Abstract
Microbial interactions are expected to be major determinants of microbiome structure and function. Although fungi are found in diverse microbiomes, their interactions with bacteria remain largely uncharacterized. In this work, we characterize interactions in 16 different bacterial-fungal pairs, examining the impacts of 8 different fungi isolated from cheese rind microbiomes on 2 bacteria (Escherichia coli and a cheese-isolated Pseudomonas psychrophila). Using random barcode transposon site sequencing (RB-TnSeq) with an analysis pipeline that allows statistical comparisons between different conditions, we observed that fungal partners caused widespread changes in the fitness of bacterial mutants compared to growth alone. We found that all fungal species modulated the availability of iron and biotin to bacterial species, suggesting that these may be conserved drivers of bacterial-fungal interactions. Species-specific interactions were also uncovered, a subset of which suggest fungal antibiotic production. Changes in both conserved and species-specific interactions resulted from deletion of a global regulator of fungal specialized metabolite production. This work highlights the potential for broad impacts of fungi on bacterial species within microbiomes.
Despite growing awareness that fungi have an immense capacity to impact ecosystems, fungi are frequently overlooked in microbiome studies1,2. Recently, fungi and other microeukaryotes have received increased attention in sequencing-based studies3–6, and there is growing interest in exploring the roles that fungi and bacterial-fungal interactions play in environmental and host-associated microbiomes7–10. While specific interaction mechanisms have been elucidated for pairwise bacterial-fungal associations, including important pathogenic bacteria and fungi11–14, analyzing a greater diversity of bacterial-fungal interactions could lead to a better ability to predict when and how these interactions contribute to microbiome diversity and function. However, broader characterization of bacterial-fungal interactions has been challenging given the complexity of many microbiomes.
Cheese rind biofilms have been developed as experimentally tractable systems to study microbiomes. These multi-kingdom biofilms form on the surface of cheese during the aging process. Prior work in this system has demonstrated that fungi can impact bacterial growth15. For example, fungi were shown to cross-feed amino acids to bacteria when grown on a cheese-based medium16. Fungal hyphal networks can also alter rind microbiome community composition by providing a means of dispersal for certain community members17.
Here, we combined the high-throughput genetic screening method RB-TnSeq18 with RNA-Seq, bacterial cytological profiling, and metabolomics to investigate bacterial-fungal interactions. Building on existing tools, we created a customized computational RB-TnSeq18 pipeline that enabled us to specifically examine the differences in bacterial growth alone versus in the presence of a fungal partner to highlight pathways important during interactions. We examined pairwise combinations of eight fungal species isolated from cheese rinds (2 yeasts and 6 filamentous fungi) and two bacteria, Pseudomonas psychrophila str. JB418 and E. coli K12.
We observed broad changes in bacterial mutant fitness in the presence of fungi compared to growth alone. A consistent impact across fungal species was the alleviation of E. coli’s requirement for its own siderophore, enterobactin. Further genetic analysis suggests that this alleviation is due to uptake of siderophores produced by filamentous fungi. We observed similar alleviation when E. coli was grown with soil and skin fungi, suggesting that fungal siderophores may impact bacterial growth in other systems. In addition, we find evidence that all fungi tested increase the need for biotin biosynthesis in both E. coli and P. psychrophila. Further, multiple lines of evidence suggest antimicrobial production by several filamentous fungal species. Deletion of laeA, a gene encoding a global regulator of fungal specialized metabolite production, led to a large decrease in the number of impacted pathways in bacteria, suggesting that specialized metabolites play a significant role in bacterial-fungal interactions.
RESULTS
Characterization of bacterial genes with differential fitness in the presence of fungal partners
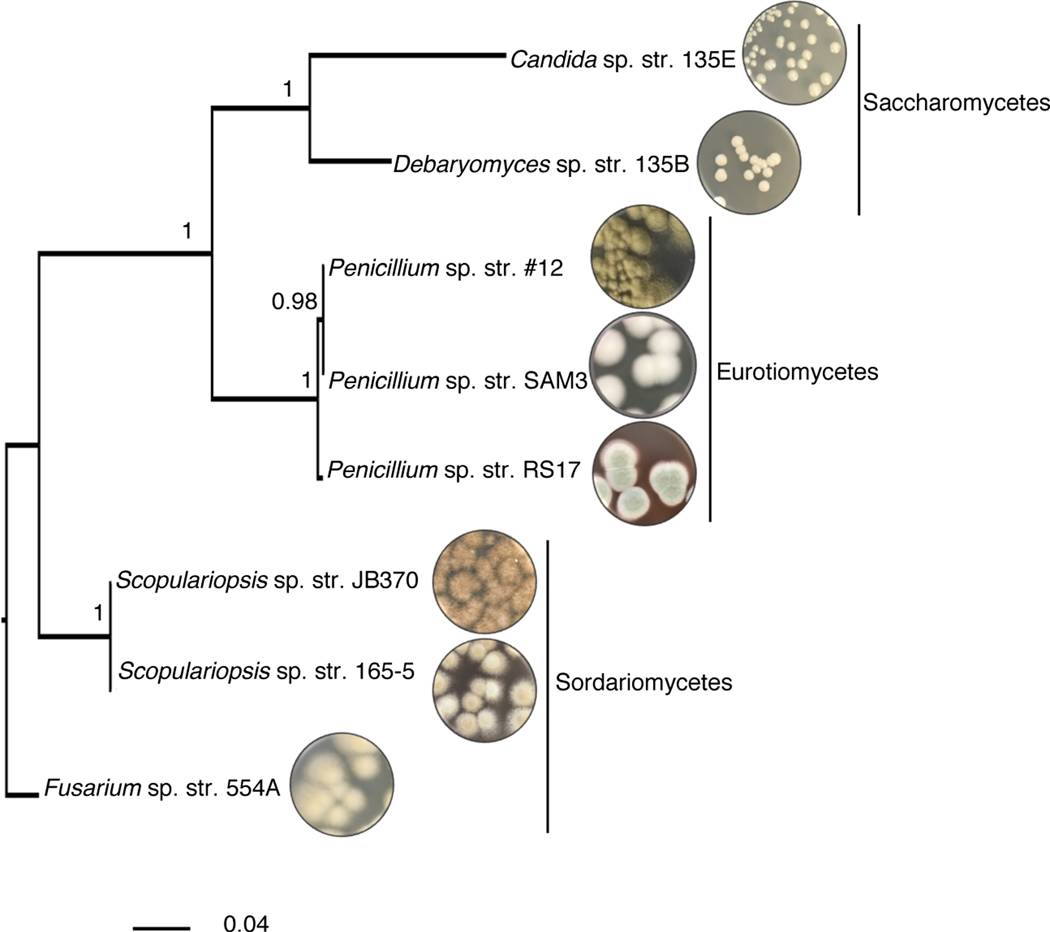
We selected a panel of 8 fungi commonly found in cheese rind microbiomes, all of which belong to the phylum Ascomycota (Figure 1). They include two yeasts, Candida sp. str. 135E and Debaryomyces sp. str. 135B, and six filamentous fungi, Penicillium sp. str. #12, Penicillium sp. str. SAM3, Penicillium sp. str. RS17, Fusarium sp. str. 554A, Scopulariopsis sp. str. JB370, and Scopulariopsis sp. str. 165–5. These genera are also found in the human gut mycobiome19, in soil microbiomes20, in other agricultural microbiomes21, and in marine environments22.
Figure 1: Fungal interaction partners span the phylogenetic and morphological diversity of the cheese ecosystem.
Phylogenetic tree based on large subunit rRNA of the cheese fungi used as interaction partners in this study. The tree was built using Bayesian phylogenetic inference with MrBayes68 and the Jukes and Cantor substitution model98. Branch labels display posterior probability.
The bacterial interaction partners selected were two species of Gammaproteobacteria, Pseudomonas psychrophila str. JB418 and Escherichia coli K12 BW25113. Proteobacteria are common inhabitants of cheese rind communities and have been shown to be responsive to the presence of fungi in experimental community conditions15 (Supplementary Figure 1 and Supplementary Figure 2). P. psychrophila str. JB418 was originally isolated from a cheese rind. E. coli K12 was selected as a bacterial partner to take advantage of the genetic resources available for this organism. Further, E. coli can be a causative agent of foodborne illness in cheese and other foods23,24.
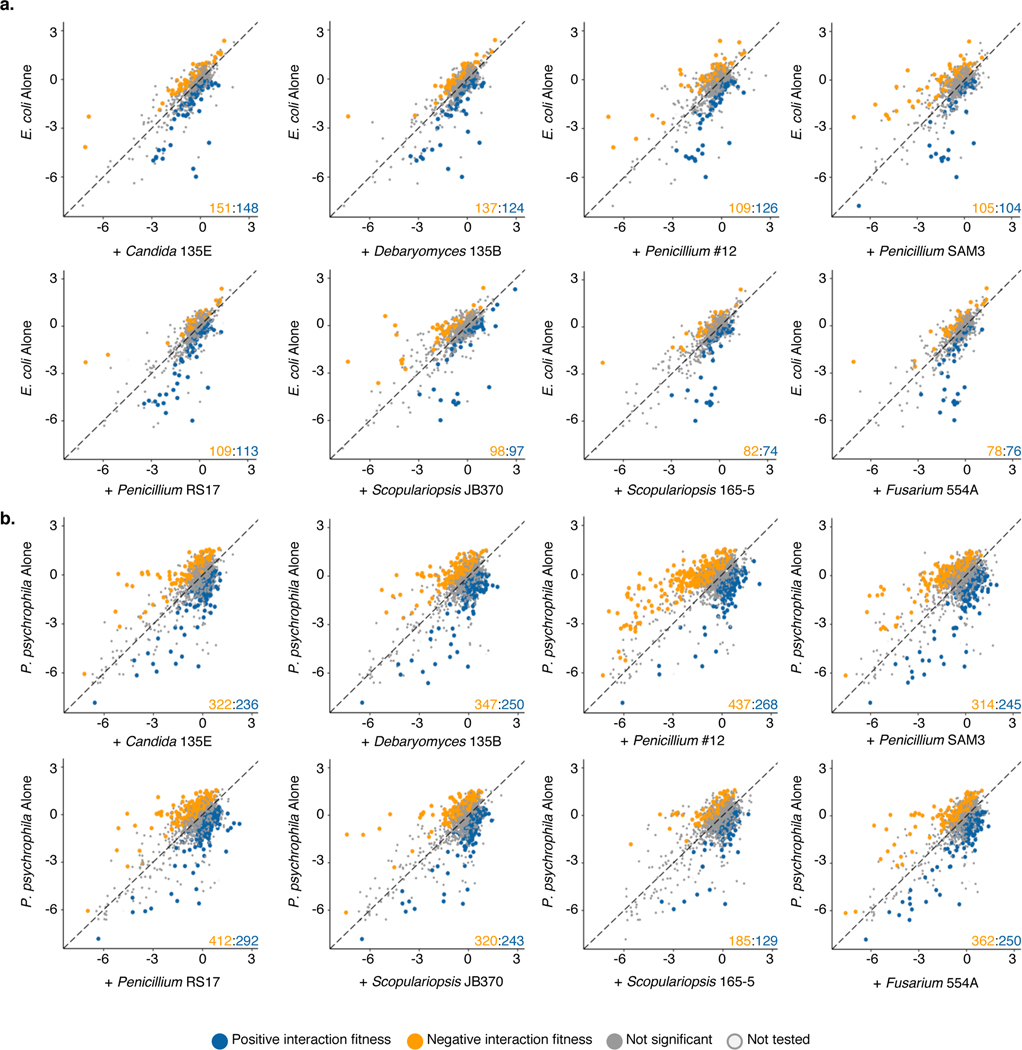
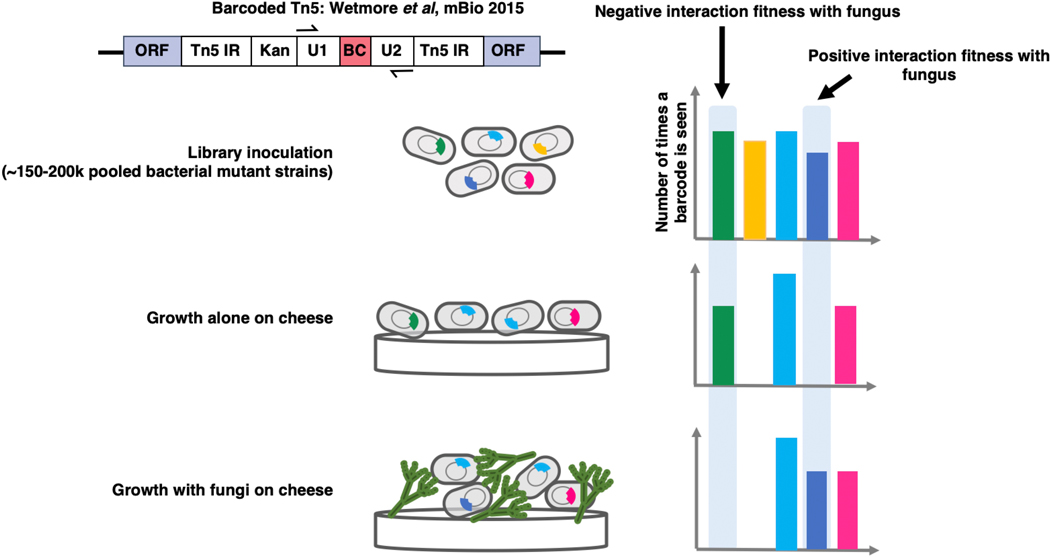
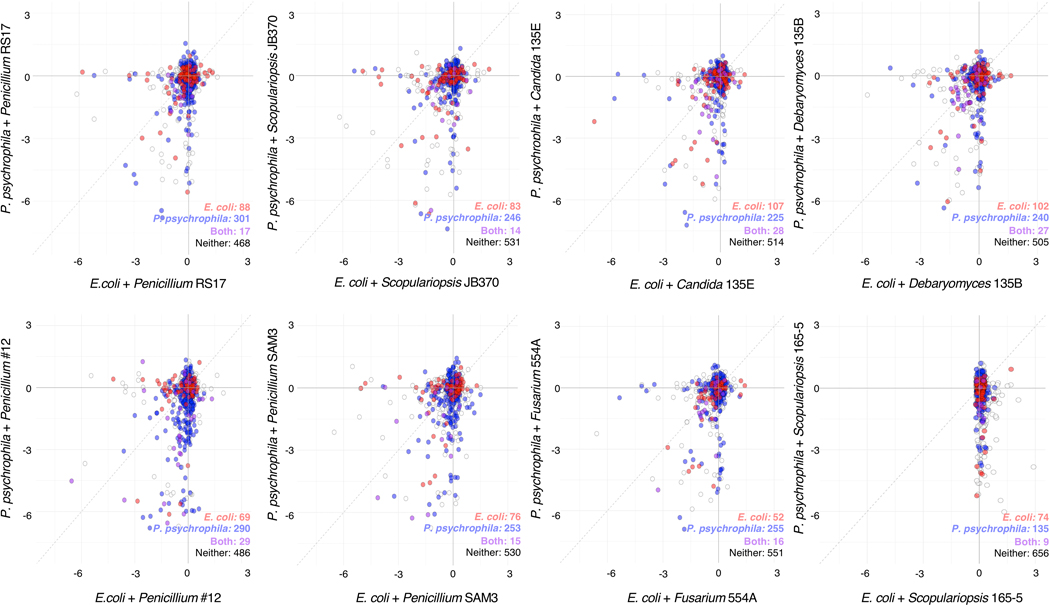
Using a pooled library of barcoded transposon-insertion mutants, RB-TnSeq18 generates a fitness value for each gene, reflecting the importance of a gene for survival in the experimental condition. A negative fitness value indicates that disruption of a given gene leads to decreased growth relative to a wild-type strain, and a positive value indicates enhanced growth, with values further from 0 indicating stronger impacts of gene disruption. To identify bacterial mutants with a significantly different fitness value in the presence of a fungal partner compared to growth alone, pooled P. psychrophila16 or E. coli18 RB-TnSeq mutant libraries were grown for seven days on solid cheese curd agar (CCA) plates25 either alone or mixed with one of the eight fungal species. As sporulation is associated with production of many fungal specialized metabolites, we selected the seven day time point in order to capture interactions related to these metabolites26. A custom computational pipeline allowed us to quantitatively compare fitness values between conditions (Extended Data 1, Supplementary Method 1, https://github.com/DuttonLab/RB-TnSeq-Microbial-interactions). The difference between these fitness values is hereafter referred to as “interaction fitness” (Figure 2, Extended Data 2). In some cases, the presence of a fungus increases the fitness of a mutant (positive interaction fitness), whereas in others the fitness of a mutant is decreased (negative interaction fitness) (Supplementary Table 1, Supplementary Table 2).
