Abstract
Hepatitis G virus (HGV) RNA was detected in 18 of 133 pregnant women from Tanzania without known risk factors for HGV infection and in 7 of 18 children born to HGV RNA-positive mothers. Molecular evidence of mother-to-infant transmission was obtained only for three of seven children. HGV RNA was also detected in 4 of 42 children born to non-HGV-infected women. Thus, mechanisms other than materno-filial may play an important role in HGV transmission during early childhood.
Infection with hepatitis G virus (HGV) (12), or GB virus C (20), is frequently detected in persons exposed to blood-borne infections and in patients with liver diseases (2, 6, 9, 11–13, 17, 20). Worldwide, the prevalence of HGV infection in blood donors ranges from 0.9 to 10% (12, 13, 17, 20, 21). However, little information is available about the epidemiologic characteristics of HGV infection on the African continent (2, 23).
Transmission of HGV by blood transfusion has already been demonstrated (18), and a role of the sexual route in the spread of HGV has also been suggested (6, 11, 21). Mother-to-infant transmission of HGV, which has been confirmed at the molecular level in a single case (7), has been previously described (5, 24). In all these reports, HGV-infected mothers were coinfected with hepatitis C virus (HCV) or human immunodeficiency virus (HIV).
We analyzed the presence of HGV RNA in 133 pregnant women from Tanzania and in 15 of the 18 children born to HGV-infected mothers. Samples from the remaining three children were not available due to migration (two cases) or death (one case). The presence of HGV RNA was tested for at 5 and 18 months of age in 11 children, at 5 months in 2, and at 18 months in 2, since only these samples were available. Forty-two children born to randomly selected HGV RNA-negative mothers were also tested for the presence of HGV RNA at the same ages. The demographic and geographical characteristics of the area of Ifakara (Tanzania) studied have been previously described (14). The prevalence of infection with hepatitis B virus, HCV, and hepatitis E virus and the risk for mother-to-infant transmission of hepatitis viruses, as well as the prevalence of antibodies to HIV, have been reported elsewhere (15).
Specific amplification of the NS3 region of HGV by reverse transcription-PCR, HGV RNA titration, and sequencing and phylogenetic analysis of HGV were performed as previously reported (8, 11, 13, 17).
Differences between groups were analyzed by use of the chi-square test and the Fisher’s exact test for categorical variables and the Mann-Whitney U test for quantitative variables.
HGV RNA was detected in 18 of the 133 women (13.5%) analyzed. Hepatitis B surface antigen was detected in 8 mothers, antibodies to HCV were found in 2, and antibodies to HIV were found in 1. The titer of HGV RNA in infected mothers varied between 102 and 106 genome equivalents per ml, but it was similar in women who transmitted the infection to their children and in those who did not (P, 0.8).
At 5 months of age, HGV RNA was detected in 4 of 13 infants (31%) born to HGV RNA-positive mothers and in 2 of 42 infants (5%) born to HGV RNA-negative mothers (odds ratio, 6.5; 95% confidence interval, 0.8 to 76.2; Fisher’s exact test; P, 0.04). At 18 months, HGV RNA was detected in 5 of 13 babies (38.5%) and in 2 of 42 babies (5%) born to HGV RNA-positive and HGV RNA-negative women, respectively (odds ratio, 8.1; 95% confidence interval, 1.1 to 90.4; Fisher’s exact test; P, 0.02).
Among children born to HGV RNA-positive mothers, HGV RNA remained detectable at 18 months of age in two of the four infants who were positive for HGV RNA at 5 months. No sample was available for retesting from the remaining two children. HGV RNA became detectable at 18 months of age in three children who had tested negative for HGV RNA at 5 months. One of these three children had received a blood transfusion, and another was hospitalized on several occasions before HGV RNA was detected. In the remaining children, no risk factors could be identified.
Among children born to HGV RNA-negative mothers, the two children with detectable HGV RNA at 5 months became negative at 18 months, whereas HGV RNA became detectable at 18 months in two children negative for HGV RNA at 5 months. It should be noted that the detection of a humoral immune response against HGV usually implies the clearance of HGV RNA from sera (22).
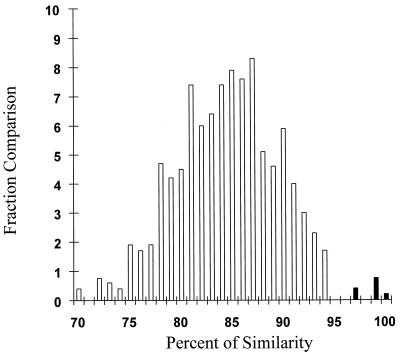
The genetic heterogeneity of HGV was analyzed by direct sequence analysis of 31 HGV RNA-positive samples from mothers and children (Fig. 1). Pairwise comparison between samples showed a close relationship in three mother-child pairs. The genetic heterogeneity ranged from 97 to 100% similarity in these three groups of samples (black bars in Fig. 1) and from 70 to 94% similarity in the remaining samples (white bars in Fig. 1). The observed intersubject NS3 sequence heterogeneity is similar to that described previously (8, 16).
