Abstract
Publishing, discussing, envisioning, modeling, designing and experimentally determining RNA three-dimensional (3D) structures involve preparation of two-dimensional (2D) drawings that depict critical functional features of the subject molecules, such as noncanonical base pairs and protein contacts. Here, we describe RiboDraw, new software for crafting these drawings. We illustrate the features of RiboDraw by applying it to several RNAs, including the Escherichia coli tRNA-Phe, the P4–P6 domain of Tetrahymena ribozyme, a −1 ribosomal frameshift stimulation element from beet western yellows virus and the 5′ untranslated region of SARS-CoV-2. We show secondary structure diagrams of the 23S and 16S subunits of the E. coli ribosome that reflect noncanonical base pairs, ribosomal proteins and structural motifs, and that convey the relative positions of these critical features in 3D space. This software is a MATLAB package freely available at https://github.com/DasLab/RiboDraw.
INTRODUCTION
Two-dimensional (2D) depictions of RNA secondary structure serve two purposes. For the authors making them, the analysis ensures thoughtful inspection of a three-dimensional (3D) molecule and distillation of its most important functional features into a human-understandable form. For readers, 2D drawings provide an entry point into complex structures, allowing for a gradual familiarization with increasing levels of detail. Over decades of publishing and presentation, a conventional ‘visual language’ for depicting RNA tertiary structure in 2D drawings has emerged, but the tooling available remains laborious.
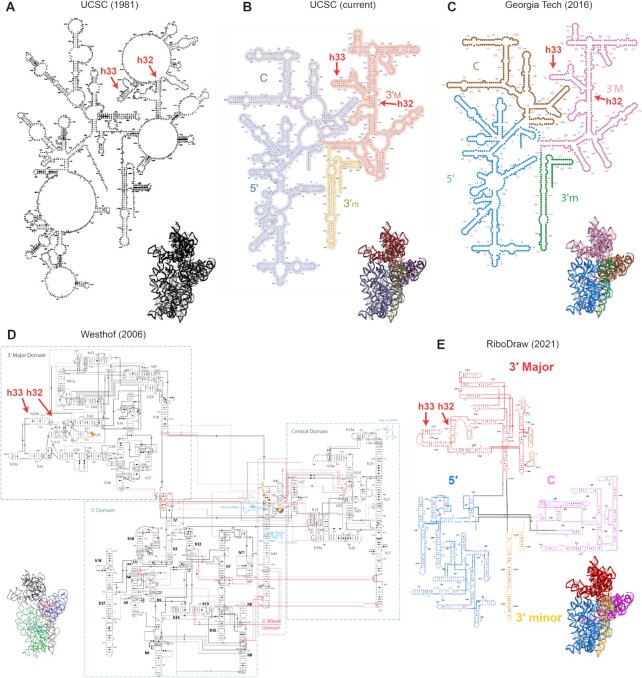
Years of researchers communicating about RNA structure have established common standards to facilitate common understanding. Helices are typically drawn in horizontal or vertical orientations; sets of shapes convey the details of base pairing geometry (Leontis–Westhof annotations) (1); numbers are provided every 10th nucleotide; and helices, or stacked sets of stems, are given names for easy reference (Figure 1). Most importantly, something about 2D layouts should convey tertiary fold, with regions in close contact in 3D also in close contact in 2D—and this remains difficult to guarantee unless the layouts are hand drawn. As an illustration, Figure 1 shows previously available drawings of the 16S rRNA of the Escherichia coli ribosome small subunit. We indicate with arrows the depiction of h33 in each drawing, which bends back in the 3′ major domain to form a tertiary contact with h32; the resulting shape forms a beak-like 3D structure in the molecule’s ‘head’. None of the 2D layouts in Figure 1A–C conveys that shape. Although there are multiple packages in wide use that implement several of the conventions for drawing 2D layouts, such as XRNA (2) and VARNA (3), no previous software directly implements each one, with the conveyance of tertiary arrangements being an almost universal omission. Consequently, a secondary structure depiction that reflects each of these principles to date, such as that in Figure 1D (4), has required tedious manual refinement in general packages like Adobe Illustrator or Microsoft PowerPoint.
Figure 1.
