Abstract
Understanding the pathogenesis of the SARS-CoV-2 infection is key to developing preventive and therapeutic strategies against COVID-19, in the case of severe illness but also when the disease is mild. The use of appropriate experimental animal models remains central in the in vivo exploration of the physiopathology of infection and antiviral strategies. This study describes SARS-CoV-2 intranasal infection in ferrets and hamsters with low doses of low-passage SARS-CoV-2 clinical French isolate UCN19, describing infection levels, excretion, immune responses and pathological patterns in both animal species. Individual infection with 103 p.f.u. SARS-CoV-2 induced a more severe disease in hamsters than in ferrets. Viral RNA was detected in the lungs of hamsters but not of ferrets and in the brain (olfactory bulb and/or medulla oblongata) of both species. Overall, the clinical disease remained mild, with serological responses detected from 7 days and 10 days post-inoculation in hamsters and ferrets respectively. The virus became undetectable and pathology resolved within 14 days. The kinetics and levels of infection can be used in ferrets and hamsters as experimental models for understanding the pathogenicity of SARS-CoV-2, and testing the protective effect of drugs.
Keywords: experimental model, ferret, hamster, infection, SARS-CoV-2, severity
Introduction
Coronavirus Disease 2019 (COVID-19) is induced by the Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) [1] and has become a pandemic causing extensive economic, social, medical and scientific disruption around the world. Since 2002, SARS-CoV-2 is the third epidemic coronavirus transmitted from animals to humans after SARS-CoV [2] and Middle East Respiratory Syndrome coronavirus (MERS-CoV) [3]. The magnitude of the COVID-19 pandemic in terms of the number of human cases is by far the greatest. Understanding the pathogenesis of the SARS-CoV-2 infection is key to maximize prevention and to develop therapeutic solutions, and this is possible through susceptible animal models such as non-human primates [cynomolgus (Macacca fascicularis) and rhesus (Macacca mulatta) macaques], cats, hamsters and ferrets [4–13], the latter two species being the most economic and the easiest to house and handle. To our knowledge, all experimental infection studies of ferrets and hamsters [9, 13] published so far, apart from two studies [9, 13], were performed with high doses of SARS-CoV-2 (ranging from 8×104 TCID50 to 105.5 TCID50 or 106 p.f.u. per animal), mainly with the objective of inducing severe clinical infections. Here, we chose to explore the effects of a lower viral infection dose, considered closer to natural and more common infection conditions with SARS-CoV-2 in humans, probably leading to mild clinical disease, but with potential viral excretion.
The main objective of the study was to characterize the kinetics of the disease (clinical signs, pathogenicity and immune responses) in ferrets and hamsters infected with low doses of a low-passage SARS-CoV-2 clinical isolate, in order to develop suitable animal models for further preclinical therapeutic and vaccine studies.
Methods
SARS-CoV-2 virus
SARS-CoV-2 viral strain UCN19 was isolated in March 2020, during the course of the active epidemic, from naso-pharyngeal flocked swabs obtained at the University Hospital of Caen, Normandy, France, from patients who were suffering from respiratory infection and were confirmed infected by SARS-CoV-2 by routine molecular diagnosis. The swabs were eluted in UTM media (Copan) at 4 °C for less than 48 h. Vero CCL-81 cells (passage 32, from ATCC), grown at 80 % confluence were inoculated with 200 µl micro-filtered elution for passage 0 (P0). Cells were visually checked for cytopathic effect on a daily basis using an inverted microscope. Cell supernatants (12 ml) were harvested on day 3 after inoculation and immediately used for passage 1 (P1) produced in T75 culture flasks containing Vero cells as previously described for P0. P1 was used for stock production of UCN19, aliquoted and stored at −80 °C before titration, genomic quantification, sequencing (GISAID access number EPI_ISL_666870) and experimental infections to ferrets and hamsters. The use of the low-passage SARS-CoV-2 clinical isolate P1 reduces the risk of cell-culture-induced genetic modification. All animals in this study were inoculated with inocula from the same stock.
Animal experimental design
The experimental protocols complied with the regulation 2010/63/CE of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes [14] and as transposed into French law [15]. These experiments were approved by the Anses/ENVA/UPEC ethics committee and the French Ministry of Research (Apafis no. 24818-2020032710416319).