FIG. 1.
Distribution of nucleotide similarities within HGV sequences. For this analysis, the percentage of nucleotide distances was rounded off to the nearest percentage, and the total number of pairwise comparisons (y axis) with a given similarity was represented as a histogram. Each sequence was compared to every other sequence. Black bars represent the percent similarity among mother-child pairs (7, 11, and 14) in which vertical transmission occurred.
Sequential samples from the two children showing the persistence of HGV RNA in serum were studied. Three nucleotide changes between isolates were detected in one child, and no nucleotide substitutions were detected in the other. Thus, the rate of nucleotide substitution in the NS3 region observed in this study is also in agreement with previous observations (8).
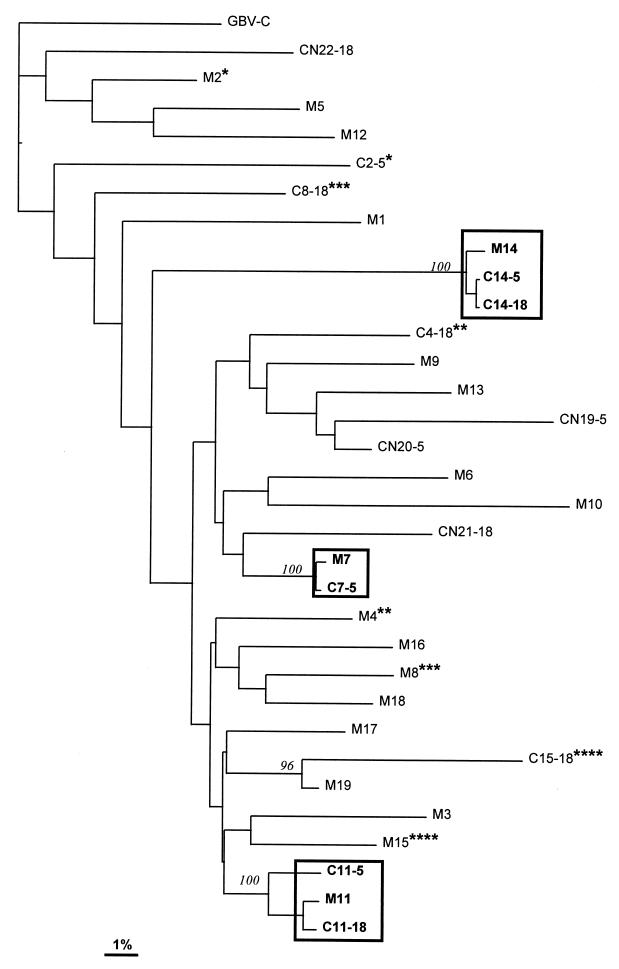
Phylogenetic reconstruction of the 31 isolates showed that three clusters of sequences (corresponding to the three mother-infant pairs mentioned above) had 100% bootstrap values (Fig. 2). None of the remaining clusters reached an 80% bootstrap value. Analysis of isolates from the remaining four children born to HGV RNA-positive mothers showed that their sequences were unique, and the level of sequence divergence between isolates from these children and isolates from their mothers was similar to that observed among isolates in the whole series analyzed. These data suggest that materno-filial transmission of HGV did not occur in these four cases.
FIG. 2.
Neighbor-joining tree for the HGV NS3 region. GB virus C (GBV-C) (20) was used as an outgroup to root the tree. Branch lengths are drawn to scale. The scale bar corresponds to 1% nucleotide sequence divergence. Numbers in italics represent bootstrap proportions in support of the adjacent node based on 100 resampling iterations. Only bootstrap values greater than 80% are shown. Boxes represent mother-child pairs for which phylogenetic analysis supports vertical transmission, and asterisks denote mother-child pairs for which mother-to-infant transmission is not supported. (Different numbers of asterisks correspond to different mother-child pairs.) For isolates from children born to HGV RNA-positive (C) and -negative (CN) mothers (M), the sample time points in months after birth are shown after the subject numbers.
Four of 42 children born to HGV RNA-negative women had detectable HGV RNA in serum, suggesting that HGV may spread through mechanisms other than vertical transmission. HGV RNA has been isolated from saliva (1) and semen (19); therefore, crowding might facilitate horizontal transmission of the virus.
