Abstract
Because current mainstream anti-glycolipid GD2 therapeutics for neuroblastoma (NB) have limitations, such as severe adverse effects, improved therapeutics are needed. In this study, we developed a GD2 aptamer (DB99) and constructed a GD2-aptamer-mediated multifunctional nanomedicine (ANM) with effective, precise, and biocompatible properties, which functioned both as chemotherapy and as gene therapy for NB. DB99 can bind to GD2+ NB tumor cells but has minimal cross-reactivity to GD2− cells. Furthermore, ANM is formulated by self-assembly of synthetic aptamers DB99 and NB-specific MYCN small interfering RNA (siRNA), followed by self-loading of the chemotherapeutic agent doxorubicin (Dox). ANM is capable of specifically recognizing, binding, and internalizing GD2+, but not GD2−, NB tumor cells in vitro. Intracellular delivery of ANM activates Dox release for chemotherapy and MYCN-siRNA-induced MYCN silencing. ANM specifically targets, and selectively accumulates in, the GD2+ tumor site in vivo and further induces growth inhibition of GD2+ tumors in vivo; in addition, ANM generates fewer or no side effects in healthy tissues, resulting in markedly longer survival with fewer adverse effects. These results suggest that the GD2-aptamer-mediated, targeted drug delivery system may have potential applications for precise treatment of NB.
Keywords: Neuroblastoma, aptamer, GD2, self-assembling nanomedicine, targeted multiple treatments
Graphical abstract
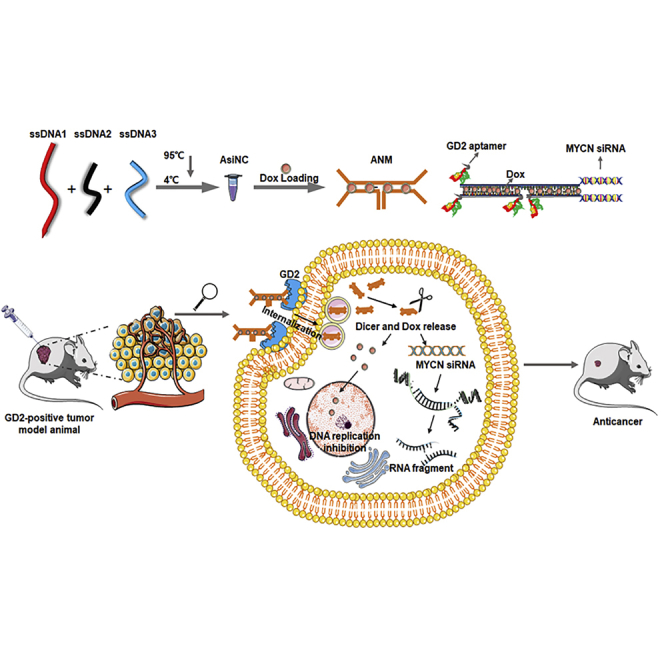
Multifunctional nanomedicine (ANM), with effective, precise, and biocompatible properties based on a GD2 aptamer, to achieve both chemotherapy and gene therapy of neuroblastoma was constructed by Zhang et al. This GD2-aptamer-mediated, targeted, multifunctional nanomedicine system may open a new avenue and have potential applications for precision treatment of NB.
Introduction
Neuroblastoma (NB), the most common extracranial solid tumor in infancy and childhood, which arises from the sympathetic nervous system, accounts for ∼15% of childhood cancer deaths and is a leading cause of death in pediatric medicine around the world.1,2 NB is classified into risk groups (low, intermediate, and high) according to the International Neuroblastoma Risk Group (INRG) classification.3 Although high-risk NB (HRNB) is the most prevalent, when compared with the low-risk and intermediate-risk groups, outcomes in the high-risk group are considerably poorer.3, 4, 5 Clinical HRNB primarily comprises children older than 18 months with distant metastatic spread and/or amplification of the MYCN oncogene.6 At present, despite multiple combination treatments, including conventional or high-dose chemotherapy, surgical resection, radiotherapy, differentiation therapy, and immunotherapy,1,7 the 5-year overall survival (OS) from HRNB has not yet changed substantially (<50%).8 Intense and high doses of chemotherapy usually cause severe sequelae, such as hearing loss, cognitive deficits, and endocrinopathies.9 Further, one-half of patients with HRNB may relapse, and survival rates after 5 years are even more dismal, at less than 10%.10 The high mortality and treatment-related high morbidity associated with current standards-of-care treatments indicate the urgent need for less-toxic and more-effective treatments for this disease.11 Hence, new drugs that exploit the molecular pathology of NB and improve the targeting ability to reduce long-term toxicities are urgently needed to treat high-risk NB at diagnosis as well as in patients with relapsed or refractory disease.
Despite the increased number of promising drugs in early clinical trials that focused on NB, the development of new agents for use in pediatric NB remains extremely limited.12 Glycolipid GD2 is one of the promising targets for NB therapy, but, to date, only one class of immunotherapies with the anti-GD2 monoclonal antibody dinutuximab has been incorporated into front-line therapy (2015 approval by the U.S. Food and Drug Administration [FDA]) for pediatric patients with HRNB since the 1980s.13,14 Dinutuximab binds to glycolipid GD2, which is overexpressed in NB cells and neuroectoderm-derived normal cells, including the central nervous system and peripheral nerves.15,16 Dinutuximab binds to GD2 on the cell surface and induces antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC). Although dinutuximab has a powerful effect, challenges still exist.17 First, more than 50% of patients will relapse, and very few relapsed patients can be cured.18,19 Second, although GD2 is an ideal target for NB therapy because it is highly expressed on NB tumor cells, it also exists in other normal tissues.20,21 Therefore, dinutuximab stimulates normal GD2+ cells, causing severe pain, fever, low platelet count, infusion reactions, hypotension, elevated liver enzymes, anemia, diarrhea, hypokalemia, capillary-leak syndrome, neutropenia, and lymphopenia. Dinutuximab treatment may result in nerve damage, infections, eye problems, electrolyte abnormalities, and bone marrow suppression.22,23 Therefore, there is an urgent need to develop an alternative GD2-targeted agent to overcome those barriers.
In recent years, aptamers, another highly clinically translatable and favorable therapeutic, have exhibited promise for tumor-targeted therapy.24 Aptamers are single-stranded DNA or RNA molecules selected via an iterative selection process called systematic evolution of ligands by exponential enrichment (SELEX).25,26 Aptamers can display complex secondary motifs, such as loops, stems, or G-quadruplexes, which may further adopt three-dimensional structures to recognize and bind targets with high affinity (dissociation constants in the low nanomolar/high picomolar range) and specificity (discrimination by only a few amino acid changes).27,28 The diverse nature of aptamers makes them not only exhibit targeting ability themselves but also enables them to be used as tools to deliver different cargos to specific cells or tissues.29,30 Because of the several substantial benefits of aptamers, compared with conventional therapeutics, such as antibodies, aptamers are more suitable for NB targeted therapy for several reasons:
-
(1)
Easy permeation and lower antigenicity. Thanks to their small size, aptamers that carry delivery cargos, such as small interfering RNA (siRNA), chemical drugs, or nanoparticles, can rapidly penetrate into cells and tissues.31 They can even infiltrate the blood-brain barrier to address metastasis in the brain. In addition, their small size decreases the chances of inducing unwanted humoral T cell-dependent immune responses.32
-
(2)
Antidotes exist. Because targets may also be expressed at low levels on normal cells, if an aptamer generates side effects, antidotes—oligonucleotides with a sequence complementary to the aptamer—can be used to bind the aptamer and further disrupt its structure and function in vivo.33,34 This makes aptamers a unique class of therapeutic agents that convey safer drug design and have an important safety advantage over other targeted therapeutics.
-
(3)
Large-scale production and lower cost. Because aptamers are chemically synthesizable, good manufacturing practices (GMP)-grade production is much more controllable, and the cost of production is less than that for antibodies.35 Therefore, development of a GD2-aptamer-targeting system is both feasible and desirable.
In this study, we attempted to construct a GD2-aptamer-targeting system. We successfully selected a DNA aptamer (termed DB99) against GD2. It has been validated that DB99 can bind to GD2+ cells with high specificity and affinity. Further, we constructed a multifunctional, biodegradable aptamer nanomedicine (ANM) based on the GD2 aptamer. Because amplification of the MYCN oncogene is observed in approximately 20% of all NB cases and in approximately 50% of high-risk cases, MYCN amplification is defined as one of the strongest unfavorable prognostic markers.5 A reduction in MYCN mRNA expression may inhibit proliferation, prevent multidrug resistance (MDR), and induce apoptosis of NB tumor cells.36 In addition, doxorubicin (Dox) is a standard chemotherapeutic for NB, and Dox can intercalate into cytosine/guanine (CG) base pairs.37 Therefore, a biostable and biocompatible ANM, containing the GD2 aptamer, MYCN siRNA, and Dox, was constructed. We hypothesized that ANM would recognize and bind with GD2+ cells because of the GD2 aptamer. Next, ANM is internalized by GD2+ cells, and Dox and MYCN siRNA are released to perform their functions. Therefore, ANM inhibits GD2+ NB tumor growth. Several functional studies in vitro and in vivo have demonstrated that ANM achieved both GD2+ cell-targeted chemotherapy and MYCN-gene-specific therapy, resulting in a combination therapy that not only achieved greater therapeutic efficacy but also had fewer nonspecific, toxic side effects on normal cells and tissues.