Fifteen 10-month-old ferrets (Mustela putorius furo, ten neutered males and five females; Euroferrets) and 21 8-week-old female hamsters (Mesocricetus auratus, strain RjHan:AURA; Janvier Labs) were used. Ferrets and hamsters were kept in cages with environmental enrichment, allocated into groups of two to five ferrets and two hamsters per cage, with non-infected animals kept in a separate room from the infected animals. Food and water were provided ad libitum. Weight, body temperature (measured once a day in the morning at about the same time for each animal by subcutaneous chips: IPTT300; Plexx) and activity levels of all animals were recorded on a daily basis throughout the duration of the experiment.
Twelve ferrets and 15 hamsters were anaesthetized with isofluorane and inoculated by the intranasal route with 2×103 p.f.u. and 1.8×103 p.f.u. of SARS-CoV-2 strain UCN19 respectively. Ferrets received the inoculum in 250 µl and hamsters in 20 µl in each nostril. At day 2 (D2) post-inoculation, D4, D7, D10 and D14, infected animals were anaesthetized with isoflurane for the collection of samples submitted to viral analysis (RT-PCR and cellular isolation): oro-pharyngeal swabs (plain swab rayon-tipped; Copan), nasal washes in ferrets only (by administering 500 µl PBS to each nostril), hamsters being too small for this sampling technique, rectal swabs in ferrets (plain swab rayon-tipped; Copan) and faecal pellets spontaneously produced in hamsters. The samples were stored at −80°C until used. At the same time-points, blood was collected from the cranial vena cava in ferrets using a vacutainer (SST tubes; BD) and from the retro-orbital vein in hamsters to isolate and store serum (at −20 °C) for immunological tests. Hamster serum was tested by RT-PCR and viral culture. The number of animals sampled at each time-point decreased over time with six inoculated animals (three ferrets, including one female ferret and three hamsters) killed sequentially at D2, D4, D7, D10 (hamsters only on that date) and D14 and submitted to post-mortem (PM) examination. At PM, the following samples were collected, homogenized and stored frozen at −80 °C for viral RNA measurement and viral culture or in 10 % buffered formalin for histology: nasal turbinates, tonsils (ferrets only/tissue absent in the hamster anatomy), proximal trachea, lung lobes (left lobes for RNA extraction and viral titration and right lobes for histopathology), liver, spleen, large intestine, kidneys, olfactory bulb and medulla oblongata. In addition, broncho-alveolar lavage (collected with PBS in ferrets only, for practical reasons) and urine were collected at PM. Three ferrets and six hamsters were inoculated with PBS as naive controls and were sampled at the end of the study to confirm their negative status and provide background data for immunological studies.
Virus titration
Viral load was determined for viral inoculum and for a subset of samples (Table S1, available in the online version of this arricle) by plaque assay on VeroE6 cells. A 12-well plate was seeded with a VeroE6 cell suspension 24 h before virus inoculation. Organs were harvested, weighed and homogenized in 1 ml of Dulbecco's modified Eagle medium (DMEM) with stainless steel beads (Qiagen) for 3 min at 30 Hz using a TissueLyserII (Qiagen). Homogenates were then clarified by centrifugation (2000 g at 4 °C for 10 min), aliquoted and stored at −80 °C. A ten-fold serial dilution of the samples (clinical samples and tissue homogenates) was performed in DMEM supplemented with FCS and penicillin/streptomycin. Inoculum was added to each well and the plate was incubated at 37 °C in 5 % CO2 for 1 h. At least one uninfected well was used as an independent negative control. One hour later, plaque assays were overlaid with carboxy-methylcellulose mix (CMC: 3.2 % DMEM, 5 % FBS, v/v) and after 5 days were fixed and stained with a crystal violet solution (3.7 % formaldehyde, 0.2 % crystal violet). The viral titre, expressed as p.f.u ml−1 [number of plaques/(dilution × volume of diluted virus added to the well)], was determined by dividing the number of plaques for the adequate dilution by the total dilution factor.