(A) Diagram of the 16S rRNA from (6), hand drawn. (B) Same diagram updated for University of California, Santa Cruz lab website that emphasizes four domains. (C) Diagram from Center for the Origin of Life website at Georgia Tech that shares a layout and domain coloring scheme with that from (7). (D) Diagram adapted from Lescoute and Westhof, currently hosted at https://eric-westhof.ibmc.cnrs.fr/teaching/, that conveys tertiary interactions. (E) New (2021) diagram generated in this study using RiboDraw. Additional detail on the h33 conformation, which is indicated in each panel here, is provided in Figure 4A.
This shortcoming has been amplified by the increasing throughput of structure determination efforts and, at the same time, the scale of the structures under study. In particular, the development of structure determination workflows like Ribosolve, which use chemical mapping and cryogenic electron microscopy, permits individual researchers to obtain numerous new structures in a year (5); to facilitate that work, we relied on an early version of the method we report here. It is telling that current secondary structure diagrams of large RNA molecules like the ribosomal RNAs dispense with these conventions, typically preferring a radial layout and omitting Leontis–Westhof symbols (see Figure 1A–C) (6–10). Although there has been significant growth in excellent software to lay out RNA secondary structures automatically (3,11–14), including methods that can guarantee outerplanarity as long as the structure lacks pseudoknots (14) and methods that take advantage of existing templates (15), tools to remodel and refine these layouts, in particular to match a known tertiary structure, are absent. Consequently, despite dozens of newly solved structures per year and enormous research interest, there has been no layout of a bacterial 23S rRNA that accurately expresses its tertiary structure.
In designing this software, we followed three guiding principles. First, the creator must still actively step through the 2D placement of helices, motifs and noncanonical pairs; the software must aid systematic and contemplative visual inspection, but it must not replace it. Second, the software must automate the tedious actions that would otherwise be necessary when refining a layout in Illustrator or PowerPoint, e.g. the dozens of operations needed to rotate a helix. Third, the software ought to encode the conventions, such as a limited set of allowed helix orientations, that have been established over the years. Figure 1D shows an example of the resulting 2D layout for the 16S rRNA, with the h32–h33 ‘beak’ more visually apparent than prior layouts, and this article presents further examples throughout. In the short term, we hope RiboDraw fills a need for the production of human-readable layouts of large RNA machines and of moderate-size RNA molecules at increased throughput. In the long term, we hope that these principles, along with a thorough description of the RiboDraw data model, will motivate a new generation of automated RNA layout software.
MATERIALS AND METHODS
Software availability
The RiboDraw software is available under an open-source MIT license on GitHub at https://github.com/ribokit/RiboDraw. The repository also contains multiple tutorials providing step-by-step walkthroughs for different drawing modes and functions, as well as example drawings for different RNA systems.
The Rosetta software suite is freely available to academics and may be commercially licensed (https://rosettacommons.org). The rna_info web server hosted on ROSIE (Rosetta Online Server that Includes Everyone), which permits easy generation of RiboDraw input files, is available at https://rosie.rosettacommons.org/rna_info/.
Ultra-high-resolution versions of the 16S and 23S ribosome drawings in Figures 5 and 6 are available in Supplementary Data, as are X, Y coordinates for each nucleotide in the 16S, 23S and P4–P6 drawings.
Figure 5.
A 2D secondary structure depiction of the 16S rRNA, as laid out and colored using RiboDraw. Note the close correspondence between the 3D orientation of domains and helices and the 2D layout. Supplementary Data include a high-resolution rendering of the secondary structure, as well as an SVG file.