In the three instances of documented vertical transmission analyzed here, the NS3 sequences in isolates obtained from children 5 months after birth differed from those of the maternal HGV isolates in 2, 5, and 5 nucleotides. In spite of being in equilibrium, an RNA viral population exists as a distribution of mutant genomes termed quasispecies (10). Viral genetic variability is generated mainly through random mutational events, but changes in the consensus sequence will be dictated by environmental selective forces, by the stochastic generation of mutants more fit than their parental counterpart, or by random sampling events (bottleneck transmission [3]), which often produce a population with a genetic composition different from that of the parental population (3, 4). Our data suggest that random sampling events during intersubject viral transmission can modify the distribution of variants in the HGV quasispecies population.
In conclusion, our study shows that the prevalence of HGV RNA among a population of pregnant women from Tanzania without known risk factors for parenterally or sexually transmitted infections is remarkably high. HGV can be transmitted from infected mothers to their children, as demonstrated at the molecular level, but mechanisms other than vertical transmission may also play a role in HGV infection in young children.
Nucleotide sequence accession numbers.
HGV sequences have been submitted to GenBank under accession no. AF099442 to AF099472.
Acknowledgments
M.G.-B. was supported by Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS). This work was supported in part by grants from CICYT (SAF 97-0103), FIS (99/0277), Generalitat de Catalunya, the Spanish Agency for International Cooperation, and the UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases. The Ifakara Health Research and Development Centre and the St. Francis Designated District Hospital receive major core funding from the Swiss Agency for Development and Cooperation.
We thank the parents and guardians of all study children. We are also grateful to the staff of the Ifakara Health Research and Development Centre and the St. Francis Designated District Hospital. We thank M. A. Martínez for valuable discussions during preparation of the manuscript.
REFERENCES
- 1.Chen M, Sonnerborg A, Johansson B, Sallberg M. Detection of hepatitis G virus (GB virus C) RNA in human saliva. J Clin Microbiol. 1997;35:973–975. doi: 10.1128/jcm.35.4.973-975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson G J, Schlauder G G, Pilot-Matias T J, Thiele D, Leary T P, Murphy P, Rosenblatt J E, Simons J N, Martinson F E A, Gutiérrez R A, Lentino J R, Pachucki C, Muerhoff A S, Widell A, Tegtmeier G, Desai S, Mushahwar I K. Prevalence studies of GB virus-C infection using reverse transcriptase-polymerase chain reaction. J Med Virol. 1996;50:97–103. doi: 10.1002/(SICI)1096-9071(199609)50:1<97::AID-JMV16>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.Duarte E, Clarke D, Moya A, Domingo E, Holland J J. Rapid fitness losses in mammalian RNA virus clones due to Muller’s ratchet. Proc Natl Acad Sci USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escarmis C, Dávila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller’s ratchet in an RNA virus. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 5.Feucht H H, Zollner B, Polywka S, Laufs R. Vertical transmission of hepatitis G. Lancet. 1996;347:615–616. [PubMed] [Google Scholar]
- 6.Fiordalisi G, Bettinardi A, Zanella Y, Stellini R, Paraninfo G. Parenteral and sexual transmission of GB virus C and hepatitis C virus among human immunodeficiency virus-positive patients. J Infect Dis. 1996;175:1025–1026. [PubMed] [Google Scholar]
- 7.Fischler B, Lara C, Chen M, Sonnerborg A, Nemeth A, Salberg M I. Genetic evidence for mother-to-infant transmission of hepatitis G virus. J Infect Dis. 1997;176:281–285. doi: 10.1086/517267. [DOI] [PubMed] [Google Scholar]
- 8.Giménez-Barcons M, Ibañez A, Tajahuerce A, Sánchez-Tapias J M, Rodés J, Martínez M A, Saiz J C. Genetic evolution of hepatitis G virus in chronically infected individual patients. J Gen Virol. 1998;79:2623–2629. doi: 10.1099/0022-1317-79-11-2623. [DOI] [PubMed] [Google Scholar]
- 9.Guilera M, Sáiz J C, López-Labrador F X, Olmedo E, Ampurdanés S, Forns X, Bruix J, Parés A, Sánchez-Tapias J M, Jiménez de Anta M T, Rodés J. Hepatitis G virus infection in chronic liver disease. Gut. 1998;42:107–111. doi: 10.1136/gut.42.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland J J. Genetic diversity of RNA viruses. Curr Top Microbiol Immunol. 1992;176:1–10. [PubMed] [Google Scholar]