RNA extraction and TaqMan RT-qPCR
Tissue homogenates and clinical samples were treated following the same protocol. A volume of 15 µl of Triton X-100 (MP Biomedicals) was added to 560 µl of AVL Lysis buffer (Qiagen) to reach a concentration of 2.7 % (required to inactivate SARS-CoV-2 in infectious samples). A negative RNA extraction control was performed for each set of 12 samples tested. TaqMan RT-qPCR was performed with the SuperScript III Platinum One-Step qRT-PCR kit (Invitrogen, Fischer Scientific) using the protocol described by Corman et al. [16]. Coronavirus primers (E_Sarbeco_F and E_Sarbeco_R) and probe (E_Sarbeco_P1 labelled with the fluorescent dye FAM-BHQ1) targeting the envelope protein gene (E gene) were used for this study. Primers and probe were provided by Eurogentec. All TaqMan RT-qPCR assays were performed on a Rotor Gene Q MDx thermocycler (Qiagen) and Lightcycler LC480 (Roche). Negative and positive controls were included in each RT-qPCR assay. Positive controls consisted of a calibrated SARS-CoV-2 RNA stock extracted from the UCN19 SARS-CoV-2 strain supernatant. The titre of the calibrated SARS-CoV-2 RNA stock was determined (3×106 E gene copies µl–1) using a serial 10-fold dilution of an E gene transcript standard control obtained from the EVAg network (https://www.european-virus-archive.com/ - ref: 026 N-03866) and a qRT-PCR system [16]. The determination of SARS-CoV-2 RNA titre in number of copies µl–1 was determined for each sample by testing a standard curve with the positive control as described above. RNA titre was expressed in number of copies mg–1 for tissue samples after application of the conversion factor calculated from the sample weight and the volumes of the different buffers used. A threshold setting (Ct) of 0.05 was used for each RT-qPCR assay. The efficiency, slope and correlation coefficient (R 2) were determined with the Rotor Gene software. All reactions were carried out as technical duplicates. A cut-off >35 was defined for low positive results (<300 copies per µl of RNA). The limit of detection of the TaqMan RT-qPCR analysis was assessed by using a serial dilution of the positive control tested as previously described [16]. This limit of detection was 15 copies per µl of RNA or 60 copies per reaction.
Histological analyses
Samples from the right cranial and caudal lung lobes together with tonsil (in ferrets only) and trachea were fixed by immersion in 10 % neutral-buffered formalin and processed routinely into paraffin wax. Sections of 4µm were cut and stained with haematoxylin and eosin (H&E) and examined microscopically. In addition, samples were stained using the RNAscope in situ hybridization (ISH) technique to identify the SARS-CoV-2 virus RNA as previously described [9]. Briefly, tissues were pre-treated with hydrogen peroxide for 10 min (room temperature), target retrieval for 30 min (98–101 °C) and protease plus for 30 min (40 °C) (Advanced Cell Diagnostics). A V-nCoV2019-S probe (Advanced Cell Diagnostics, Biotechne) was incubated on the tissues for 2 h at 40 °C. Amplification of the signal was carried out following the RNAscope protocol using the RNAscope 2.5 HD Detection kit – Red (Advanced Cell Diagnostics, Biotechne).
Serological analyses
The sera of six different animals (three ferrets and three hamsters) were sampled at each time-point, except at D10 and D14 when the same ferrets were tested consecutively. The plates were coated with SARS-CoV-2-infected Vero E6 cell lysates at 4 °C overnight or with non-infected Vero E6 lysate to provide sample background optical density (OD) values. This SARS-CoV-2 antigen was previously validated with negative and positive human sera as provided by an independent medical analysis laboratory. For each ELISA test of the study, sera already tested were used as positive and negative controls. Hamster and ferret serum samples were diluted 1 : 100 and incubated for 1 h at 37 °C. Specific antibody binding was detected by peroxidase-labelled goat anti-hamster IgG (H+L) (Invitrogen) and peroxidase-labelled goat anti-ferret IgG (H+L) (KPL), diluted 1 : 5000 and 1 : 100 respectively, for 1 h at 37 °C. Substrate (TMB single solution; Fisher Scientific) was then added and the reaction was stopped with 10.6 % H3PO4. The optical density was determined at 450 nm for both species. Net OD values were calculated by removing the sample background OD value from OD sample values.