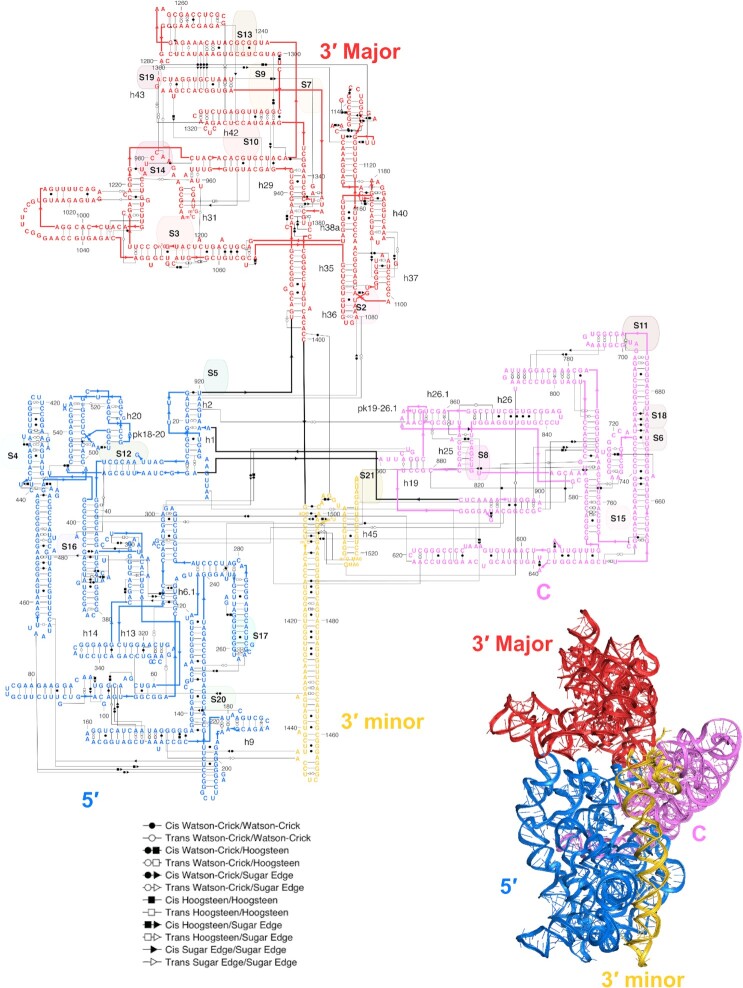
Figure 6.
A 2D layout of the 23S rRNA and 5S rRNA. While the complexity of the large subunit prevents a perfect reproduction of the 3D orientations for the whole molecule, an ‘exploded’ depiction preserves the local orientations of helices within separate layers of the ribosome while long arrows between the layers convey the covalent connections. The peptidyl transferase center has been redrawn compared to Figure 4 to better show noncanonical pairs and connections to other domains. Supplementary Data include a high-resolution rendering of the secondary structure, as well as an SVG file.
RESULTS
The RiboDraw software was written using graphical manipulation toolboxes available in MATLAB (16). As a reference for users and prospective developers, an overview of all current RiboDraw functions is given in Supplementary Table S1, and a description of the input/output data format is given in Supplementary Table S2. In the following, we illustrate the general steps to use RiboDraw on a ∼200-nucleotide model system, the P4–P6 RNA (17); then describe alternative display modes and workflows useful for preparing visualizations of RNA–protein complexes, depicting chemical mapping data or creating custom layouts for puzzles on the Eterna RNA design platform and secondary structure design tool; and finally present RiboDraw drawings of entire E. coli small and large ribosomal subunits.
Steps to make a drawing
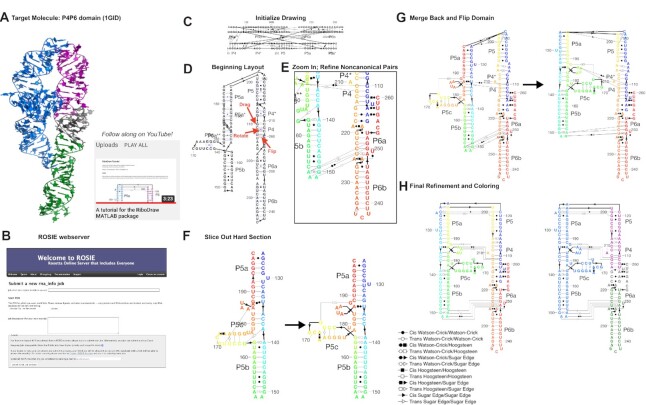
Figure 2 illustrates the steps that a user can apply to convert a set of 3D coordinates from an RNA crystal structure in PDB format to a 2D layout. The model molecule is the P4–P6 domain of the Tetrahymena group I self-splicing intron, based on its first crystal structure by Cate et al. (17) (PDB ID: 1GID; Figure 2A). Descriptions of specific commands, along with example images and files, are provided in the tutorial/ directory of RiboDraw, and we provide a video walkthrough for the essential steps in Supplementary Data. Here, we focus on our rationale for each step rather than the details of their implementation or access functions in the software.
