Abstract
Chromosomal rearrangements are important resources for genetic studies. Recently, a Cre-loxP-based method to introduce defined chromosomal rearrangements (deletions, duplications, and inversions) into the mouse genome (chromosome engineering) has been established. To explore the limits of this technology systematically, we have evaluated this strategy on mouse chromosome 11. Although the efficiency of Cre-loxP-mediated recombination decreases with increasing genetic distance when the two endpoints are on the same chromosome, the efficiency is not limiting even when the genetic distance is maximized. Rearrangements encompassing up to three quarters of chromosome 11 have been constructed in mouse embryonic stem (ES) cells. While larger deletions may lead to ES cell lethality, smaller deletions can be produced very efficiently both in ES cells and in vivo in a tissue- or cell-type-specific manner. We conclude that any chromosomal rearrangement can be made in ES cells with the Cre-loxP strategy provided that it does not affect cell viability. In vivo chromosome engineering can be potentially used to achieve somatic losses of heterozygosity in creating mouse models of human cancers.
Specific chromosomal rearrangements can be engineered in mice to model human chromosomal disorders, such as those associated with deletions or duplications of chromosomal segments (for example, Smith-Magenis syndrome, Downs syndrome, and Charcot-Marie-Tooth type 1A) (5, 7, 10). Chromosomal rearrangements also facilitate genetic studies (2, 14). Inversion chromosomes can be used to establish balanced lethal systems to facilitate stock maintenance. Deletions can be used for mapping and in genetic screens for recessive mutations.
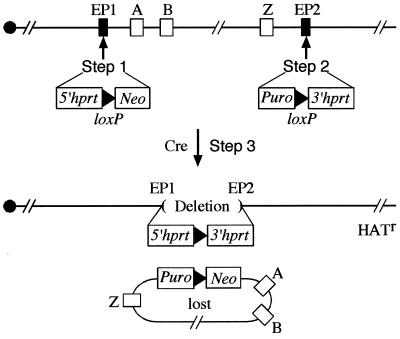
In Drosophila melanogaster there is a wealth of chromosomal rearrangements that are widely used as genetic tools. In particular, chromosomal deletions (deficiencies) which collectively cover approximately 60 to 70% of the genome have been indispensable in mapping recessive mutations and in region-specific mutagenesis screens. The use of deletions in mice, however, has been much more limited because of the paucity of chromosomal deletions which, until recently, were restricted to a few regions of the mouse genome flanking visible genetic markers (14). The application of the Cre-loxP recombination system over large distances in mouse embryonic stem (ES) cells has made it possible to engineer specific chromosomal rearrangements in the mouse (13, 17). This chromosome engineering strategy involves three manipulation steps in ES cells (see Fig. 1): (i) one loxP site is targeted to one endpoint along with the 5′ half of an Hprt selectable marker gene (5′ hprt); (ii) another loxP site is targeted to a second endpoint with the 3′ half of the Hprt gene (3′ hprt); and (iii) transient expression of Cre recombinase catalyzes loxP site-specific recombination, leading to the desired rearrangement. Reconstitution of a full-length Hprt gene provides selection for ES cells with the recombination products in culture in HAT (hypoxanthine-aminopterin-thymidine) medium. By using this technology, deletions, duplications, inversions, or translocations can be generated depending upon the relative position and orientation of the two loxP sites and selection cassettes (13, 17).
FIG. 1.
The Cre-loxP based chromosome engineering strategy. 5′ hprt was previously named hprtΔ3′; 3′ hprt was previously name hprtΔ5′. A neomycin (Neo) or a puromycin (Puro) resistance gene is linked to the first or the second loxP site, respectively, for positive selection during gene targeting. In this case, Cre recombination between two loxP sites targeted in the same orientation in cis (on the same chromosome) leads to a deletion that is neomycin and puromycin sensitive due to the loss of the Neo- and Puro-carrying reciprocal product, a ring chromosome in G1 (shown) or a duplication sister chromatid in G2 (not shown). If the two loxP sites are on the two different chromosome homologues (in trans), a deletion and a duplication will be produced. The rearrangements can then be transmitted through the mouse germline if viable.
The Cre-loxP chromosome engineering strategy provides a unique and unprecedented opportunity to manipulate the mouse genome. However, several critical questions remain to be answered in order to explore fully the potential of this technology. First, is there any limit as to the kind and size of rearrangements that can be made with this technology? While there are likely to be biological limits in mice, ES cells harboring large chromosomal deletions offer an opportunity to perform haploid genetic screens in vitro. For such applications, the larger the deletion, the more powerful the screen. Second, what is the efficiency of Cre-mediated recombination for substrates of different genetic distances? This will be pertinent to the scope and applicability of this technology. Third, can this strategy be used to engineer chromosomes somatically, that is, in a tissue- or cell-type-specific manner without the strong positive selection schemes that are used in cell culture? Tissue-specific deletions also enable recessive genetics to be employed somatically, for instance, to induce loss of heterozygosity (LOH) to model genetic changes in human cancers or to conduct screens for novel tumor suppressor genes in combination with mutagenesis strategies. Somatically induced deletions may avoid the developmental problems associated with larger germline deletions and consequently a larger chromosomal region can be studied in a single animal.
