Abstract
We present a genome assembly from a clonal population of Eimeria tenella Houghton parasites (Apicomplexa; Conoidasida; Eucoccidiorida; Eimeriidae). The genome sequence is 53.25 megabases in span. The entire assembly is scaffolded into 15 chromosomal pseudomolecules, with complete mitochondrion and apicoplast organellar genomes also present.
Keywords: Eimeria tenella, Apicomplexa, parasite, protist, genome sequence, chromosomal
Species taxonomy
Eukaryota; Apicomplexa; Conoidasia; Eucoccidiorida; Eimeriidae; Eimeria; Eimeria tenella Tyzzer 1929 (NCBItxid:5802).
Introduction
The genome of Eimeria tenella (Houghton strain) was sequenced as part of the Darwin Tree of Life Project, a collaborative effort to sequence all of the named eukaryotic species in Britain and Ireland. Here we present a chromosomally complete genome sequence based on a clonal specimen maintained initially at the Houghton Poultry Research Station (HPRS) and more recently at the Royal Veterinary College, Hertfordshire, UK, where it was collected from experimentally infected Gallus gallus domesticus. This apicomplexan parasite is a major cause of coccidiosis in farmed chickens in the UK.
Genome sequence report
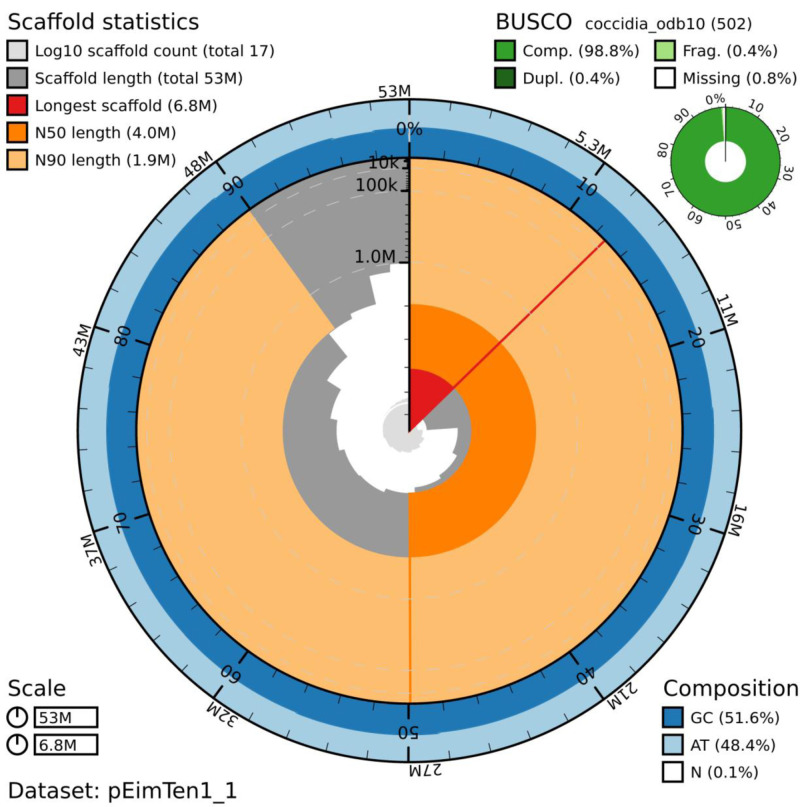
The genome was sequenced from a clonal specimen of E. tenella collected from experimentally infected G. gallus domesticus at the Royal Veterinary College, UK. A total of 41-fold coverage in Pacific Biosciences single-molecule long reads (N50 8 kb) and 107-fold coverage in 10X Genomics read clouds were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 200 missing/misjoins, reducing the scaffold number by 77.9%, increasing the scaffold N50 by 0.1% and decreasing the assembly length by 1.85%. The final assembly has a total length of 53.25 Mb in 15 chromosomal scaffolds, one mitochondrial scaffold and one apicoplast scaffold. The total scaffold N50 was 4.01 Mb ( Table 1). The chromosomal scaffolds are numbered by sequence length, 1 being the smallest and 15 the largest, as is typical for Apicomplexa ( Figure 1– Figure 3; Table 2). The organellar mitochondrial and apicoplast genome sequences were each assembled into single contigs and circularized to remove redundancy. The assembly has a BUSCO v5.1.2 ( Simao et al., 2015) completeness of 98.8% and duplication rate of 0.2% using the coccidia_odb10 reference set.