Figure 2.
The steps to make a RiboDraw drawing based on 3D coordinates: identifying a PDB structure of interest (A); running the rna_info web server to annotate structural features (B); initializing the drawing using the initialize_drawing command (C); laying out individual stems and loops using mouse controls to move helices and nucleotides as well as rotate and flip helices using handles displayed via show_helix_controls (D); refining the positions of particular noncanonical pairs, which may be shown via show_noncanonical_pairs as well as important linkers between nucleotides via show_linker_controls (E); extracting particular challenging sections for focused attention using slice_drawing (F); merging the subdrawing back using merge_drawing, flipping its orientation using tools enabled by show_domain_controls and recoloring (G); and finalizing linker geometries and coloring to match the 3D structure using color_drawing and setup_domain (H). This step-by-step tutorial is available online in the RiboDraw GitHub repository as well as on YouTube at https://youtu.be/gLcxN6HxEjQ.
In order to convey an RNA structure, a 2D secondary structure drawing should minimally show the residue numbers; RNA double helices, or stems (secondary structure); and noncanonical pairs. To avoid human error, this information may be extracted from the PDB coordinate file using a Rosetta application. To avoid having to install and compile Rosetta, we have provided a server called rna_info to accomplish this step at ROSIE (Figure 2B) (18,19). If the user prefers local operation, the Rosetta application rna_motif produces the same output text files, which describe each base pair, each ligand, each motif, each tertiary contact, each nucleobase stack and each stem. Conventionally, layout diagrams for RNA structure label helices with names like ‘P4’ (for ‘paired region 4’) or similar, with the exact helix names and numbers typically depending on conventions developed by researchers who have previously studied these molecules. Since these conventions are not embedded in the PDB coordinate file, the RiboDraw user may use a text editor to add the conventional helix names (e.g. ‘P4’, ‘P5’, ‘P6’, etc.) to each line of a text file produced by rna_info that lists the residue numbers corresponding to each stem (see Supplementary Data). RiboDraw treats pseudoknotted helices just as it does ordinary stems. For RNAs of unknown 3D structure, RiboDraw also provides alternative functions that set up these files based on sequence, numbering and expected secondary structure.
Next, the user must compose the drawing itself. To ensure that the user makes conscious choices for how to lay out each important element in the RNA, the objects are not pre-laid out in a specific order. Within MATLAB, a single command loads all the required files into RiboDraw (Figure 2C). At this point, all the information needed for the drawing is present, and the user now refines how to lay out each helix, each non-helix residue, and then each drawing label and linker corresponding to a strand connection or pairing. These steps, depicted in Figure 2D–F, are accelerated through single-click operations for helix flipping or rotation that would require numerous tedious rearrangements in graphical software such as Illustrator or PowerPoint. Through the process, the user can guide their choices in laying out elements by inspecting how helices are arranged in three dimensions by simultaneously visualizing the 3D structure in PyMOL (20,21). Coloring nucleotides in RiboDraw to match those in PyMOL makes it easier to see correspondences; to make that simpler, RiboDraw supports PyMOL’s color names. Functions to select and manipulate subdomains of the drawing, such as the P5abc subdomain of the P4–P6 RNA, enable refinement of those parts, including flipping (Figure 2G). Final refinement of helix label positions, tick numbers and linkers gives the final drawing (Figure 2H).
During the editing process, the drawing may be saved for subsequent work as JavaScript Object Notation or as a MATLAB’s Hierarchical Data Format (HDF5)-based .mat file; all the data that are saved in order to reproduce a drawing are documented in Supplementary Table S2. Once ready for external visualization, the drawing can be exported in standard image file formats, including PNG (Portable Network Graphics), SVG (Scalable Vector Graphics) and PDF (Portable Document Format) files that are further editable in applications like Adobe Illustrator. Example output files are included in Supplementary Data as well as in the online tutorial.
Alternative drawing modes
Figure 3 shows a gallery of alternative drawing styles that have been implemented in RiboDraw to meet demands for high-quality drawings that have arisen in a number of RNA systems. Accessory tutorials for accessing each of these features are available in the RiboDraw repository.