Results
Selection and characteristics of GD2 aptamers
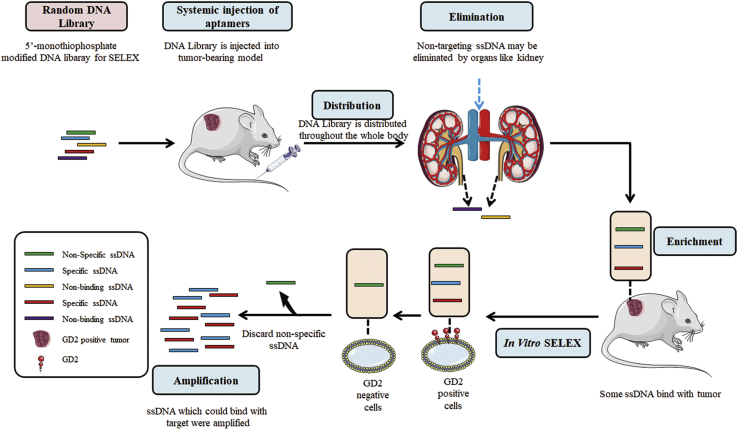
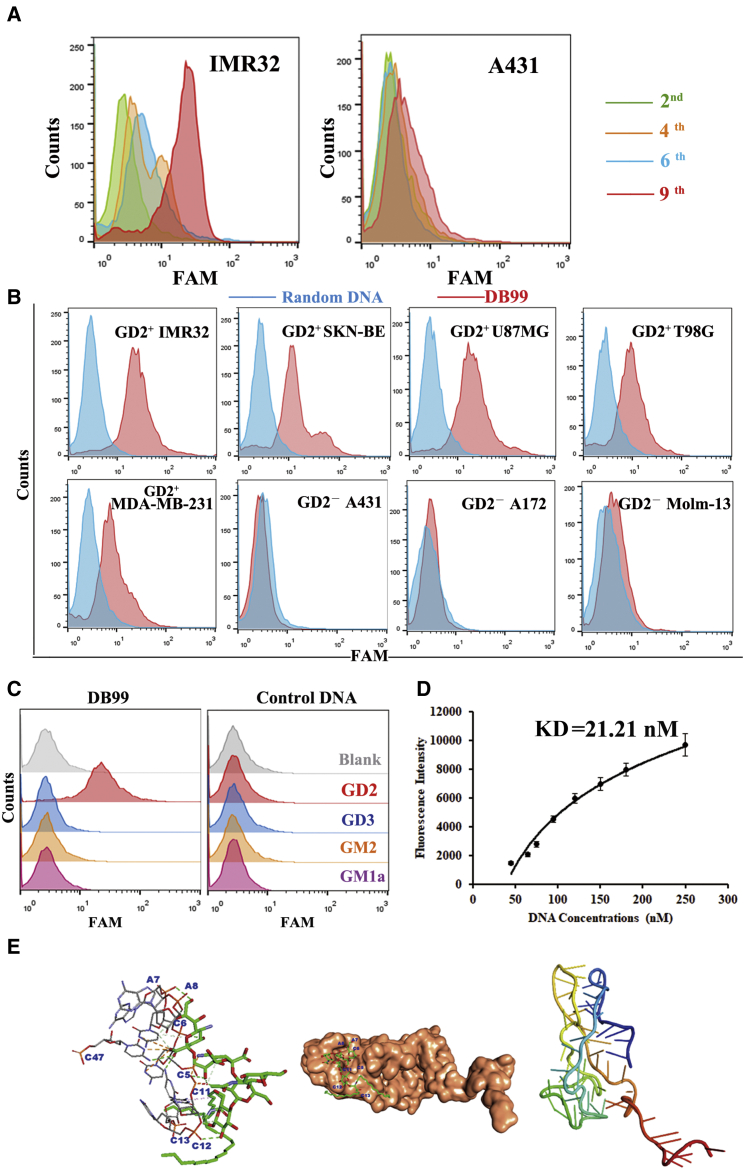
The GD2 aptamer was selected via a combined SELEX, which consisted of an in vivo part and an in vitro part (Figure 1). IMR32-cell-bearing mice were used for in vivo SELEX. In total, nine SELEX rounds were completed. Flow cytometry was applied to monitor the selection effect of combined SELEX in each round. As shown in Figure 2A, when compared with the GD2− cell line A431, there was an increasing amount of single-stranded DNA (ssDNA) bound to GD2+ IMR32 cells during the fourth, sixth, and ninth rounds when compared with the second round, indicating that ssDNA binding increased after round 2. Fluorescence values were highest in IMR32 at the ninth selection round, indicating full enrichment. The DNA pool was subsequently cloned. Ninety-nine clones were used for further functional evaluation. Sequences were analyzed and divided into families according to alignment. Interestingly, those sequences revealed two dominant clones, termed clone A and clone B (Table S1). Of those clones, the representation of clone A, named DB99, was slightly greater than that of the other clone and so was chosen for further identification and characterization. First, to evaluate binding specificity, DB99 was amplified with a carboxyfluorescein (FAM)-labeled forward primer and a biotin-labeled reverse primer by PCR. FAM-labeled DB99 was separated by streptavidin beads, and 40 pmol was incubated with several cell lines (GD2+ cell lines: IMR32, SKN-BE, MDA-MB-231, U87MG, and T98G cells; GD2− cell lines: A431, A172, and Molm-13 cell lines). Cells were evaluated by flow cytometry. As shown in Figure 2B, when compared with the random DNA pool, DB99 generated a relatively stronger signal in GD2+ cell lines. This result indicates that DB99 has relatively high specificity and sensitivity to GD2. Further, because DB99 generated a relatively stronger signal in GD2+ cells and we processed an in vitro selection part to exclude the influence of other molecules on cell membranes, it was necessary to confirm and validate whether the binding target of DB99 was GD2. First, expression of GM2/GD2 synthase in IMR32 cells was knocked down using siRNA. GD2 expression was observed by confocal microscopy (Figure S1A), and protein expression was evaluated by western blot (Figure S1B). The expression of GM2/GD2 synthase was relatively less in KD-IMR32 cells, whereas the expression of the reference protein, tubulin, was the same in IMR32 and KD-IMR32 cells. Cells were incubated with FAM-DB99 and analyzed by flow cytometry. As shown in Figure S1C, with decreasing GD2 expression, binding between DB99 and cells was also decreased, indicating the binding specificity of DB99 to GD2. Furthermore, FAM-labeled DB99 was incubated with GD2-coated beads and assessed by flow cytometry. As shown in Figure 2C, when compared with control ganglioside (ganglioside sugar-β-aminopropyl GM1a, ganglioside sugar GD3, and ganglioside sugar GM2), there was relatively stronger binding of DB99 to GD2, whereas control DNA did not generate obvious binding signals in all gangliosides. Using nonlinear regression analysis, DB99 was found to have a KD of 21.21 nM (Figure 2D). In addition, molecular docking was applied to evaluate the binding between GD2 and aptamer DB99. As shown in Figure 2E, DB99 generated two quadruplets, including domain 1 and domain 2. Aptamer DB99 could bind with GD2 with a binding energy of −11.600 kcal/mol. The primary interaction elements were nucleotides C5, C6, A7, A8, C11, C12, and C13.
Figure 1.
Schematic representation of combined SELEX selection system
Figure 2.
Characterization of GD2 aptamer DB99
(A) Monitoring of SELEX efficacy by flow cytometry. (B) Binding specificity of DB99 to GD2+ cell lines. (C) Binding specificity of DB99 to GD2 molecules. (D) Flow cytometry evaluation of the binding affinity of DB99 to GD2+ cells. (E) Molecular docking of DB99 to the GD2 molecule.
Construction of ANM
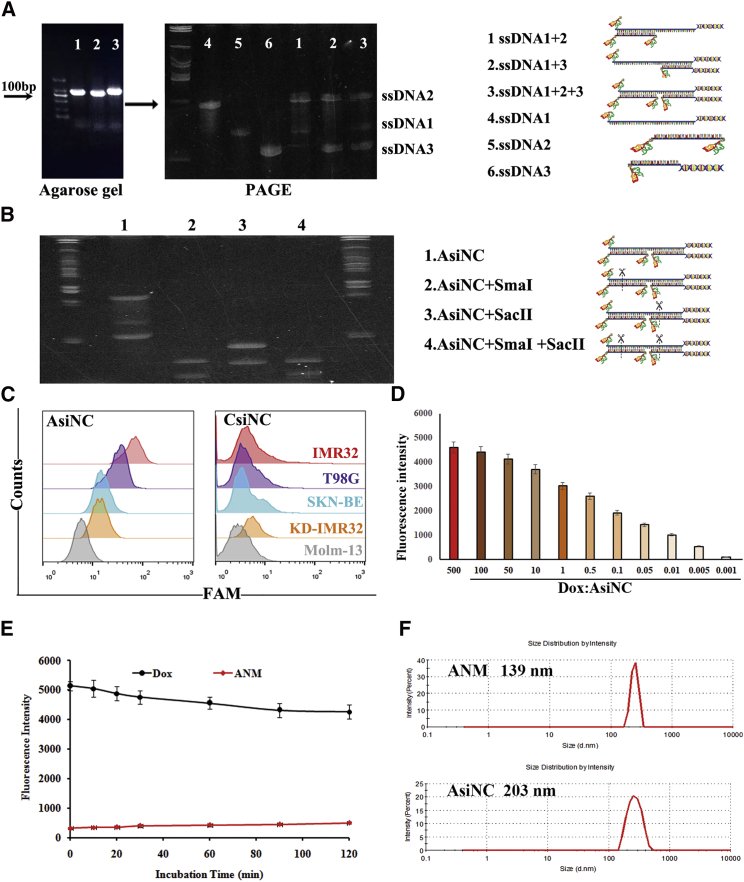
To construct the ANM, the aptamer/siRNA nanocomplex (AsiNC) was first formulated through a self-assembly process (Figure S2). To that end, three complementary, functional, ssDNAs (ssDNA1, ssDNA2, and ssDNA3) were synthesized. Equimolar amounts of ssDNA1, ssDNA2, and ssDNA3 were mixed to generate aptamer/siRNA complexes. To validate whether the aptamer/siRNA nanocomplex AsiNC was formed, PAGE was applied. As presented in Figure 3A, when compared with single ssDNA1, ssDNA2, or ssDNA3, the ssDNA1+ssDNA2, ssDNA1+ssDNA3, and ssDNA1+ssDNA2+ssDNA3 generated different mobility shifts on a gel. The ssDNA1+ssDNA2+ssDNA3 group generated a single band, indicating successful construction. In addition, to further confirm AsiNC formation, AsiNC was exposed to restriction endonucleases SmaI and/or SacII, and the resultant products were analyzed by PAGE (Figure 3B). The digested fragments were assessed after treatment with the endonucleases SmaI and SacII. Thus, AsiNC formation through self-assembly was validated because the endonucleases SmaI and SacII only cleave double-stranded DNA (dsDNA) nanostructures.
Figure 3.
Construction and characterization of AsiNC and ANM
(A) Formation of AsiNC. Markers between 50 and 1,000 bp. (B) Endonuclease digestion of AsiNC for AsiNC forming confirmation. AsiNC incubated with endonuclease were assessed by PAGE (1, AsiNC; 2, mixture of AsiNC+SmaI; 3, mixture of AsiNC+SacII; and 4, mixture of AsiNC+SmaI+SacII). (C) Selective reorganization and binding of AsiNC to GD2+ cells. (D) Formation of ANM. (E) Stability of ANM. (F) Size of AsiNC and ANM was examined with a Zeta-sizer nanodetector.
Subsequently, to evaluate the binding specificity of AsiNC, FAM-labeled AsiNC (Figure S2) or CsiNC (Figure S3) was mixed with several GD2+ or GD− cell lines, and the cells were analyzed by flow cytometry. CsiNC replaced the GD2 aptamer in AsiNC with control ssDNA. This control ssDNA was also generated from SELEX but could not recognize GD2. As shown in Figure 3C, compared with CsiNC, there was an obvious signal in AsiNC, indicating that AsiNC maintained binding ability to GD2.
To construct ANM, given that the fluorescence of free Dox will be quenched after intercalation into dsDNA, AsiNC was incubated with an aqueous solution of Dox and monitored by fluorescence spectrometry. As shown in Figure 3D, sequential decreases in the native fluorescence spectrum of Dox were observed with increasing concentrations of AsiNC. When the AsiNC/Dox molar ratio was below 1,000:1, the fluorescence signal reached its lowest level and was not obviously altered, indicating that AsiNC was fully incorporated up to the Dox payload. The same construction method was also suitable for Dox loading for CsiNC and ACsiNC (Figure S4). ACsiNC replaced MYCN siRNA in AsiNC with control siRNA, which could not downregulate MYCN expression. ACM and CNM were constructed in the same way (Figure S5A). Furthermore, to explore the stability of ANM, the ANM was incubated with fetal bovine serum (FBS) or at different pH levels, and the mixtures were examined for Dox fluorescence. There was no obvious increase in Dox fluorescence, indicating the anti-nuclease activity and stability of ANM (Figures 3E and S5B). The physical properties of the formulated ANM were assessed using a dynamic light-scattering assay. As shown in Figure 3F, ANM was smaller than AsiNC, which makes it more suitable for theranostics. The diameters of single ssDNAs are shown in Figure S6.
ANM selectively binds and delivers drugs to GD2+ cells
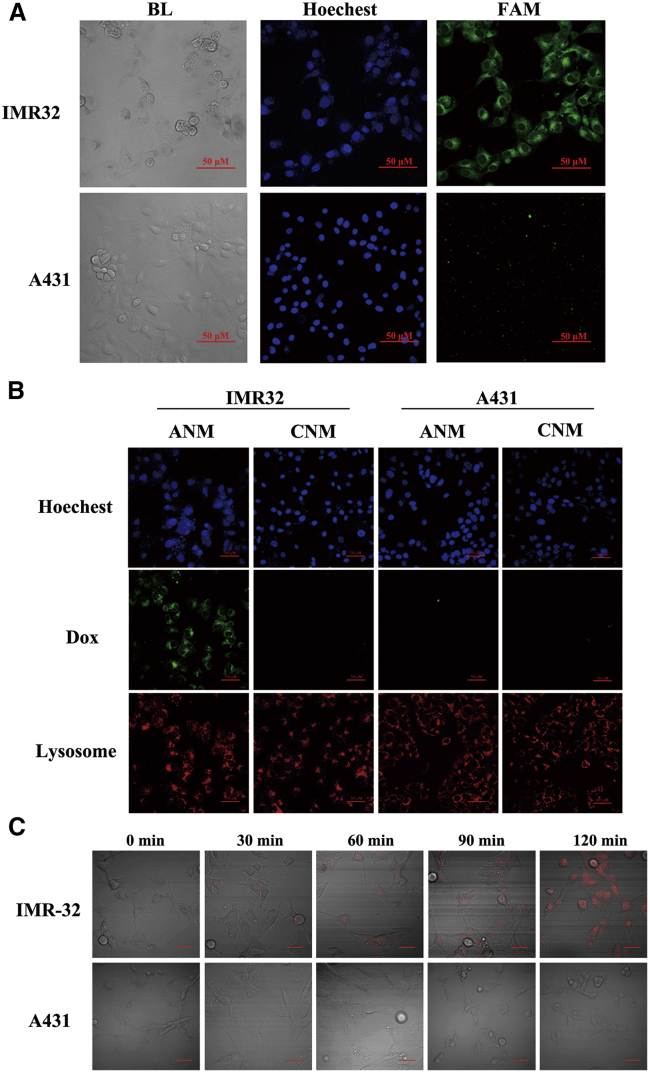
First, to confirm whether ANM maintains GD2 binding specificity, FAM-modified ANM was incubated with IMR32 or A431 cells and evaluated by confocal microscopy. As shown in Figure 4A, there was significant binding to IMR32 cells but little cross-binding to A431 cells. These results demonstrated that ANM still recognized GD2 molecules or GD2+ cells.