Table 1. Genome data for Eimeria tenella, pEimTen1.1.
| Project accession data | |
|---|---|
| Assembly identifier | pEimTen1.1 |
| Species | Eimeria tenella |
| Specimen | pEimTen1 |
| NCBI taxonomy ID | NCBI:txid5802 |
| BioProject | PRJEB43184 |
| BioSample ID | SAMEA7524401 |
| Isolate information | Clonal specimen, Houghton strain |
| Raw data accessions | |
| PacificBiosciences SEQUEL I | ERR6447337 |
| 10X Genomics Illumina | ERX5693366-ERX5693369 |
| Hi-C Illumina | ERX5693901 |
| Genome assembly | |
| Assembly accession | GCA_905310635.1 |
| Span (Mb) | 381 |
| Number of contigs | 35 |
| Contig N50 length (Mb) | 14 |
| Number of scaffolds | 33 |
| Scaffold N50 length (Mb) | 14 |
| Longest scaffold (Mb) | 16 |
| BUSCO * genome score | C:98.8%[S:98.4%,D:0.4%],F:0.4%,
M:0.8%,n:502 |
*BUSCO scores based on the coccodia_odb10 BUSCO set using v5.1.2. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/Eimeria%20tenella/dataset/pEimTen1_1/busco.
Figure 1. Genome assembly of Eimeria tenella Houghton, pEimTen1.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/pEimTen1.1/dataset/pEimTen1_1/snail.
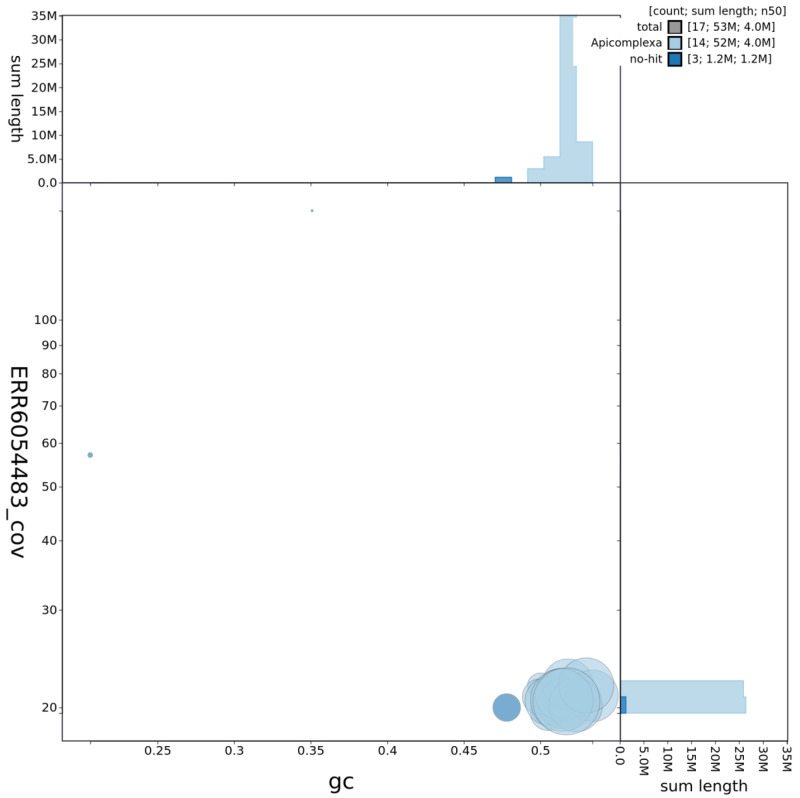
Figure 2. Genome assembly of Eimeria tenella Houghton, pEimTen1.1: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of chromosome length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/pEimTen1.1/dataset/pEimTen1_1/blob.