To address these questions, we applied the Cre-loxP chromosome engineering strategy to various parts of mouse chromosome 11 (Chr 11) in ES cells and in vivo. With an improved selection cassette, we obtained an 11% deletion efficiency for a two-centimorgan (2-cM; equivalent to 4 Mb) deletion substrate in murine ES cells. Rearrangements of up to three-quarters of Chr 11 have been made, demonstrating that there appears to be no recombination-based restriction as to what type of rearrangements can be made provided that ES cells tolerate the genetic change. We found that the efficiency of Cre-mediated recombination between two loxP sites on the same chromosome (cis) decreases with increasing genetic distance. We found that large chromosomal deletions may be deleterious to ES cells and that deletions which were lethal to developing embryos could be engineered somatically at high efficiencies, breaking ground for somatic chromosome engineering.
MATERIALS AND METHODS
Construction of targeting vectors.
The Hsd17b1 targeted cell line has been described elsewhere (13). The targeting vectors for Wnt3 (modified from a previous version [8]) and p53 have also been described elsewhere (24). All microsatellite markers were targeted with insertion vectors. The targeting vectors for D11Mit199 and D11Mit69 were modified from previous versions (8), replacing the mutant 3′ hprt cassette with the wild-type sequence.
The D11Mit142 and D11Mit71 loci were targeted with insertion vectors generated from a targeting-ready genomic library that contains the puromycin resistance gene, a loxP site, 3′ hprt cassette, and an agouti coat color transgene in the vector backbone (23). Clones isolated from this library were restriction mapped, and a gap was created in the region of homology which was used as the probe to detect targeting by Southern analysis. The targeting vectors for D11Mit142 have been described (23). A clone with a 10.9-kb genomic insert at D11Mit71 was isolated from the 3′ hprt library and mapped with several restriction enzymes. The insert consists of two flanking (2.3 and 3.9 kb) and three internal (0.8, 3.3, and 0.6 kb) NcoI fragments. The internal fragments were deleted from the clone to create a gap in the region of homology, resulting in targeting vector pTVD11Mit71F. The insert was then flipped by using rare cutter AscI sites that flank the insert, resulting in targeting vector pTVD11Mit71R which was used to deliver the loxP site to the D11Mit71 locus with the reverse orientation. The 3.3-kb internal NcoI fragment was used as the probe in mini-Southern analysis to detect gap repair-dependent targeting events (20, 23). This probe hybridizes to a 6-kb and a (weak) 2.8-kb wild-type EcoRI fragment and, in targeted clones, an additional 18.6-kb targeted fragment resulting from the insertion of the vector sequence into the targeted locus.
Generation and analysis of chromosomal rearrangements.
ES cell cultures, gene targeting, and germ line transmission were performed as described previously (12). AB2.2 ES cells were used in most experiments except in a few cases where a hybrid ES cell line (between 129S7 and C57BL/6-Tyrc-Brd), ER3.4, was used (E. Regel and A. Bradley, unpublished data). Electroporation of the Cre expression plasmid pOG231 (11), selection of Cre recombination products with HAT medium, and drug (neomycin and puromycin) resistance tests were performed as described earlier (8, 13) with some modifications. ES cells (80% confluent) were passaged 1 day before electroporation and fed with medium 2 h before electroporation. The cells were then trypsinized and resuspended in phosphate-buffered saline (PBS), and cell counting was performed with a Coulter Counter. The cells were again suspended in PBS to make the final cell density of 1.1 × 107 cells/ml. In a typical transient Cre expression experiment, 25 μg of pOG231 (prepared by CsCl centrifugation, unlinearized) was electroporated into 107 ES cells in 0.9 ml of PBS. The electroporation was conducted with a Bio-Rad GenePulser and a Gene Pulser cuvette with a 0.4-cm electrode gap at 230 V and 500 μF. Cells (in PBS) were then mixed with M15 medium and plated on two to three plates at different densities. For the cis 2-cM substrates, electroporated cells were subject to serial dilution before plating to enable counting of the HAT-resistant colonies. HAT selection was initiated about 48 h after electroporation, maintained for 8 days and released in hypoxanthine thymidine (HT) for 2 days before the colonies were counted and picked. In all experiments a 104 dilution was also plated for each cell line under no selection to count and calculate the number of colonies that survived electroporation. Assessed by this procedure, usually about one-third of the cells undergoing electroporation survived and formed colonies on feeder plates in M15 medium. To control between different experiments, a 2-cM double-targeted cell line was included in each experiment as a control for the Cre recombination efficiency, and this efficiency (∼11%) has been consistent throughout all of the experiments.
FISH.
Metaphase chromosome spreads from ES cells were prepared as described previously (15). Fluorescence in situ hybridization (FISH) was performed with phage or BAC probes according to a standard protocol (3). The mPer1 phage clone, BAC 330H2, was labeled with digoxigenin and detected by anti-digoxigenin-rhodamine antibody. BAC 293C22 and BAC 330P14 were labeled with biotin and detected with fluorescein isothiocyanate-avidin. BAC 232M23 was labeled with a mixture of digoxigenin and biotin. The chromosomes were stained with DAPI (4′,6′-diamidino-2-phenylindole). The images were taken as monotonic pictures, and the composites were made with artificial coloration for clarity.
PCR and sequence analysis.
Primer Pa, 5′-AGG ATG TGA TAC GTG GAA GA (Hprt intron, forward), and primer Pb, 5′-GCC GTT ATT AGT GGA GAG GC (polymerase II promoter in the neomycin resistance gene, reverse), were used to specifically amplify by PCR a fragment containing exon 3 sequence in the 5′ hprt cassette. Primer Pa and primer Pc, 5′-CCA GTT TCA CTA ATG ACA CA (Hprt exon 9, reverse), were used to specifically amplify exon 3 sequence in the 3′ hprt cassette. Primer Pd, 5′-GCA TTG TTT TGC CAG TGT C (Hprt exon 6, reverse), was used to sequence exon 3 in the PCR products.