Figure 4.
Selective binding and drug delivery of ANM to GD2+ cells
(A) Selective binding of ANM to GD2+ cells. Scale bars: 50 μM. (B) Monitoring of intracellular drug unloading via endocytosis/lysosome pathway. Scale bars: 50 μM. (C) Drug uptake of IMR32 and A431 cells. Scale bars: 50 μM.
Second, to assess whether ANM delivers Dox to GD2+ NB cells in a targeted manner, IMR32 or A431 cells were seeded on glass slides and incubated with ANM (20 nM) or CNM (20 nM). CNM was constructed via Dox interacting in CsiNC. It has been reported that aptamer-mediated drug delivery systems can be internalized into endosomal or lysosomal pathways.38,39 Therefore, to validate the potential mechanism of intracellular delivery, lysosomes were also located. As shown in Figure 4B, there was strong Dox fluorescence in IMR32 cells incubated with ANM but not in IMR32 cells incubated with CNM or A431 cells incubated with either ANM or CNM. This result confirmed that ANM delivers Dox to target cells with specificity. In addition, there was colocalization of Dox and lysosomes, which again confirmed the endocytosis/lysosome pathway as the mechanism for internalization of the aptamer-mediated drug delivery system.
Third, to observe the Dox delivery process of ANM to GD2+ cells, IMR32 or A431 cells were seeded into 6-well plates and incubated with ANM (20 nM). Cells were maintained at 37°C for 0 min, 30 min, 60 min, 90 min and 120 min, and fluorescence signals of Dox were subsequently observed. As illustrated in Figure 4C, the content of Dox in IMR32 cells increased with longer incubation times. Meanwhile, although the incubation time increased, there was hardly any Dox fluorescence signal in GD2− cells. These results indicate the accurate and effective drug delivery of ANM to GD2+ cells.
ANM specifically inhibits GD2+ NB cells in vitro
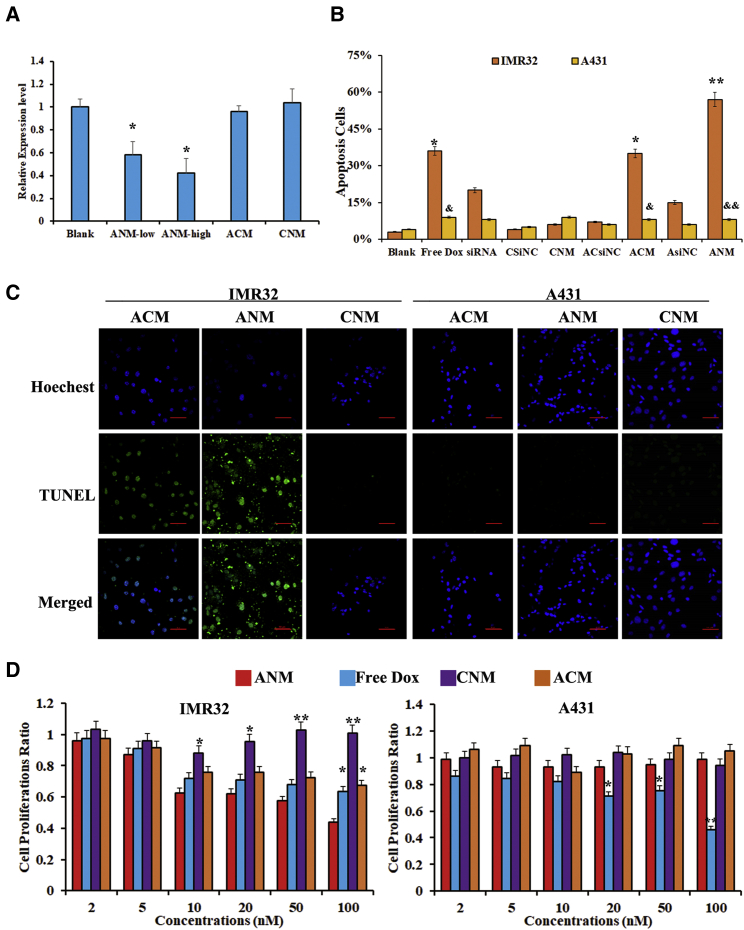
To address whether ANM could silence MYCN expression in GD2+ cells, IMR32 cells were incubated with ANM, ACM, or CNM. ACM was constructed via Dox interacting in ACsiNC. Total RNA was extracted, and qRT-PCR was applied to evaluate the expression of MYCN. As presented in Figure 5A, after incubation with ANM, MYCN expression levels in IMR32 cells were significantly decreased when compared with that of the control group (p < 0.05). In addition, MYCN expression levels in IMR32 treated with a higher concentration of ANM was lower than that treated with a lower concentration of ANM. However, MYCN expression in IMR32 cells treated with ACM or CNM did not markedly change. This result verified that ANM specifically recognizes GD2+ cells and delivers gene-editing agents.
Figure 5.
Inhibition ability of ANM to GD2+ cells for chemotherapy and gene therapy in vitro
(A) MYCN gene silencing by ANM in GD2+ IMR32 cells. ANM-high indicates IMR32 cells incubated with 20 nM ANM, and ANM-low indicates IMR32 cells incubated with ANM. The difference compared with the blank is labeled. ∗p < 0.05. (B) Rates (%) of apoptotic cells detected by TUNEL staining The difference compared with the blank is labeled. ∗p < 0.05, ∗∗p < 0.01. The difference compared with the IMR32 cells with the same treatment is labeled &p < 0.05, &&p < 0.01. (C) Fluorescence microscopy imaging of ANM-induced apoptotic cells, detected by TUNEL staining. Apoptosis cells were stained in green. Scale bars: 50 μM. (D) MTS assays in vitro after the cells were treated with ANM or ACM. The difference compared with the ANM group at the same concentration is labeled. ∗p < 0.05, ∗∗p < 0.01.
According to the results above, ANM specifically delivers agents to GD2+ NB cells. Thus, to further investigate whether ANM could inhibit GD2+ cells and avoid damaging GD2− cells, apoptosis and cell proliferation were assessed. As shown in Figures 5B and 5C, IMR32 cells incubated with ANM presented increased apoptotic cells compared with the PBS group (p < 0.01), whereas IMR32 cells incubated with CNM did not exhibit obvious apoptosis. In addition, A431 cells incubated with either ANM or CNM did not change appreciably. ACM and ANM generated obvious inhibition in IMR32 cells compared with A431 (ACM: p < 0.05, ANM: p < 0.01). Interestingly, a free Dox group generated a similar apoptosis ratio when compared with ACM, which contained GD2 aptamer and free Dox; In addition, the siRNA group generated a similar apoptosis ratio when compared with the AsiNC group, which contained a GD2 aptamer and siRNA. These results further suggested successful targeted delivery to GD2+ cells because of the GD2 aptamer. Next, cell proliferation was assessed, and the data are shown in Figure 5D. In IMR32 cells, both ANM and free Dox inhibited cell proliferation with no significant difference. ACM also inhibited cell growth because it contains a GD2 aptamer. ACM also delivered Dox to cells. However, the lack of MYCN siRNA resulted in a smaller inhibitory effect than observed in response to ANM (2 nM, 5 nM, 10 nM, and 20 nM: p < 0.05; 50 nM and 100 nM: p < 0.01). CNM exhibited almost no inhibition in cells because there was no GD2 aptamer, and CNM was unable to bind with IMR32 cells. In A431 cells, because A431 does not express GD2, neither ANM nor ACM inhibited cells compared with free Dox (20 nM and 50 nM: p < 0.05; 100 nM: p < 0.01). Meanwhile. CNM also exhibited almost no inhibition of A431 cells. To assess the specificity of ANM toward GD2+MYCN+ cells compared with GD2+MYCN− cells, GD2+MYCNhi IMR32, and GD2+MYCNlow, SH-SY-5Y and SK-N-SH cells were used.40 As shown in Figure S7, ANM generated increased inhibition of SK-N-SH, SH-SY-5Y, and IMR32 cells, consistent with MYCN expression levels in those three cell lines (p < 0.05). Compared with ACM and CNM, ANM generated greater inhibition than either ACM or CNM (p < 0.05). These results suggest that ANM tends to reduce damage to GD2− cells and retain the efficacy of Dox and gene-editing agents against GD2+ cells.
ANM treatment inhibits GD2+ NB tumor growth and improves mouse survival
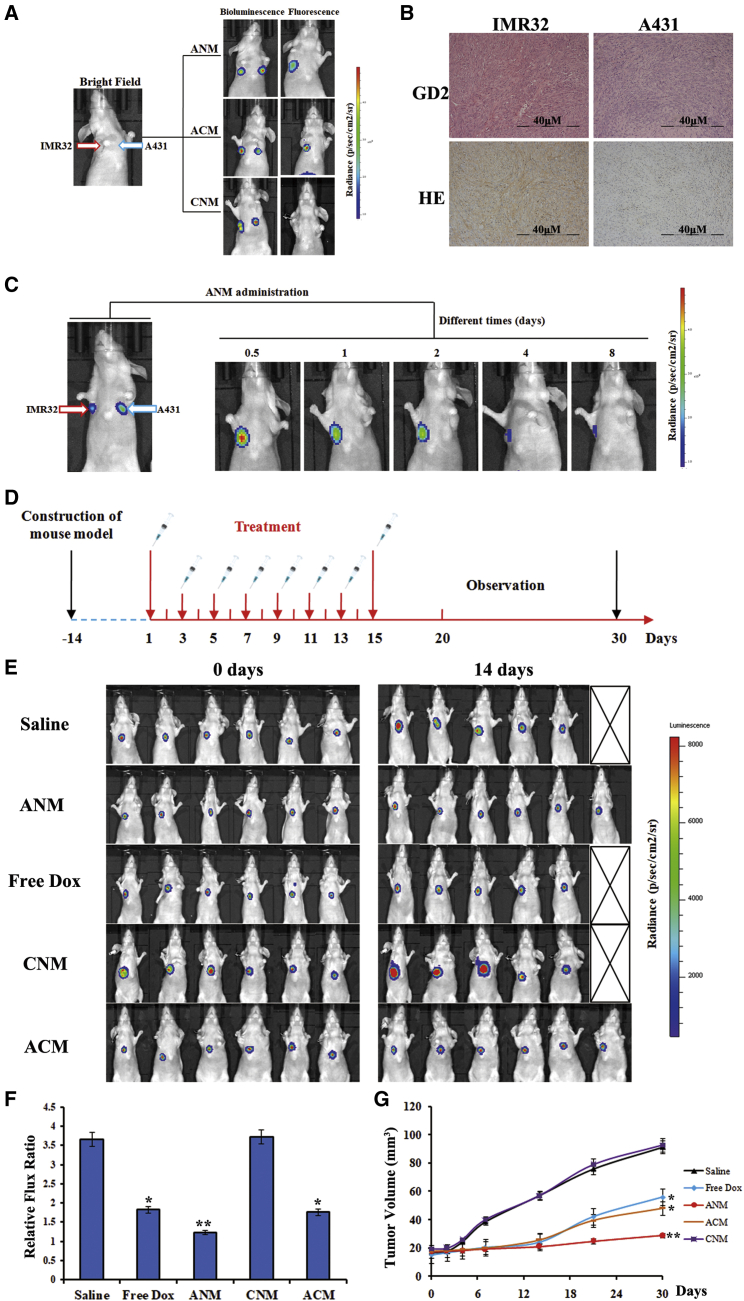
To evaluate whether ANM inhibits GD2+ NB tumors effectively and avoids damaging GD2− cells, first, the ANM targeting ability was evaluated in GD2+ NB tumors. GD2+ IMR32 and GD2− A431 cells stably expressing GFP and luciferase were subcutaneously inoculated into BALB/c mice. The IMR32 cell line was injected into the right shoulder and A431 into the left shoulder. To observe a clear signal view, we chose 50 mM, which was a very large concentration for an in vivo experiment. In brief, ANM, ACM, and CNM (50 mM per mouse) were systemically administered through the tail veins and were observed using an IVIS 200 Imaging System after administration. Because Dox fluoresces, the location of the ANM could be observed. Tumors were monitored by bioluminescence imaging, given that the tumor cells stably expressed luciferase. As presented in Figure 6A, compared with A431, ANM fully highlighted the IMR32 cells expressing GD2, and quantitative imaging analysis indicated a greater signal in IMR32 cells than in A431 cells. In addition, because ACM also contains the GD2 aptamer, it was also located in the IMR32 tumor tissues. CNM exhibited no signal in either IMR32 or the A431 tumors.