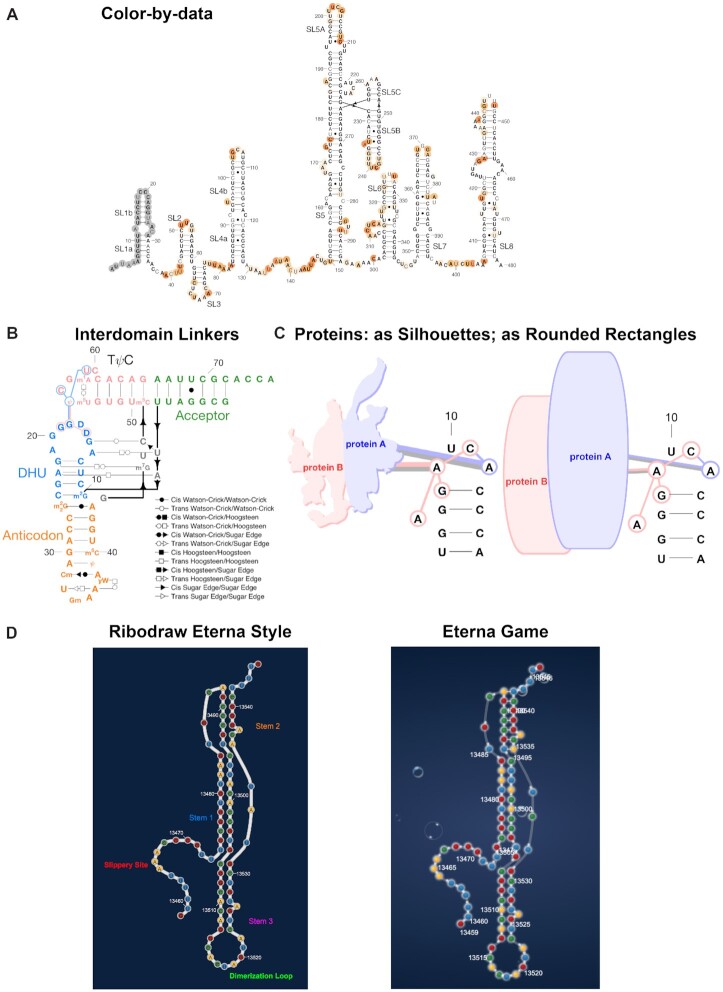
Figure 3.
RiboDraw offers diverse structural depictions that each implement the same fundamental conventions. (A) The simplest depiction, as in this structure of the SARS-CoV-2 5′ untranslated region, only includes ordinary secondary structure, although here we show how that depiction can be colored according to chemical mapping reactivity. (B) Noncanonical pairs may be annotated with Leontis–Westhof symbols, and chemically modified nucleotides may be given their IUPAC names like m22G, as in this tRNA-Phe. (C) Protein binding, such as the MS2 coat protein/hairpin interaction, may be depicted via colored linkers to the binding nucleotides, and the proteins themselves may be shown as silhouettes (left) or as rounded rectangles (right). (D) RiboDraw may also be used to create layouts for the online video game and secondary structure design tool Eterna, such as this layout for the frameshift stimulation element of SARS-CoV-2 (left). These layouts correspond closely to the visual style of the game itself (right). Play this puzzle at https://eternagame.org/game/puzzle/9837790/!
Most simply, it is straightforward to remove noncanonical pairs and most labels to give streamlined drawings with just secondary structures (Figure 3A). For RNAs with complex tertiary contacts with numerous noncanonical pairs, the contacts can be grouped into ‘interdomain’ tertiary linkers to further simplify the drawing, as is shown in Figure 3B for tRNA (phenylalanine). This drawing also highlights RiboDraw’s ability to recognize chemical modifications (e.g. by showing the letter ψ for pseudouridine) and a functionality to identify and depict some standard tertiary motifs, including U-turns and intercalating T-loop motifs.
Proteins, metal ions and small-molecule ligands are important cofactors for most functional RNA molecules (22–25). RiboDraw provides a number of ways to depict these factors, illustrated for the MS2 viral coat protein dimer bound to an RNA hairpin (PDB ID: 1ZDH). The most detailed representation gives ‘silhouettes’ of proteins and linkers of the protein to each RNA residue it contacts (Figure 3C), while simpler representations such as rounded rectangles without linkers are also available (Figure 3C; see below for ribosome drawings).