The primers used to detect the cardiac specific 2-cM deletion were P1, 5′-CCT CAT GGA CTA ATT ATG GAC (Hprt exon 2, forward), and P2, the same as Pc (Hprt exon 9, reverse). The primer pair used to detect the αMyHC-Cre (Cre coding sequence under the control of α-myosin heavy chain promoter) transgene has been described elsewhere (1).
RESULTS
High-efficiency Cre-loxP based chromosomal engineering with an improved vector in mouse ES cells.
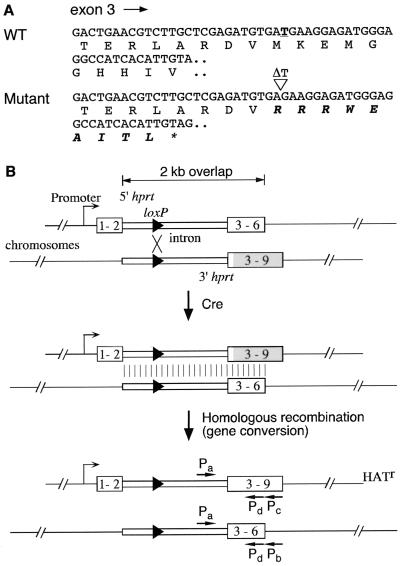
Sequence analysis identified a frameshift mutation in the coding portion of the 3′ hprt selection cassette (Fig. 2) previously successfully used for chromosome engineering (13), leading to a translation stop codon nine codons downstream of the mutation (Fig. 2A). This mutation should render a reconstituted Hprt minigene nonfunctional, yet HAT-resistant colonies were obtained with this cassette. These may have resulted from a repair event during or following Cre recombination (see below). Since the events we have scored to date required selection, the efficiency of Cre-mediated loxP site-specific recombination on multimegabase substrates may be greater than that scored by the number of selected HAT-resistant clones. Because the recombination efficiency is pertinent in applications of the Cre-loxP-based chromosome engineering strategy, we reassessed this efficiency for a 2-cM interval between Hsd17b1 (E2DH) and D11Mit199 on Chr 11 (8) by using cassettes without the frameshift mutation. The D11Mit199 locus was retargeted with the corrected 3′ hprt cassette in an ES cell line that had been targeted at the Hsd17b1 locus with the 5′ hprt cassette (13) so that the loxP sites were in the same orientation (8). The double-targeted cell lines were electroporated with a Cre expression plasmid (pOG231) (11) or a control plasmid (TyBS) (22), and the recombination efficiency was assessed (defined here as the number of HAT-resistant colonies per cell surviving electroporation). No HAT-resistant colony was obtained with the control plasmid. With the Cre expression plasmid, approximately half of the double-targeted clones yielded recombination efficiencies of approximately 11%, while the rest had efficiencies of approximately 0.047% (Table 1). Cre recombination products from the former group are puromycin sensitive (indicating that these cell lines have a deletion), while those from the latter group are puromycin resistant (indicating that these cell lines have a deletion and a duplication). Therefore, the two different efficiencies reflect the configuration of the targeted loxP sites on the Chr 11 homologues in the parental cell lines, which is resolved into a deletion in cis and which is resolved into a deletion and a duplication in trans (8, 13). The cis events occurred several hundred times more efficiently than the trans events. To rule out any effect of HAT selection on Cre-mediated recombination, we transfected three cis double-targeted cell lines with the Cre expression plasmid and randomly picked colonies grown under no drug selection. We then determined the percentage of the colonies that were recombined by both drug resistance test and Southern analysis on individual clones. Of 279 colonies picked for all three double-targeted cell lines, 24 had undergone the 2-cM Cre-mediated deletion, yielding a Cre recombination efficiency of ∼9%, which is not significantly different from that assessed by HAT selection. As a control, the 2-cM substrate with the mutant 3′ hprt cassette gave Cre recombination efficiencies of 0.007% for cis and 0.0001% for trans (8). Thus, the Cre recombination efficiency is improved by approximately 3 orders of magnitude after correction of the frameshifted 3′ hprt selection cassette.
FIG. 2.
A frameshift mutation in the original 3′ hprt cassette used in chromosome engineering. (A) Partial exon 3 sequence of the wild-type (WT) and mutant 3′ hprt cassette with conceptual translation. The thymidine residue in the wild-type sequence that is deleted in the mutant is in boldface and underlined. The altered amino acid residues affected by the mutation is in italicized boldface. ∗, Stop codon. (B) A proposed mechanism by which HAT-resistant colonies were obtained by a combination of Cre-loxP site-specific recombination and homologous recombination, as shown for Cre recombination between two loxP sites in trans in the same orientation that leads to a deletion and a duplication. 1–2, 3–6, and 3–9 refer to Hprt exons. Pa, Pb, Pc, and Pd, primers for PCR and sequence analysis of exon 3 of the Hprt gene. The portion of the coding sequence affected by the mutation in exons 3–9 is shaded. The polyadenylation signal and neomycin and puromycin resistance genes are not shown for simplicity.
TABLE 1.