Figure 6.
Targeted delivery and anti-cancer of ANM in vivo
(A) Specific targeting ability of ANM to GD2+ tumor site in mouse model detected by IVIS 200 imaging. (B) Immunohistochemistry and H&E staining of tumor tissues. Scale bars: 40 μM. (C) Imaging of ANM versus time to evaluate retention in tumor area. (D) Flow chart of drug administration in mice. (E) Anti-cancer ability of ANM in vivo. Whole-body imaging of mice was to assess tumor volume change after agent administration. (F) Fluorescence signal value ratio of tumor site. ∗p < 0.05, ∗∗p < 0.01. (G) Tumor volume. The significant difference compared with the saline group is labeled. ∗p < 0.05, ∗∗p < 0.01.
Furthermore, tumor tissues were assessed using histological and immunohistochemical studies for expression of GD2, and fresh tumor cells were observed under a fluorescence microscope. As demonstrated in Figures 6B and S8, IMR32 tumor cells overexpressing GD2 were highlighted by Dox, but no GD2 or Dox signals were detected in A431 cells.
Because ANM was designed to inhibit GD2+ tumor cells, to explore the drug delivery and the potential for therapeutic use of ANM, biostability and retention time were assessed. Mice with both IMR32 and A431 tumors were injected with ANM, and signals were measured at different time points (Figure 6C). ANM selectively accumulated in GD2+ cells after administration, and the signal could be observed for 8 days, demonstrating the promising capacity of ANM for therapeutic use. These results indicate that ANM selectively recognizes GD2+ tumor cells with relatively high specificity and affinity.
To evaluate the inhibitory ability of GD2+ cells in vivo, a tumor model was generated by subcutaneously injecting 2 × 107 in vitro propagated GD2-expressing IMR32 cells into the flanks of BALB/c mice. Tumor-bearing mice were randomly assigned to one of three groups that received different treatments (six mice per group). Dox dose used in this section was the common 20 μg. As shown in Figure 6D, mice were administered ANM, ACM, and CNM (carrying the 20-μg Dox payload), equimolar amounts of free Dox (20 μg), or placebo saline (untreated controls). To assess inhibition ability, tumor growth was monitored by whole-body bioluminescence (Figure 6E). ANM significantly decreased tumor growth compared with the other groups. ACM showed little inhibition ability even though it also contained DB99 and could deliver Dox. However, ACM lacked MYCN siRNA. Therefore, the anticancer ability of ACM was weaker than that of ANM. One mouse each in the saline/CNM and free Dox groups died from rapid tumor growth (saline group and CNM) and severe side effects (free Dox), respectively. To further validate anti-cancer ability, the change in fluorescence intensity was calculated (Figure 6F). When compared with the saline group (almost 3.6-fold increase in signal at 14 days when compared with 0 days), the ANM group showed almost a 1.3-fold increase in signal at 14 days when compared with 0 days (p < 0.01). The CNM group showed almost a 3.7-fold increase in signal at 14 days when compared with 0 days, which was the same with the saline group. In addition, the ACM and free Dox group showed a similar ratio because of Dox, indicating effective delivery ability of the GD2 aptamer (p < 0.05). Meanwhile, the ANM group showed smaller change than the ACM and free Dox group did, because ANM not only contained Dox but also contained MYCN siRNA. The average tumor volume in the ANM group was much lower than that in the other groups and inhibited tumor growth more effectively than free Dox did. In addition, the ACM group showed slower tumor growth than that of the free Dox group, which may be due to the selective Dox delivery of ACM and higher agent concentrations in tumor tissues, findings that support the high therapeutic efficacy of ANM (Figure 6G).
GD2 aptamer-mediated ANM reduces side effects in normal tissues and exhibits biosafety in vivo
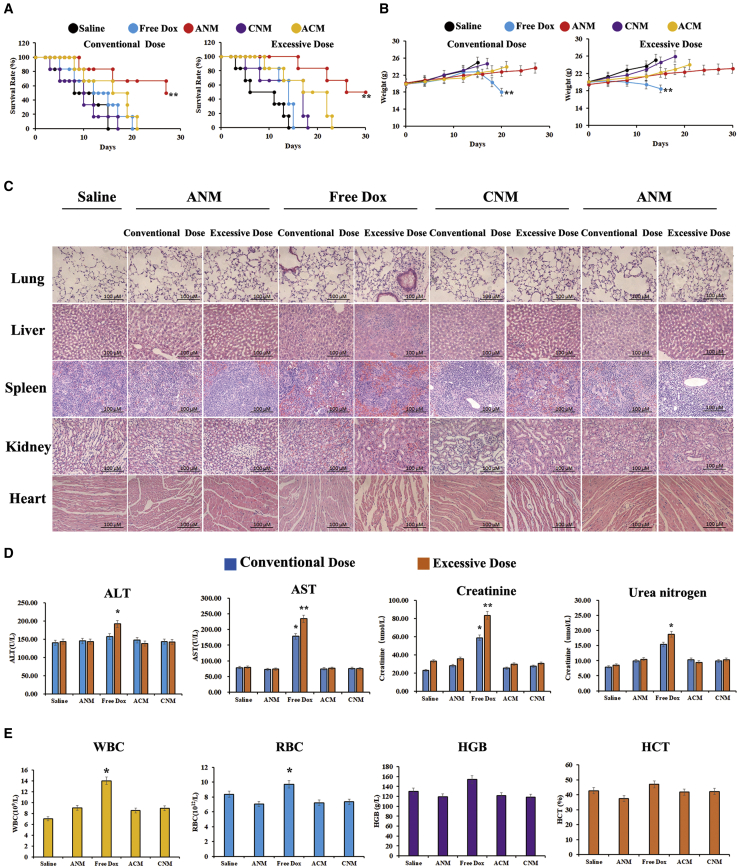
We evaluated whether ANM attenuated side effects in normal tissues, as shown in Figures 7A and 7B, and under both conventional and excessive doses (twice the conventional doses), the survival time in the ANM and free Dox groups was longer than that in the saline, ACM, and CNM groups (p < 0.01). The ANM group exhibited increased survival times compared with the free Dox group. In addition, excessive doses exhibited a longer survival period than conventional doses because of the much higher concentration of Dox (p < 0.01). The weights of each group are presented in Figure 7B. Regardless of whether mice received a conventional or excess dose, mouse weights in ANM group changed little, indicating minimal toxicity in mice in response to the treatment. In contrast, the weight in treatment with free Dox under the conventional dose and the excess dose remained stable at the beginning, ultimately significantly decreasing because of the severe side effects from Dox (p < 0.01). Consistent with the prediction, body weight in the excess-dosage free Dox declined faster than in regular-dosage free Dox. Body weight in the saline, ACM, and CNM groups (conventional and excessive doses) increased rapidly because of the quick growth of tumor masses. Furthermore, mice were euthanized, and the blood, heart, liver, kidney, spleen, and lung tissues were collected for further study. The hematoxylin and eosin staining (H&E staining) of heart, liver, kidney, spleen, and lung tissues revealed that there was bleeding, inflammation, and cell damage in the free Dox group compared with that of the ANM, saline, ACM, and CNM groups. In addition, the damage in excessive-dose free Dox group was severer than that of the conventional group (Figure 7C). Dox at excessive doses caused far more severe damage than the conventional dose did. Serum biomarkers were consistent with histological and immunohistochemical results, as shown in Figures 7D&7E. These values in the ANM, ACM, and CNM group were quite similar to those in the saline group at both conventional and excess dosages, whereas they were much higher in the free Dox group, indicating obvious tissue damage. With an increasing amount of free Dox, the damage to normal tissues and cells was much more severe. Therefore, we concluded that the GD2 aptamer binds to GD2+ cells and that ANM reduces cytotoxicity to normal cells and tissues, indicating that GD2-aptamer-mediated ANM exhibits promising anticancer applications and biosafety.
Figure 7.
Potential reduced side effects of ANM in vivo
(A) Survival rate of mice after treatment initiation. ∗p < 0.05, ∗∗p < 0.01. (B) Body weight of mice after treatment initiation. ∗p < 0.05, ∗∗p < 0.01. (C) Histochemistry of mice organs. Scale bars: 100 μM. (D) Serology assessment. Each bar represents means with SD of six replicates. ∗p < 0.05, ∗∗p < 0.01. (E) Blood routine examination of mice peripheral blood. ∗p < 0.05, ∗∗p < 0.01.
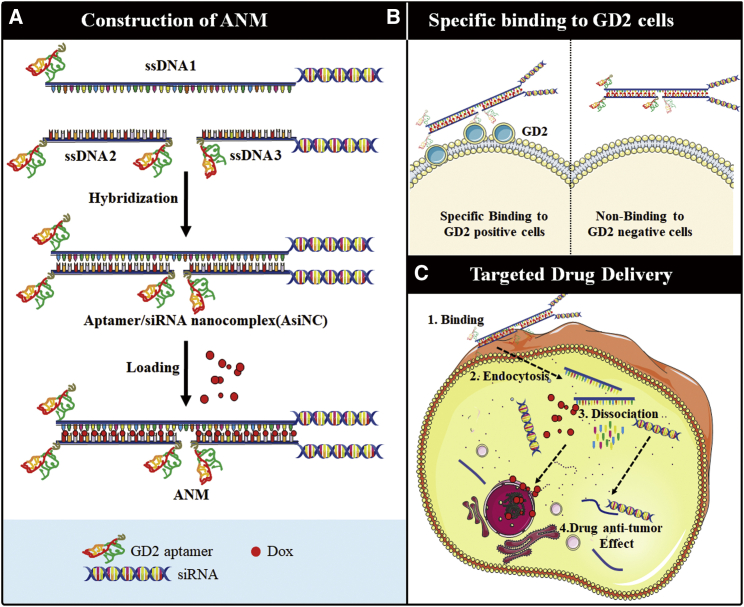
In conclusion, ANM was constructed by self-assembly of ssDNA1, ssDNA2, and ssDNA3 and further Dox intercalation (Figure 8A), and ANM was internalized by GD2+ cells because of the GD2 aptamer (Figure 8B). Further, ANM was degraded, and anti-cancer agents were released to inhibit GD2+ NB tumor growth (Figure 8C). This biostable and biocompatible complex may have potential application for precise treatment of NB.
Figure 8.
Self-assembly schematics of GD2-aptamer-mediated nanostructure for targeting transportation of chemotherapeutics and gene therapy in NB theranostic applications
(A) Self-construction of ANM from ssDNA. (B) Specific recognition of ANM to GD2+ cells. (C) Targeted drug delivery of ANM to GD2+ cells: (1, ANM first bind with GD2+ cells specially via the binding specificity of GD2 aptamer to GD2 molecule; 2, ANM was transferred by endocytic/lysosome vacuole; 3, ANM was degraded, chemotherapeutics Dox and siRNA were released; and 4, Dox and siRNA were released, further inducing cell death and gene silencing).
