranscriptome. Mol Cell (2016) 62(3):323–4. doi: 10.1016/j.molcel.2016.04.024 [DOI] [PubMed] [Google Scholar]
- 25. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a Mammalian RNA Demethylase That Impacts RNA Metabolism and Mouse Fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. M(6)a Potentiates Sxl Alternative Pre-mRNA Splicing for Robust Drosophila Sex Determination. Nature (2016) 540(7632):301–4. doi: 10.1038/nature20577 [DOI] [PubMed] [Google Scholar]
- 30. Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The Critical Role of RNA M(6)a Methylation in Cancer. Cancer Res (2019) 79(7):1285–92. doi: 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- 31. Muller S, Glass M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 Promotes SRF-Dependent Transcription in Cancer in a M6a- and Mirna-Dependent Manner. Nucleic Acids Res (2019) 47(1):375–90. doi: 10.1093/nar/gky1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ben-Haim MS, Moshitch-Moshkovitz S, Rechavi G. FTO: Linking M6a Demethylation to Adipogenesis. Cell Res (2015) 25(1):3–4. doi: 10.1038/cr.2014.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. M(6)a Regulator-Mediated Methylation Modification Patterns and Tumor Microenvironment Infiltration Characterization in Gastric Cancer. Mol Cancer (2020) 19(1):53. doi: 10.1186/s12943-020-01170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X, Zhang S, Yan X, Shan Y, Liu L, Zhou J, et al. M6a Regulator-Mediated RNA Methylation Modification Patterns are Involved in Immune Microenvironment Regulation of Periodontitis. J Cell Mol Med (2021) 25(7):3634–45. doi: 10.1111/jcmm.16469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li M, Zha X, Wang S. The Role of N6-Methyladenosine mRNA in the Tumor Microenvironment. Biochim Biophys Acta Rev Cancer (2021) 1875(2):188522. doi: 10.1016/j.bbcan.2021.188522 [DOI] [PubMed] [Google Scholar]
- 36. Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, et al. High-Dimensional Single-Cell Analysis Predicts Response to Anti-PD-1 Immunotherapy. Nat Med (2018) 24(2):144–53. doi: 10.1038/nm.4466 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between the levels of IGF2BP1 and m6A in peripheral blood of CRC patients. Absence of correlation between the m6A levels and IGF2BP1 expression.
The m6A levels of monocytes isolated from peripheral blood of CRC patients and normal subjects. The m6A levels of monocytes isolated from CRC patients was higher than those in monocytes from normal subjects.
IGF2BP1 and IGF2BP3 expression are negatively associated with several immune response pathways. (A, B) EnrichmentMap pathways network exhibited connectivity among IGF2BP1 (A) and IGF2BP3 (B) high-expressed phenotype enriched pathways relating to immunity response in peripheral blood of CRC patients. (C, D) GSEA indicated that IGF2BP1 (C) and IGF2BP3 (D) were negatively correlated with the immune response of monocytes.
Expressions of FTO and ALKBH5 in peripheral blood RNA of CRC patients. (A, B) Q-PCR analysis of FTO (A) and ALKBH5 (B) mRNA expression levels in peripheral blood of NCs and CRC patients.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.