Figure 2: Comparison of bacterial gene fitness with fungi against growth alone and identification of bacterial genes with significant interaction fitness across fungal partners.
a, E. coli. b, P. psychrophila. Gene fitness has been calculated for each gene during growth with a fungal partner (x-axis) and during growth alone (y-axis). Each point represents a gene, with colored points indicating genes with a significant difference between gene fitness during growth alone versus with a fungal partner identified by a two-sided t-test and an adjusted p-value lower than 5% using Benjamini-Hochberg correction for multiple comparison testing75. This difference is hereafter referred to as interaction fitness. Exact p-values are available in Supplementary Tables 1 and 2. The colored numbers in the lower right-hand corner indicate how many genes have either positive (blue) or negative (orange) interaction fitness. Positive interaction fitness indicates that a gene fitness value is significantly higher in the presence of the fungal partner compared to growth alone while negative interaction fitness indicates a lower fitness value in the presence of the fungal partner. Non-significant points are plotted smaller to aid in visualization of significant genes. Genes not included in the t-test are labeled as not tested.
In total, we found 731 E. coli and 1606 P. psychrophila genes whose disruption leads to a change in fitness in the presence of at least one of the fungal partners (Supplementary Table 3, Supplementary Table 4). This represents an average of 216 ± 50 E. coli genes per fungal condition and 576 ± 122 P. psychrophila genes per fungal condition that have an interaction fitness. For E. coli, interaction fitness values range from −5.66 to 5.71, and for P. psychrophila, −6.18 to 5.74, highlighting the large positive and negative impacts of fungi on bacteria.
To assess conservation of fitness effects, we identified homologous genes between E. coli and P. psychrophila (Supplementary Table 5 and Supplementary Table 6, Extended Data 3). The set of 88 genes with interaction fitness for both E. coli and P. psychrophila in the same set of fungal conditions is enriched for genes in amino acid biosynthesis, including isoleucine/valine biosynthesis. The isoleucine/valine biosynthesis genes have negative interaction fitness with both bacteria, suggesting that fungi may be competing for amino acids available from cheese (Supplementary Table 7). We have previously seen that these genes are important for E. coli growth alone on cheese, suggesting that these amino acids may be limited in this medium16.
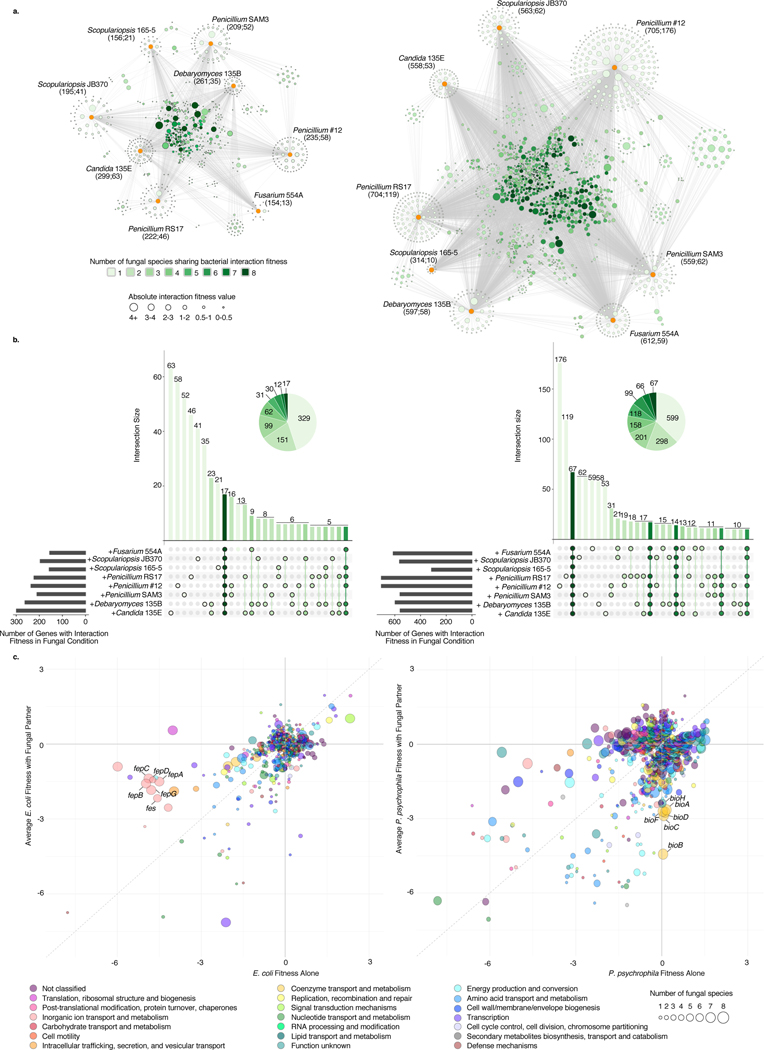
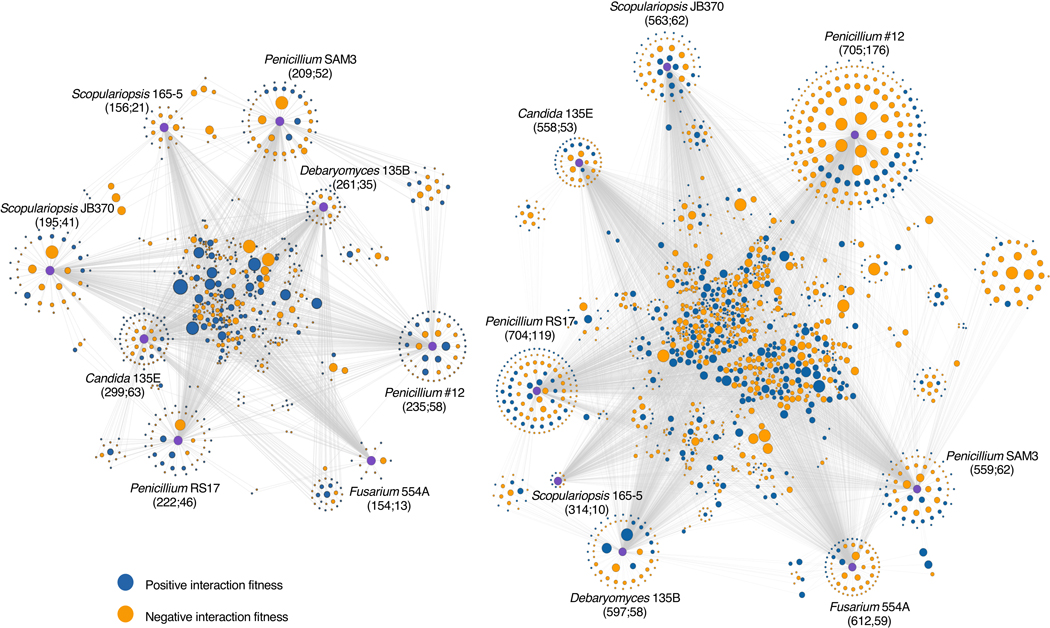
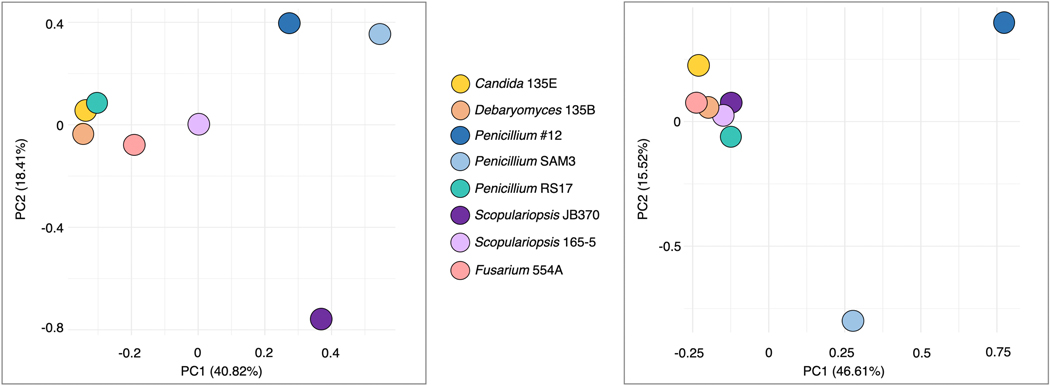
To assess the specificity of fungal impacts on bacteria, we evaluated the intersections of gene sets across the entire set of fungal interaction conditions (Figure 3a, Extended Data 4, Supplementary Table 8, Supplementary Table 9). Around 21 percent (n=152) of the interaction-related genes for E. coli and 32 percent (n=508) for P. psychrophila were common to at least four of the eight fungal interaction conditions (Figure 3b). In addition to conserved effects, we observed a large number of fungal species-specific effects. For E. coli, 45 percent of the genes with interaction fitness are specific to a single fungus (n=329), and for P. psychrophila, 37 percent (n=599). For both E. coli and P. psychrophila, growth with Penicillium sp. str. #12 and Penicillium sp. str. SAM3 results in a large number of the same genes with significant interaction fitness (n=83 and n=318, Figure 3b). These species also cluster away from the other fungi in principal component analysis (Extended Data 5).
Figure 3: Cross-comparison and functional characterization of bacterial genes with interaction fitness in the presence of fungi.
a, Network of E. coli (left) or P. psychrophila (right) genes with an interaction fitness based on RB-TnSeq. Each orange node represents a fungal partner and is labeled as follows: fungal partner (number of genes with interaction fitness; number of genes with interaction fitness unique to this condition). Each green node represents a bacterial gene. Green nodes are shaded by the number of fungal conditions in which this gene has an interaction fitness, as shown in the legend below, and are sized by average strength of interaction fitness across partners. b, UpSet99 plots showing the intersections of E. coli (left) or P. psychrophila (right) gene sets with interaction fitness across fungal partners. These UpSet plots are conceptually similar to Venn Diagrams. The connected circles indicate which fungal conditions are included in the intersection, and the size of the intersection (the number of genes that have an interaction fitness in all the highlighted conditions) is displayed in the main bar chart. The horizontal bar chart displays the number of genes with significant interaction fitness per fungal condition. Intersections <5 genes are not shown for E. coli and <10 genes are not shown for P. psychrophila. For example, in the E. coli panel, 17 genes have an interaction fitness with all partners (all fungi circles are connected), while 16 other genes have an interaction fitness with Penicillium sp. str. #12 and with Penicillium sp. str. SAM3 (only Penicillium sp. str. #12 and Penicillium sp. str. SAM3 circles are connected). Intersections are green color-coded based on the number of fungal partners sharing the interaction as in Figure 3a. c, Comparison of E. coli (left) or P. psychrophila (right) gene fitness values alone compared to fitness values with a fungal partner, colored by COG category and sized by the conservation of effect among fungal partners (1–8 fungal species). Genes discussed further in the text related to E. coli enterobactin uptake and P. psychrophila biotin biosynthesis are labeled.
Penicillium sp. str. #12 and Penicillium sp. str. SAM3 induce bacterial envelope stress
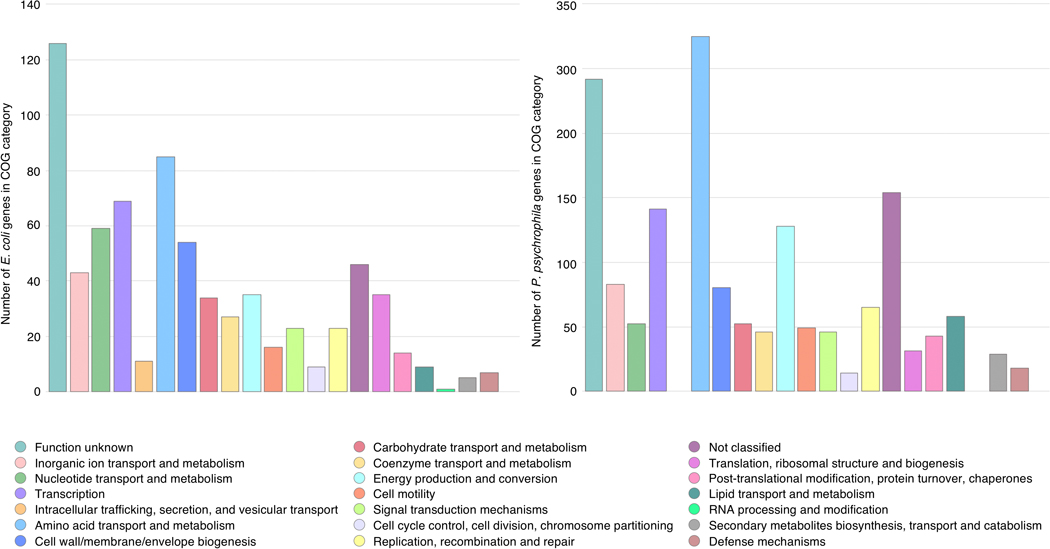
To identify potential mechanisms underlying bacterial-fungal interactions, we used a combination of methods: clusters of Orthologous Genes (COG) categorization, functional enrichment analysis, and analysis of conservation of the effect across fungal species (Figure 3c, Supplementary Table 10, Supplementary Table 11, Extended Data 6). Penicillium sp. str. #12 and Penicillium sp. str. SAM3 consistently shared effects on bacterial mutant fitness, as seen by their large number of network connections (Figure 3a). The gene sets impacted by these fungi suggest that these two fungal species are creating bacterial envelope stress, potentially through production of antibiotic molecules, as they include drug efflux pumps, envelope stress response systems, penicillin-binding proteins, and lipopolysaccharide/peptidoglycan biosynthesis genes (Supplementary Table 12). For example, disruption of the multidrug efflux pump MdtK results in a decreased fitness specifically in the presence of these two fungi (gene fitness alone = 0.34, with Penicillium sp. str. #12= −2.49, with Penicillium sp. str. SAM3 = −1.92).