The presence of neutralizing antibodies was detected by performing a plaque reduction neutralization test (PRNT) as described previously [17] using P2 of SARS-CoV-2 strain UCN19. Hamster and ferret sera were serially diluted and mixed with an equal volume containing 100 p.f.u. of virus per 125 µl. The mixtures were incubated for 1 h and subsequently inoculated into wells of 12-well tissue plates containing confluent Vero E6 cells. After adsorption for 1 h, the wells were overlaid with a mixture of CMC (carboxymethylcellulose sodium salt of low viscosity) and 5 % FCS DMEM medium. Plates were incubated for 5 days and finally coloured by using crystal violet. A 50 % reduction in the number of plaques was used as the criterion for virus neutralization titres. Endpoint titres of neutralizing antibodies were determined.
Statistical analyses
Wilcoxon’s signed-rank test was performed via the package stat in R (version 3.3.3) to compare viral RNA quantities in nasal washes between male and female ferrets and with GraphPad Prism v6 to compare the areas under the curves for the kinetics of viral presence in clinical samples and of weight changes from D0. Two-way ANOVA was used (significance level fixed at 0.05) to analyse the effect of ‘species’ and ‘time’ on RNA levels recovered in tissue samples collected from ferrets and hamsters using GraphPad Prism v6.
Results
Clinical observations
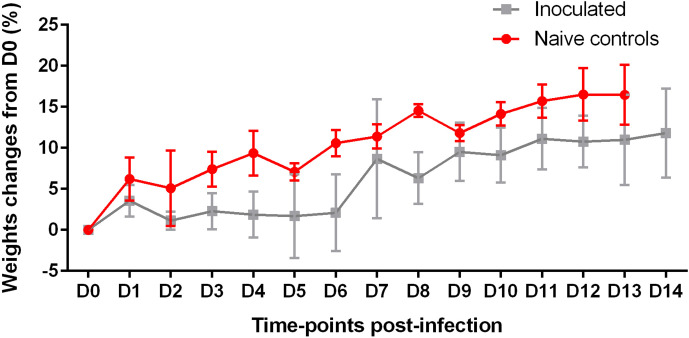
The experimental SARS-CoV-2 infection did not result in any death and no clinical endpoint was reached in any animal. Lethargy was observed on days 7 and 8 post-infection in three ferrets. Snoring was also reported in one ferret between D7 and D14. Hyperthermia was not recorded at any time point measured in any infected animal. Weight remained stable in ferrets. In hamsters, an average weight gain was observed in animals over time, and on average was reduced, although not significantly, in the inoculated group (Fig. 1).
Fig. 1.
Hamster weight changes from D0 (%) with mean and standard deviation after inoculation with SARS-CoV-2 or PBS (in the naïve controls). At all time-points, n=6 for naïve controls. For inoculated hamsters, n=15 between D0 and D2, n=12 between D3 and D4, n=9 between D5 and D7, n=6 between D8 and D10, and n=3 between D11 and D14.