Last, RiboDraw offers a tool to preview custom layouts of structured RNAs for the online Eterna RNA design project, as illustrated for the pseudoknotted frameshift stimulation element from the SARS-CoV-2 genome in Figure 3D. Functionality to export and display these RiboDraw layouts in the EternaJS online game (24) and secondary structure design tool (26) has been implemented, such as for the challenge found at https://eternagame.org/game/puzzle/9837790/.
Ribosome drawings
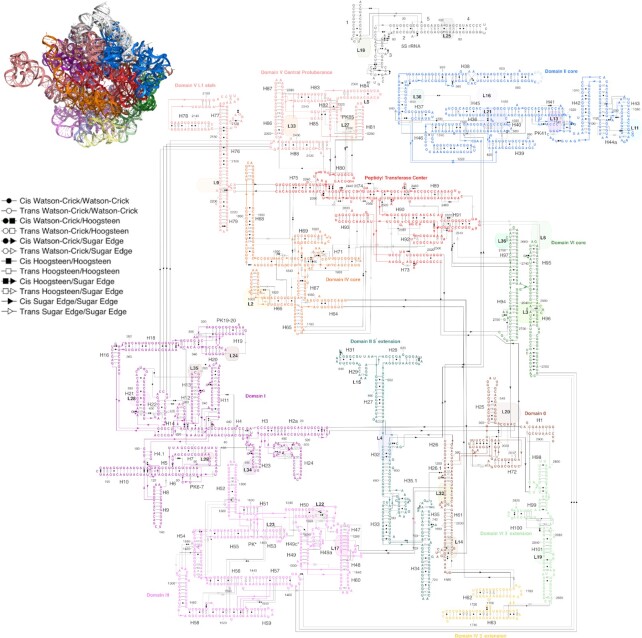
We were able to use the RiboDraw software to prepare drawings of the E. coli ribosome in a 2D arrangement following the 3D organization found in experimental structures, such as that in (27) (PDB ID: 4YBB). As a small-scale illustration of the value of secondary structure diagrams that reflect a 3D structure, we show the conformation of h33 in the 16S rRNA (Figure 4A) and the peptidyl transferase center of the 23S rRNA (Figure 4B). The complexity of both regions is more manageable when approached through the lens of a 2D diagram than a single view of the 3D structure, while interaction details are preserved.
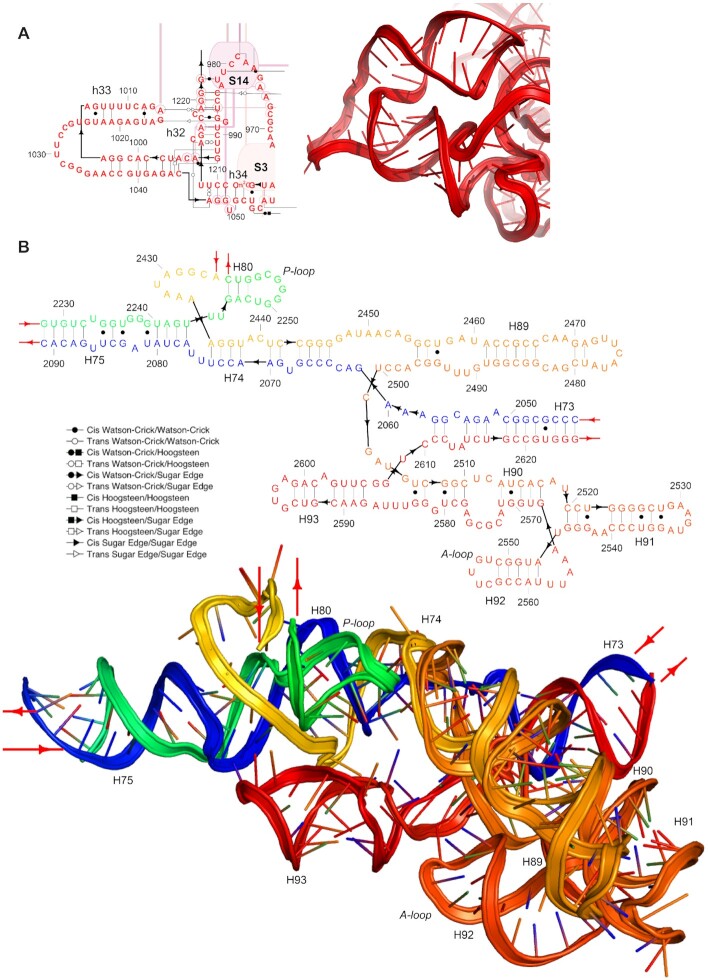
Figure 4.