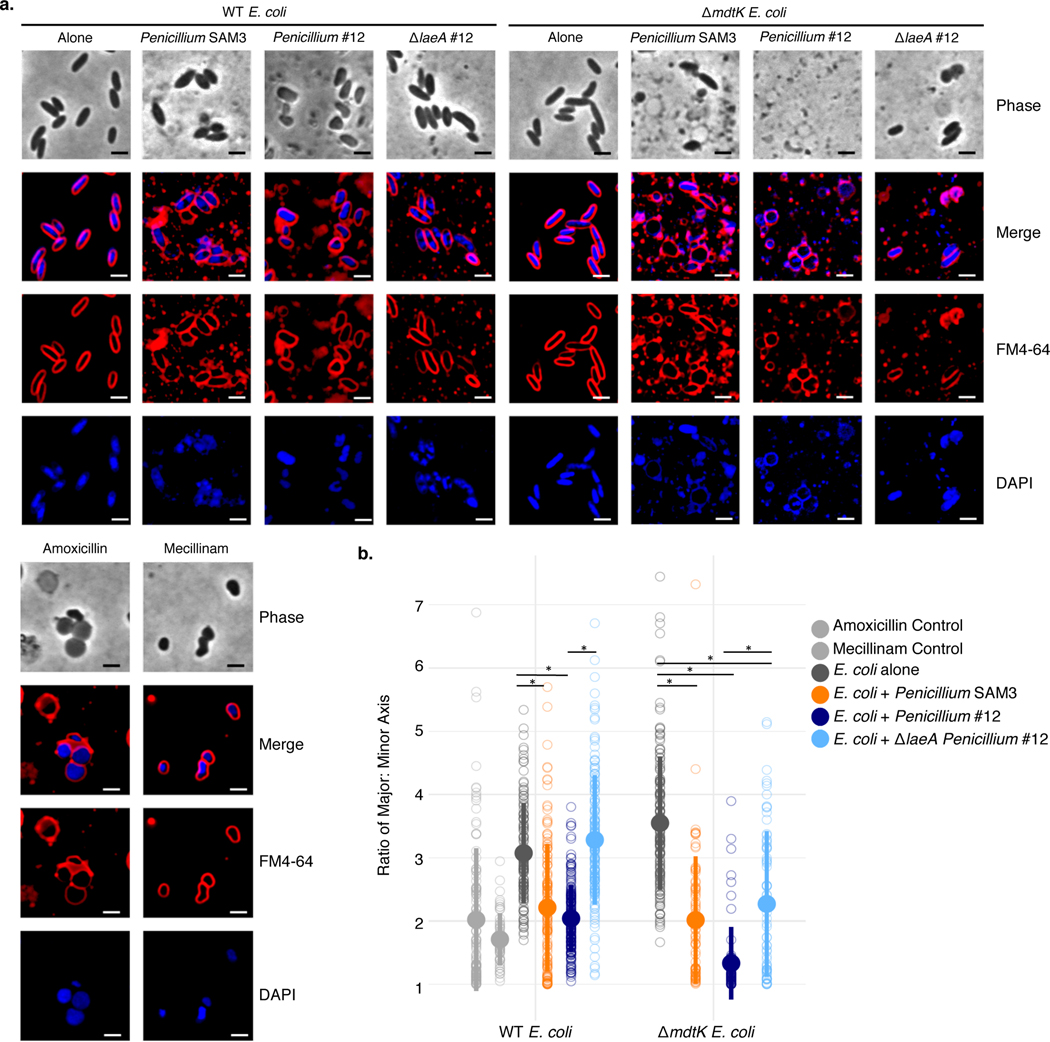
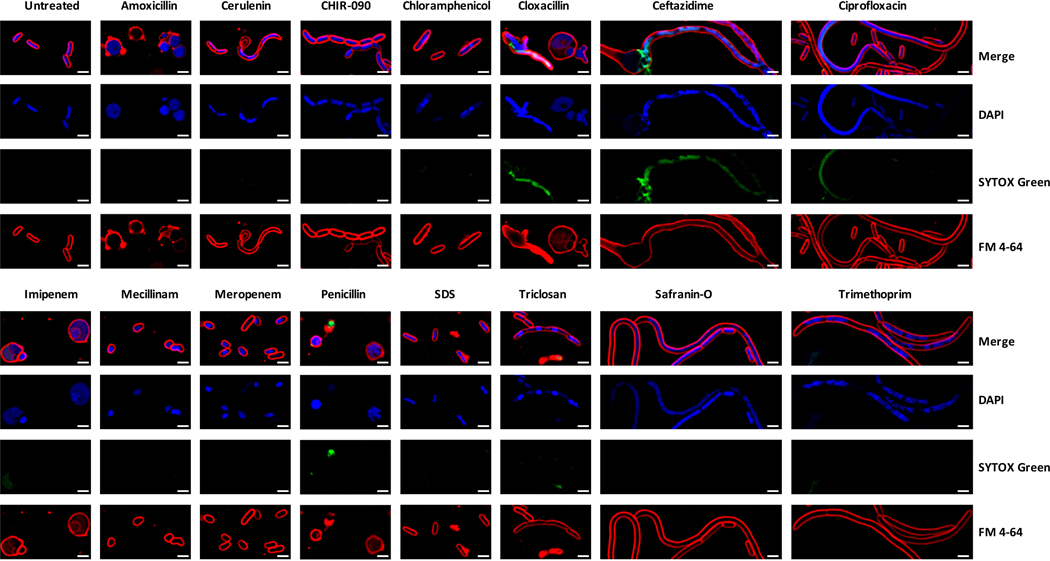
We performed bacterial cytological profiling (BCP)27, a microscopy-based method used to predict the mechanism of action for antibiotics, on WT or ΔmdtK E. coli grown alone or in a mixed biofilm with Penicillium sp. str. #12 or Penicillium sp. str. SAM3. Microscopy showed a strong change in cell morphology for both WT and ΔmdtK E. coli when grown with Penicillium compared to growth alone (Figure 4a). When cultured with these fungi, E. coli cells are significantly more round, consistent with a reduction in cell wall integrity (Figure 4b). ΔmdtK cells are strongly affected and have spheroplasted, indicative of the complete loss of structural integrity. Control experiments with known antibiotic compounds show that this effect is similar to that of antibiotics that target cell wall biosynthesis, such as mecillinam and amoxicillin (Figure 4, Extended Data 7).
Figure 4: Bacterial cytological profiling (BCP) of E. coli grown with Penicillium sp. str. SAM3, Penicillium sp. str. #12, and ΔlaeA Penicillium sp. str. #12 on CCA plates.
a, The phenotype of E. coli grown with these fungi is similar to that seen when E. coli is exposed to antibiotics targeting cell wall biosynthesis. This effect is more dramatic in E. coli lacking the mdtK multidrug efflux pump. Representative fields of deconvoluted images are displayed. DAPI dye stains DNA and FM4–64 dye stains bacterial membranes. Scale bars represent 2 μm. b, Quantification of microscopy results. The major and minor axes of individual cells were measured (all cells in the image for multiple images), and the ratio of these measurements was used as an indicator of cell roundness. Each empty circle represents an individual cell (from left to right, n=110, 53, 121, 136, 181, 144, 153, 79, 70, and 73 cells examined from one independent experiment per condition). The filled circle displays the mean, and the thick bar extending from the mean displays standard deviation. WT E. coli has a ratio of about 3, and the cells become rounder as the ratio approaches 1. Asterisks indicate significantly different roundness in the presence of a fungus relative to growth alone or significantly different roundness in the presence of WT Penicillium sp. str. #12 relative to ΔlaeA Penicillium sp. str. #12 (unpaired two-sample Wilcoxon test p-value < 0.05). Exact p-values: E. coli/Penicillium SAM3- E. coli alone=3.05e-13; E. coli/Penicillium #12- E. coli alone=3.55e-28; E. coli/Penicillium #12- E. coli/ΔlaeA Penicillium #12=5.21e-37; ΔmdtK E. coli/Penicillium SAM3- ΔmdtK E. coli alone=4.13e-25; ΔmdtK E. coli/Penicillium #12- ΔmdtK E. coli alone=1.14e-41; ΔmdtK E. coli/ΔlaeA Penicillium #12- ΔmdtK E. coli alone=2.68e-13; ΔmdtK E. coli/Penicillium #12- ΔmdtK E. coli/ΔlaeA Penicillium #12=1.16e-07.
Previous studies have shown that fungal specialized metabolite production in other ascomycete fungi is controlled by the global regulator, LaeA28,29. To test the contribution of this gene to the potential antibiotic activity, we generated a ΔlaeA mutant in Penicillium sp. str. #12. WT E. coli cells are significantly less rounded when grown with Penicillium sp. str. #12 ΔlaeA, suggesting that loss of this global regulator has potentially decreased fungal antibiotic production (Figure 4). Neither of these two fungal strains are known producers of penicillin, and analysis of the Penicillium sp. str. #12 draft genome failed to detect penicillin biosynthesis gene clusters30. However, both fungi are causing consistent morphological and genetic effects suggesting cell envelope stress similar to that seen with beta-lactam antibiotics.
Fungi increase bacterial need for biotin biosynthesis
P. psychrophila RB-TnSeq data showed that the disruption of genes associated with biotin biosynthesis (bioB, bioD, bioF, bioA, bioH and bioC) results in a negative interaction fitness with most fungi (average fitness alone: 0.08, average fitness across fungi:−2.97) (Figure 3c). This gene set represents all genes needed to synthesize biotin from pimeloyl-CoA. Biotin is present in our cheese curd agar medium at 73 nmol/mg and represents an essential cofactor for enzymes involved in key cellular functions like amino acid metabolism and lipid synthesis31. In E. coli, biotin biosynthesis genes have a neutral fitness alone and do not show interaction fitness in our RB-TnSeq experiments. However, RNA-Seq of WT E. coli grown either alone or in the presence of Penicillium sp. str. #12, a predicted biotin prototroph, showed that bioA, bioB, bioC, bioD, and bioF were all significantly upregulated in the presence of Penicillium sp. str. #12 with an average fold change of 4.4 (Supplementary Table 13). This highlights an increased need for bacterial biotin synthesis in both P. psychrophila and E. coli, suggesting competition for available biotin in the medium, or potentially that bacteria have higher biotin requirements in the presence of fungi.
Fungi increase iron availability for bacterial partners
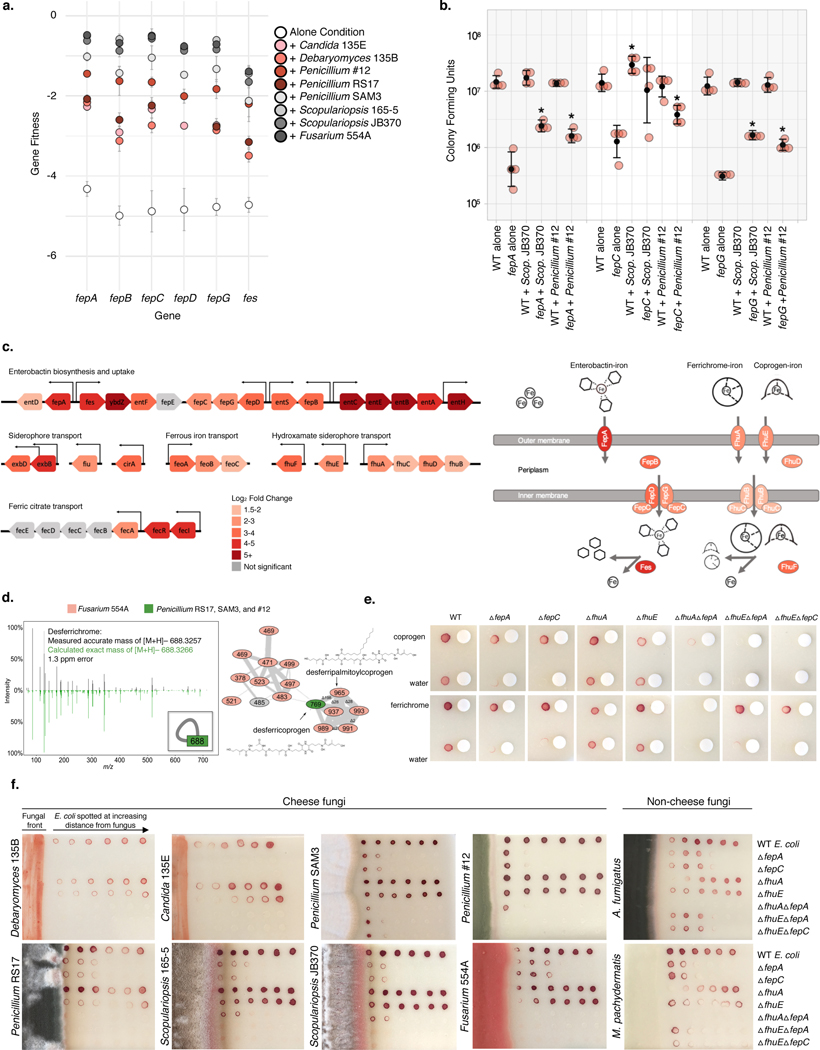
Because cheese is an iron-limited environment (free iron levels approximately 3 ppm32), microbial species require iron chelators such as siderophores to grow16,32,33. Our RB-TnSeq fitness data revealed that E. coli mutants defective in transport of its siderophore, enterobactin, grow poorly in the alone condition (fitness <−4). However, the presence of any fungal partner significantly improves the growth of mutants in the fep operon (fepC, fepG, fepA, fepB), which encodes enterobactin transport functions (average positive interaction fitness 3.11) (Figure 3c, Figure 5a, Supplementary Table 10). The positive effect of fungi on growth was further supported by competitive mutant fitness assays with isolated enterobactin uptake mutants (Figure 5b).
Figure 5: Utilization of fungal siderophores by E. coli.
a, RB-TnSeq fitness values for fep operon genes in alone or with-fungi conditions, showing an increase in fitness in the presence of fungal species. Fitness values are not shown for non-significant differences from alone. Error bars show standard deviation of fitness values from the mean. b, Colony forming units of WT E. coli and Δfep mutants after 7 days of 1:1 competitive growth on CCA, pH 7. Competitions between the two E. coli strains were performed either with no fungus present (“alone”) or with Penicillium sp. str. #12 or Scopulariopsis sp. str. JB370. N=4 biologically independent experiments, error bars show standard deviation from the mean (black circle). Asterisks indicate significantly different growth in the presence of a fungus relative to growth without the fungus (“alone”) based on a two-sided two-sample t-test p-value < 0.05. Exact p-values associated with asterisks (from left to right): 0.001, 0.007, 0.034, 0.022, 0.0001, 0.0006. c, E. coli iron-related genes upregulated in the presence of Penicillium sp. str. #12. Significance cutoff made at abs(log2(fold-change)) >1.5 and adjusted p-value <0.05. Differential expression analysis was performed using the default function DESeq87 which performs a Wald test with Benjamini-Hochberg75 correction for multiple comparison testing. Exact p-values are available in Supplementary Table 14. d, Fungal siderophores identified by mass spectrometry. Inset in the left box shows the node that represents the desferrichrome fragmentation pattern depicted while the network on the right represents coprogen-related molecules. Coprogen B and ferrichrome were found by matching fragmentation patterns to library spectra. Both identifications were confirmed using retention time and fragmentation matching to a purchased standard. e, Visual assays of Δfep mutant growth with purified siderophores coprogen and ferrichrome. f, Visual assays of E. coli mutant growth at varying distances from pre-cultured cheese fungi, A. fumigatus (soil, human pathogen), and M. pachydermatis (skin commensal). For e, and f, growth was performed on CCA. Tetrazolium chloride, a red indicator of cellular respiration, was added to the medium to visualize colony growth on opaque CCA.
Although siderophore production and uptake have not been previously characterized in P. psychrophila str. JB418, three putative iron-related genes have an effect similar to that seen with E. coli fep genes, having a fitness defect alone, but a positive interaction fitness with any fungus: Ga0212129_113525, Ga0212129_115698, and Ga0212129_114260. These genes are annotated as iron complex outer membrane receptor protein, putative iron-dependent peroxidase, and uncharacterized iron-regulated membrane protein, respectively. Immediately upstream of Ga0212129_114260, we find a ferric enterobactin receptor (FepA) and the PfeR-PfeS two-component regulatory system required for the ferric enterobactin receptor34. Although pyochelin and pyoverdine are two well-known Pseudomonas siderophores35, antiSMASH does not predict these siderophores in P. psychrophila.
RNA-Seq analysis of WT E. coli revealed 34 genes (out of a total of 348 significantly upregulated genes) involved in iron acquisition that are specifically upregulated in the presence of Penicillium sp. str. #12 (Supplementary Table 14). We observe upregulation of enterobactin biosynthesis and uptake (ent and fep-associated genes), suggesting that even in the presence of fungi, E. coli still produces and utilizes its native siderophore (Figure 5c). In addition to uptake of enterobactin, E. coli possesses the Fhu system, which enables uptake of hydroxamate-type siderophores, including those produced by fungi, such as ferrichrome and coprogen36,37. Notably, our RNA-Seq data show upregulation of fhuA (outer membrane receptor for ferrichrome) and fhuE (outer membrane receptor for coprogen) in the presence of Penicillium, suggesting that this fungus may alleviate the growth defects seen in the fep mutants through the provision of hydroxamate-type siderophores taken up by the Fhu system.
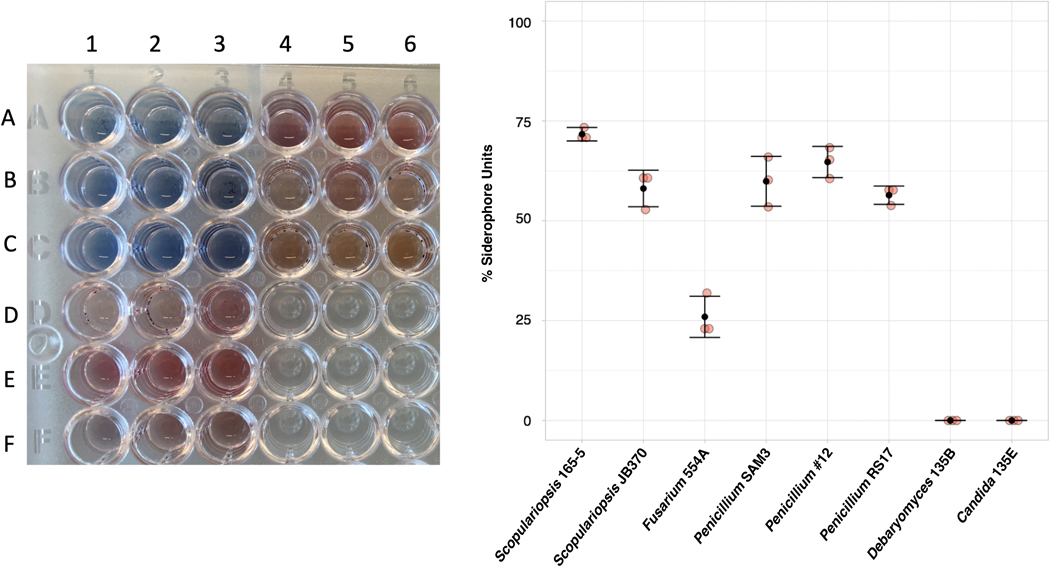
All filamentous fungi in this study, but not yeast, produce siderophores detectable by Chrome Azurol S (CAS) assay (Extended Data 8). In addition, liquid chromatography mass spectrometry (LC-MS and LC-MS/MS) showed evidence of the hydroxamate fungal siderophores coprogen and ferrichrome in Fusarium and Penicillium species (Figure 5d). Although not detected in these extracts, Scopulariopsis sp. str. JB370 is predicted to make dimethylcoprogen based on antiSMASH analysis of the draft genome38.