Local viral RNA presence measured by RT-PCR
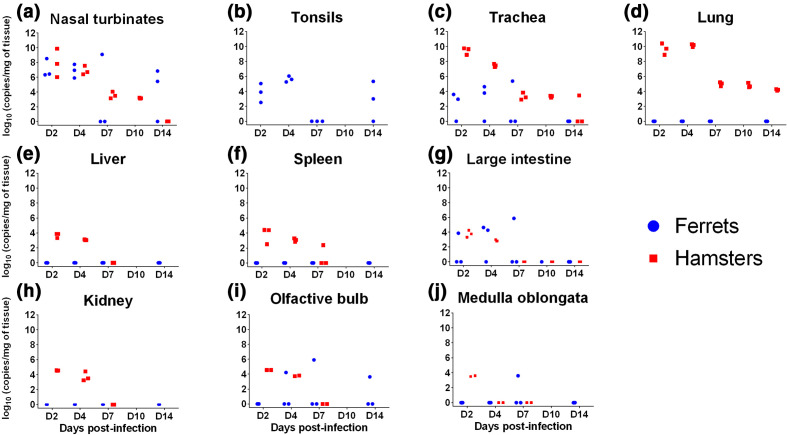
Viral RNA was recovered from a large range of tissues in both species from 2 days post-infection and tended to decrease over time, with almost all tissues negative by D14, except in nasal turbinates (Fig. 2a), tonsils (tissue absent in the hamster anatomy) (Fig. 2b) and medulla oblongata (Fig. 2j) in ferrets, and the trachea (Fig. 2c) and the lungs (Fig. 2d) in hamsters. More variability was observed in RNA levels within ferret groups than hamster groups at each time-point. The most heavily infected tissues were: nasal turbinates with no significant difference between average infection levels measured in ferrets and hamsters; the trachea, being significantly more infected in hamsters than in ferrets; and the lungs, with high RNA levels in hamsters and no detection in ferrets. Viral RNA was recovered at lower levels than in the respiratory tissues in the liver (Fig. 2e), the spleen (Fig. 2f) and the kidneys (Fig. 2h) on D2 and D4 in hamsters only. In the large intestine (Fig. 2g), average viral RNA levels were similar in the ferret and hamster groups, although individual ferrets presented values higher than hamsters at D2, D4 and D7. In the olfactiory bulb (Fig. 2i), viral RNA was recovered in both species, with higher average levels in hamsters at D2 and D4, but a longer persistence in ferrets (up to D14). All three hamsters were positive in the medulla oblongata (Fig. 2j) at D2 but subsequently became negative, while only one ferret was positive at D7. Urine samples were negative at all time-points in both species, as well as retro-pharyngeal lymph nodes (tested in ferrets only) and hamster serum (data shown in Table S1).
Fig. 2.
Viral RNA loads in three ferrets and three hamsters inoculated with UCN19 SARS-CoV-2. (a) Nasal turbinates, (b) tonsils, (c) trachea, (d) lung, (e) liver, (f) spleen, (g) large intestine, (h) kidney, (i) olfactory bulb, (j) medulla oblongata.
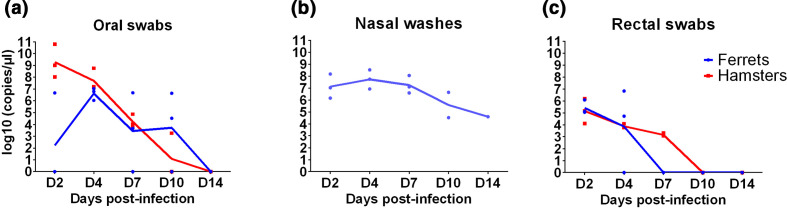
Viral RNA was found in oral swabs of both species from D2 to D10 (Fig. 3a) with no oral swab positive at D14 in either species. The areas under the curves did not differ between hamsters and ferrets. Nasal wash fluids were positive from D2 to D14 (Fig. 3b) (collected in ferrets only), consistent with the high levels of viral RNA found in nasal turbinates in this species (Fig. 2a). The viral RNA levels in nasal washes were not different between male and female ferrets independently of the experimental time point (P=0.28 on D2, 0.55 on D4, and 1 on D7, D10 and D14). RNA levels decreased faster in rectal swabs (Fig. 3c) than in the other clinical samples and were all negative by D10, with no difference between areas under the curves for ferrets and hamsters.
Fig. 3.
Viral RNA loads in clinical samples. (a) Oral swabs, (b) nasal washes and (c) rectal swabs collected from to D2 to D14 in three ferrets and three hamsters inoculated intranasally with UCN19 SARS-CoV-2 (nasal washes were only collected in ferrets and olfactory bulbs and medulla oblongata were collected in two hamsters rather than three from D2 to D14).
Virus titration
In hamsters, infectious virus was detected from nasal turbinates, trachea and lungs in all the necropsied hamsters at D2 and D4 with 2.7×100 to 1.32×104 p.f.u. ml−1 tissue and from oral swabs for two out of three hamsters at D2 (4×101 and 2.4×102 p.f.u. ml−1) and one at D4 (8.0×101 p.f.u. ml−1). In ferrets, infectious virus was detected in turbinates and nasal washes, but not in the lungs. Nasal turbinates were found to be infectious (in four out of nine samples) between D2 and D7 with a maximal titre of 1.28×104 p.f.u. ml−1 at D7. Almost all nasal washes (five out of six samples) were infectious between D2 and D4 with a maximal titre of 3.04×103 p.f.u. ml−1 at D2.