Efficiency of Cre-mediated loxP site-specific recombination over different genetic distances
| Interval (5′ hprt-3′ hprt) | Genetic distance (cM) | Cre recombination efficiencya (no. of colonies in HAT/no. of colonies with no drug)
|
||
|---|---|---|---|---|
| loxP sites in opposite orientation: cis (inversion)b (n) |
loxP sites in the same orientation
|
|||
| cis (deletion) (n) | trans (deletion-duplication) (n) | |||
| Hsd17b1-D11Mit199 | 2 | ND | (1.1 ± 0.5) × 10−1 (8) | (4.7 ± 0.7) × 10−4 (5) |
| Hsd17b1-D11Mit69 | 22 | ND | (3.7 ± 2.4) × 10−5 (4) | (3.1 ± 1.1) × 10−5 (11) |
| Wnt3-p53 | 24 | (2.2 ± 0.6) × 10−3 (5) | 2.9 × 10−5 (1)c | (8.2 ± 0.9) × 10−5 (5) |
| Hsd17b1-D11Mit142 | 30 | (3.2 ± 0.7) × 10−4 (3) | (9.8 ± 1.7) × 10−6 (2)c | (5.9 ± 4.7) × 10−5 (5) |
| Hsd17b1-D11Mit71 | 60 | (8.3 ± 0.8) × 10−5 (2) | (9.5 ± 2.1) × 10−7 (2)c | (1.4 ± 0.2) × 10−5 (4) |
Numbers are the means ± the standard deviations. ND, not determined.
No colonies were obtained with the trans configuration because dicentric and acentric chromosomes are produced.
Not confirmed by FISH analysis.
Coupled Cre-loxP recombination and gene conversion.
The Hprt cassette reconstructed by Cre-loxP recombination from the mutant 3′ hprt selection cassette should be nonfunctional. However, HAT-resistant colonies were readily obtained (8, 13). This raised the question as to the nature of the event that leads to the HAT-resistant colonies in these experiments. The frequency of spontaneous reversion is too low to explain the observed frequency of HAT-resistant clones from the mutant cassette. The frameshift mutation is located in a 2-kb overlap between the 5′ and the 3′ hprt cassettes, and therefore the mutation in the 3′ cassette may be corrected by homologous recombination with sequences in the 5′ cassette. We hypothesized that Cre brings the two loxP sites together to promote site-specific recombination and that during or immediately after this process the endogenous homologous recombination machinery repairs the mutation (Fig. 2B). This notion would predict that all recombination products would have the wild-type exon 3 sequence rather than a correcting single nucleotide insertion resulting from a spontaneous reversion. Sequence analysis demonstrated that all HAT-resistant colonies had acquired a wild-type sequence in the reconstituted full-length Hprt minigene (n = 10) (see Fig. 2B and Materials and Methods).
trans recombination events also generate an Hprt+ deletion chromosome and the reciprocal product, a duplication chromosome, which retains the recombined overlapping region between the 5′ and 3′ cassettes (Fig. 2B). Sequence analysis of PCR products from exon 3 in the reciprocal product revealed that this exon 3 remained wild type in all cases analyzed (n = 17), indicating that the repair results from a gene conversion event (see Fig. 2B and Materials and Methods).
Long-range chromosomal rearrangements can be made in ES cells.
Our chromosome engineering strategy has primarily focused on deletions, duplications, and inversions of a few centimorgans (8, 13). The ability to manipulate a larger region of the chromosome is desirable in many instances. For example, large inversions, when marked with a recessive lethal mutation, can be used as balancer chromosomes (2). ES cells with a large deletion may be useful in screens for recessive mutations in vitro. Since the apparent Cre recombination efficiency was dramatically increased with the corrected 3′ hprt cassette, we tested whether long-range (defined here as tens of megabases) deletions can be made in ES cells.
A deletion of 22 cM between Hsd17b1 and D11Mit69 on Chr 11 was used for this test. Previous attempts to generate this deletion in ES cells with the mutant 3′ hprt cassette had failed (8). The D11Mit69 locus was targeted with the 3′ hprt cassette oriented for a deletion in an ES cell line that had been targeted at Hsd17b1 (8). Fifteen double-loxP-targeted cell lines were transiently transfected with a Cre expression plasmid, and HAT-resistant colonies were counted after 12 days. Drug resistance tests indicated that four parental cell lines were double targeted in cis and eleven were double targeted in trans. The recombination events were confirmed to be Cre dependent because a mock transfection with a control plasmid (TyBS) yielded no HAT-resistant colonies for one cis and one trans double-targeted parental cell line. Unlike previous cis-trans tests, however, Cre recombination for both cis and trans configurations occurred at a similar efficiency of approximately 3 × 10−5 (Table 1 and see below). We further successfully generated a number of long-range rearrangements on Chr 11 (Fig. 3; Table 1). The most dramatic example is illustrated in Fig. 4D, where Cre recombination between two loxP sites targeted in trans to Hsd17b1 and D11Mit71 that are 60 cM away from each other on Chr 11 leads to a minideletion chromosome and a large duplication chromosome. Therefore, long-range chromosomal rearrangements, including deletions and deletion-duplications, can be generated with the improved selection cassette.
FIG. 3.