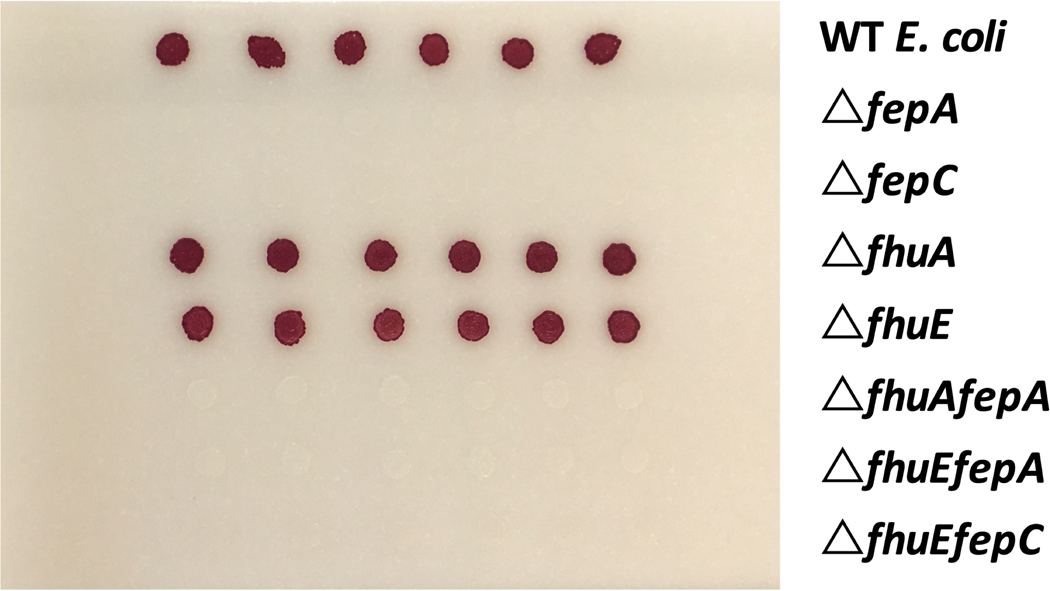
To confirm that hydroxamate siderophores could rescue fep mutants via the Fhu pathway, we verified that purified coprogen and ferrichrome rescue the growth defect of ΔfepA or ΔfepC on CCA (Figure 5e). As expected, mutants of fhuA or fhuE alone do not show a growth defect on CCA, likely because the enterobactin system is intact. Thus, to specifically examine whether these genes are required for uptake of the purified siderophores, we constructed mutants of fhuA or fhuE in an enterobactin-uptake defective background (ΔfepC or ΔfepA). Combined loss of enterobactin uptake and fhuA eliminates the alleviation seen with ferrichrome, whereas loss of either fhuA or fhuE in the ΔfepA background seems to eliminate the alleviation seen with coprogen (Figure 5e). This suggests that E. coli requires fhuA for ferrichrome uptake, and both fhuA and fhuE for coprogen uptake.
We next examined whether the presence of fungal species changed the growth of strains defective in siderophore uptake (Figure 5f, Extended Data 9). Growth of ΔfepA and ΔfepC is restored closest to the filamentous fungi, but not when grown near yeasts. For filamentous fungi, this effect is either fhuA or fhuE dependent. Thus, E. coli is likely to use and benefit from fungal hydroxamate siderophores produced by filamentous fungi that are taken up by the Fhu system independently of the enterobactin uptake system.
Because iron limitation is a common challenge across many environments, we wanted to examine whether fungal species from other ecosystems could also be producing siderophores that are accessible to neighboring bacterial species. We performed similar assays with Aspergillus fumigatus, a soil-dwelling filamentous ascomycete, and Malassezia pachydermatis, a basidiomycete yeast that is a commensal resident on animal skin. Results suggest that A. fumigatus produces a siderophore capable of being imported through FhuA (Figure 5f). We see a similar effect using M. pachydermatis, suggesting that bacteria are able to utilize siderophores from a yeast species using the Fhu system (Figure 5f). We performed AntiSMASH38 analysis on a previously published genome of this Malassezia pachydermatis strain and were able to identify an NRPS biosynthetic gene cluster containing a ferrichrome peptide synthetase38,39. In sum, our results suggest that cheese-associated filamentous fungi, and select fungi from other environments, can reduce bacterial dependence on their own siderophores.
Loss of a fungal secondary metabolite regulator alters the profile of interaction fitness
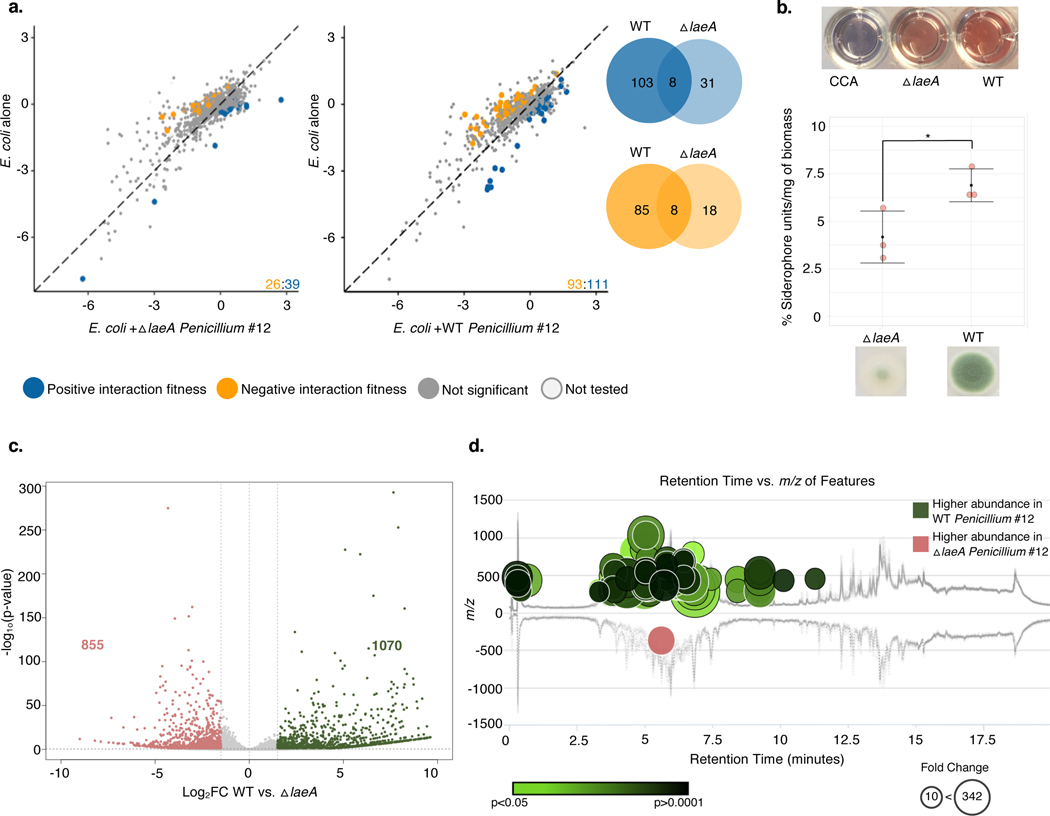
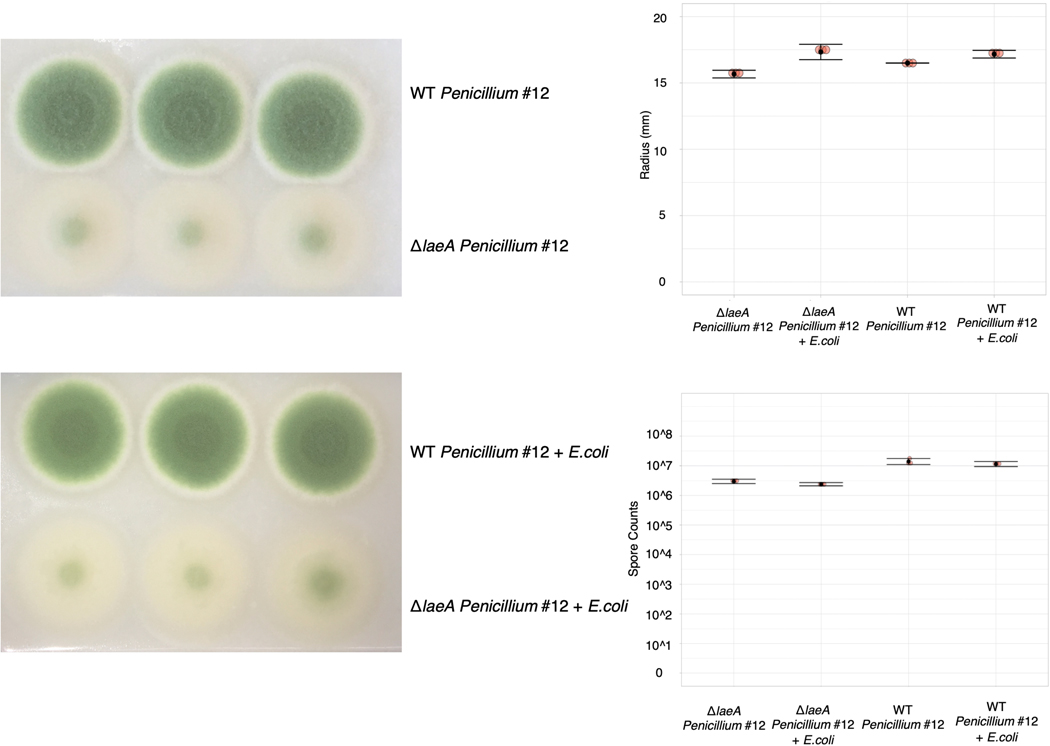
The cases above show that bacterial gene fitness is impacted by the production of fungal specialized metabolites, including siderophores and potentially antibiotics. To determine the extent to which the global regulator LaeA is responsible for fungal-induced changes in bacterial fitness, we performed RB-TnSeq experiments with the Penicillium sp. str. #12 ΔlaeA mutant. Despite comparable fungal growth between WT and ΔlaeA, we see only 65 E. coli genes with interaction fitness when grown with ΔlaeA compared to 204 with WT, suggesting that many of the fitness effects we see may be due to fungal specialized metabolite production (Figure 6a, Extended Data 10).
Figure 6: Fungal metabolite production impacts bacterial-fungal interactions.
a, E. coli genes with significant interaction fitness with ΔlaeA or WT Penicillium sp. str. #12. Each point represents a gene, with colored points indicating genes with interaction fitness. X and Y values (+fungal partner on x-axis; alone on y-axis) indicate gene fitness values in each condition, and the numbers in the lower right hand corner indicate how many genes have either positive (blue) or negative (orange) interaction fitness. Venn diagrams display the overlap of these gene sets. b, Liquid CAS assay of supernatants from blank control CCA medium, ΔlaeA, or WT Penicillium sp. str. #12 normalized to fungal biomass. N=3 biologically independent experiments, error bars show standard deviation from the mean. Asterisk indicates significantly different siderophore production (two-sided two-sample t-test p-value 0.04). c, Differential expression of WT Penicillium sp. str. #12 relative to ΔlaeA after three days of growth on CCA. Labeled on the volcano plot are the number of genes with a log2 fold change of >1.5 (green) or <−1.5 (red) and adjusted p-value of <0.05. Differential expression analysis was performed using the default function DESeq87 which performs a Wald test with Benjamini-Hochberg75 correction for multiple comparison testing. d, The metabolomics data analysis platform XCMS100 was used to compare features detected by LC-MS analyses of ΔlaeA and WT Penicillium sp. str. #12 extracts (two-sided Welch’s t-test for unequal variances). Features of higher abundance in WT relative to ΔlaeA are depicted as green nodes on the top of the mirror plot and features of lower abundance in WT relative to ΔlaeA are depicted as red nodes on the bottom. Node radius is proportional to the fold change of the detected features and color intensity is dependent on p-value. The graph displays only those features with a p-value less than or equal to 0.05, fold change higher than or equal to 10, m/z between 200 and 2000 Da, and intensity higher than 500. Exact p-values are available in Supplementary Table 18.
Given that siderophore production in other fungi is controlled by LaeA, we would expect that E. coli enterobactin uptake mutants would not have positive interaction fitness with the ΔlaeA mutant. Indeed, we no longer see a positive interaction fitness for fes, fepA, fepB, fepC, fepD and fepG genes when E. coli is grown with ΔlaeA Penicillium sp. str. #12 (Supplementary Table 15). Additionally, we see negative interaction fitness for hydroxamate siderophore transport genes fhuB and fhuC with WT Penicillium sp. str. #12 but not with ΔlaeA. Liquid CAS assays demonstrate that ΔlaeA Penicillium sp. str. #12 produces less siderophores than WT on cheese media (Figure 6b). Overall, these results demonstrate that loss of the LaeA regulator decreases siderophore production in Penicillium sp. str. #12 and abolishes the positive interaction effect seen for fep genes grown with WT fungus.
We next examined whether there were changes in the fitness of genes related to responses to antibiotics. A number of genes involved in cell envelope maintenance have a negative interaction fitness with WT but not ΔlaeA Penicillium sp. str. #12 (Supplementary Table 12). These genes include dacA, which encodes penicillin-binding protein 5; loss of this gene has been shown to increase E. coli’s susceptibility to β-lactam antibiotics40. The gene encoding the MdtK efflux protein has improved fitness with ΔlaeA relative to WT Penicillium sp. str. #12. As seen in our BCP analysis, maintenance of cell envelope integrity is important for E. coli growing with WT Penicillium sp. str. #12, but less so in the absence of LaeA.
Based on RNA-Seq, fourteen percent of the Penicillium sp. str. #12 genome was differentially expressed between WT and ΔlaeA (Figure 6c, Supplementary Table 16). This is consistent with previous findings in A. fumigatus that LaeA influenced expression of around 10 percent of the fungal genome41. GO term enrichment analysis identified a number of specialized metabolite biosynthesis pathways overrepresented in the genes more expressed in WT (Supplementary Table 17). Genes in eleven antiSMASH38-predicted biosynthetic gene clusters are downregulated in ΔlaeA, including one terpene cluster, two type I polyketide synthase clusters, and eight non-ribosomal peptide synthetase (NRPS) clusters. Consistent with our findings of decreased siderophore production in ΔlaeA, one NRPS cluster contains four genes with homology to sidD, sidF, sidH, and sitT; these genes are associated with siderophore biosynthesis and transport in Aspergillus42.
LC-MS comparison of WT and ΔlaeA Penicillium sp. str. #12 showed differential production of many metabolites, 94 of which have a >10-fold change between the two (Supplementary Table 18). Of these, 93 are less abundant in the ΔlaeA mutant, which is consistent with the loss of secondary metabolite production in the ΔlaeA mutant (Figure 6d). Cyclopenol, a biosynthetic intermediate for viridicatol43, was the only molecule reported to have antibiotic activity identified in the LC-MS data; it is produced by the WT Penicillium sp. str. #12 in >10-fold higher quantity than ΔlaeA. However, further work is needed to determine if this molecule is related to the antibacterial activity seen in BCP. In sum, these data highlight an important diminution of specialized metabolite production in the ΔlaeA strain.
DISCUSSION
Fungi have the potential to strongly impact bacterial neighbors in diverse systems, from soil to polymicrobial infections44–48. We combined the high-throughput genetic screen RB-TnSeq with BCP, RNA-Seq, and metabolomics to identify a diversity of bacterial genes involved in, and the associated fungal contributors to, bacterial-fungal interactions in our system. This study provides new insight into the wide range of fungal impacts on bacteria that can occur even in a relatively simple system with pairwise combinations of bacteria and fungi. Our study focused on fungi from the Ascomycota because this phylum is abundant in cheese rinds, and therefore represents a small sampling of the full genomic and functional diversity across fungi. It remains to be seen how these interactions might change with a more diverse set of partners, in conditions with varying ratios of bacteria and fungi, in varied environmental conditions, or with increasing community complexity16.