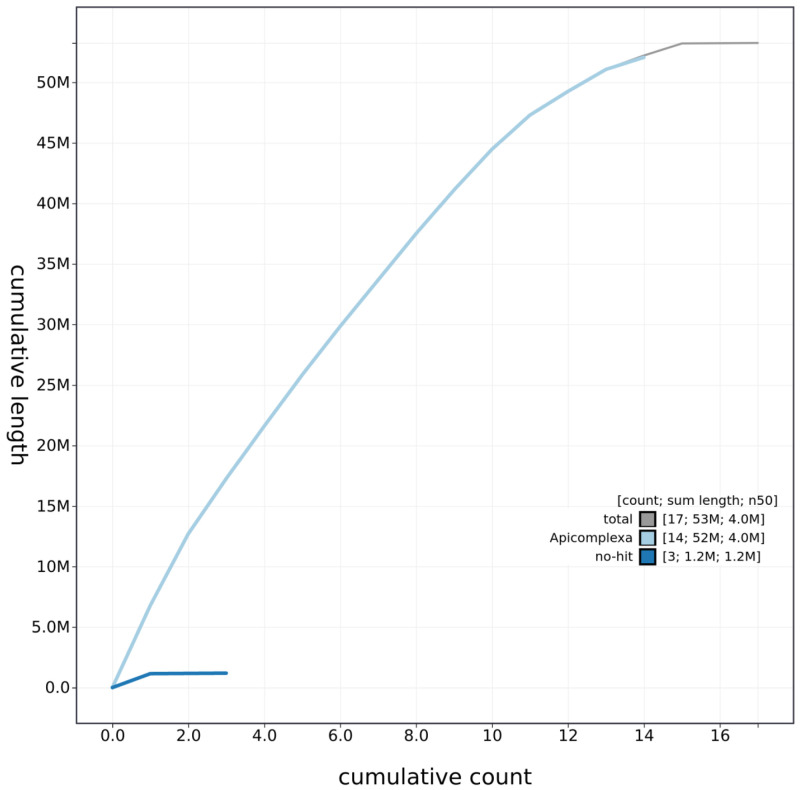
Figure 3. Genome assembly of Eimeria tenella Houghton, pEimTen1.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all chromosomes. Coloured lines show cumulative lengths of chromosomes assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/pEimTen1.1/dataset/pEimTen1_1/cumulative.
Table 2. Chromosomal pseudomolecules in the genome assembly of Eimeria tenella, pEimTen1.1.
The numbering of chromosomes is based on ordering the pEimTen1.1 assembly scaffolds by size in reverse order, so the chromosome names do not necessarily correspond to chromosome names in previously existing literature on Eimeria tenella.
| INSDC
accession |
Chromosome | Size (kb) | GC% | Gaps | Putative centromeric
region (bp) |
|---|---|---|---|---|---|
| HG994961 | 1 | 998.4 | 50.0 | 0 | 837838-871939 |
| HG994962 | 2 | 1,151.2 | 47.8 | 1 | 530700-562938 |
| HG994963 | 3 | 1,819.2 | 50.5 | 1 | 1605419-1629071 |
| HG994964 | 4 | 1,948.7 | 50.0 | 2 | 1130403-1164585 |
| HG994965 | 5 | 2,810.7 | 52.0 | 2 | 2256341-2281694 |
| HG994966 | 6 | 3,367.5 | 51.8 | 3 | 2700201-2728956 |
| HG994967 | 7 | 3,616.8 | 50.8 | 9 | 1871503-1901473 |
| HG994968 | 8 | 3,810.9 | 51.5 | 1 | 1320955-1355380 |
| HG994969 | 9 | 3,854.2 | 51.8 | 2 | 2305986-2344056 |
| HG994970 | 10 | 4,007.2 | 53.4 | 1 | 2379713-2394995 |
| HG994971 | 11 | 4,218.1 | 51.4 | 4 | 747612-790704 |
| HG994972 | 12 | 4,348.4 | 52.3 | 0 | 418148-444959 |
| HG994973 | 13 | 4,564.6 | 53.0 | 7 | 830432-888266 |
| HG994974 | 14 | 5,913.3 | 51.4 | 4 | 3126091-3200449 |
| HG994975 | 15 | 6,779.9 | 51.7 | 9 | 346670-377612 |
| HG994976 | MT | 6.2 | 35.0 | 0 | N/A |
| HG994977 | Apicoplast | 34.8 | 20.5 | 0 | N/A |
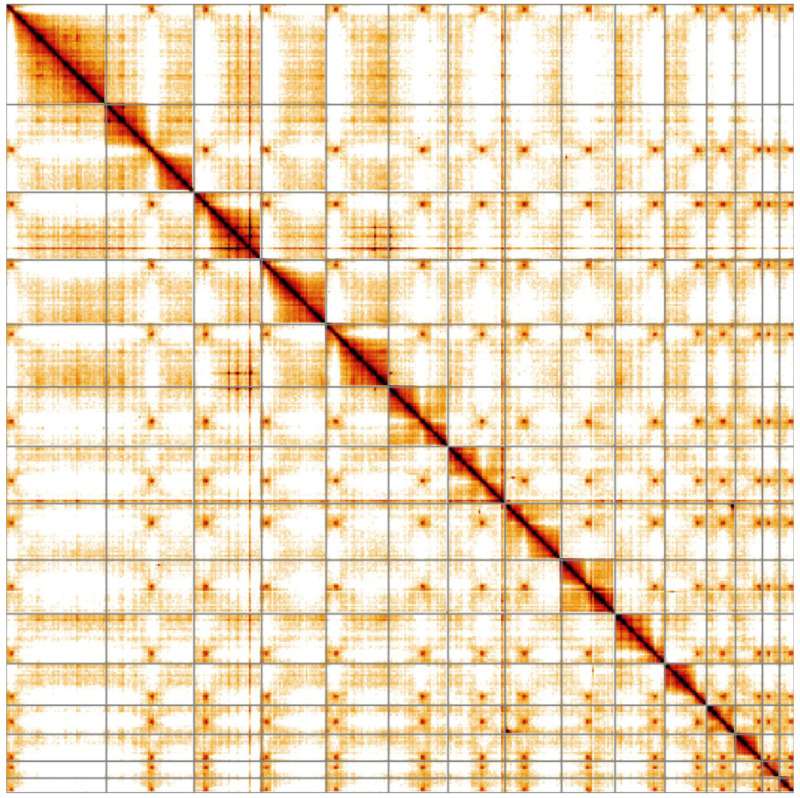
Of particular note is that 15 chromosomal scaffolds were identified, each with telomeres attached to both ends. This calls into question previous reports which suggested a haploid chromosome number of 14 for this species ( del Cacho et al., 2005). The Hi-C map ( Figure 4) shows that each of the 15 chromosomal scaffolds has a single contact region with each of the others. It has been shown in the coccidian relative Toxoplasma gondii that centromeres are sequestered together within the nucleus throughout the cell cycle ( Brooks et al., 2011). The Hi-C map suggests that this also occurs in E. tenella and if true, further supports the existence