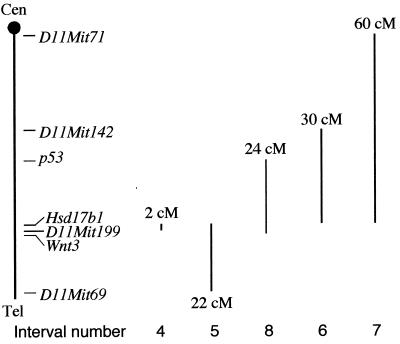
Genetic intervals of rearrangements made on mouse Chr 11 in this study. 2 cM, Hsd17b1-D11Mit199, deletion, deletion-duplication; 22 cM, Hsd17b1-D11Mit69, deletion, deletion-duplication; 24 cM, Wnt3-p53, inversion, deletion-duplication; 30 cM, Hsd17b1-D11Mit142, inversion, deletion-duplication; 60 cM, Hsd17b1-D11Mit71, inversion, deletion-duplication. The total genetic distance from centromere (Cen) to telomere (Tel) is about 80 cM.
FIG. 4.
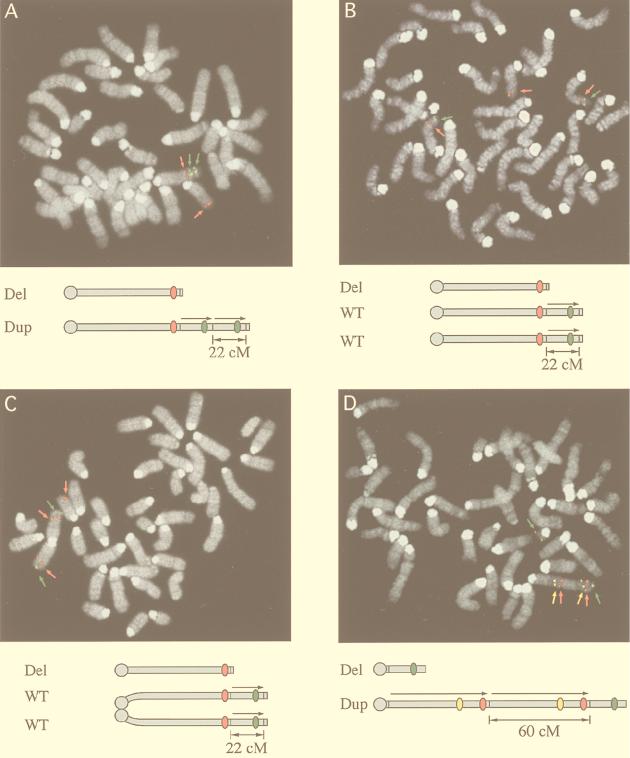
FISH analysis of long-range Cre recombination products on Chr 11. (A) Del(11)5Brd-Dp(11)5Brd, a 22-cM deletion chromosome and a 22-cM duplication chromosome produced by a trans event between Hsd17b1 and D11Mit69. (B) Del(11)5Brd-WT-WT, a 22-cM deletion chromosome produced by a cis event between Hsd17b1 and D11Mit69, while the remaining wild-type chromosome is duplicated to survive. (C) The same as in panel B except that the duplicated wild-type chromosomes are in a Robertsonian (or iso-chromosome) configuration. (D) Del(11)7Brd-Dp(11)7Brd, a 60-cM deletion chromosome and a 60-cM duplication chromosome produced by a trans event between Hsd17b1 and D11Mit71. Colors: yellow, BAC 232M23 (D11Mit320); red, BAC 330H2 (D11Mit263); green, BAC 330P14 (D11Mit11). Two or more probes were differentially labeled and artificially colored.
Large chromosomal deletions may cause ES cell lethality.
The Cre-mediated deletion efficiency for the cis configuration differs by more than 3 orders of magnitude between a 2-cM (Hsd17b1-D11Mit199) and a 22-cM (Hsd17b1-D11Mit69) substrate (Table 1). The reduced Cre recombination efficiency for a larger substrate may simply reflect a lower efficiency of Cre-loxP juxtaposition with greater physical separation. However, it is also possible that ES cells with larger deletions may be selected against if the deletion has deleterious effects on cell viability or growth. In this scenario, only cells that have undergone a compensatory genetic change would survive. To test this, the deletion cell lines were analyzed by FISH with probes both internal and external to the deletion interval. Intriguingly, of five recombination products derived from three independent cis double-targeted parental cell lines, all were trisomy 11, with two wild-type and one deletion chromosome. The two wild-type chromosomes were found to exist as two separate chromosomes (as in Fig. 4B, three of five analyzed) or as a Robertsonian fusion in other cases (as in Fig. 4C, two of five analyzed, both of which derived from independent double-targeted parental cell lines). In contrast, the majority (three of four) of the trans recombination products analyzed contain the expected single deletion and duplication chromosomes. The remaining trans product contained a duplication chromosome and two deletion chromosomes in the Robertsonian configuration. All double-targeted parental cell lines analyzed, irrespective of the cis or trans configuration, contain two wild-type chromosomes (data not shown). These results indicate that the deletion in cis, which leads to a single copy of the 22-cM region of Chr 11, is haploinsufficient in ES cells. Consequently, rare variants are selected in which the remaining wild-type chromosome is duplicated. Thus, the hemizygous 22-cM deletion causes ES cell lethality or a severe growth disadvantage.
Cre-loxP recombination efficiency decreases over increasing genetic distances.