Our study highlighted several important areas that contribute to bacterial-fungal interactions. First, multiple lines of evidence suggest that cheese-isolated Penicillium species exhibit antibiotic-like activity, although the mechanism behind this activity is currently unknown. Although Penicillium species are known to produce a wide-range of specialized metabolites, detection of known antibiotics such as penicillin in food products is limited30. Second, we found an increased need for biotin biosynthesis in the presence of fungi, suggesting fungal competition for available biotin. Previous studies have pointed to roles of B vitamins in bacterial-fungal and even plant-bacterial interactions49,50. Third, the strongest and most widespread bacterial-fungal interaction that we observed suggests that fungal species can dramatically modulate access to iron through the provision of fungal siderophores, such as ferrichrome and coprogen. It has long been known that bacteria grown in isolation are able to uptake purified fungal siderophores, but the ecological relevance of this putative interaction has not been demonstrated36,51. Our results demonstrate that this exchange takes place between bacteria and filamentous fungi growing in a biofilm and that this exchange can have impacts on the competitive fitness of bacteria. Our data suggest that cheese-associated yeast species may alleviate bacterial iron limitation through a different mechanism, as we did not detect siderophore production by these species.
Due to the importance of iron in bacterial physiology and the prevalence of fungi in microbial ecosystems, we expect that iron-based bacterial-fungal interactions are important in other microbiomes. For example, growth of non-siderophore producing mutants of soil-dwelling Streptomyces coelicolor was restored by the presence of siderophores from airborne contaminant Penicillium52. Moreover, many filamentous fungi outside of the genera studied here can produce siderophores53. In addition to filamentous fungi, we showed that the basidiomycete skin yeast Malassezia pachydermatis alleviated bacterial iron limitation. Human skin microbiome yeasts Malassezia restricta and Malassezia globosa have previously been found to possess genes for siderophore biosynthesis54,55. Fungal growth may also be affected by interkingdom siderophore exchange, as some fungal species have evolved mechanisms of utilizing bacterial siderophores while others are inhibited by bacterial siderophores56–58.
FhuE and FhuA receptors are widespread in Proteobacteria, further suggesting that interkingdom siderophore exchange could play an important role in diverse systems. Hydroxamate siderophore uptake systems have also been identified in Gram-positive bacterial pathogens59,60. Additionally, hydroxamate siderophore uptake systems impact bacterial fitness in a murine infection model61. Bacteroides fragilis, a human gut symbiont, is able to use ferrichrome to grow in iron-limiting conditions, and fhu genes are expressed by E. coli in colonic mucus62,63. Fermented foods are known to contain fungal siderophores, which could be a source of fungal siderophores in the gut in addition to potential siderophore production by gut-resident species64,65.
We anticipated that by looking for fungal impacts on E. coli, we could leverage the genetic information available for this species. However, even in this well-characterized organism, 38% of genes identified as having interaction fitness are annotated as hypothetical, uncharacterized, or putative. For P. psychrophila, 27% of genes with interaction fitness are hypothetical proteins. Similarly, the chemical identity and ecological relevance of most of the specialized metabolites we identify in our Penicillium species is unknown. This highlights that many genes and molecules involved in interspecies interactions have yet to be characterized, and that studying microbes in the context of their interactions with other species, and not just in monoculture, provides an avenue for uncovering new areas of biology.
METHODS
Source Information for Strains and Libraries
Source information for strains and libraries used in this study is provided in Supplementary Table 19.
Fungal Ribosomal RNA Sequencing
Genomic DNA was extracted with phenol-chloroform (pH 8) from cultures of the eight cheese fungal species used in this study. For each extraction: 125 μL of 425–600 μm acid-washed beads and 125 μL of 150–212 μm acid-washed beads were poured in a screw-capped 2 mL tube. 500 μL of 2X buffer B (200 mM NaCl, 20 mM EDTA) and 210 μL of SDS 20% were added to the tube containing fungal material and 500 μL of Phenol:Chloroform (pH 8). Cells were lysed by vortexing the tubes for 2 min at maximum speed. Aqueous and organic phases were separated by centrifugation at 4°C, 8,000 RPM for 3 min and 450 μL of the aqueous phase (upper phase) were recovered in a 1.5 mL Eppendorf tube. 45 μL of sodium acetate 3M and 450 μL of ice-cold isopropanol were added before incubating the tubes at −80°C for 10 min. The tubes were then centrifuged for 5 min at 4°C at 13,000 RPM. The pellet was then washed in 750 μL of 70% ice cold ethanol and re-suspended in 50 μL of DNase/RNase free water. Following DNA extraction, LROR (ACCCGCTGAACTTAAGC) and LR6 (CGCCAGTTCTGCTTACC)66 primers were used to amplify the large subunit of the ribosomal RNA and for Penicillium species, Bt2a (GGTAACCAAATCGGTGCTGCTTTC) and Bt2b (ACCCTCAGTGTAGTGACCCTTGGC)67 primers were used to amplify the beta-tubulin gene. PCR was performed in a final volume of 50 μL: 25 μL of Q5 polymerase master mix (New England Biolabs), 2.5 μL of the forward primer at 10 μM, 2.5 μL of the reverse primer at 10 μM, 100 ng of genomic DNA, and water using the following PCR programs: LSU-(i) 98°C - 30 s, (ii) 35 cycles of: 98°C – 10 s; 52°C – 30 s; 72°C – 1.5 min, (iii) 72°C – 5 min; Beta-tubulin-(i) 98°C - 30 s, (ii) 35 cycles of: 98°C – 10 s; 57°C – 30 s; 72°C – 1 min, (iii) 72°C – 5 min. PCR products were purified with the QIAquick PCR purification kit (Qiagen) and sequenced using the forward and reverse primer by Eton Bioscience Inc. (San Diego, USA). Consensus sequences from forward and reverse sequencing reactions of the LROR/LR6 PCR product were aligned using Geneious version R9 9.1.8 (http://www.geneious.com). The MrBayes68 plugin for Geneious was used to build the phylogenetic tree with the following parameters: Substitution model- JC69; Rate variation- gamma; Outgroup- Fusarium sp. str. 554A; Gamma categories-4; Chain Length- 1100000; Subsampling freq- 200; Heated chains-4; Burn-in length- 100000; Heated chain temp- 0.2; Random seed-1160; Unconstrained branch lengths- 1, 0.1, 1, 1. FigTree v1.4.4 was used for visualization (https://github.com/rambaut/figtree/releases).
Bacterial-Fungal Growth Assays
To approximate a 1:1 ratio of bacteria: fungi based on cell size, we inoculated 60,000 bacterial cells alone or with 6,000 fungal spores per well on 10% CCA medium25 adjusted to pH 7 in a 96-well plate. Each bacterial or bacterial-fungal assay was done in triplicate. After 7 days of growth at room temperature, the entire well was harvested and homogenized in 1xPBS-Tween 0.05% prior to dilution and plating on LB with 20 μg/ml cycloheximide (for bacterial counts) or milk plate count agar with 50 μg/ml chloramphenicol (for fungal spore counts). Counts were done at inoculation and after harvest. Final growth counts were then compared in co-culture condition relative to growth alone to identify interaction impacts. Significant growth impacts were determined based on Dunnett’s test69, p-value < 0.05. Plots were made with R package ggplot2 3.2.170.
Microbial Culturing for LC-MS/MS extraction
All fungal cultures were grown on plate count agar milk salt (PCAMS; 1 g/L whole milk powder, 1 g/L dextrose, 2.5 g/L yeast extract, 5 g/L tryptone, 10 g/L sodium chloride, 15 g/L agar). Plates were kept at room temperature and spores were harvested at 7 days of growth (or after sporulation was observed) for subsequent experiments. Spores harvested from fungi were put into PBS and normalized to an OD600 of 0.1 for working stock.
Extraction of cultures
Three biological replicates of each condition were plated (distinct samples) and extracted from solid agar. For extraction from solid agar plates, 5 μL of fungal working stock were spotted onto 10% CCA medium adjusted to pH 7. Following 7 days of growth, agar was removed from the Petri dish and placed into 50 mL falcon tubes. Acetonitrile (10 mL) was added to each tube and all were sonicated for 30 minutes. All falcon tubes were centrifuged, and liquid was removed from the solid agar pieces and transferred to 15 mL falcon tubes. The 15 mL falcon tubes containing liquid were then centrifuged and liquid was again removed from any residual solid debris and transferred to glass scintillation vials. These liquid extractions were then dried in vacuo. Dried extracts were weighed and diluted with MeOH to obtain 1 mg/mL solutions which were stored at −20°C until analysis via LC-MS/MS.
LC-MS/MS data collection
High resolution LC-MS and LC-MS/MS data were collected on a Bruker impact II qTOF in positive mode with the detection window set from 50 to 1500 Da, on a 2.1×150mm C18 cortecs UPLC column with a flow rate of 0.5mL/min for a gradient of 10–100% ACN with 0.1% formic acid over 16 minutes. For each sample, 10 μL of a 1mg/mL solution was injected. The ESI conditions were set with the capillary voltage at 4.5 kV. For MS2, dynamic exclusion was used, and the top nine precursor ions from each MS1 scan were subjected to collision energies scaled according to mass and charge state for a total of nine data dependent MS2 events per MS1. MS2 data for pooled biological replicates is deposited under MassIVE accession number MSV000085070. MS1 and MS2 data for ΔlaeA and WT Penicillium sp. str. #12 is deposited under MassIVE accession number MSV000085054 and was collected under identical conditions on a Bruker compact qTOF.
Molecular Networking
For all extractions, all precursor m/z’s that were found in solvent and agar controls (based on both retention time and mass tolerance) were removed prior to input into GNPS using the BLANKA algorithm71. A molecular network ( https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=464b331ef9d54de9957d23b4f9b9db14) was created using the online workflow in GNPS. The data was filtered by removing all MS/MS peaks within +/− 17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top 6 peaks in the +/− 50Da window throughout the spectrum. The data was then clustered with MS-Cluster with a parent mass tolerance of .02 Da and a MS/MS fragment ion tolerance of .02 Da to create consensus spectra. Further, consensus spectra that contained less than 2 spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than 6 matched peaks. Further edges between two nodes were kept in the network only if each of the nodes appeared in each other’s respective top 10 most similar nodes. The spectra in the network were then searched against GNPS’ spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 6 matched peaks. Solvent and agar control files were also loaded into the networks in order to perform removal based on fragmentation patterns. All nodes with precursor masses less than 200Da were also removed. The extensive background and low m/z Da removal was done to more accurately reflect the metabolomic profiles of fungal genera in an attempt to represent only true metabolites. The DEREPLICATOR was used to annotate MS/MS spectra72,73. The molecular networks were visualized using Cytoscape software74.
RB-TnSeq Assays
All RB-TnSeq assays were performed on 10% CCA medium adjusted to pH 7. Prior to inoculation, one aliquot of each library was thawed and inoculated into 25 mL of liquid LB-kanamycin (50 μg/mL). This is the same media used for initial library creation and is expected to be non-selective. Once the culture reached mid-log phase (OD = 0.6–0.8), 5 mL of that pre-culture was pelleted and stored at −80°C for the T0 reference in the fitness analysis. The remaining cells were used to inoculate the fitness assay conditions. For each RB-TnSeq fitness assay, we aimed to inoculate 7,000,000 cells of the bacterial library (on average 50 cells per insertion mutant). For fitness assays including a fungal partner, 700,000 fungal cells were inoculated based on spore counts. We inoculated 10X more bacterial cells than fungal spores in order to approximate a 1:1 volume ratio of bacteria: fungi, as fungal cells are approximately 10 times larger than bacterial cells. Pre-cultured cells were washed in 1xPBS-Tween 0.05%, mixed with appropriate volumes of quantified fungal spore stocks, and then inoculated by spreading evenly on a 100 mm petri dish containing 10% CCA medium, pH 7. For each condition, assays were performed in triplicate (3 distinct samples). After seven days, each plate was flooded with 1.5 mL of 1xPBS-Tween 0.05% and cells were scraped off, taking care to not disturb the CCA. The liquid was then transferred into a 1.5 mL microfuge tube and cells were pelleted by centrifugation. After removing the supernatant, the cells were washed in 1 mL of RNAprotect solution (Qiagen, Hilden, Germany), pelleted and stored at −80°C until genomic DNA (gDNA) extraction. gDNA was extracted with phenol-chloroform (pH 8) using the same protocol used for fungal gDNA extraction described above. Samples were stored at −80°C until further analysis.
After gDNA extraction, extracts containing Penicillium sp. str. #12 DNA were purified using the OneStep PCR Inhibitor Removal Kit (Zymo Research, CA, USA). Then, the 98°C BarSeq PCR as described in Wetmore et al., 201518 was used to amplify only the barcoded region of the transposons. PCR was performed in a final volume of 50 μL: 25 μL of Q5 polymerase master mix (New England Biolabs, MA, USA), 10 μL of GC enhancer buffer (New England Biolabs), 2.5 μL of the common reverse primer (BarSeq_P1 – Wetmore et al., 2015) at 10 μM, 2.5 μL of a forward primer from the 96 forward primers (BarSeq_P2_ITXXX) at 10 μM and either 200 ng of gDNA for alone conditions, or 2 μg of gDNA for fungal interaction conditions. For E. coli analysis, we performed 84 PCRs (T0 sample and 28 harvest samples in triplicate) involving 28 different multiplexing indexes. For P. psychrophila str. JB418 analysis, we performed 84 PCRs (T0 sample and 28 harvest samples in triplicate) involving 28 different multiplexing indexes. We used the following PCR program: (i) 98°C - 4 min, (ii) 30 cycles of: 98°C – 30 s; 55°C – 30 s; 72°C – 30 s, (iii) 72°C – 5 min. After the PCR, for both E. coli and P. psychrophila, 10 μL of each of the PCR products were pooled together to create the BarSeq sequencing library and 200 μL of the pooled library were purified using the MinElute purification kit (Qiagen, Germany). The final elution of the BarSeq library was performed in 30 μL in DNase and RNase free water. The BarSeq libraries were then quantified using the Qubit dsDNA HS assay kit (Invitrogen, CA, USA) and sequenced on a HiSeq4000 (75 bp, single-end reads), by the IGM Genomics Center at the University of California, San Diego. The sequencing depth for each condition varied between 6.1 and 11.7 million reads for E. coli and 5.8 and 13.3 million reads for P. psychrophila.