No infectious virus was isolated in any other tested sample (Tables S1 and S2).
Histopathology
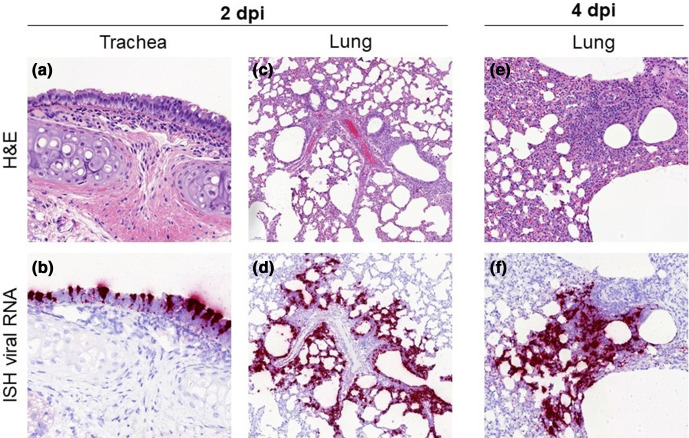
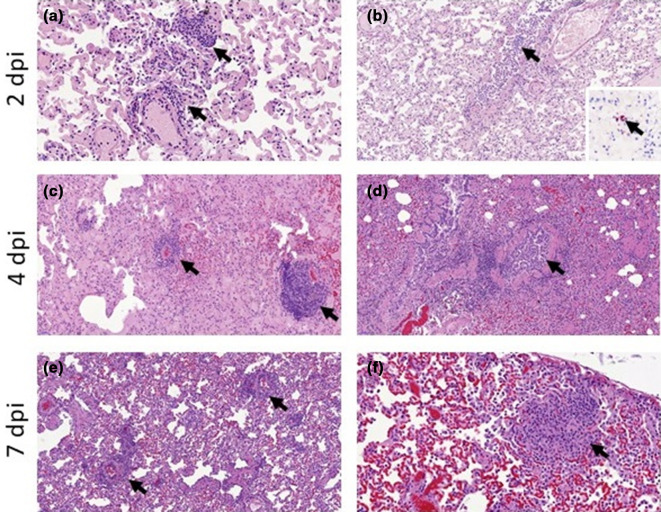
In hamsters, inflammatory infiltration was observed in the lung from D2 to D7, with the presence of macrophages, lymphocytes and neutrophils, usually surrounding airways and within the alveolar walls (Fig. 4). These inflammatory infiltrates showed signs of cell death and a high amount of viral RNA detected by ISH, mostly at D2 and D4 (Fig. 4), and at much lower level at D7. Animals killed at D10 and D14 did not show any remarkable changes in the lungs and no presence of viral RNA. In the trachea, mild inflammatory infiltration of the epithelium was observed together with minimal to mild epithelial necrosis at D2, D4 and D7. Viral RNA was very abundant within the tracheal epithelial cells at D2, and decreased from D4 with a complete absence from D7 onwards (Fig. 4).
Fig. 4.
Histopathological findings in hamsters inoculated with SARS-CoV-2 strain UCN19. Mild inflammatory cell infiltration was observed in the trachea at D2 (a, 400×) with high levels of viral RNA by ISH within respiratory epithelial cells (b, 400×). Inflammatory infiltrates with the lung parenchyma, mostly within the bronchial and bronchiolar mucosa but also surrounding airways and blood vessels, were observed at D2 (c, 100×) and D4 (e, 200×). The presence of inflammatory infiltrates is correlated with the viral RNA staining in sequential sections at D2 (d, 100×) and D4 (f, 200×).