Discussion
In this study, using combined SELEX technology, we developed a GD2 aptamer, DB99, which recognizes and binds to GD2+ cells with relatively high specificity and affinity. Further, a drug-targeted delivery system termed the “drug-targeted delivery ANM system,” based on DB99, bound to GD2+ tumor cells and elicited selective chemotherapy of intracellular delivery of Dox and MYCN siRNA. The data indicate that ANM achieved growth-inhibition rates of GD2+ NB tumor cells and exhibited negligible toxicity to normal tissues compared with equimolar amounts of free Dox. Aptamers exhibit obvious advantages compared with that of antibodies, such as a high tissue penetration rates, easy modification, limited cost of synthesis, nonimmunogenicity, and inter-batch uniformity.31,41,42 However, to date, although aptamers have been widely used as diagnostic tools (biosensors) or as therapeutic agents, they have lagged behind in reaching the clinical arena despite their potential clinical translation.43, 44, 45 Most aptamers may not function in vivo, primarily because aptamers used as therapeutics were selected in vitro, and the spatial structure of targets in the physiological environment may be different when compared with that of recombinant proteins, as well as in cells in vitro. To solve that obstacle, scientists developed in vivo SELEX. Mi et al.43 performed whole-organism in vivo SELEX in a model of intrahepatic colorectal carcinoma (CRC) metastases, generating an aptamer that bound to and blocked activity of the oncogenic helicase p68 after 14 rounds of in vivo selection. Wang et al.44 performed whole-organism in vivo SELEX using human xenografts of NCI-H460 cells to generate aptamers. The selection pool was a 2′-fluoropyrimidine- and polyethylene glycol (PEG)-modified RNA pool. After 11 rounds of selection, the aptamer RA16 demonstrated high affinity and binding selectively to NCI-H460 cells in vivo. Thus, to generate a potentially transformative GD2 aptamer, we used in vivo SELEX. In addition, to improve the efficacy and ensure that the target of the aptamer was GD2, but not other molecules, we performed an in vitro section. After nine rounds, we selected an ideal aptamer. This combined SELEX procedure may represent a more-promising method of aptamer selection for therapeutic applications.
Aptamers have been selected as appealing candidates for the fabrication of various functional systems, including diagnosis and bioimaging, especially drug delivery.45,46 Aptamer-mediated nanoparticles (NPs) can assist NPs by precisely directing the target cells to the targeted sites. Generally, many types of NPs have been used for drug-delivery systems, including calcium carbonate NPs, mesoporous silica NPs (MSNs), poly(lactic-co-glycolic acid) (PLGA) NPs, and lipid-based NPs.47,48 However, these systems have limitations for clinical applications, such as hepatotoxicity caused by NP aggregation. In addition, these materials are foreign to the human body and may pose unknown risks for in vivo use, potentially causing side effects in normal cells. To solve those issues, scientists have recently been inclined to use nucleic acids in the construction of nanoparticles. Wu et al.9 designed a CD123-aptamer-mediated targeted drug train (TDT) with effective, economical, biocompatible, and high-drug-loading capacity. The TDT is self-assembled from two short primers by ligand-modified ZW25, which acts as an initiation position for elongation, and intercalated by Dox. The TDT has been validated as being capable of transporting a high capacity of Dox to target cells and retaining the efficacy of Dox. Wang et al.28 generated a mesenchymal stem cell (MSC) aptamer and developed feasible, economical, biocompatible, and functional MSC-aptamer-directed NPs without complex manufacturing to recruit MSC for bone-defect regeneration. This nano-aptamer ball (NAB) was constructed using an N-hydroxysuccinimide/1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (NHS/EDC) reaction and healed bone defects significantly better. Many reports have also validated the efficacy of those systems.49, 50, 51 Therefore, in this study, we developed a completely biocompatible and biodegradable system, termed ANM. ANM was designed to logically combine multiple therapeutic modalities for additional or synergistic efficacy, including chemotherapeutics and gene therapy. Our data also confirmed that ANM significantly downregulated MYCN expression in GD2+ NB cells. ANM exhibited better inhibition ability and negligible toxicity in normal tissues than equimolar amounts of free Dox. The current blood tests and serum tests indicates relative protection of ANM in normal cells. Although free Dox generated inflammation (increased white blood cells [WBCs]) and organ damage (increased aspartate transaminase [AST] and alanine transaminase [ALT]), ANM did not generate obvious increases in those parameters. The aforementioned findings reveal a promising GD2 aptamer and an ANM drug delivery system of new-precision nanomedicines for NB-targeted therapy. ANM shows promising application, and additional follow-up studies should focus on areas that would increase its likelihood of clinical translation, such as pharmacokinetics, in vivo stability, biosafety, and improving efficacy.
Materials and methods
Reagents
The random DNA library and primers were synthesized by Sangon Biotech (Shanghai, China). DMEM and RPMI 1640 medium were obtained from HyClone (Thermo Scientific, USA). FBS was purchased from Gibco (Invitrogen, USA). Dox was purchased from Sigma-Aldrich (St. Louis, MO). Streptavidin-coated magnetic beads were obtained from Promega (Madison, WI). The CRISPR-Cas9 knockout (KO) plasmid transfection kit was purchased from Santa Cruz Biotechnology. GD2 ganglioside sugar-β-NAc-spacer3-biotin (GalNAcβ1-4(Neu5Acα2-8Neu5Acα2-3) and Galβ1-4Glcβ-NAc-spacer3-biotin [C47H72N4O32Na2]), GD3-β-N-acetyl–spacer3-biotin (ammonium salt), GM3-β-N-acetyl-spacer3-biotin, GD1a ganglioside sugar-β-NAc-sp3-biotin, and GM2 ganglioside sugar-β-NAc-sp3-biotin were purchased from ELICITYL (France). FAM anti-human ganglioside GD2 antibody and Alexa Fluor 647 anti-human ganglioside GD2 antibody were obtained from BioLegend (San Diego, CA). MYCN siRNA was synthesized and purchased from GenePharma (Shanghai, China). Lentiviruses stably expressing GFP and luciferase were obtained from OBiO (Shanghai, China). The MTS cell proliferation colorimetric assay kit was purchased from BioVision (USA).
Cell lines and culture
The human NB cell line IMR32 (ATCC: CCL-127), the NB cell line SKN-BE (ATCC: CRL-2271), and the human glioma cell lines U87MG (ATCC: HTB-14), T98G (ATCC: CRL-1690), and A172 (ATCC: CRL-1620) were all purchased from ATCC (Manassas, VA, USA). The human acute myelocytic leukemia cell line Molm-13, human epidermal cancer cell line A431, and human breast cancer cell line MDA-MB-231 were all obtained from Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences/Peking Union Medical College (IBMS, CAMS/PUMC). Adherent cells were cultured in DMEM, and cells were cultured in suspension with RPMI 1640 medium with 10% FBS and a mixture of penicillin/streptomycin. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2. All experiments were performed on cells in the exponential growth phase.
GM2/GD2 synthase knockdown of the IMR32 cell line and cell labeling
IMR32 cells were seeded into six-well culture plates at a density/well of 1.5 × 105–2.5 × 105 cells in 3 mL of antibiotic-free standard growth medium 24 h before transfection. The confluence degree of cells at the time of transfection was 30%–50%. Before transfection, the siRNA/microRNA (miRNA) oligo storage solution was diluted to 1 μM. The siRNA/miRNA oligo solution was added to 100 μL Opti-MEM at room temperature for 5 min. Then, 2 μL siRNA-Mate was immediately added. After incubating for 5 min, this complex was added dropwise to the cells. Cells were incubated for 24–96 h until suitable for target gene analysis. Expression of GM2/GD2 synthase mRNA was evaluated at 24–48 h, and GM2/GD2 synthase protein was assessed at 48–72 h by western blot. Tubulin protein was treated as a reference protein. IMR32 cells with GM2/GD2 synthase knockdown were termed KD-IMR32 cells. 1.0 × 106 IMR32 or KD-IMR32 cells were incubated with FAM-labeled GD2 aptamer at 37°C for 30 min. Cells were washed with PBS and assessed by flow cytometry. 1.0 × 104 cells were determined.
IMR32 and A431 cells stably expressing GFP and luciferase were constructed by transfection of GFP adenovirus and subsequently selected. Adherent cells were cultured with DMEM with 10% FBS and a mixture of penicillin/streptomycin. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Tumor-bearing model
The protocol of the animal study in this paper was reviewed and approved by the Ethics Committee of Xi’an Jiaotong University Affiliated Children’s Hospital (Xi’an Children’s Hospital, Xi’an, China), no. C2018004. Six- to eight-week-old female immunodeficient BALB/c (CAnN. Cg-Foxn1nu/Crl) mice were purchased from the Xi’an JiaoTong University Laboratory Animal Centre (Xi’an) and raised under pathogen-free conditions. For the targeting-ability-evaluation assay, mice were anesthetized with 2% vaporized isoflurane, and 2 × 107 GD2+ IMR32 and GD2− A431 cells stably expressing GFP and luciferase were subcutaneously inoculated into each mouse at different anatomic sites at the same time (IMR32 in the right armpit and A431 in the left armpit). Agents were systemically administered via tail vein injection when the tumor reached a diameter ≥5 mm. For SELEX in vivo and anticancer assays, mice were anesthetized with 2% vaporized isoflurane and administered a subcutaneous injection of 2 × 107 IMR32 cells. To avoid post-injection bleeding and tumor cell leakage, brief local compression was executed. A single tumor nodule in each mouse was allowed to grow for several days. All animal procedures were performed according to the standards of the Institutional Animal Care and Use Committee.
Combined SELEX (in vivo and in vitro) selection procedure
The random DNA pool thiophosphorylated by 5′-monothiophosphate substitutions of all dA was synthesized with the sequence 5′-CCGCCCAAATCCCTAAGAG-(N28)-CAGACACACTACACACGCA-3′, where N28 represents 28 random nucleotides. A 20-nmol DNA-aptamer pool in 200 μL of saline was administered through tail vein injection to IMR32 tumor-bearing mice. Mice injected with PBS were used as controls. Considering that aptamers can escape the vasculature after injection, we chose to sacrifice mice between 20 and 30 min after injection. To remove blood from the tumor tissues, mice were sacrificed by heart perfusion. Tumor tissues were collected and frozen in liquid nitrogen. Tissues were homogenized, and DNA was extracted from tumor tissues by All-In-One DNA/RNA/protein mini-preps kits (Sangon Biotech, China). DNA extracted was amplified using a forward primer (5′-TGCGTGTGTAGTGTGTCTG-3′) and a biotin-labeled reverse primer (5′-CCGCCCAAATCCCTAAGAG-3′). The PCR amplification procedure was performed as follows: 5 min 95°C for initial denaturation; 25 cycles of 30 s at 95°C, 30 s at 67°C, and 40 s at 72°C; and 10 min 72°C for extension. PCR products were assessed by 2% agarose gel electrophoresis; PCR products were mixed with streptavidin-coated magnetic beads to separate ssDNA. In brief, 20 pmol of PCR products were incubated with 5 μL beads for 15 min. Beads were washed, and 10 μL NaOH (1 M) was added for 2 min to separate ssDNA. The supernatant was added to 10 μL HCl (1 M) to neutralize the pH. Then, 20 pmol of separated ssDNA was incubated with 1 × 105 IMR32 cells at 37°C for 30 min. Cells were washed twice with PBS, centrifuged (without removal of the supernatant), and further heated at 95°C for 10 min. It is known that, when in 95°C, DNA can denature and lose its three-dimensional structures. As a result, aptamers may lose GD2 binding ability and be released to the supernatant. Therefore, ssDNA that could bind to GD2+ cells was released to the supernatant and further collected. The supernatant fluid was collected and incubated with 1 × 105 KD-IMR32 cells at 37°C for 30 min and subsequently amplified by PCR. ssDNA was separated by streptavidin-coated magnetic beads, and 20 nM ssDNA was used for the next round.
Monitoring the SELEX effect
To monitor the selection efficiency of SELEX and the enrichment of aptamers, forward primers were labeled with FAM, and reverse primers were labeled with biotin. ssDNA enriched from each round was amplified with labeled primers by PCR. FAM-ssDNA was separated from PCR products by streptavidin-coated magnetic beads, and 20 pmol was incubated with 1 × 105 IMR32 or A431 cells scraped off the plates at 37°C for 30 min. Cells were washed twice with PBS, and FAM fluorescence was determined by fluorescence-activated cell sorting (FACS) caliber cytometer (BD Biosciences). 1 × 104 cells were determined. When the fluorescence signal in IMR32 cells was significantly stronger than that in A431 cells or random DNA, SELEX was ended, and the selected ssDNA pool was PCR-amplified using unmodified primers and cloned into Escherichia coli using a TA cloning kit (cat. no. CT101) for DNA sequencing. Flow cytometry was performed three times.