RB-TnSeq Data Processing
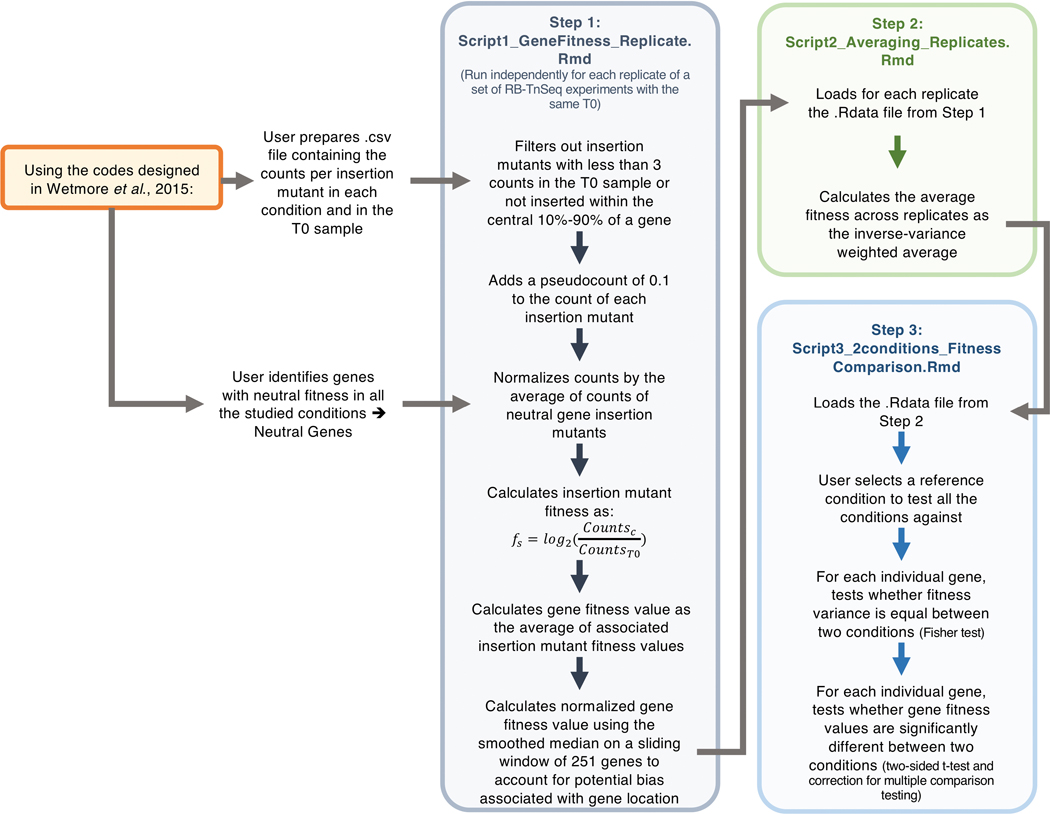
Custom R scripts were used to determine the average fitness scores for each gene across three RB-TnSeq assay replicates. These scripts are available at https://github.com/DuttonLab/RB-TnSeq-Microbial-interactions. The Readme document provides an in-depth explanation of all data processing steps performed in these scripts. Insertion mutants that did not have a sufficient T0 count (3) in each condition or that are not centrally inserted (10–90% of gene) were removed from analysis. Counts determined by Wetmore et al., 201518 scripts were then normalized using a set of 5 reference genes (glgP, acnA, modE, leuA and idnK in E. coli (average of 52 strains each) and respective closest protein BLAST match Ga0212129_11488, Ga0212129_114557, Ga0212129_111416, Ga0212129_112128 and Ga0212129_112491(average of 74 strains each) in P. psychrophila) to be able to compare across conditions and to account for differences in sequencing depth. These genes have an absolute fitness effect <0.6 in all conditions for all replicates in any condition based on former fitness determination developed by Wetmore et al., 201518. Strain fitness (fs) was then calculated per insertion mutant as the log2 of the ratio of the normalized counts in the condition and the normalized counts in the T0 sample (Equation (1)).
| (1) |
with Cc: normalized counts in condition C and Ct0: normalized counts in T0
Gene fitness and variance were next calculated by averaging insertion mutants within a gene. These values were then normalized based on the position of the gene along the chromosome using a smoothed median on a window of 251 genes as described in Wetmore et al., 201518. These steps were all done on individual replicates. For all conditions, replicates were highly correlated with an averaged Pearson correlation coefficient of 0.85 for E. coli and 0.84 for P. psychrophila. Next, the average gene fitness (fg) (Equation (2)) and associated standard deviation () (Equation (3)) were calculated using the inverse of variance weighted average of the fitness values across the three different replicates.
| (2) |
| (3) |
With wi the inverse of the gene fitness variance for each replicate, n the number of replicates and the weighted gene fitness average across the n replicates.
Final fitness values were then compared for fungal interaction conditions compared to bacterial alone conditions using two-sided t-tests (when equality of variance was verified by Fisher test) and correction for multiple comparison (Benjamini-Hochberg method75). Comparisons associated with an adjusted p-value lower than 5% were considered a significant interaction fitness (alphaF parameter=0.002 and alphaT parameter=0.05 in Script3_2conditions_FitnessComparison.Rmd code). The overall pipeline is described in Extended Data 1, and Supplementary Method 1 provides an example for a complete run. Networks of fitness values were visualized in Cytoscape v. 3.5.174 and PCA plots were made with R package ggplot2 3.2.170 and ggfortify 0.4.776. COG category mapping of E. coli and P. psychrophila protein sequences was done by eggNOG-mapper v277.
Functional Enrichment Analysis of Bacterial Gene Sets
ClusterProfiler78 was used for GO ontology functional enrichment analysis of bacterial gene sets with FDR p-value adjustment cutoff of 0.1. For E. coli, the E. coli K12 database (org.EcK12.eg.db) was used79. For P. psychrophila, a custom annotation database was created using eggNOG-mapper v277 GO assignments using AnnotationForge80 in R.
Bacterial Cytological Profiling
Approximately 7,000,000 WT E. coli K-12 strain BW25113 or Keio collection mdtK mutant cells81 were inoculated alone or co-inoculated with 700,000 Penicillium sp. str. #12, Penicillium sp. str. #12 ΔlaeA, or Penicillium sp. str. SAM3 spores on 10% CCA pH 7. After 7 days of growth, 1 mL of T-Base buffer was added to the surface of the biofilms, and biofilms were scraped into the buffer. For co-culture conditions, the sample was filtered through a 0.5 μm filter to specifically remove fungal material. 2 μL of concentrated dye mix (1 μL 1 mg/mL FM4–64, 1 μL 2 mg/mL DAPI in 48 μL T-Base) were added to 20 μL of filtrate. The dye-filtrate mix was spotted onto agarose-LB pads (1% agarose, 20% LB liquid medium, 80% ddH2O) and imaged by fluorescence and phase contrast using an Applied Precision Deltavision Spectris imaging system with an Olympus UPLFLN100XO2PH objective. Control compound references on CCA medium were obtained by spotting and drying 30 μL of 5x, 10x, 25x, and 100x MIC dilutions of antibiotics onto quadrants on CCA medium pH 7 plates and then spread-plating 200 μL of log-phase (OD 0.1) E. coli cultures. After two days of growth, cells near the edge of the zone of inhibition on appropriate dilution spots were resuspended in 10 μL of prediluted dye mix (1 μL 1 mg/mL FM4–64, 1 μL 2 mg/mL DAPI in 998 μL T-Base) and spotted onto agarose-LB pads and imaged as described above. Resulting images were deconvoluted using Deltavision SoftWorx software (Applied Precision, Inc., WA, USA), analyzed using Fiji82, and assembled in Adobe Photoshop (Adobe, CA, USA). Brightness was altered linearly in Fiji to aid visualization. For quantification of cell roundness, we defined the cell major axis as the longest possible line along the cell, and the cell minor axis was measured as the longest possible line orthogonal to the cell major axis. Cell measurements were obtained via the measure tool in Fiji82, and single-cell major and minor axes measurements were collated. Pixel to micron ratio was set as 15.6 as per the microscope specifications. Major/minor axis ratio was calculated for all cells in the field. Number of fields was chosen to ensure measurement of at least 50 cells for each experimental condition. Individual ratio values for each cell were plotted via R package ggplot2 3.2.170, and differences in major:minor ratios in the presence of a fungus relative to growth alone or with WT Penicillium sp. str. #12 relative to ΔlaeA Penicillium sp. str. #12 were determined based on an unpaired two-sample Wilcoxon test p-value < 0.05.
CCA medium biotin quantification
Biotin quantification of CCA medium was performed on three replicate samples by Creative Proteomics (NY, USA) as follows: 100 mg of each sample was homogenized in water (10 μL/mg) for 1 minute three times with the aid of 5-mm metal balls on a MM400 mill mixer. Methanol at 10 μL/mg was then added. Water-soluble vitamins were extracted by vortex-mixing for 2 min and sonication in a water batch for 5 min. After centrifugation, the clear supernatants were cleaned up by solid-phase extraction on a Strata-X (60 mg/mL) cartridge. The eluted fractions containing water-soluble vitamins were collected, pooled and then dried under a gentle nitrogen gas flow in a nitrogen evaporator. The residues were dissolved in 200 μL of 10% methanol. Twenty μL aliquots were injected to run UPLC-MRM/MS with the use of a C18 UPLC column and with (+) ion detection and (−) ion detection, respectively. Calibration curves were prepared by injection of serially-diluted mixed standard solutions of water-soluble vitamins. Concentrations of detected vitamins were calculated by interpolating the linear calibration curves.
Δfep mutant competitive growth assays
Approximately 60,000 bacterial cells (a 1:1 ratio of WT cells and either ΔfepA, ΔfepC, or ΔfepG Keio collection81 mutant cells) were inoculated either alone (no fungus) or co-inoculated with approximately 6,000 Penicillium sp. str. #12 or Scopulariopsis sp. str. JB370 spores on 10% CCA pH 7 in a 96-well plate in four replicates each (4 distinct samples). After seven days of growth, the entire well was harvested and homogenized in 1xPBS-Tween 0.05% prior to dilution and plating on LB with 20 μg/ml cycloheximide (total bacterial counts) or with 20 μg/ml cycloheximide and 50 μg/ml kanamycin (bacterial mutant counts). Final growth counts were then compared in fungal co-culture conditions relative to bacterial growth alone to identify interaction impacts. Significant growth impacts were determined by significantly different growth in the presence of a fungus relative to growth alone based on a two-sided two-sample t-test p-value < 0.05. Plots made with R package ggplot2 3.2.170.
Siderophore Detection with Chrome Azurol S (CAS) Assay
The following methods were adapted from those described by Schwyn and Neilands83 and Payne84. All glassware, caps, and stir bars were cleaned with 6M HCl and rinsed with deionized water. Plastic spatulas and doubly deionized water were used for solution preparation. A 2 mM CAS stock solution was prepared and stored in the dark at room temperature and a 1 mM FeCl3 stock solution was prepared. Piperazine buffer was prepared by dissolving 4.3095 g of anhydrous piperazine in 30 mL of water and adding 5 M HCl until the pH reached 5.635. To prepare the CAS reagent, 1.1202 mL of 0.05 M hexadecyltrimethylammonium bromide (HDTMA) was added to 50 mL of water. 1.5 mL of 1 mM FeCl3 stock solution was mixed with 7.5 mL of 2 mM CAS solution and added to the HDTMA solution. Lastly, the piperazine buffer was added to the solution and stirred. The resulting CAS assay solution was stored in the dark at room temperature. 0.2 M shuttle solution was prepared with 5-sulfosalicylic acid dihydrate in water. The shuttle solution was stored in the dark at room temperature.
For detection of siderophore presence, fungal species were inoculated in triplicate into liquid 2% CCA pH 7 and cultures were grown at room temperature for twelve days. For filamentous fungi, cultures were left standing without shaking. After twelve days, supernatants were filtered through a 0.22 micron filter. Prior to use, the CAS assay solution was vortexed until all precipitates were resuspended. CAS assay incubations were performed in the dark at room temperature. For each fungal supernatant or CCA filtrate, 100 μL of CAS assay solution was added to 100 μL of supernatant. The resulting solution was mixed by pipetting and incubated for 15 minutes. After incubation, 2 μL of shuttle solution was added to the solution and mixed by pipetting. The solution was incubated for an additional 30 minutes. Sample absorbance of 630 nm light was measured in a 96-well plate using an Epoch 2 plate reader (BioTek, Winooski, VT).
For CAS assay comparisons of relative siderophore production in WT and ΔlaeA Penicillium sp. str. #12, 200,000 spores of WT or ΔlaeA were inoculated in triplicate in 3 mL of liquid 2% CCA pH 7. After seven days of growth at room temperature without shaking, the biomass of the fungal mat was removed from the top of the culture and the entire supernatant was filtered through a 0.22 micron filter. Total filtrate was measured. Fungal mats were dried in a 60°C drying oven for 2 days before being weighed. Filtrates were concentrated 3X in a SpeedVac Vacuum Concentrator and 100 μL of three replicates each of WT, ΔlaeA, and 2% liquid CCA were added to 100 μL of CAS solution and CAS assays were performed as described above. Following CAS measurements, % siderophore units were normalized to the entire volume of 1X filtrate and expressed per mg of dried fungal biomass.
RNA-Seq and differential expression analysis of E. coli with Penicillium sp. str. #12
Approximately 7,000,000 E. coli cells were inoculated in triplicate (3 distinct samples) either alone or with approximately 700,000 Penicillium sp. str. #12 spores on 10% CCA pH 7. After 3 days, the biofilms were harvested for RNA extraction and washed with 1 mL of RNAprotect. RNA was extracted by a phenol-chloroform extraction (pH 8) using the same extraction protocol as for gDNA. Extractions were then purified with the OneStep PCR Inhibitor Removal Kit (Zymo Research, CA, USA).
Sequencing libraries were prepared as follows: RNA samples were treated with DNase using the ‘Rigorous DNase treatment’ for the Turbo DNA-free kit (AMBION, Life Technologies, Waltham, MA, USA) and RNA concentration was measured by nucleic acid quantification in Epoch Microplate Spectrophotometer (BioTek, Winooski, VT, USA). Transfer RNAs and 5S RNA were then removed using the MEGAclear Kit Purification for Large Scale Transcription Reactions (AMBION, Life Technologies) following manufacturer instructions. Absence of tRNA and 5S RNA was verified by running 100 ng of RNA on a 1.5% agarose gel and RNA concentration was quantified by nucleic acid quantification in an Epoch Microplate Spectrophotometer. Also, presence of gDNA was assessed by PCR using universal bacterial 16S PCR primers (Forward primer: AGAGTTTGATCCTGGCTCAG, Reverse Primer: GGTTACCTTGTTACGACTT). The PCR was performed in a final volume of 20 μL: 10 μL of Q5 polymerase master mix (New England Biolabs), 0.5 μL of forward primer 10 μM, 0.5 μL of reverse primer 10 μM, and 5 μL of non-diluted RNA. PCR products were run on a 1.7% agarose gel and if gDNA was amplified, another DNase treatment was performed as well as a new verification of absence of gDNA.
Ribosomal RNA depletion was performed using the RiboMinus Transcriptome Isolation Kit (Yeast and Bacteria) for the E. coli alone samples and using both the RiboMinus Transcriptome Isolation Kit (Yeast and Bacteria) and the RiboMinus Eukaryote Kit v2 for the mixed E. coli/Penicillium sp. str. #12 samples (ThermoFisher Scientific). For the E. coli alone samples, each sample was divided into two for treatment and then repooled for RNA recovery with ethanol precipitation. For the E. coli/Penicillium sp. str. #12 samples, an equal volume of the eukaryotic probe and RiboMinus Bacterial Probe Mix were used to deplete both bacterial and fungal ribosomal RNA and RNA was recovered with ethanol precipitation. Concentrations after ribosomal RNA depletion were measured using Qubit RNA HS Assay Kits (Invitrogen). The RNA-Seq library was produced using the NEBNext Ultra RNA Library Prep Kit for Illumina for purified mRNA or ribosome-depleted RNA. We prepared a library with a fragment size of 300 nucleotides and used the 10 μM NEBNext Multiplex Oligos for Illumina (Set 1, NEB #E7335 lot 0091412) and the NEBNext multiplex Oligos for Illumina (Set 2, NEB #E7500 lot 0071412). We performed PCR product purification with 0.8X Agencourt AMPure XP Beads. Library samples were quantified with Qubit DNA HS Assay Kits before the quality and fragment size were validated by TapeStation (HiSensD1000 ScreenTape). Library samples were pooled at a concentration of 15 nM each and were sequenced on a HiSeq4000 (50 bp, single-end). TapeStation assays and sequencing were performed by the IGM Genomics Center at the University of California, San Diego.
Following sequencing, reads were mapped to the concatenated genome of E. coli K-12 BW2511385 and Penicillium sp. str. #12 using Geneious version R9 9.1.8 (http://www.geneious.com). Only the reads that uniquely mapped to a single location on the E. coli genome section were kept. E. coli expression analysis was performed using the following R packages: Rsamtools (R package version 2.0.3), GenomeInfoDb (R package version 1.20.0), GenomicFeatures86 (R package version 1.36.4), GenomicAlignments86 (R package version 1.20.1), GenomicRanges86 (R package version 1.36.1) and DESeq287 (R package version 1.20.1). We followed the workflow described by Love et al. 2014 and performed the differential expression analysis using the package DESeq2. Differentially expressed genes between conditions were selected with an adjusted p-value lower than 5% (Benjamini-Hochberg correction for multiple testing75) and an absolute log2 fold change equal to or greater than 1.5.
Construction of E. coli mutants and visual interaction assays
Visual assays for purified hydroxamate siderophore stimulation:
Antibiotic assay discs (Whatman) were placed on CCA medium pH 7 with .005% tetrazolium chloride (an indicator of cellular respiration) and 20 μL of water, 10 μM coprogen or ferrichrome (EMC Microcollections GmbH) solutions (in water) were slowly pipetted onto the disc and allowed to absorb. 2.5 μL of 37°C overnight LB cultures of E. coli K-12 BW25113 WT, ΔfepA, ΔfepC, ΔfhuE, ΔfhuA, ΔfepAΔfhuE, ΔfepCΔfhuE, or ΔfepAΔfhuA mutants81 were spotted next to the discs. Double mutants were constructed as described below. Plates were left at room temperature until development of red color resulting from tetrazolium chloride, an indicator of respiration.