- 11.Ibañez A, Giménez-Barcons M, Tajahuerce A, Tural C, Sirera G, Clotet B, Sánchez-Tapias J M, Rodés J, Matínez M A, Saiz J C. Prevalence and genotypes of GB virus C/hepatitis G virus (GBV-C/HGV) and hepatitis C virus (HCV) among human immunodeficiency virus (HIV) infected patients: evidence of GBV-C/HGV sexual transmission. J Med Virol. 1998;55:293–299. doi: 10.1002/(sici)1096-9071(199808)55:4<293::aid-jmv7>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 12.Linnen J, Wages J, Zhoug-Keck Z Y, Fry K E, Krawczynski K, Alter H J, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W K, Young L M P, Jr, Hoover C, Fernández J, Chen S, Zou J C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and association of GB virus C/hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 13.Masuko K, Mitsui T, Iwano K, Yamazaki C, Okuda K, Meguro T, Murayama M, Inoue T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. Infection with hepatitis GB virus C in patients on maintenance hemodialysis. N Engl J Med. 1996;334:1485–1490. doi: 10.1056/NEJM199606063342301. [DOI] [PubMed] [Google Scholar]
- 14.Menendez C, Kahigwa E, Hirt R, Vounatsou P, Aponte J J, Font F, Acosta C J, Schellenberg D M, Galindo C M, Kimario J, Urassa H, Brabin B, Smith T A, Kitua A Y, Tanner M, Alonso P L. Randomised placebo-controlled trial of iron supplementation and malaria chemoprophylaxis for prevention of severe anemia and malaria in Tanzanian infants. Lancet. 1997;350:844–850. doi: 10.1016/S0140-6736(97)04229-3. [DOI] [PubMed] [Google Scholar]
- 15.Menendez, C., J. M. Sánchez-Tapias, E. Kahigwa, H. Mshinda, J. Costa, J. Vidal, C. Acosta, F. X. López-Labrador, E. Olmedo, M. Navia, M. Tanner, J. Rodés, and P. L. Alonso. Prevalence and mother-to-infant transmission of hepatitis viruses B, C and E in southern Tanzania. J. Med. Virol., in press. [DOI] [PubMed]
- 16.Pickering J M, Thomas H C, Karayiannis P. Genetic diversity between hepatitis G virus isolates: analysis of nucleotide variation in the NS-3 and putative “core” peptide genes. J Gen Virol. 1997;78:53–60. doi: 10.1099/0022-1317-78-1-53. [DOI] [PubMed] [Google Scholar]
- 17.Saiz J C, Ampurdanés S, Olmedo E, López-Labrador F X, Forns X, Guilera M, Tassies D, Costa J, Sánchez-Tapias J M, Jiménez de Anta M T, Rodés J. Hepatitis G virus infection in chronic hepatitis C: frequency, features and response to interferon. J Hepatol. 1997;26:787–793. doi: 10.1016/s0168-8278(97)80243-7. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt B, Korn K, Fleckenstein B. Molecular evidence for transmission of hepatitis G virus by blood transfusion. Lancet. 1996;347:909. doi: 10.1016/s0140-6736(96)91396-3. [DOI] [PubMed] [Google Scholar]
- 19.Semprini A E, Persico T, Thiers V, Oneta M, Tuveri R, Serafini P, Boschini A, Giuntelli S, Pardi G, Brechot C. Absence of hepatitis C virus and detection of hepatitis G virus/GB virus C RNA sequences in the semen of infected men. J Infect Dis. 1998;177:848–854. doi: 10.1086/515257. [DOI] [PubMed] [Google Scholar]
- 20.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of a novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 21.Stark K, Bienzle U, Hess G, Engel A M, Hagenscheid B, Schlüter V. Detection of the GB virus C/hepatitis G virus genome among injecting drug users, homosexual and bisexual men, and blood donors. J Infect Dis. 1996;174:1320–1323. doi: 10.1093/infdis/174.6.1320. [DOI] [PubMed] [Google Scholar]
- 22.Tacke M, Schmolke S, Schleuter V, Sauleda S, Esteban J I, Tanaka E, Kiyosawa K, Alter H J, Schmitt U, Hess G, Ofenloch-Haehnle B, Engel A M. Humoral immune response to E2 protein of hepatitis G virus is associated with long-term recovery from infection and reveals a high frequency of hepatitis G exposure among healthy blood donors. Hepatology. 1997;26:1626–1633. doi: 10.1002/hep.510260635. [DOI] [PubMed] [Google Scholar]
- 23.Tucker T J, Louw S J, Robson S C, Isaacs S, Kirsch R E. High prevalence of GBV-C hepatitis G virus infection in a rural South African population. J Med Virol. 1997;53:225–228. [PubMed] [Google Scholar]
- 24.Viazov S, Riffelmann M, Sarr S, Ballauff A, Meisel H, Roggendorf M. Transmission of GBV-C/HGV from drug-addicted mothers to their babies. J Hepatol. 1997;27:85–90. doi: 10.1016/s0168-8278(97)80284-x. [DOI] [PubMed] [Google Scholar]