Aptamer-binding specificity assays
To evaluate the binding specificity of aptamers, aptamers were amplified with FAM-labeled forward primers and biotin-labeled reverse primers by PCR. Adherent cells were removed from the plates using a scraper. FAM-labeled aptamers were separated by streptavidin beads, and 40 pmol was incubated with 1 × 106 IMR32, A431, SKN-BE, MDA-MB-231, T98G, U87MG, A172, or Molm-13 cells at 37°C for 30 min. Cells were washed twice with PBS, resuspended in 0.1 mL of PBS buffer, and analyzed by flow cytometry. A random DNA pool was treated as a negative control. Cells were analyzed by flow cytometry using three replicates. 1 × 104 cells were determined for each test.
Target validation
To confirm whether the target of the aptamer was GD2, but not other molecules, 10 mM biotin-GD2, GD3, GM1a, and GM2 were mixed with 20 μL of streptavidin-coated beads for 20 min to fix GD2 on beads. Then, 40 pmol of FAM-modified GD2 aptamer was incubated with those beads for 30 min. Beads were washed twice with PBS and assessed by flow cytometry three times. 5 × 103 beads were determined. Random DNA was used as a negative control. GD3, GM1a, and GM2 were also treated as negative controls.
Molecular docking
The GD2 aptamer structure was predicted using the Discovery Studio program, and the energy was optimized using the Sybyl program. The optimized structure was introduced into the Maestro program for blind docking (taking the central coordinate of the nucleic acid aptamer as the active site, the docking range includes the whole nucleic acid aptamer region, coordinates 0.198109, 0.004182, 1.998088, radius 36) to form a lattice for molecular docking. The compound ganglioside GD2 was prepared according to the pH range of 5∼9, and a conformational search was performed to obtain the 3D structure for docking. Docking was performed in extra precision (XP) and flexible mode. After docking, the conformation of the complex was submitted for molecular dynamics simulation at 50 ns using soft Desmond.
Aptamer-binding affinity assays
To assess the binding specificity of aptamers, gradient concentrations (45, 65, 75, 95, 120, 150, 180, and 250 pM) of FAM-labeled aptamer were incubated with 2 × 105 GD2-coated beads at 37°C for 30 min. Beads were washed three times, resuspended in 0.2 mL PBS buffer and subjected to flow cytometric analysis. 5 × 103 beads were determined. Random DNA was used as a negative control to measure nonspecific binding.50 The equilibrium dissociation constants (KD) of the aptamer were obtained by fitting the dependence of the fluorescence intensity of specific binding on the concentration of the aptamers to the equation Y = BmaxX/(KD + X), where Y represents the reciprocal of the average fluorescence intensity (average fluorescence intensity = fluorescence intensity in DB99-fluorescence intensity in DB99 control DNA), X represents the reciprocal of the aptamer’s concentration, and Bmax represents the maximum binding capacity of the aptamer bound to the target. Three replicates were performed.
Formation of aptamer/siRNA nanocomplex
To construct the aptamer/siRNA nanocomplex, three oligonucleotides were designed and synthesized. This ANM not only targeted GD2+ cells but also delivered siRNA and Dox to specifically targeted cells. Thus, three oligonucleotides were designed. ssDNA1 was the principal part, containing the GD2 aptamer, Dox loading linker 1 for Dox loading, and MYCN siRNA 1 for oncogene silencing; ssDNA2 and ssDNA3 were designed for accessories of targeting ability enhancement, Dox loading, and oncogene silencing. The orders of three oligonucleotides were as follows: ssDNA1: 5′-GD2 aptamer/Dox loading linker 1/MYCN siRNA 1 (5′-GD2 aptamer-CCCGGGCGCGCGCCCGC GGCCCGGGCGCGCGCCCGCGG-CGGAGTTGGTAAAGAATGA-3′); ssDNA2: 5′-GD2 aptamer/Dox loading linker 2/GD2-aptamer-3′ (5′-GD2 aptamer-GGGCCCGCGCGCGGGCGCC-GD2 aptamer-3′); and ssDNA3: 5′-GD2 aptamer/Dox loading linker 2/MYCN siRNA 2–3′ (5′-GD2 aptamer-GGGCCCGCGCGCGGGCGCC-CGGAGATGCTGCTTGAGAA-3′). To validate whether aptamer/siRNA nanocomplex was formed, two endonuclease restriction sites, SmaI (underlined nucleotides in sequences) and SacII (font in italics in sequences), were introduced into three oligonucleotides. Nanocomplexes were formed by programmed hybridization of complementary ssDNA sequences. Three oligonucleotides were mixed at an equal molar ratio (1:1:1) in Tris EDTA buffer (Sigma-Aldrich, St. Louis, MO). The mixture was heated at 95°C for 5 min and cooled at 4°C for programmed hybridization. Control complexes, termed CsiNC and ACsiNC, were constructed in the same way.
Evaluation of AsiNC
First, to validate whether AsiNC was formed, a mixture of oligonucleotides (ssDNA1+ssDNA2, ssDNA1+ssDNA3, and ssDNA1+ssDNA2+ssDNA3) was incubated. After incubation, the ssDNA mixture was assessed by 1% agarose gel electrophoresis, and the bands were cut and extracted from the gel using a DiaSpin DNA gel extraction kit (Sangon Biotech, China) according to the manufacturer’s instructions. Furthermore, SmaI and/or SacII restriction endonuclease (1 U) were mixed with extracted DNA (1 μg) at 37°C for 3 h. The products were also analyzed on a 5% denaturing PAGE gel.
Second, to evaluate the binding specificity of AsiNC, the GD2 aptamer in ssDNA1 and ssDNA3 was modified with FAM at the 5′ end. Adherent cells were removed from the plates with a scraper. FAM-labeled AsiNC (20 mM) was mixed with 1 × 106 cells (IMR32, KD-IMR32, Molm-13, T98G, and SKN-BE) at 37°C for 30 min. Cells were washed twice with PBS, resuspended in 0.1 mL PBS buffer, and analyzed by flow cytometry. Twenty millimolar FAM-labeled CsiNC was treated as a control group. Cells were analyzed by flow cytometry, and 1 × 104 cells were determined. The fluorescence intensity was determined using three replicates.
Construction of ANM
To determine the loading ability of AsiNC for Dox, AsiNC was incubated in an aqueous solution of Dox (5 nM) for 30 min in a black 96-well plate at a range of aptamer/Dox molar ratios. The system was kept in the dark at room temperature for 1 h. Saturation of Dox loading was monitored by fluorescence spectrometry by a Synergy 4 analyzer (BioTek UK) (Eex = 488 nm, Eem = 594 nm) using three replicates. The same construction method was also suitable for Dox loading for CsiNC and ACsiNC. After Dox loading, the complexes were termed CNM and ACM, respectively.
Measurement of size and zeta potential
A total of 5 × 106 AsiNC and ANM were diluted in double-distilled water (ddH2O) to 1 mL. The scale distribution of nanoparticles was measured using a dynamic light-scatter meter at 25°C. The physical properties of ssDNA1, ssDNA2, and ssDNA3 were assessed as described above.
Stability of ANM
ANM (100 nM) was diluted in 100 μL of PBS and incubated with FBS or different pH levels for 2 h. The mixtures were assessed for Dox fluorescence by fluorescence spectrometry using a Synergy 4 analyzer (Eex = 488 nm, Eem = 594 nm). PBS mixed with 5% FBS was considered a negative control. Three replicates were performed.
Binding specificity of ANM
IMR32 or A431 cells (1 × 105) were seeded on glass slides, and the GD2 aptamer in ssDNA1 and ssDNA3 was modified with FAM at the 5′ end. FAM-modified ANM (10 mM) was incubated with cells at 37°C for 1 h. Cells were washed twice with PBS and evaluated by confocal microscopy.
Selective intracellular drug delivery of ANM observed by confocal microscopy
IMR32 or A431 cells (1 × 105) were seeded on glass slides and incubated with ANM (20 nM) or CNM (20 nM). Cells were kept in the dark at 37°C for 1 h. Then, cells were washed with PBS for lysosomal staining. In brief, 20 μL of LysoNIR Indicator was diluted in 10 mL of Live Cell staining buffer to prepare a dye working solution. Equal volumes of the dye working solution were added to the cells and incubated at 37°C for 1 h. Cells were washed with prewarmed Hank’s balanced salt solution (HBSS) buffer, fixed, and observed by confocal microscopy. Fluorescence signals were detected at different wavelengths (Eex/Eem = 630–650 nm for lysosomes; Eex = 488 nm, Eem = 565 nm for Dox; Eex = 350 nm, Eem = 460 nm for Hoechst).
Drug uptake
IMR32 or A431 cells (1 × 106) were seeded into 6-well plates and incubated with ANM (20 nM). Cells were kept at 37°C for 0 min, 30 min, 60 min, 90 min and 120 min. Then, cells were washed with PBS and subjected to confocal microscopy to observe the fluorescence signals of Dox.
Gene silencing of the MYCN gene by ANM
IMR32 cells (1 × 105) were seeded onto glass slides and incubated with ANM (low: 10 nM; high: 20 nM), ACM (20 nM), or CNM (20 nM). Cells were kept in the dark at 37°C for 1 h. Then, cells were washed with PBS and incubated for an additional 24 h. Cells were collected, and RNA was extracted for RT-PCR. The RNeasy Mini Kit (QIAGEN) was used to extract total RNA from cell lines. The TaKaRa PrimeScript RT reagent kit was used to transcribe cDNA for qRT-PCR analysis. Relative gene expression was measured using SYBR Green qPCR Master Mix (Perkin-Elmer Applied Biosystems). The primers applied were as follows: forward primer: 5′-ACTGTAGCCATCCGAGGACA-3′; reverse primer: 5′-TCGGAAGCAGAAACAGTCCC-3′. β-actin was treated as an internal reference gene. The primers applied were as follows: forward, 5′-CAGCCTTCCTTCTTGGGTAT-3′; reverse, 5′-TGGCATAG AGGTCTTTACGG-3′. Three replicates were performed. Data were analyzed using Applied Biosystems QuantStudio real-time PCR software.
Inhibition ability of ANM analyzed by apoptosis in vitro
IMR32 and A431 (1 × 105) were seeded and incubated with free Dox, MYCN siRNA, CsiNC, CNM, ACsiNC, ACM, AsiNC, and ANM, respectively. CsiNC, CNM, ACsiNC, ACM, AsiNC, and ANM were 5 nM. The groups of free Dox and siRNA alone were added with the same concentration of Dox or siRNA in the ANM group. Cells were incubated at 37°C for 1 h, washed with PBS, and incubated for an additional 24 h. 54 μL TDT buffer was added to cells at 37°C for 1 h. Then, cells were incubated with termination solution and labeled with peroxidase-labeled anti-digoxin antibody. Cells were evaluated under a microscope and the apoptosis ratio (%) = apoptosis cell number/total cell number.
IMR32 or A431 cells (1 × 105) were seeded onto glass slides and incubated with ANM (20 nM), CNM (20 nM), or ACM (20 nM). Cells were kept in the dark at 37°C for 1 h. Then, cells were washed with PBS and incubated for an additional 24 h. Then, cells were fixed in 4% neutral formalin at room temperature for 10 min. Cells were washed twice with PBS and incubated in PBS containing 2% H2O2 at room temperature for 5 min. Cells were washed with PBS, and 54 μL TDT buffer was added to cells at 37°C for 1 h. Then, cells were incubated with termination solution and labeled with peroxidase-labeled anti-digoxin antibody. To observe cells, 0.05% 3,3′-diaminobenzidine (DAB) was added. Cells were evaluated under a microscope.