(A) The stem h33 of the E. coli 16S ribosomal RNA exhibits an unusual 13/2 two-way junction that induces a nearly complete turn between its two Watson–Crick helices, permitting the hairpin loop that terminates it to form noncanonical base pairs with nucleotides from h32. This bent geometry is absent from prior depictions but captured in a RiboDraw depiction. (B) The peptidyl transferase center is essential to ribosomal function and comprises domain V along with the L1 stalk and central protuberance. The RiboDraw layout captures the central stack of H75, H74 and H89; the P-loop capping H80; and the lower domain featuring H73, H90, H91 and H93 as well as the A-loop capping H92.
Although there have been inspiring precedents, in particular for the 16S rRNA (Figure 1D) (4), the depictions of the whole ribosomal subunits prepared for this manuscript (Figure 5, the 16S rRNA; Figure 6, the 23S rRNA and 5S rRNA) are the first layouts of the entire bacterial ribosomal RNA to our knowledge that reflect the 3D structure and fully depict noncanonical pairs. These layouts have been useful for our lab and Eterna’s efforts to re-engineer ribosomes (28) and illustrate the applicability of RiboDraw to large and complex RNA molecules.
DISCUSSION
RiboDraw enables users to create publication-quality secondary structure diagrams with tools specialized to the task: instead of manipulating geometric shapes and text boxes, the user may perform fundamental operations on objects that are structurally meaningful, e.g. stems, bound proteins and noncanonical base pairs. This abstraction enables a qualitative change in efficiency, permitting the creation of high-quality depictions of complex RNAs like the ribosome that reflect its 3D structure.
The RiboDraw software faces some limitations. MATLAB is not primarily intended for manipulating diagrams with thousands of graphical elements, as a ribosomal RNA demands, nor is it a common setting for biochemistry software development. Improving performance, adding a graphical user interface, direct integration with 3D molecular visualization and reaching a broader audience through the Web may demand a port to a language like JavaScript and is under exploration. GNU Octave is compatible with most MATLAB code but conflicts with MATLAB’s current graphics handling scheme and thus cannot run RiboDraw. Any port will be accelerated by starting from the design principles and open-source code established here.
Future development of RiboDraw may lead to accelerations through further automation. Any layout operation that could benefit from expert judgment (e.g., the relative position of two helices) ought to be performed manually, but an automated method to, for example, rapidly draw linkers connecting contiguous residues or denoting noncanonical pairs may be helpful. Operations like structure threading, transferring the 2D layout for one RNA structure to a homologous structure, could also be automated, while producing a report of any discrepancies or ambiguities for manual expert inspection. We propose that, in future development, the overall philosophy should remain to augment and assist expert composition rather than to replace it.
Supplementary Material
ACKNOWLEDGEMENTS
We dedicate this paper to Neocles Leontis, whose passion for RNA structure and social justice will be dearly missed. We acknowledge Luc Jaeger for inspiring discussions of ribosome structure, Sergey Lyskov for maintenance of the ROSIE web server platform, Ramya Rangan for advice and contributions to chemical mapping visualization code, Kalli Kappel for test drawings for SAM-IV and other Ribosolve models, and Jonathan Romano and Guy Geva for advice on how to use RiboDraw to create custom Eterna layouts.
Contributor Information
Rhiju Das, Department of Physics, Stanford University, Stanford, CA 94305, USA; Department of Biochemistry, Stanford University School of Medicine, Stanford, CA 94305, USA.
Andrew M Watkins, Department of Biochemistry, Stanford University School of Medicine, Stanford, CA 94305, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Institutes of Health [R35 GM 122579 and R21 CA 219847].
Conflict of interest statement. None declared.