Visual assays for fungal stimulation of bacterial mutants:
Fungal spores were inoculated on CCA pH 7 with .005% tetrazolium chloride. After fungal pre-culturing at room temperature (cheese fungal isolates) or 30°C (A. fumigatus and M. pachydermatis), 2.5 μL of E. coli overnight cultures grown in LB medium at 37°C were spotted at increasing distances from the fungal front. Plates were left at room temperature until red color developed. A. fumigatus isolate AF293 was received from Nancy Keller, University of Wisconsin-Madison. M. pachydermatis was originally isolated from the ear of a dog in Sweden (ATCC14522).
Creation of ΔfepAΔfhuE and ΔfepCΔfhuE:
Chemically competent cells for ΔfepA or ΔfepC mutants were created. An overnight culture of ΔfepA or ΔfepC mutants was diluted 1:100 and grown at 37°C until OD 0.4–0.6. The culture was placed on ice for 20 minutes and then centrifuged at 4°C for 10 minutes at 6000 rpm to collect the cells. Supernatant was removed and cells were resuspended in half the previous volume of pre-cooled 0.1M CaCl2. After being left on ice for 30 minutes, centrifugation was repeated, and supernatant was removed before resuspension in a quarter of the original volume of pre-cooled 0.1M CaCl2/15% glycerol. Cells were aliquoted and stored at −80°C until transformation. These cells were transformed with the pKD46 plasmid88, recovered at 30°C, and plated on LB plates with 100 μg/mL Ampicillin and grown at 30°C. Overnight cultures were started from individual colonies for creation of electrocompetent cells. Overnight cultures of ΔfepC-pkD46 or ΔfepA-pkD46 were diluted 1:100 in fresh LB-100 μg/mL Ampicillin and grown at 30°C until an OD of 0.1. 20 μL of fresh 1 M L-arabinose were added, and growth was continued at 30°C until OD 0.4–0.6. Cells were then chilled on ice for 15 minutes and then centrifuged for ten minutes at 4000 rcf 4°C. Cells were resuspended in 1 mL of ice water and centrifuged for ten minutes at 4000 rcf at 4°C. Cells were resuspended in 0.5 mL of ice water and centrifuged for ten minutes at 4000 rcf 4°C. Cells were resuspended in 50 μL of ice water and kept on ice until transformation. The chloramphenicol resistance cassette was amplified from the pKD3 plasmid88 using the following custom primers: FhuEcatF (CAGATGGCTGCCTTTTTTACAGGTGTTATTCAGAATTGATACGTGCCGGTAATGGCGCGCCTTACGCCCC) and FhuEcatR (CCTCCTCCGGCATGAGCCTGACGACAACATAAACCAAGAGATTTCAAATGCTGGGCCAACTTTTGGCGAA) and the following PCR conditions (i) 98°C - 30 sec, (ii) 30 cycles of: 98°C – 10 s; 70°C – 20 s; 72°C – 30 s, (iii) 72°C – 5 min. Amplification was performed on 4 ng of pKD3 plasmid using Q5 High-Fidelity 2X Master Mix (New England Biolabs). The PCR product was digested for 1 hour with the restriction enzymes DpnI and ClaI at 37°C and then the PCR product was run on a 1% agarose gel. The PCR product was extracted using the QIAquick Gel Extraction Kit (Qiagen) and then dialyzed for 4 hours with TE buffer. 1.5 μL of dialyzed PCR product was used to transform the electrocompetent ΔfepC-pkD46 or ΔfepA-pkD46 cells. After 2 hours of recovery in SOC medium with 1 mM arabinose at 37°C, the transformation was plated on LB with 50 mg/mL kanamycin and chloramphenicol. Transformants were confirmed to be ΔfhuE with Eton Bioscience Inc. sequencing of the chloramphenicol cassette.
Creation of ΔfepAΔfhuA:
Creation was done as with ΔfepAΔfhuE, except that the chloramphenicol resistance cassette was amplified from pKD388 using FhuAcatF (ATCATTCTCGTTTACGTTATCATTCACTTT ACATCAGAGATATACCAATGAATGGCGCGCCTTACGCCCCAATGGCGCGCCTTACGCCCC) and FhuAcatR (GCACGGAAATCCGTGCCCCAAAAGAGAAATTAGAAACGGAAGGTTGCGGTCTGGGCCAACTTTTGGCGAACTGGGCCAACTTTTGGCGAA) custom primers.
Penicillium sp. str. #12 genome sequencing, assembly, and annotation
Genomic DNA was extracted from Penicillium sp. str. #12 using the genomic DNA extraction protocol described above. High molecular weight DNA (average 16 kilobases) was sequenced on the Oxford Nanopore MinION with a R.9.5 flow cell using 1D2 sequencing adaptors from kit SQK-LSK308 (Oxford Nanopore Technologies, Oxford, United Kingdom). Raw data was basecalled using guppy 3.3.0 (Oxford Nanopore Technologies, Oxford, United Kingdom)(guppy_basecaller -config dna_r9.5_450bps.cfg --fast5_out) for 1D basecalls and these were used to also obtain higher accuracy 1D^2 basecalls (guppy_basecaller_1d2 -i 1Dbasecall/workspace/ --config dba_r9.5_450bps_1d2_raw.cfg --index_file 1Dbasecall/sequencing_summary.txt) These reads were assembled by canu 1.889 and polished by racon 1.4.390 four times and by pilon 1.2391 once. The final assembly is 38 Mbp and consists of 52 contigs.
Penicillium sp. str. #12 genome annotations were obtained by combining genomic and transcriptomic information from RNA-Seq. To obtain the gene expression profile of Penicillium sp. str. #12, approximately 700,000 WT Penicillium sp. str. #12 spores were inoculated in triplicate on 10% CCA pH 7. After 3 days, the biofilms were harvested for RNA extraction and washed with 1 mL of RNAprotect. RNA was extracted and RNA-Seq libraries were prepared as described above with the following modification: ribosomal RNA depletion was performed using the RiboMinus Eukaryote Kit v1 and RNA was recovered with ethanol precipitation. After sequencing, the RNA-Seq reads from these Penicillium sp. str. #12 alone cultures were concatenated with RNA-Seq reads from the previously described E. coli/Penicillium sp. str. #12 co-culture conditions that uniquely mapped to a single location on the Penicillium sp. str. #12 genome. The full set of transcriptomic reads was then used as input into the FunGAP annotation pipeline and 77 million of these reads mapped92. This pipeline predicted 13261 protein-coding genes in the Penicillium sp. str. #12 genome. Interproscan93 was used within the FunGAP pipeline for function prediction of genes. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAASRZ000000000. The version described in this paper is version JAASRZ010000000.
Creation and confirmation of Penicillium sp. str. #12 laeA deletion mutant
Deletion cassette design strategy: In order to knockout laeA in Penicillium sp. str. #12, the isolate was first screened for hygromycin and phleomycin resistance. Penicillium sp. str. #12 showed a confirmed sensitivity to both antibiotics. A three round PCR deletion strategy was used to replace the laeA ORF with the hph gene, whose expression confers selection on hygromycin94. The schematic representation of the laeA gene replacement with the hph gene is depicted in Supplementary Figure 3. The deletion cassette (5’flank- hph- 3’flank) was constructed using three sequential PCR reactions. In the first PCR round, about 1 kilobase genomic sequence flanking either the 5’ or 3’ end of the laeA ORF were amplified using the primer sets P12_KOlaeA_5’ F (CTCCGTTGGGCCCTCAC) and 5’R (GCAATTTAACTGTGATAAACTACCGCATTAAAGCTGTTGATATCGGCAATCAATCAATG) or P12_KOlaeA_3’F (GGTGGGCCTTGACATGTGCAGCCGGTGGAGCGGCGCCTGGTGAATCCTACCCACATGG) and 3’R (CGTTGGGAGGAAAAGCTTCTGCG), respectively. The hph gene was amplified from plasmid pUCH2–8 using primers hph_F (AGCTTTAATGCGGTAGTTTATCACAG) and hph_R (CTCCACCGGCTGCACATGTC). A second PCR reaction was performed to assemble the three individual fragments from the first round PCR by homologous recombination. The deletion cassettes were finally amplified using the nested primer set, P12_KOlaeA_NestedF (CAGACGGTCCGCATCCCG) and P12_KOlaeA_NestedR (GGTCCAGGTGCAGTAGTACTG).
To generate the deletion strains, a protoplast-mediated transformation protocol was employed. Briefly, 109 fresh spores were cultured in 500 mL of liquid minimal medium (LMM) for 12 h at 25°C and 280 rpm. Newly born hyphae were harvested by centrifugation at 8000 rpm for 15 min and hydrolyzed in a mixture of 30 mg Lysing Enzyme from Trichoderma (Sigma-Aldrich) and 20 mg Yatalase (Fisher Scientific) in 10 mL of Osmotic Medium (1.2 M MgSO4, 10 mM NaPB, pH 5.8). The quality of the protoplasts was monitored under the microscope after four hours of shaking at 28 °C and 80 rpm. The protoplast mixture was later overlaid with 10 mL of trapping buffer (0.6 M sorbitol, 100 mM Tris-HCl pH 7.0) and centrifuged for 15 min at 4°C and 5000 rpm. Protoplasts were collected from the interface, overlaid with an equal volume of STC (1.2 M sorbitol, 10 mM Tris-HCL pH 7.5,10 mM CaCl2) and decanted by centrifugation at 6000 rpm for 8 min. The protoplast pellet was resuspended in 500 μL STC and used for transformation. After 5 days of incubation at 25 °C, colonies grown on stabilized minimal medium (SMM) plates supplemented with hygromycin were subjected to a second round of selection on hygromycin plates. In total, 25 hygromycin-resistant transformants were isolated after a rapid screening procedure on SMM supplemented with hygromycin. Single-spored transformants were later tested for proper homologous recombination at the ORF locus by PCR and Southern blot analysis.
The correct replacement of laeA with the hph gene was first verified by PCR analysis of gDNA from the transformant strains using primer set P12_laeA_F (CACAATGGCTGAACACTCTCGG) and P12_laeA_R (GGGATATGGAGCATCGAAGTTGC) that amplify the laeA ORF. About 12% (3/25) of the monoconidial lines generated from primary transformants of Penicillium sp. str. #12 were PCR-positive for the absence of the laeA ORF. The positive deletion strains were further checked for a single insertion of the deletion cassette by Southern blot analysis and revealed single-site integration of the deletion cassette in one transformant (Supplementary Figure 3). Probes corresponding to the 5’ and 3’ flanks of the laeA gene in each strain were labeled using [α32P] dCTP (PerkinElmer, USA) following the manufacturer’s instructions.
RNA-Seq analysis of WT and ΔlaeA Penicillium sp. str. #12
To characterize the effect of the laeA deletion on the Penicillium sp. str. #12 gene expression profile, we performed RNA-Seq analysis for ΔlaeA Penicillium sp. str. #12. As for WT Penicillium sp. str. #12, 700,000 ΔlaeA Penicillium sp. str. #12 spores were inoculated in triplicate (3 distinct samples) on 10% CCA pH 7 and biofilms were harvested after 3 days. Harvest, RNA extraction, and library preparation were performed identically to WT Penicillium sp. str. #12. Then, Penicillium sp. str. #12 and ΔlaeA differential expression analysis was performed as described for E. coli/Penicillium sp. str. #12 above. To look for enrichment of functions in the set of differentially expressed genes, we input the protein sequences of the genes into the gene-list enrichment function of KOBAS 3.095. Sequences were searched against the Gene Ontology (GO) database96,97 using A. fumigatus as a reference for GO assignment before conducting a hypergeometric test with Benjamini-Hochberg correction. Functions with a corrected p-value <.05 were considered enriched.
WT and ΔlaeA Penicillium sp. str. #12 Growth Assays
For radial growth assays, 2,000 WT or ΔlaeA Penicillium sp. str. #12 fungal spores were inoculated in triplicate on 10% CCA pH 7 either alone or with 20,000 E. coli cells. The radius of fungal growth was measured after 7 days. For spore counting assays, we aimed to inoculate 6,000 WT or ΔlaeA Penicillium sp. str. #12 fungal spores per well on 10% CCA pH 7 in a 96-well plate either alone or co-inoculated with 60,000 E. coli cells. Each assay was done in triplicate. After 7 days of growth, the entire well was harvested and homogenized in PBS1X-Tween 0.05% prior to dilution and plating on milk plate count agar with 50 μg/ml chloramphenicol (for fungal spore counts).
DATA AVAILABILITY
Sequence data that support the findings of this study (RB-TnSeq, RNA-Seq) have been deposited in the NCBI SRA database with SRA accession codes SRR11514793-SRR11514872 and BioProject PRJNA624168. Mass spectrometry data is available in the MassIVE database under accession numbers MSV000085070 and MSV000085054. The GNPS molecular network is available at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=464b331ef9d54de9957d23b4f9b9db14. The E. coli annotation database used for gene ontology functional enrichment is available at http://bioconductor.org/packages/release/data/annotation/html/org.EcK12.eg.db.html. The Whole Genome Shotgun project for Penicillium sp. str. #12 including reads, genome assembly, and annotation has been deposited at DDBJ/ENA/GenBank under the accession JAASRZ000000000 in BioProject PRJNA612335 (BioSample SAMN14369290 and SRA SRR11536435). In addition to these sources, data used to create figures 2,3,5, and 6 is available in the Supplementary Tables provided with the paper. Uncropped Southern blots associated with Supplementary Figure 3 are provided with the manuscript as Source Data.
CODE AVAILABILITY
The R scripts developed for processing RB-TnSeq data described in this manuscript are available at https://github.com/DuttonLab/RB-TnSeq-Microbial-interactions along with usage instructions. The perl scripts needed for initial processing of RB-TnSeq data published in Wetmore et al. 2015 are available at https://bitbucket.org/berkeleylab/feba/src/master/.
AVAILABILITY OF BIOLOGICAL MATERIALS
All unique materials, including described fungal strains isolated from cheese, the P. psychrophila JB418 strain and RB-TnSeq library, Penicillium sp. str. #12 laeA deletion mutant, and E. coli siderophore-uptake double mutants, are readily available from the authors upon request. The E. coli RB-TnSeq library and Keio strains can be requested from the groups that created these resources (PMID references provided in Supplementary Table 19).
Extended Data
Extended Data Fig. 1. RB-TnSeq R data processing pipeline for gene fitness comparison across multiple conditions.
The pipeline is divided into three main scripts. Script 1 calculates the normalized gene fitness for each replicate of an RB-TnSeq experiment. This script has to be run for each replicate independently. Then, the .Rdata files from Script 1 are loaded in Script 2. Script 2 calculates for each RB-TnSeq condition the average gene fitness across replicates (inverse-variance weighted average). Finally, Script 3 compares gene fitness values of each RB-TnSeq condition against a chosen reference condition.
Extended Data Fig. 2. RB-TnSeq assay for fungal impacts on bacterial gene fitness.
Characterized pooled bacterial mutant libraries were grown in a biofilm either alone or with a fungal partner. After seven days of growth, mutant abundances were compared to the starting library abundances for each condition. Changes in barcode abundances were used to calculate gene fitness values. Genes with fitness values that differed significantly between co-culture and alone conditions (significant interaction fitness) were identified as potentially relevant to fungal interaction.
Extended Data Fig. 3. Comparison of E. coli and P. psychrophila interaction fitness values for the 874 genes found in both bacteria.
BLAST comparison (e-value cutoff of 1e-2) of protein sequences from P. psychrophila to those from E. coli and comparison of eggNOG gene assignments were used to find the best cross-species gene match for all genes with significant interaction fitness for at least one of the two bacterial species. A match was found for 874 genes. For each fungal condition, the fitness value of these genes with E. coli is on the x-axis and with P. psychrophila on the y-axis. In each condition, the genes are colored according to whether they have significant interaction fitness in the condition for E. coli (red), P. psychrophila (blue), both (purple), or neither (white).
Extended Data Fig. 4. Network of E. coli (left) or P. psychrophila (right) genes with positive and negative RB-TnSeq interaction fitness.
Each purple node represents a fungal partner and is labeled as follows: fungal partner (number of genes with interaction fitness; number of genes with interaction fitness unique to this condition). Each blue or orange node represents a bacterial gene. Nodes are colored by whether the average interaction fitness is positive (blue) or negative (orange) as shown in the legend below and are sized by average strength of interaction fitness across partners.