Inhibition ability of ANM analyzed by cell proliferation in vitro
IMR32 or A431 cells (1 × 105) were seeded into 96-well plates and incubated with various concentrations (2, 5, 10, 20, 50, and 100 nM) of ANM, free Dox, ACM, or CNM. Cells treated with PBS were used as controls. Cells were kept in the dark at 37°C for 1 h. Then, cells were washed with PBS and incubated for an additional 24 h. Then, 10 μL of CCK-8 was added to each well and incubated at 37°C for 4 h. The absorption value of each well was evaluated at 450 nm by an enzyme-linked immunodetector. Cell proliferation is presented as the cell-proliferation ratio, which was calculated by the Absorption value in agent group/Absorption value in the PBS group.
IMR32, SH-SY-5Y, and SK-N-SH cells were seeded into 96-well plates (5 × 104 cells per well). Cells were treated with PBS, ANM, CNM, and ACM (20 nM). Cells were kept in the dark at 37°C for 1 h. Then, cells were washed with PBS and incubated for an additional 24 h. For the cytotoxicity study assessed by MTS, 20 μL MTS reagent was added to each well for a 4-h incubation. The absorbance (490 nm) of each well was determined to evaluate cell viability. Cell proliferation was assessed by 5-bromo-2′-deoxyuridine (BrdU) ELISA: the BrdU buffer was added and incubated for 24 h. After removing the incubation buffer, 100 μL of fixed buffer was added for 30 min. Then, 100 μL of BrdU antibody buffer was added for 1 h followed by 300 μL of washing buffer three times. Horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody was added and incubated for 1 h. Cells were washed three times, and 100 μL of 3,3′5,5′-tetramethylbenzidine (TMB) was finally added. After 15 min, 50 μL of terminating buffer was added, and the absorbance (450 nm) of each well was measured. Cell proliferation is presented as the cell proliferation ratio, which was calculated by the Absorption value in agent group/Absorption value in the PBS group. All tests were performed three times.
Targeting ability of ANM in vivo
The protocol of the animal study in this paper was reviewed and approved by the Ethics Committee of Xi’an Jiaotong University Affiliated Children’s Hospital (Xi’an Children’s Hospital, Xi’an, China); 4–8-weeks-old BALB/c (CAnN. Cg-Foxn1nu/Crl) mice were purchased from the Fourth Military Medical University (Xi’an, ShaanXi, China); 2 × 107 GD2+ IMR32 and GD2− A431 cells stably expressing GFP and luciferase were subcutaneously inoculated into each mouse at different anatomic sites at the same time (IMR32 in the right armpit and A431 in the left armpit). When the tumor reached a diameter ≥5 mm, 50 mM ANM and the control group (ACM and CNM) were systemically administered via the tail vein injection (N = 6). Tumor tissues were observed by GFP. Because Dox fluoresces (Eex = 488 nm, Eem = 595 nm), the agent location could be identified. Therefore, the targeting ability of agents was evaluated using the IVIS 200 imaging system to image the entire body at different times (0.5, 1, 2, 4, and 8 days) (N = 3).
Anticancer efficacy of ANM in vivo
To evaluate the anticancer effect of ANM, a mouse xenograft tumor model was generated by subcutaneously injecting 2 × 107 in vitro propagated GD2-expressing IMR32 cells into the abdomen of BALB/c (CAnN; Cg-Foxn1nu/Crl) mice. Dorsal tumor nodules were allowed to grow to a volume of ∼20 mm3 before initiation of treatment. Tumor-bearing mice were randomly assigned to one of five groups, with six mice in each group, as follows: (1) mice treated with saline, (2) mice treated with ANM carrying 20 μg Dox to each mouse, (3) mice treated with 20 μg free Dox to each mouse, (4) mice treated with CNM carrying 20 μg Dox to each mouse, and (5) mice treated with ACM carrying 20 μg Dox to each mouse. Drugs were administered through tail vein injection once a day, and the treatment was continued for 15 days. Tumor length and width were measured using calipers, and tumor volume was calculated using the following equation: Tumor volume = (Tumor length × [Tumor width2])/2. Animals were evaluated using the IVIS 200 Imaging System (N = 6). The fluorescence intensity change was calculated using the following equation: Relative ratio = (Fluorescence intensity at 14 days)/(Fluorescence intensity at 0 days).
Systemic toxicity and safety evaluation
To assess the therapeutic safety and systemic toxicity of ANM, mouse xenograft tumor models were divided into nine groups, with six mice in each group. Mice were euthanized by cervical ligation when tumor volume exceeded 2,000 mm3. Group administrations (N = 6) were performed as follows: (1) saline, (2) ANM carrying 20 μg Dox, (3) twice ANM, (4) 20 μg free Dox, (5) twice Dox, (6) CNM carrying 20 μg Dox, (7) twice CNM, (8) ACM carrying 20 μg Dox, and (9) twice ACM. Treatments were applied every 2 days and for 10 days. Mice were observed for survival rate and weight. All mice were euthanized, and blood samples were collected in BD Microtainer blood collection tubes with K2-EDTA (Becton Dickinson, USA). Blood samples were subjected to hematologic analysis, including WBCs, red blood cells (RBCs), platelets (PLTs), hemoglobin (HGB), and hematocrit (HCT). In addition, serum markers of organ damage (creatinine, AST, ALT, and blood urea nitrogen [BUN]) were assessed. The body weight of each mouse was also measured weekly to monitor potential drug toxicity. After confirmation of death, the heart, liver, kidney, spleen, and lung were collected and subjected to H&E staining to evaluate any adverse effects of ANM on normal tissues.
Statistical analysis
SPSS 11.0 software was applied at Xi’an Jiaotong University. All numerical data are expressed as means ± SD. Differences among groups were examined using Student’s two-tailed t test, except for tumor-growth curves and mouse-survival curves. The statistical significance for the tumor-growth curves was calculated using a one-way ANOVA with Dunnett’s multiple comparison test. For the mouse-survival curves, log-rank tests were applied. A p < 0.05 was identified as a significant difference.
Acknowledgments
The authors would like to thank Dr. Dan Tan and Dr. Jiping Zhao for their assistance with the chemistry experiments at the Xi’an Jiaotong University. This work was supported by Shaanxi Provincial Program (grants 2019SF-207 and 2017SF-280).
Author contributions
All authors listed made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data. The detailed contributions are as follows: conceived and designed the experiments, H.L. and L.Z.; performed the experiments, L.Z., M.W., and Z.Z.; analyzed the data, L.Z. and S.C.; contributed reagents/materials/analytic tools, H.W., Y.Y., F.C., and Q.L.; wrote the paper: L.Z. and H.L. All authors were also involved in drafting the manuscript and have given final approval of the version to be published.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.08.021.
Supplemental information
References
- 1.McNerney K.O., Karageorgos S.A., Hogarty M.D., Bassiri H. Enhancing neuroblastoma immunotherapies by engaging iNKT and NK cells. Front. Immunol. 2020;11:873. doi: 10.3389/fimmu.2020.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J., Nelson C., Wong M., Tee A.E., Liu P.Y., La T., Fletcher J.I., Kamili A., Mayoh C., Bartenhagen C. Targeted therapy of TERT-rearranged neuroblastoma with BET bromodomain inhibitor and proteasome inhibitor combination therapy. Clin. Cancer Res. 2021;27:1438–1451. doi: 10.1158/1078-0432.CCR-20-3044. [DOI] [PubMed] [Google Scholar]
- 3.Southgate H.E.D., Chen L., Curtin N.J., Tweddle D.A. Targeting the DNA damage response for the treatment of high risk neuroblastoma. Front. Oncol. 2020;10:371. doi: 10.3389/fonc.2020.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coorens T.H.H., Farndon S.J., Mitchell T.J., Jain N., Lee S., Hubank M., Sebire N., Anderson J., Behjati S. Lineage-independent tumors in bilateral neuroblastoma. N. Engl. J. Med. 2020;383:1860–1865. doi: 10.1056/NEJMoa2000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borenäs M., Umapathy G., Lai W.Y., Lind D.E., Witek B., Guan J., Mendoza-Garcia P., Masudi T., Claeys A., Chuang T.P. ALK ligand ALKAL2 potentiates MYCN-driven neuroblastoma in the absence of ALK mutation. EMBO J. 2021;40:e105784. doi: 10.15252/embj.2020105784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George S.L., Parmar V., Lorenzi F., Marshall L.V., Jamin Y., Poon E., Angelini P., Chesler L. Novel therapeutic strategies targeting telomere maintenance mechanisms in high-risk neuroblastoma. J. Exp. Clin. Cancer Res. 2020;39:78. doi: 10.1186/s13046-020-01582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucianò A.M., Perciballi E., Fiore M., Del Bufalo D., Tata A.M. The combination of the M2 muscarinic receptor agonist and chemotherapy affects drug resistance in neuroblastoma cells. Int. J. Mol. Sci. 2020;21:E8433. doi: 10.3390/ijms21228433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonso Cadenas J.A., Almodóvar Martín J.L., Iglesias Bouzas M.I., Jiménez García R., Serrano González A. Adrenergic crisis as a debut form of a neuroblastoma. Rev. Chil. Pediatr. 2020;91:767–772. doi: 10.32641/rchped.vi91i5.1828. [DOI] [PubMed] [Google Scholar]
- 9.Wu H., Wang M., Dai B., Zhang Y., Yang Y., Li Q., Duan M., Zhang X., Wang X., Li A., Zhang L. Novel CD123-aptamer-originated targeted drug trains for selectively delivering cytotoxic agent to tumor cells in acute myeloid leukemia theranostics. Drug Deliv. 2017;24:1216–1229. doi: 10.1080/10717544.2017.1367976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heczey A., Courtney A.N., Montalbano A., Robinson S., Liu K., Li M., Ghatwai N., Dakhova O., Liu B., Raveh-Sadka T. Anti-GD2 CAR-NKT cells in patients with relapsed or refractory neuroblastoma: an interim analysis. Nat. Med. 2020;26:1686–1690. doi: 10.1038/s41591-020-1074-2. [DOI] [PubMed] [Google Scholar]
- 11.Holmes K., Pötschger U., Pearson A.D.J., Sarnacki S., Cecchetto G., Gomez-Chacon J., Squire R., Freud E., Bysiek A., Matthyssens L.E., International Society of Paediatric Oncology Europe Neuroblastoma Group (SIOPEN) Influence of surgical excision on the survival of patients with stage 4 high-risk neuroblastoma: a report from the HR-NBL1/SIOPEN study. J. Clin. Oncol. 2020;38:2902–2915. doi: 10.1200/JCO.19.03117. [DOI] [PubMed] [Google Scholar]
- 12.Moreno L., Barone G., DuBois S.G., Molenaar J., Fischer M., Schulte J., Eggert A., Schleiermacher G., Speleman F., Chesler L. Accelerating drug development for neuroblastoma: summary of the Second Neuroblastoma Drug Development Strategy forum from Innovative Therapies for Children with Cancer and International Society of Paediatric Oncology Europe Neuroblastoma. Eur. J. Cancer. 2020;136:52–68. doi: 10.1016/j.ejca.2020.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Cheung I.Y., Cheung N.V., Modak S., Mauguen A., Feng Y., Basu E., Roberts S.S., Ragupathi G., Kushner B.H. Survival impact of anti-GD2 antibody response in a phase II ganglioside vaccine trial among patients with high-risk neuroblastoma with prior disease progression. J. Clin. Oncol. 2021;39:215–226. doi: 10.1200/JCO.20.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladenstein R., Pötschger U., Valteau-Couanet D., Luksch R., Castel V., Yaniv I., Laureys G., Brock P., Michon J.M., Owens C. Interleukin 2 with anti-GD2 antibody ch14.18/CHO (dinutuximab beta) in patients with high-risk neuroblastoma (HR-NBL1/SIOPEN): a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19:1617–1629. doi: 10.1016/S1470-2045(18)30578-3. [DOI] [PubMed] [Google Scholar]