REFERENCES
- 1. Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001; 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. XRNA 2004; (1 September 2021, date last accessed)http://rna.ucsc.edu/rnacenter/xrna/xrna.html.
- 3. Darty K., Denise A., Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lescoute A., Westhof E The interaction networks of structured RNAs. Nucleic Acids Res. 2006; 34:6587–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kappel K., Zhang K., Su Z., Watkins A.M., Kladwang W., Li S., Pintilie G., Topkar V.V., Rangan R., Zheludev I.N. et al. Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods. 2020; 17:699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Noller H.F., Woese C.R. Secondary structure of 16S ribosomal RNA. Science. 1981; 212:403. [DOI] [PubMed] [Google Scholar]
- 7. Gulen B., Petrov A.S., Okafor C.D., Vander Wood D., O’Neill E.B., Hud N. V., Williams L.D. Ribosomal small subunit domains radiate from a central core. Sci. Rep. 2016; 6:20885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Noller H.F. 2017; Ribosome images(28 August 2021, date last accessed)http://rna.ucsc.edu/rnacenter/ribosome_images.html. [Google Scholar]
- 9. Bernier C.R., Petrov A.S., Waterbury C.C., Jett J., Li F., Freil L.E., Xiong X., Wang L., Migliozzi B.L.R., Hershkovits E. et al. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 2014; 169:195–207. [DOI] [PubMed] [Google Scholar]
- 10. Weinberg Z., Breaker R.R. R2R: software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kerpedjiev P., Hammer S., Hofacker I.L. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015; 31:3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elias R., Hoksza D. TRAVeLer: a tool for template-based RNA secondary structure visualization. BMC Bioinformatics. 2017; 18:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shabash B., Wiese K.C. jViz.RNA 4.0: visualizing pseudoknots and RNA editing employing compressed tree graphs. PLoS One. 2019; 14:e0210281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wiegreffe D., Alexander D., Stadler P.F., Zeckzer D. RNApuzzler: efficient outerplanar drawing of RNA-secondary structures. Bioinformatics. 2019; 35:1342–1349. [DOI] [PubMed] [Google Scholar]
- 15. Sweeney B.A., Hoksza D., Nawrocki E.P., Ribas C.E., Madeira F., Cannone J.J., Gutell R., Maddala A., Meade C.D., Williams L.D. et al. R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat. Commun. 2021; 12:3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MathWorks MATLAB Using MATLAB Graphics. MathWorks, Natick, MA. 2000; [Google Scholar]
- 17. Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996; 273:1678–1685. [DOI] [PubMed] [Google Scholar]
- 18. Moretti R., Lyskov S., Das R., Meiler J., Gray J.J. Web-accessible molecular modeling with Rosetta: the Rosetta Online Server that Includes Everyone (ROSIE). Protein Sci. 2018; 27:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lyskov S., Chou F.C., Conchúir S.Ó., Der B.S., Drew K., Kuroda D., Xu J., Weitzner B.D., Renfrew P.D., Sripakdeevong P. et al. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One. 2013; 8:e63906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeLano W.L. PyMOL: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002; 1–9. [Google Scholar]
- 21. DeLano W.L. The PyMOL molecular graphics system, version 2.3. Schrödinger LLC. 2020;
- 22. Hampel K.J., Tinsley M.M. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006; 45:7861–7871. [DOI] [PubMed] [Google Scholar]
- 23. Hampel A., Cowan J.A. A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997; 4:513–517. [DOI] [PubMed] [Google Scholar]
- 24. Lee J., Kladwang W., Lee M., Cantu D., Azizyan M., Kim H., Limpaecher A., Yoon S., Treuille A., Das R. RNA design rules from a massive open laboratory. Proc. Natl Acad. Sci. U.S.A. 2014; 111:2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bokinsky G., Nivón L.G., Liu S., Chai G., Hong M., Weeks K.M., Zhuang X. Two distinct binding modes of a protein cofactor with its target RNA. J. Mol. Biol. 2006; 361:771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jain S., Laederach A., Ramos S.B.V., Schlick T. A pipeline for computational design of novel RNA-like topologies. Nucleic Acids Res. 2018; 46:7040–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.D. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnaud, C.H. Engineered ribosomes could make new polymers. C&EN. 2021; 99:26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.