of 15 chromosomes. We examined the putative centromeric regions as identified by Hi-C in the Artemis genome browser ( Carver et al., 2012) and found almost all to be in intergenic regions of, on average, 35 kb (min=15 kb, max=74 kb). The exception was chromosome 1, where it was adjacent to a repeat near to the end of the chromosome. The data suggest that E. tenella chromosomes have single, well-localised centromeres which occupy acrocentric and sub-metacentric positions ( Table 2).
Figure 4. Genome assembly of Eimeria tenella, pEimTen1.1: Hi-C contact map.
Hi-C contact map of the pEimTen1.1 assembly, visualised in HiGlass.
The GC content of the genome was 58.6%.
Genome annotation report
We identified 7268 protein coding genes. Around 2000 gene models were manually corrected. The average exon length was 350.1, average intron length 298.1, with an average of 6.34 exons per gene. We annotated 44 pseudogenes, 32 degraded LTR retrotransposons (currently not included in GFF annotation), 140 rRNAs, 31 repeat regions, 28 ncRNAs and 345 tRNAs.
Methods
A clonal specimen of E. tenella was collected from experimentally infected G. gallus domesticus at the Royal Veterinary College, Hertfordshire, UK. Four-week-old Lohmann Valo chickens reared under specific pathogen-free conditions were used to propagate oocysts of the E. tenella Houghton strain as described previously ( Long et al., 1976). Standard methods were used to purify and sporulate oocysts and to purify sporozoites through nylon wool and DE-52 columns ( Pastor-Fernández et al., 2019; Shirley et al., 1995). Animals were raised in strict accordance with the Animals (Scientific Procedures) Act 1986, an Act of Parliament of the United Kingdom. All animal studies and protocols were approved by the Royal Veterinary College Animal Welfare & Ethical Review Body (AWERB) and the UK Government Home Office under specific project licence.
DNA was extracted from the clonal specimen using the Qiagen MagAttract HMW DNA kit according to the manufacturer’s instructions. Pacific Biosciences CLR long read and 10X Genomics read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Hi-C data were generated using the Arima Hi-C kit. Sequencing was performed by the Scientific Operations DNA Pipelines at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL I (long read), Illumina HiSeq (10X) and Illumina MiSeq (Hi-C) instruments.