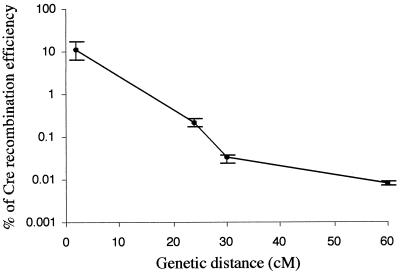
The Cre recombination efficiency is an important consideration in designing Cre-loxP-based chromosome engineering experiments. To provide a framework for future experiments, we determined this efficiency for cis events at different genetic distances. Since a 22-cM deletion had been observed to cause cell death or a growth disadvantage, we assessed the efficiency of inversions as the indicator of Cre recombination efficiency for the larger intervals. Four rearrangements were included in this analysis: (i) a 2-cM deletion between Hsd17b1 and D11Mit199, Del(11)4Brd; (ii) a 24-cM inversion between p53 and Wnt3, In(11)8Brd; (iii) a 30-cM inversion between Hsd17b1 and D11Mit142, In(11)6Brd; and (iv) a 60-cM inversion between Hsd17b1 and D11Mit71, In(11)7Brd (Fig. 3). When the two loxP sites are in opposite orientations, approximately half of the independent double-targeted cell lines give HAT-resistant colonies (interpreted as loxP sites in cis), and the other half do not give any colonies (interpreted loxP sites in trans), presumably due to the formation of dicentric and acentric chromosomes. FISH analysis confirmed that the relevant inversions had occurred in representative clones from all three large genetic intervals (data not shown). As shown in Fig. 5, between 2 and 60 cM, the logarithm of the Cre recombination efficiency is inversely proportional to the genetic distance between the loxP sites.
FIG. 5.
Efficiency of Cre recombination over genetic distance. The percentage of Cre recombination efficiency (y axis, in log10 scale) is plotted against the genetic distance (x axis, in linear scale). The first datum point represents a deletion. The other three points with larger genetic distances represent inversions. Error bars indicate the standard deviations. The numbers of independent experiments were as indicated in Table 1.
Tissue-specific chromosome engineering.
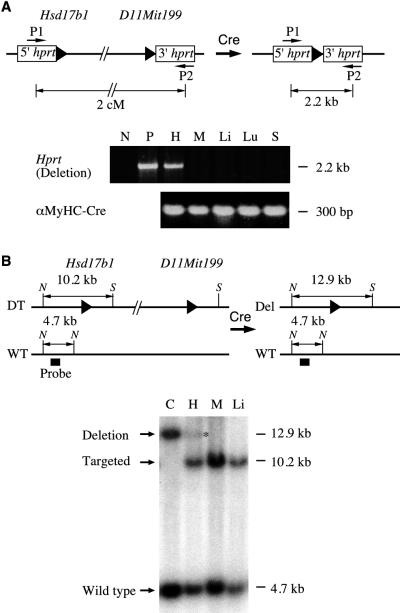
Several deletions of a few centimorgans around the Hsd17b1 locus on Chr 11 are heterozygous lethal (8). Although this underscores the developmental importance of this chromosomal region, lethal deletions cannot be used for genetic screens. However, if the deletion can be made somatically, for instance, in a tissue- or cell-type-specific manner, the problem of heterozygous lethality can be partially circumvented. To test this possibility, we generated a 2-cM Hsd17b1-D11Mit199 double-targeted mouse line (deletion substrate) and crossed it to a cardiac-specific Cre (i.e., αMyHC-Cre) (1). The αMyHC-Cre line had previously been used to make cardiac-specific deletions of several kilobases with an efficiency of up to 90% (1). Tissue DNA was isolated from two progeny that inherited both the αMyHC-Cre transgene and the 2-cM substrate. PCR analysis with primers specific to the reconstituted Hprt minigene was performed to determine whether the Cre-mediated recombination had occurred (Fig. 6A). This analysis demonstrated that the Cre recombination occurred in heart, but not in skeletal muscle, liver, lung, or spleen (Fig. 6A). To provide a more quantitative measure of Cre recombination, Southern analysis was performed on two animals by using restriction digestions and a probe at Hsd17b1 that would distinguish the wild-type allele, the (double) targeted allele and the Cre-recombined allele (Fig. 6B). Deletion occurred exclusively in the heart but not in the other organs tested (Fig. 6B). Based on the ratio of intensity of the recombined fragment and the predeletion allele for both animals tested, the deletion efficiency in the heart is about 10%.
FIG. 6.
Cardiac-specific 2-cM deletion between Hsd17b1 and D11Mit199. (A) PCR analysis of the deletion products. P1 and P2, primers used to specifically amplify the full-length Hprt. PCR reactions on αMyHC-Cre serve as a control. N, negative control; P, positive control; H, heart; M, skeletal muscle; Li, liver; Lu, lung; S, spleen. (B) Southern analysis of the deletion products. C, control cell line that contains a deletion and a wild-type chromosome. The cardiac-specific deletion band is indicated by an asterisk. N, NheI; S, SfiI; solid triangle, loxP site.
DISCUSSION
The organism that the Cre-loxP system is derived from, bacteriophage P1, evolved the system to resolve its ∼100-kb genome into monomeric circular forms (18). The Cre-loxP site-specific recombination system has been extensively used for conditional genetic technology, namely, the temporal and spatial control of gene expression in mice (16). In these applications, the genetic material involved (as determined by the distance between the two loxP sites) is usually a few kilobases. We have previously shown that this system can be adapted for substrates of several megabases by incorporating a positive selection scheme (8, 13). In the present study, we redefined the Cre recombination efficiency for a 4-Mb substrate, after we corrected a mutation in the selection cassette. Surprisingly, the efficiency for this substrate is approximately 11% by transient Cre expression. This efficiency approaches that obtained with substrates of several kilobases and indicates that at between several kilobases and several megabases the Cre-loxP recombination occurs at comparable efficiencies. This might reflect aspects of chromatin domain organization such that sequences that are 1-kb to 1-Mb apart may have similar separations in three-dimensional space. In this aspect, the fact that the 2-cM cardiac-specific deletion can be detected by Southern analysis is of particular significance. In many cancers, interstitial deletions are the dominant mode for loss of the remaining allele of a tumor suppressor gene (6). Therefore, in vivo chromosomal deletions can be used to mimic somatic LOH in human cancers and in searches for novel tumor suppressor genes in combination with point mutagenesis.