Extended Data Fig. 5. Principal Component Analysis of RB-TnSeq data.
Analysis was done on the fitness values in each fungal condition for all E. coli (left) or P. psychrophila (right) genes with an interaction fitness in at least one fungal condition. Each colored circle represents a fungal condition.
Extended Data Fig. 6. Clusters of Orthologous Genes (COG) categories of genes with interaction fitness.
Charts display the number of genes with interaction fitness that fall into each COG category for E. coli (left) or P. psychrophila (right).
Extended Data Fig. 7. Bacterial Cytological Profiling of ΔtolC E.coli treated with known antibiotic compounds on cheese curd agar.
DAPI dye stains DNA and FM4–64 dye stains bacterial membranes. SYTOX green stains nucleic acids but cannot penetrate live cells. Scale bars represent 2 μm. Testing of each antibiotic at four concentrations was performed once, and cells from the edges of zones of clearing were imaged for at least 5 fields from each condition to ensure consistency in phenotype.
Extended Data Fig. 8. Siderophore production by filamentous fungi.
Liquid CAS assay was performed on filtered and concentrated fungal supernatants from three replicates grown in 2% liquid cheese pH 7 for 12 days. Row A) 1–3: Liquid cheese control 4–6: Penicillium SAM3. Row B) 1–3: Debaryomyces 135B 4–6: Penicillium #12. Row C) 1–3: Candida 135E. 4–6: Penicillium RS17. Row D) 1–3: Scopulariopsis 165–5 Row E) 1–3: Scopulariopsis JB370. Row F) 1–3: Fusarium 554A. % Siderophore units calculated as [(Ar - As )/(Ar )]*100, where Ar is the absorbance of the cheese curd agar supernatant blank and As is the absorbance of the sample. N=3 biologically independent samples, error bars show standard deviation and black point is the mean.
Extended Data Fig. 9. Fitness defect of Δfep mutants on iron-limiting CCA.
Visual assays of E. coli mutant growth spotted alone on CCA pH 7 supplemented with tetrazolium chloride, an indicator of respiration.
Extended Data Fig. 10. Comparison of Penicillium sp. str. #12 WT and laeA deletion mutant growth on CCA.
Radial growth assay, including quantification, of Penicillium sp. str. #12 WT and laeA deletion mutant grown alone or with E. coli on CCA pH 7 (N=3 biologically independent experiments, error bars show standard deviation and black point is the mean). Spore counts from Penicillium sp. str. #12 WT and laeA deletion mutant grown alone or with E. coli for 7 days on CCA are also shown (N=3 biologically independent samples, error bars show standard deviation and black point is the mean).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank: the Arkin lab and the Deutschbauer lab at UC Berkeley for the E. coli Keio_ML9 library, Kristen Jepsen at the IGM Genomics Center at the University of California San Diego for assistance with sequencing, Dr. Sergey Kryazhimskiy (UCSD) for his input on RB-TnSeq data processing, Cong Dinh (UCSD) for assistance with fungal genome assembly, William Bushnell (UCSD) for assistance with fitness validation experiments, Dr. Sinem Beyhan (JCVI) for advice on fungal genome annotation, Lisa Marotz (UCSD) for assistance with non-cheese yeasts, and members of the Dutton lab, especially Brooke Anderson and Christina Saak, for constructive comments on the manuscript. This work was supported by National Institutes of Health grants T32-AT007533 (J.C.L.) and F31-AT010418 (J.C.L.), National Institutes of Health grant R01-AI117712 (R.B.L), National Science Foundation grant MCB-1817955 (L.M.S.), NSF grant MCB-1817887 (R.J.D. and L.M.S.), National Science Foundation grant MCB-1715553 (B.E.W.), the UCSD Center for Microbiome Innovation (E.C.P.), the UCSD Ruth Stern Award (E.C.P.), NIH Institutional Training grant 5 T32 GM 7240-40 (E.C.P.), and National Institutes of Health grant R01GM112739-01 (N.P.K.).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Laforest-Lapointe I. & Arrieta M-C Microbial Eukaryotes: a Missing Link in Gut Microbiome Studies. mSystems 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huseyin CE, O’Toole PW, Cotter PD & Scanlan PD Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol. Rev 41, 479–511 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Bergelson J, Mittelstrass J. & Horton MW Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep 9, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huffnagle GB & Noverr MC The emerging world of the fungal microbiome. Trends Microbiol. 21, 334–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford LL & Ravel J. The vaginal mycobiome: A contemporary perspective on fungi in women’s health and diseases. Virulence 8, 342–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Filippis F, Laiola M, Blaiotta G. & Ercolini D. Different Amplicon Targets for Sequencing-Based Studies of Fungal Diversity. Appl. Environ. Microbiol 83, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang TT et al. Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host Microbe 22, 809–816.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagg C, Schlaeppi K, Banerjee S, Kuramae EE & van der Heijden MGA Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun 10, 4841 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durán P. et al. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 175, 973–983.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tourneroche A. et al. Bacterial–Fungal Interactions in the Kelp Endomicrobiota Drive Autoinducer-2 Quorum Sensing. Frontiers in Microbiology vol. 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsay AK & Hogan DA Candida albicans: Molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus. Fungal Biol. Rev 28, 85–96 (2014). [Google Scholar]
- 12.Xu X-L et al. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 4, 28–39 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Spraker JE et al. Conserved Responses in a War of Small Molecules between a Plant-Pathogenic Bacterium and Fungi. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalid S. et al. NRPS-Derived Isoquinolines and Lipopetides Mediate Antagonism between Plant Pathogenic Fungi and Bacteria. ACS Chem. Biol 13, 171–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe BE, Button JE, Santarelli M. & Dutton RJ Cheese Rind Communities Provide Tractable Systems for In Situ and In Vitro Studies of Microbial Diversity. Cell 158, 422–433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morin M, Pierce EC & Dutton RJ Changes in the genetic requirements for microbial interactions with increasing community complexity. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Kastman EK, Guasto JS & Wolfe BE Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat. Commun 9, 336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wetmore KM et al. Rapid Quantification of Mutant Fitness in Diverse Bacteria by Sequencing Randomly Bar-Coded Transposons. MBio 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallen-Adams HE & Suhr MJ Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frąc M, Hannula SE, Bełka M. & Jędryczka M. Fungal Biodiversity and Their Role in Soil Health. Front. Microbiol 9, 707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dukare AS et al. Exploitation of microbial antagonists for the control of postharvest diseases of fruits: a review. Crit. Rev. Food Sci. Nutr 59, 1498–1513 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Richards TA, Jones MDM, Leonard G. & Bass D. Marine fungi: their ecology and molecular diversity. Ann. Rev. Mar. Sci 4, 495–522 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Choi K-H, Lee H, Lee S, Kim S. & Yoon Y. Cheese Microbial Risk Assessments - A Review. Asian-australas. J. Anim. Sci 29, 307–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrin F. et al. Quantitative Risk Assessment of Haemolytic and Uremic Syndrome Linked to O157:H7 and Non-O157:H7 Shiga-Toxin Producing Escherichia coli Strains in Raw Milk Soft Cheeses : Quantitative Risk Assessment of HUS Linked to Pathogenic STEC in Cheese. Risk Anal. 35, 109–128 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Cosetta CM & Wolfe BE Deconstructing and Reconstructing Cheese Rind Microbiomes for Experiments in Microbial Ecology and Evolution. Curr. Protoc. Microbiol 56, e95 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Calvo AM, Wilson RA, Bok JW & Keller NP Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev 66, 447–59, table of contents (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nonejuie P, Burkart M, Pogliano K. & Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. U. S. A 110, 16169–16174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bok JW & Keller NP LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527–535 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosalková K. et al. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 91, 214–225 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Laich F, Fierro F. & Martín JF Production of penicillin by fungi growing on food products: identification of a complete penicillin gene cluster in Penicillium griseofulvum and a truncated cluster in Penicillium verrucosum. Appl. Environ. Microbiol 68, 1211–1219 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streit WR & Entcheva P. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol 61, 21–31 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Kastman EK et al. Biotic Interactions Shape the Ecological Distributions of Staphylococcus Species. MBio 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonham KS, Wolfe BE & Dutton RJ Extensive horizontal gene transfer in cheese-associated bacteria. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dean CR & Poole K. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol. Microbiol 8, 1095–1103 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Schalk IJ, Rigouin C. & Godet J. An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environ. Microbiol 22, 1447–1466 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Fecker L. & Braun V. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J. Bacteriol 156, 1301–1314 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer M, Hantke K. & Braun V. Ferric-coprogen receptor FhuE of Escherichia coli: processing and sequence common to all TonB-dependent outer membrane receptor proteins. J. Bacteriol 169, 2044–2049 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blin K. et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Triana S. et al. Draft Genome Sequence of the Animal and Human Pathogen Malassezia pachydermatis Strain CBS 1879. Genome Announc. 3, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar SK, Chowdhury C. & Ghosh AS Deletion of penicillin-binding protein 5 (PBP5) sensitises Escherichia coli cells to beta-lactam agents. Int. J. Antimicrob. Agents 35, 244–249 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Perrin RM et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3, e50 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep 31, 1266–1276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luckner M. [On the synthesis of quinoline alkaloids in plants. 2. Fermentativ conversion of the penicillin alkaloids cyclopenin and cyclopenol to viridicatin and viridicatol]. Eur. J. Biochem 2, 74–78 (1967). [DOI] [PubMed] [Google Scholar]
- 44.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW & Shirtliff ME Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev 25, 193–213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scherlach K, Graupner K. & Hertweck C. Molecular bacteria-fungi interactions: effects on environment, food, and medicine. Annu. Rev. Microbiol 67, 375–397 (2013). [DOI] [PubMed] [Google Scholar]
- 46.de Boer W. & Folman LB Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS microbiology (2005). [DOI] [PubMed]
- 47.Johansson JF, Paul LR & Finlay RD Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol 48, 1–13 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Tarkka MT, Sarniguet A. & Frey-Klett P. Inter-kingdom encounters: recent advances in molecular bacterium-fungus interactions. Curr. Genet 55, 233–243 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Taga ME & Walker GC Sinorhizobium meliloti requires a cobalamin-dependent ribonucleotide reductase for symbiosis with its plant host. Mol. Plant. Microbe. Interact 23, 1643–1654 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deveau A. et al. Role of fungal trehalose and bacterial thiamine in the improved survival and growth of the ectomycorrhizal fungus Laccaria bicolor S238N and the helper bacterium Pseudomonas fluorescens BBc6R8. Environ. Microbiol. Rep 2, 560–568 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Hantke K. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol. Gen. Genet 191, 301–306 (1983). [DOI] [PubMed] [Google Scholar]
- 52.Arias AA et al. Growth of desferrioxamine-deficient Streptomyces mutants through xenosiderophore piracy of airborne fungal contaminations. FEMS Microbiol. Ecol 91, (2015). [DOI] [PubMed] [Google Scholar]
- 53.Haas H, Eisendle M. & Turgeon BG Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol 46, 149–187 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Park M, Cho Y-J, Lee YW & Jung WH Understanding the Mechanism of Action of the Anti-Dandruff Agent Zinc Pyrithione against Malassezia restricta. Sci. Rep 8, 12086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gründlinger M. et al. Fungal siderophore biosynthesis is partially localized in peroxisomes. Mol. Microbiol 88, 862–875 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heymann P, Ernst JF & Winkelmann G. A gene of the major facilitator superfamily encodes a transporter for enterobactin (Enb1p) in Saccharomyces cerevisiae. Biometals 13, 65–72 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Sass G. et al. Studies of Pseudomonas aeruginosa Mutants Indicate Pyoverdine as the Central Factor in Inhibition of Aspergillus fumigatus Biofilm. J. Bacteriol 200, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Briard B. et al. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep 5, 8220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clancy A. et al. Evidence for siderophore-dependent iron acquisition in group B streptococcus. Mol. Microbiol 59, 707–721 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Jin B. et al. Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes. Mol. Microbiol 59, 1185–1198 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Mishra RPN et al. Staphylococcus aureus FhuD2 is involved in the early phase of staphylococcal dissemination and generates protective immunity in mice. J. Infect. Dis 206, 1041–1049 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Rocha ER & Krykunivsky AS Anaerobic utilization of Fe(III)-xenosiderophores among Bacteroides species and the distinct assimilation of Fe(III)-ferrichrome by Bacteroides fragilis within the genus. Microbiologyopen 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H. et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun 6, 8292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ong SA & Neilands JB Siderophores in microbially processed cheese. J. Agric. Food Chem 27, 990–995 (1979). [DOI] [PubMed] [Google Scholar]
- 65.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rehner SA & Samuels GJ Molecular systematics of the Hypocreales: a teleomorph gene phylogeny and the status of their anamorphs. Can. J. Bot 73, 816–823 (1995). [Google Scholar]
- 67.Glass NL & Donaldson GC Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol 61, 1323–1330 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ronquist F. & Huelsenbeck JP MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Dunnett CW A Multiple Comparison Procedure for Comparing Several Treatments with a Control. J. Am. Stat. Assoc 50, 1096–1121 (1955). [Google Scholar]
- 70.Wickham H. ggplot2: Elegant Graphics for Data Analysis. (2009).
- 71.Cleary JL, Luu GT, Pierce EC, Dutton RJ & Sanchez LM BLANKA: an Algorithm for Blank Subtraction in Mass Spectrometry of Complex Biological Samples. J. Am. Soc. Mass Spectrom 30, 1426–1434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohimani H. et al. Dereplication of microbial metabolites through database search of mass spectra. Nature Communications vol. 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohimani H. et al. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol 13, 30–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamini Y. & Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol 57, 289–300 (1995). [Google Scholar]
- 76.Tang Y, Horikoshi M. & Li W. ggfortify: Unified Interface to Visualize Statistical Result of Popular R Packages. The R Journal vol. 8 (2016). [Google Scholar]
- 77.Huerta-Cepas J. et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu G, Wang L-G, Han Y. & He Q-Y clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carlson M. org.EcK12.eg.db: Genome wide annotation for E coli strain K12. (2019) doi: 10.18129/B9.bioc.org.EcK12.eg.db. [DOI]
- 80.Carlson M. & Pagès H. AnnotationForge: Tools for building SQLite-based annotation data packages. R package version 1.26.0. [Google Scholar]
- 81.Baba T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwyn B. & Neilands JB Universal chemical assay for the detection and determination of siderophores. Anal. Biochem 160, 47–56 (1987). [DOI] [PubMed] [Google Scholar]
- 84.Payne SM Detection, isolation, and characterization of siderophores. Methods Enzymol. 235, 329–344 (1994). [DOI] [PubMed] [Google Scholar]
- 85.Grenier F, Matteau D, Baby V. & Rodrigue S. Complete Genome Sequence of Escherichia coli BW25113. Genome Announc. 2, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love MI, Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koren S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vaser R, Sović I, Nagarajan N. & Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walker BJ et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Min B, Grigoriev IV & Choi I-G FunGAP: Fungal Genome Annotation Pipeline using evidence-based gene model evaluation. Bioinformatics 33, 2936–2937 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Jones P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lim FY, Sanchez JF, Wang CCC & Keller NP Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 517, 303–324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie C. et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ashburner M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jukes TH & Cantor CR Evolution of protein molecules. Mammalian protein metabolism (1969).
- 99.Conway JR, Lex A. & Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Patti GJ et al. A view from above: cloud plots to visualize global metabolomic data. Anal. Chem 85, 798–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study (RB-TnSeq, RNA-Seq) have been deposited in the NCBI SRA database with SRA accession codes SRR11514793-SRR11514872 and BioProject PRJNA624168. Mass spectrometry data is available in the MassIVE database under accession numbers MSV000085070 and MSV000085054. The GNPS molecular network is available at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=464b331ef9d54de9957d23b4f9b9db14. The E. coli annotation database used for gene ontology functional enrichment is available at http://bioconductor.org/packages/release/data/annotation/html/org.EcK12.eg.db.html. The Whole Genome Shotgun project for Penicillium sp. str. #12 including reads, genome assembly, and annotation has been deposited at DDBJ/ENA/GenBank under the accession JAASRZ000000000 in BioProject PRJNA612335 (BioSample SAMN14369290 and SRA SRR11536435). In addition to these sources, data used to create figures 2,3,5, and 6 is available in the Supplementary Tables provided with the paper. Uncropped Southern blots associated with Supplementary Figure 3 are provided with the manuscript as Source Data.