The assembly pEimTen1.1 is based on 41x PacBio data, 10X Genomics Chromium data, and Arima Hi-C data generated by the Darwin Tree of Life Project. PacBio subreads were assembled with Canu 1.6 ( Koren et al., 2017). After running Canu, some deduplication of contigs was performed using GAP5 v1.2.14-r3753M ( Bonfield & Whitwham, 2010). The assembly was scaffolded with scaff10x 4.2 using E. tenella 10x Chromium Illumina reads. This was then broken with break10x 3.1 and re-scaffolded using SALSA2 (October 2019 version) ( Ghurye et al., 2019) and E. tenella Hi-C reads. Juicebox 1.9.1 ( Robinson et al., 2018) and Tigmint 1.1.2 ( Jackman et al., 2018) were used to break scaffolds. RaGOO 1.1 ( Alonge et al., 2019) was then used to re-scaffold, using another assembly generated from the same PacBio reads using wtdbg2 2.5 (20190621) ( Ruan & Li, 2020). The assembly was then polished with Arrow (gcpp 1.0.0-SL-release-8.0.0, with pbmm2 version 1.1.0). Further polishing of the assembly was done with Pilon 1.19 ( Walker et al., 2014), using 10x Chromium Illumina reads from which 10x bar codes and linkers had been removed. The assembly was checked for contamination and analysed using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation was performed using gEVAL, HiGlass ( Kerpedjiev et al., 2018) and Pretext, before final polishing with Pilon. The genome was analysed and BUSCO v5.1.2 scores generated using BlobToolKit 2.6.1 ( Challis et al., 2020). The software tools used, with versions, are summarised in Table 3.
Table 3. Software tools used.
| Software
tool |
Version | Source |
|---|---|---|
| Canu | 1.6 | ( Koren et al., 2017) |
| GAP5 | v1.2.14-r3753M | ( Bonfield & Whitwham, 2010) |
| scaff10x | 4.2 | https://github.com/wtsi-hpag/Scaff10X |
| break10x | 3.1 | https://github.com/wtsi-hpag/Scaff10X |
| SALSA2 | October 2019 | ( Ghurye et al., 2019) |
| Juicebox | 1.9.1 | ( Durand et al., 2016) |
| Tigmint | 1.1.2 | ( Jackman et al., 2018) |
| RaGOO | 1.1 | ( Alonge et al., 2019) |
| Wtdbg2 | 2.5 (20190621) | ( Ruan & Li, 2020) |
| Arrow | gcpp 1.0.0-SL-release-8.0.0 | https://github.com/PacificBiosciences/GenomicConsensus |
| Pilon | 1.19 | ( Walker et al., 2014) |
| STAR | 2.5.3a | ( Dobin et al., 2013) |
| Cufflinks | 2.2.1 | ( Trapnell et al., 2010) |
| HISAT2 | 2.2.0 | ( Kim et al., 2019) |
| Companion | May 2020 | ( Steinbiss et al., 2016) |
| gEVAL | N/A | ( Chow et al., 2016) |
| HiGlass | 1.11.8 | ( Kerpedjiev et al., 2018) |
| PretextView | 0.1 | https://github.com/wtsi-hpag/PretextView |
| BlobToolKit | 2.6.1 | ( Challis et al., 2020) |
An initial annotation was performed using Companion ( Steinbiss et al., 2016) with the previous Eimeria tenella strain Houghton assembly and annotation as the reference ( Ling et al., 2007). Eimeria tenella RNA-seq reads (from project PRJEB3308 in the European Nucleotide Archive, runs ERR178634, ERR178635, ERR178636, ERR178637 and ERR178638 ( Reid et al., 2014)) were mapped to the assembly using 2-pass mapping method with STAR RNA-seq aligner version 2.5.3a ( Aunin et al., 2020; Dobin et al., 2013). The mapped reads were processed with Cufflinks v2.2.1 ( Trapnell et al., 2010) to produce a GTF file, which was then used as an input for Companion. Companion (May 2020 version) was run with Augustus threshold set to 0.2, alignment of proteins to the target genome enabled and other settings left as default. The annotations were then manually curated using Artemis v18.1.0 ( Rutherford et al., 2000) and the Artemis Comparison Tool v18.1.0 ( Carver et al., 2005) with the help of previously published RNA-seq data ( Reid et al., 2014). For viewing in Artemis, the RNA-seq data ( Reid et al., 2014) was mapped to the assembly with HISAT2 2.2.0 ( Kim et al., 2019).