- 15.Yu J., Hung J.T., Wang S.H., Cheng J.Y., Yu A.L. Targeting glycosphingolipids for cancer immunotherapy. FEBS Lett. 2020;594:3602–3618. doi: 10.1002/1873-3468.13917. [DOI] [PubMed] [Google Scholar]
- 16.Xu X., Huang W., Heczey A., Liu D., Guo L., Wood M., Jin J., Courtney A.N., Liu B., Di Pierro E.J. NKT cells coexpressing a GD2-specific chimeric antigen receptor and IL15 show enhanced in vivo persistence and antitumor activity against neuroblastoma. Clin. Cancer Res. 2019;25:7126–7138. doi: 10.1158/1078-0432.CCR-19-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ornell K.J., Chiu B., Coburn J.M. Development of a dinutuximab delivery system using silk foams for GD2 targeted neuroblastoma cell death. J. Biomed. Mater. Res A. 2021;109:1393–1405. doi: 10.1002/jbm.a.37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diccianni M.B., Kempińska K., Gangoti J.A., Yu A.L., Sorkin L.S. Anti-GD2 induced allodynia in rats can be reduced by pretreatment with DFMO. PLoS ONE. 2020;15:e0236115. doi: 10.1371/journal.pone.0236115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H.W., Sheard M.A., Malvar J., Fernandez G.E., DeClerck Y.A., Blavier L., Shimada H., Theuer C.P., Sposto R., Seeger R.C. Anti-CD105 antibody eliminates tumor microenvironment cells and enhances anti-GD2 antibody immunotherapy of neuroblastoma with activated natural killer cells. Clin. Cancer Res. 2019;25:4761–4774. doi: 10.1158/1078-0432.CCR-18-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emberesh M., Rubinstein J.D., Young J., Benoit S.W., Dandoy C.E., Weiss B.D. Tolerance of dinutuximab therapy for treatment of high-risk neuroblastoma in two patients with end-stage renal disease on dialysis. Pediatr. Blood Cancer. 2021;68:e28852. doi: 10.1002/pbc.28852. [DOI] [PubMed] [Google Scholar]
- 21.Fletcher J.I., Ziegler D.S., Trahair T.N., Marshall G.M., Haber M., Norris M.D. Too many targets, not enough patients: rethinking neuroblastoma clinical trials. Nat. Rev. Cancer. 2018;18:389–400. doi: 10.1038/s41568-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 22.Gartrell J., Shulkin B.L., Helmig S., Caldwell K.J., Furman W., Federico S.M. Induction chemotherapy with an anti-GD2 monoclonal antibody (dinutuximab) and cytokines in children with newly diagnosed high-risk neuroblastoma: a case series. J. Pediatr. Hematol. Oncol. 2021;43:e692–e696. doi: 10.1097/MPH.0000000000001992. [DOI] [PubMed] [Google Scholar]
- 23.Anghelescu D.L., Goldberg J.L., Faughnan L.G., Wu J., Mao S., Furman W.L., Santana V.M., Navid F. Comparison of pain outcomes between two anti-GD2 antibodies in patients with neuroblastoma. Pediatr. Blood Cancer. 2015;62:224–228. doi: 10.1002/pbc.25280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangel A.E., Hariri A.A., Eisenstein M., Soh H.T. Engineering aptamer switches for multifunctional stimulus-responsive nanosystems. Adv. Mater. 2020;32:e2003704. doi: 10.1002/adma.202003704. [DOI] [PubMed] [Google Scholar]
- 25.Lv M., Zhou W., Tavakoli H., Bautista C., Xia J., Wang Z., Li X. Aptamer-functionalized metal-organic frameworks (MOFs) for biosensing. Biosens. Bioelectron. 2021;176:112947. doi: 10.1016/j.bios.2020.112947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirian M., Kouhpayeh S., Shariati L., Boshtam M., Rahimmanesh I., Darzi L., Taghizadeh R., Jahanian-Najafabadi A., Khanahmad H. Generation of HBsAg DNA aptamer using modified cell-based SELEX strategy. Mol. Biol. Rep. 2021;48:139–146. doi: 10.1007/s11033-020-05995-2. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J., Wang M., Yang Y., Wang G., Che F., Li Q., Zhang L. CD123 thioaptamer protects against sepsis via the blockade between IL-3/CD123 in a cecal ligation and puncture rat model. Nucleosides Nucleotides Nucleic Acids. 2021;40:16–31. doi: 10.1080/15257770.2020.1815770. [DOI] [PubMed] [Google Scholar]
- 28.Wang M., Wu H., Li Q., Yang Y., Che F., Wang G., Zhang L. Novel aptamer-functionalized nanoparticles enhances bone defect repair by improving stem cell recruitment. Int. J. Nanomedicine. 2019;14:8707–8724. doi: 10.2147/IJN.S223164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayramoglu G., Ozalp V.C., Dincbal U., Arica M.Y. Fast and sensitive detection of salmonella in milk samples using aptamer-functionalized magnetic silica solid phase and MCM-41-aptamer gate system. ACS Biomater. Sci. Eng. 2018;4:1437–1444. doi: 10.1021/acsbiomaterials.8b00018. [DOI] [PubMed] [Google Scholar]
- 30.Vázquez-González M., Willner I. Aptamer-functionalized micro- and nanocarriers for controlled release. ACS Appl. Mater. Interfaces. 2021;13:9520–9541. doi: 10.1021/acsami.0c17121. [DOI] [PubMed] [Google Scholar]
- 31.Safari Yazd H., Yang Y., Li L., Yang L., Li X., Pan X., Chen Z., Jiang J., Cui C., Tan W. Precise deposition of polydopamine on cancer cell membrane as artificial receptor for targeted drug delivery. iScience. 2020;23:101750. doi: 10.1016/j.isci.2020.101750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., He J., Shen L., Wang T., Yang J., Li Y., Wang Y., Quan D. Brain-targeted delivery of obidoxime, using aptamer-modified liposomes, for detoxification of organophosphorus compounds. J. Control. Release. 2021;329:1117–1128. doi: 10.1016/j.jconrel.2020.10.039. [DOI] [PubMed] [Google Scholar]
- 33.Krissanaprasit A., Key C.M., Froehlich K., Pontula S., Mihalko E., Dupont D.M., Andersen E.S., Kjems J., Brown A.C., LaBean T.H. Multivalent aptamer-functionalized single-strand RNA origami as effective, target-specific anticoagulants with corresponding reversal agents. Adv. Healthc. Mater. 2021;10:e2001826. doi: 10.1002/adhm.202001826. [DOI] [PubMed] [Google Scholar]
- 34.Dyke C.K., Steinhubl S.R., Kleiman N.S., Cannon R.O., Aberle L.G., Lin M., Myles S.K., Melloni C., Harrington R.A., Alexander J.H. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490–2497. doi: 10.1161/CIRCULATIONAHA.106.668434. [DOI] [PubMed] [Google Scholar]
- 35.Gao F., Zhou J., Sun Y., Yang C., Zhang S., Wang R., Tan W. Programmable repurposing of existing drugs as pharmaceutical elements for the construction of aptamer-drug conjugates. ACS Appl. Mater. Interfaces. 2021;13:9457–9463. doi: 10.1021/acsami.0c18846. [DOI] [PubMed] [Google Scholar]
- 36.Shao F., Wang Z., Wang S. Identification of MYCN-related gene as a potential biomarker for neuroblastoma prognostic model by integrated analysis and quantitative real-time PCR. DNA Cell Biol. 2021;40:332–347. doi: 10.1089/dna.2020.6193. [DOI] [PubMed] [Google Scholar]
- 37.Selvi S.V., Prasannan A., Chen S.M., Vadivelmurugan A., Tsai H.C., Lai J.Y. Glutathione and cystamine functionalized MoS2core-shell nanoparticles for enhanced electrochemical detection of doxorubicin. Mikrochim. Acta. 2021;188:35. doi: 10.1007/s00604-020-04642-8. [DOI] [PubMed] [Google Scholar]
- 38.Cao X., Shang X., Guo Y., Zheng X., Li W., Wu D., Sun L., Mu S., Guo C. Lysosomal escaped protein nanocarriers for nuclear-targeted siRNA delivery. Anal. Bioanal. Chem. 2021;413:3493–3499. doi: 10.1007/s00216-021-03297-5. [DOI] [PubMed] [Google Scholar]
- 39.Lv C., Yang C., Ding D., Sun Y., Wang R., Han D., Tan W. Endocytic pathways and intracellular transport of aptamer-drug conjugates in live cells monitored by single-particle tracking. Anal. Chem. 2019;91:13818–13823. doi: 10.1021/acs.analchem.9b03281. [DOI] [PubMed] [Google Scholar]
- 40.Aygun N., Altungoz O. MYCN is amplified during S phase, and c-myb is involved in controlling MYCN expression and amplification in MYCN-amplified neuroblatoma cell lines. Mol. Med. Rep. 2019;19:345–361. doi: 10.3892/mmr.2018.9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong L., Gan L., Deng Z., Liu X., Peng H., Tang H., Liu X., Fang F., Yao F., Li W. Antitumor activity of lipid-DNA aptamer modified T lymphocytes in carcinoma. J. Biomed. Nanotechnol. 2020;16:1110–1118. doi: 10.1166/jbn.2020.2954. [DOI] [PubMed] [Google Scholar]
- 42.Yang L., Liang M., Cui C., Li X., Li L., Pan X., Yazd H.S., Hong M., Lu J., Cao Y.C., Tan W. Enhancing the nucleolytic resistance and bioactivity of functional nucleic acids by diverse nanostructures through in situ polymerization-induced self-assembly. ChemBioChem. 2021;22:754–759. doi: 10.1002/cbic.202000712. [DOI] [PubMed] [Google Scholar]
- 43.Mi J., Liu Y., Rabbani Z.N., Yang Z., Urban J.H., Sullenger B.A., Clary B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010;6:22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H., Zhang Y., Yang H., Qin M., Ding X., Liu R., Jiang Y. In vivo SELEX of an inhibitory NSCLC-specific RNA aptamer from PEGylated RNA library. Mol. Ther. Nucleic Acids. 2018;10:187–198. doi: 10.1016/j.omtn.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge M.H., Zhu X.H., Shao Y.M., Wang C., Huang P., Wang Y., Jiang Y., Maimaitiyiming Y., Chen E., Yang C., Naranmandura H. Synthesis and characterization of CD133 targeted aptamer-drug conjugates for precision therapy of anaplastic thyroid cancer. Biomater. Sci. 2021;9:1313–1324. doi: 10.1039/d0bm01832e. [DOI] [PubMed] [Google Scholar]
- 46.Koudrina A., McConnell E.M., Zurakowski J.A., Cron G.O., Chen S., Tsai E.C., DeRosa M.C. Exploring the unique contrast properties of aptamer-gadolinium conjugates in magnetic resonance imaging for targeted imaging of thrombi. ACS Appl. Mater. Interfaces. 2021;13:9412–9424. doi: 10.1021/acsami.0c16666. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura K., Akagi S., Ejiri K., Yoshida M., Miyoshi T., Sakaguchi M., Amioka N., Suastika L.O.S., Kondo M., Nakayama R. Inhibitory effects of RAGE-aptamer on development of monocrotaline-induced pulmonary arterial hypertension in rats. J. Cardiol. 2021;78:12–16. doi: 10.1016/j.jjcc.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Mashreghi M., Zamani P., Karimi M., Mehrabian A., Arabsalmani M., Zarqi J., Moosavian S.A., Jaafari M.R. Anti-EpCAM RNA aptamer-conjugated liposomal doxorubicin as an efficient targeted therapy in mice bearing colon carcinoma tumor model. Biotechnol. Prog. 2020;37:e3116. doi: 10.1002/btpr.3116. [DOI] [PubMed] [Google Scholar]
- 49.Wu J., Tian Y., He L., Zhang J., Huang Z., Luo Z., Duan Y. An efficient localized catalytic hairpin assembly-based DNA nanomachine for miRNA-21 imaging in living cells. Analyst (Lond.) 2021;146:3041–3051. doi: 10.1039/d1an00001b. [DOI] [PubMed] [Google Scholar]
- 50.Taghdisi S.M., Danesh N.M., Ramezani M., Lavaee P., Jalalian S.H., Robati R.Y., Abnous K. Double targeting and aptamer-assisted controlled release delivery of epirubicin to cancer cells by aptamers-based dendrimer in vitro and in vivo. Eur. J. Pharm. Biopharm. 2016;102:152–158. doi: 10.1016/j.ejpb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 51.Boyacioglu O., Stuart C.H., Kulik G., Gmeiner W.H. Dimeric DNA aptamer complexes for high-capacity-targeted drug delivery using pH-sensitive covalent linkages. Mol. Ther. Nucleic Acids. 2013;2:e107. doi: 10.1038/mtna.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.