The 22-cM deletion between Hsd17b1 and D11Mit69 on Chr 11 appears to cause ES cell lethality or a severe growth disadvantage because deletion products for this interval exclusively carry an additional wild-type Chr 11. This may be due to a dosage effect of one or multiple genes in this interval such that a single copy of these genes cannot support the normal growth of ES cells (haploinsufficiency). The Cre-loxP-mediated deletion of this 22-cM region therefore selects for cells that have duplicated the wild-type Chr 11. This result underscores the tight control of the euploid ES cell genome. A region of haploinsufficiency has also been proposed to reside on Chr 9 in studies on a radiation-induced deletion complex (19). The observation of haploinsufficiency in ES cells is in direct contrast with many cancer cells that often carry large chromosomal deletions and chromosomal losses. Such a unique feature of ES cells may be further studied by isolating suppressors of this lethality caused by the deletions. On the other hand, these data indicate that duplications are tolerated better than deletions in ES cells. This is consistent with the notion that monosomies rarely, if ever, exist, whereas trisomy 8, 11, and 15 and several other chromosomes have been observed in ES cells (9). The relatively frequent occurrence of trisomy 11 is further suggested by our observation that one of four 22-cM deletion-duplication products analyzed by FISH contain one duplication and two deletion chromosomes where the deletion chromosome is presumably not required to be duplicated for cell survival or growth. The lethality caused by large deletions in ES cells precludes a straightforward approach of using the deletion as a partial haploid reagent in mutagenesis screens.
It is possible that the partial-trisomy ES cells selected by the 22-cM cis deletion are derived from an underlying trisomy 11 population in the ES cells transfected with Cre. Although these cells are not detected by analysis of double-targeted clones, extrapolation of the inversion recombination efficiencies suggests that either these cells are present at 10−2 frequencies in the transfected clones or that this nondisjunction event is induced by the Cre-loxP recombination event itself.
Large-deletion-associated ES cell lethality can obscure the Cre recombination efficiency. We therefore determined the Cre efficiency by using large inversion substrates. This analysis indicates that Cre recombination efficiency decreases over increasing genetic distances. However, in all cases, the recombination products (HAT-resistant colonies) are readily obtained in sufficient numbers in a single experiment except when inviable products are generated (dicentric and acentric chromosomes). For multimegabase substrates, the logarithm of the Cre recombination efficiency is approximately inversely proportional to the genetic distance (Fig. 5). This can be used as a guide for future experiments with Cre-loxP-based chromosome engineering. However, other factors, such as chromosomal locations and differences in experimental manipulations, may affect the Cre recombination efficiency. For deletions, the Cre recombination drops more precipitously as the genetic distance increases for two reasons. First, the physical barrier Cre has to overcome to bring the two loxP sites together is greater as the distance between the two loxP sites increases, as in inversions. Second, larger deletions may cause ES cell lethality or a growth disadvantage and are consequently selected against after Cre recombination. In the trans configuration where a deletion and a duplication chromosomes are the products, Cre recombination efficiency is moderately reduced with an increasing genetic distance (Table 1). This suggests that chromosome homologues may pair in a mitotic cell cycle, assisting Cre recombination by bringing the two loxP substrates on different chromosomes to the same subcellular location. Under such circumstances, the closer the two loxP sites are genetically, the closer they are physically when the two chromosomes pair, and therefore, the higher the Cre recombination efficiency. Alternatively, if trans recombination occurs mainly in G2 and recombined sister chromatids tend to segregate away from each other, as reported in Drosophila (4), the HAT-resistant deletion products will frequently contain a wild-type chromosome instead of the duplication chromosome. In this scenario, trans deletion-duplication events involving a larger distance will appear to occur at a lower frequency due to the production of haploinsufficient deletions.
The Cre recombination efficiency for large deletion-duplications is probably comparable to that for translocations between nonhomologues. In some of our experiments, we analyzed some random integration clones when targeting the second loxP site. Upon Cre expression, approximately half of these clones give HAT-resistant colonies and the other half do not give viable HAT-resistant colonies. The former group presumably yields translocations, while the latter group yields dicentric and acentric products. The efficiency of generating these translocations is about 10−5. It has been reported using a similar strategy that Cre recombination efficiency for a translocation between chromosomes 12 and 15 occurs at about 10−7 (17). The higher efficiency in our experiments may be due to the Cre plasmid, the tissue culture conditions, and/or the electroporation procedures used. It remains possible that the 2-kb homology between our 5′ hprt and 3′ hprt cassettes assists the Cre-loxP recombination by recruiting the homologous recombination machinery to help secure the loxP site recombination synapse.