Data availability
European Nucleotide Archive: Eimeria tenella (Coccidian parasite). Accession number PRJEB43184: https://identifiers.org/ena.embl:PRJEB43184
The genome sequence is released openly for reuse. The E. tenella genome sequencing initiative is part of the Darwin Tree of Life (DToL) project. All raw sequence data and the assembly have been deposited in INSDC databases. Raw data and assembly accession identifiers are reported in Table 1.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328). SAM is supported by Wellcome (207492).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- Alonge M, Soyk S, Ramakrishnan S, et al. : RaGOO: Fast and Accurate Reference-Guided Scaffolding of Draft Genomes. Genome Biol. 2019;20(1):224. 10.1186/s13059-019-1829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aunin E, Böhme U, Sanderson T, et al. : Genomic and Transcriptomic Evidence for Descent from Plasmodium and Loss of Blood Schizogony in Hepatocystis Parasites from Naturally Infected Red Colobus Monkeys. PLoS Pathogens. 2020;16(8):e1008717. 10.1371/journal.ppat.1008717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield JK, Whitwham A: Gap5—editing the Billion Fragment Sequence Assembly. Bioinformatics. 2010;26(14):1699–1703. 10.1093/bioinformatics/btq268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CF, Francia ME, Gissot M, et al. : Toxoplasma Gondii Sequesters Centromeres to a Specific Nuclear Region throughout the Cell Cycle. Proc Natl Acad Sci U S A. 2011;108(9):3767–72. 10.1073/pnas.1006741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Harris SR, Berriman M, et al. : Artemis: An Integrated Platform for Visualization and Analysis of High-Throughput Sequence-Based Experimental Data. Bioinformatics. 2012;28(4):464–69. 10.1093/bioinformatics/btr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, et al. : ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21(16):3422–23. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL — a Web-Based Browser for Evaluating Genome Assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Cacho E, Pages M, Gallego M, et al. : Synaptonemal Complex Karyotype of Eimeria Tenella. Int J Parasitol. 2005;35(13):1445–51. 10.1016/j.ijpara.2005.06.009 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, et al. : STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics. 2013;29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, et al. : Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 2016;3(1):99–101. 10.1016/j.cels.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. Gigascience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman SD, Coombe L, Chu J, et al. : Tigmint: Correcting Assembly Errors Using Linked Reads from Large Molecules. BMC Bioinformatics. 2018;19(1):393. 10.1186/s12859-018-2425-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, et al. : Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat Biotechnol. 2019;37(8):907–15. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, et al. : Canu: Scalable and Accurate Long-Read Assembly via Adaptive K-Mer Weighting and Repeat Separation. Genome Res. 2017;27(5):722–36. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KH, Rajandream MA, Rivailler P, et al. : Sequencing and Analysis of Chromosome 1 of Eimeria Tenella Reveals a Unique Segmental Organization. Genome Res. 2007;17(3):311–19. 10.1101/gr.5823007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long PL, Millard BJ, Joyner LP, et al. : A Guide to Laboratory Techniques Used in the Study and Diagnosis of Avian Coccidiosis. Folia Vet Lat. 1976;6(3):201–17. [PubMed] [Google Scholar]
- Pastor-Fernández I, Pegg E, Macdonald SE, et al. : Laboratory Growth and Genetic Manipulation of Eimeria Tenella. Curr Protoc Microbiol. 2019;53(1):e81. 10.1002/cpmc.81 [DOI] [PubMed] [Google Scholar]
- Reid AJ, Blake DP, Ansari HR, et al. : Genomic Analysis of the Causative Agents of Coccidiosis in Domestic Chickens. Genome Res. 2014;24(10):1676–85. 10.1101/gr.168955.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Turner D, Durand NC, et al. : Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Syst. 2018;6(2):256–58.e1. 10.1016/j.cels.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H: Fast and Accurate Long-Read Assembly with wtdbg2. Nat Methods. 2020;17(2):155–58. 10.1038/s41592-019-0669-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, et al. : Artemis: Sequence Visualization and Annotation. Bioinformatics. 2000;16(10):944–45. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- Shirley MW, Bushell AC, Bushell JE, et al. : A Live Attenuated Vaccine for the Control of Avian Coccidiosis: Trials in Broiler Breeders and Replacement Layer Flocks in the United Kingdom. Vet Rec. 1995;137(18):453–57. 10.1136/vr.137.18.453 [DOI] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics. 2015;31(19):3210–12. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Steinbiss S, Silva-Franco F, Brunk B, et al. : Companion: A Web Server for Annotation and Analysis of Parasite Genomes. Nucleic Acids Res. 2016;44(W1):W29–34. 10.1093/nar/gkw292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. : Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat Biotechnol. 2010;28(5):511–15. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, et al. : Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS One. 2014;9(11):e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]