The mutant 3′ hprt cassette used in previous chromosome engineering experiments provides a unique opportunity for studying a potential interaction between homologous and site-specific recombination. Sequence analysis of Cre recombination products indicates that the mutation is repaired by homologous recombination with the wild-type template in the 5′ hprt cassette. This homologous recombination cannot occur in the absence of site-specific recombination since the homology is only 2 kb, but the substrates are on different chromosomes or far away (multimegabases) from each other on the same chromosome. Therefore, it must have occurred during or immediately after the Cre-loxP recombination. It is possible that the Holliday junction structure created by Cre (21) can be resolved by homologous recombination machinery. This scenario would suggest that the two recombination events are not mutually exclusive and can be coupled under specific circumstances. The other possibility is that Cre-loxP recombination facilitates gene conversion merely by bringing the two substrates together. Immediately after site-specific recombination, homologous recombination occurs. Since HAT-resistant colonies for a 2-cM substrate are obtained with an efficiency of approximately 3 orders of magnitude higher with the wild-type 3′ selection cassette than with the mutant version, homologous recombination responsible for repairing the mutation occurs ca. 0.1% of the time after Cre recombination.
Taken together, the Cre-loxP chromosome engineering strategy provides a powerful tool for genetic studies and for genome manipulation. We explored the possibility and determined the efficiency of generating various chromosomal rearrangements on mouse Chr 11. We conclude that any desired rearrangement can be made with the Cre-loxP system provided that the rearrangement does not have any deleterious effect on the ES cells. Cre-loxP recombination is very efficient for substrates of a few centimorgans both in tissue culture and in vivo. This efficiency decreases over increasing genetic distances between the two loxP sites. The work presented here provides a framework for future applications of chromosome engineering.
ACKNOWLEDGMENTS
We thank Sandra Rivera, Sukeshi Vaishnav, and Yin-Chai Cheah for technical assistance; Hong Su for an Hsd17b1 targeted cell line; Wei Wen Cai for the BAC clones used in FISH analysis; Michael Schneider for the αMyHC-Cre mouse line; Pentao Liu for helpful discussions; Patrick Biggs, Xiaozhong Wang, and Meredith Wentland for helpful comments on the manuscript; and Sylvia Perez for secretarial assistance.
This work is partially supported by grants from the National Institutes of Health, the National Cancer Institute, and DAMD17-98-1-8280. A.B. is an investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Agah R, Frenkel P A, French B A, Michael L H, Overbeek P A, Schneider M D. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Investig. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M. Drosophila: a laboratory handbook. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 3.Baldini A, Lindsay E A. Mapping human YAC clones by fluorescence in situ hybridization using Alu-PCR from single yeast colonies. In: Choo K H A, editor. In situ hybridization protocols. Vol. 33. Totowa, N.J: Humana Press; 1994. pp. 75–84. [DOI] [PubMed] [Google Scholar]
- 4.Beumer K J, Pimpinelli S, Golic K G. Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila. Genetics. 1998;150:173–188. doi: 10.1093/genetics/150.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen K S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault A C, Lee C C, Lupski J R. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 6.Croce C M. Genetic approaches to the study of the molecular basis of human cancer. Cancer Res. 1991;51:5015s–5018s. [PubMed] [Google Scholar]
- 7.Korenberg J R, Chen X N, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu P, Zhang H, McLellan A, Vogel H, Bradley A. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse Chromosome 11. Genetics. 1998;150:1155–1168. doi: 10.1093/genetics/150.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Wu H, Loring J, Hormuzdi S, Disteche C M, Bornstein P, Jaenisch R. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev Dyn. 1997;209:85–91. doi: 10.1002/(SICI)1097-0177(199705)209:1<85::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Lupski J R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 11.O'Gorman S, Dagenais N A, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez-Solis R, Davis A C, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 14.Rinchik E M, Russell L B. Germ-line deletion mutations in the mouse: tools for intensive functional and physical mapping of regions of the mammalian genome. In: Davies K E, Tilghman S M, editors. Genome analysis. 1. Genetic and physical mapping. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 121–158. [Google Scholar]
- 15.Robertson E J. Embryo-derived stem cell lines. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford, England: IRL; 1987. pp. 71–112. [Google Scholar]
- 16.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 17.Smith A J, De Sousa M A, Kwabi-Addo B, Heppell-Parton A, Impey H, Rabbitts P. A site-directed chromosomal translocation induced in embryonic stem cells by Cre-loxP recombination. Nat Genet. 1995;9:376–385. doi: 10.1038/ng0495-376. [DOI] [PubMed] [Google Scholar]
- 18.Sternberg N L. Cloning high molecular weight DNA fragments by the bacteriophage P1 system. Trends Genet. 1992;8:11–16. doi: 10.1016/0168-9525(92)90018-y. [DOI] [PubMed] [Google Scholar]
- 19.Thomas J W, LaMantia C, Magnuson T. X-ray-induced mutations in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1998;95:1114–1119. doi: 10.1073/pnas.95.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valancius V, Smithies O. Double-strand gap repair in a mammalian gene targeting reaction. Mol Cell Biol. 1991;11:4389–4397. doi: 10.1128/mcb.11.9.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voziyanov Y, Pathania S, Jayaram M. A general model for site-specific recombination by the integrase family recombinases. Nucleic Acids Res. 1999;27:930–941. doi: 10.1093/nar/27.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokoyama T, Silversides D W, Waymire K G, Kwon B S, Takeuchi T, Overbeek P A. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 1990;18:7293–7298. doi: 10.1093/nar/18.24.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng B, Mills A A, Bradley A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 1999;27:2354–60. doi: 10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng B, Sage M, Cai W W, Thompson D M, Tavsanli B C, Cheah Y C, Bradley A. Engineering a mouse balancer chromosome. Nat Genet. 1999;22:375–378. doi: 10.1038/11949. [DOI] [PubMed] [Google Scholar]