In ferrets, histopathological findings were less severe than in hamsters. Non-inoculated ferrets showed only minimal to mild inflammatory cell infiltrates around the bronchioles and blood vessels (peribronchiolar and perivascular cuffing). In inoculated animals, mild bronchiolitis was observed at D2 in all animals with the presence of inflammatory cell infiltrates within the bronchiolar luminae, mostly neutrophils but also some eosinophils, macrophages and lymphocytes. Perivascular and peribronchiolar cuffing was also observed (mainly mononuclear cells) (Fig. 5). One of the three animals killed at D2 showed a few scattered cells positive to viral RNA within alveolar walls, not related to any histopathological lesion (Fig. 5b). At D4, D7 and D14, individual animals showed mild to moderate perivascular cuffing, together with areas of bronchiolitis similar to those seen in animals killed at D2 (Fig. 5). Small areas of consolidation (inflammatory infiltrates composed mainly of macrophages and lymphocytes within the parenchyma) were also observed (Fig. 5f). No remarkable change was observed in the tonsil or the proximal trachea, with only a few scattered positive cells to viral RNA (ISH) in the trachea at D2 not related to any histopathological lesion.
Fig. 5.
Histopathological findings in the lungs of ferrets inoculated with the UNC19 SARS-CoV-2 strain. Perivascular cuffing (a, 400×, arrows) and mild bronchiolitis (b, 200×, arrow) was observed at D2, with minimal presence of viral RNA (b, inset, 400×) within alveolar walls not related to histopathological lesions. Perivascular cuffing was also observed at D4 (c, 200×, arrows) and D7 (e, 200×, arrows). Mild bronchiolitis with presence of intraluminal inflammatory infiltrates was observed at D4 (d, 200×, arrow). Scattered foci of parenchymal inflammation were also observed at D7 (f, 400×, arrow).
Serology
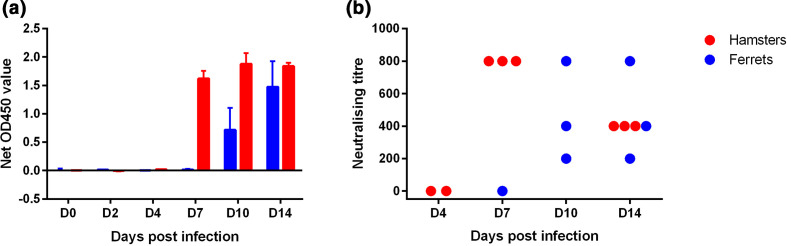
A detectable level of IgG appeared from D7 in hamster serum and from D10 in ferrets. The level of IgG increased rapidly at D7 and remained consistently high until D14 in hamsters (Fig. 6a), while in ferrets the increase appeared more gradual from D10. Neutralizing antibodies were detected from D7 and D10 in hamsters and ferrets respectively (Fig. 6b). The IgG levels did not correlate with the neutralizing titres. Indeed, in hamsters, the neutralizing titres halved between D7 and D14 whereas the level of detected IgG remained stable. More variability was observed in ferret responses at D10 and D14 than in hamsters. For one ferret, the level of neutralizing antibodies was halved at D14 compared with D10, as observed for the hamsters. For another ferret, the response doubled from D10 to D14, while the last ferret of the group presented persistently the highest responses on D10 and D14 (Fig. 6b).
Fig. 6.
Antibody responses (measured at each time-point in inoculated hamsters and ferrets. (a) Net OD450 for IgG ELISA [n=3 animals for each time-point except for ferrets at D0 (n=2) and (b) sero-neutralization, with one dot per individual blood sample analysis (in red, hamsters with n=2 at D4, n=3 at D7 and n=3 at D14/in blue, ferrets with n=1 at D7, n=3 at D10 and n=3 at D14). Sero-neutralization was not carried out for the hamster samples of D0 and D2 and for the ferret samples of D0, D2 and D4 because of the ELISA-negative results.
Discussion
A number of studies have already been published on the experimental infection of ferrets or hamsters with SARS-CoV-2, but none has compared the two models with the same strain. Differences between protocols and strains may impact the clinical, immunological and pathological outcomes, and therefore impair the comparison of independent investigations. Moreover, most studies have focused on high inoculation doses in an attempt to induce severe forms of COVID-19 and understand their pathogenicity. In this study, we have focused on milder forms of COVID-19, more common than the severe clinical course in human patients, that also require preventive and therapeutic approaches. Only one other study [13] used a similarly low SARS-CoV-2 dose of 103 p.f.u. inoculated to young female hamsters, but it was delivered simultaneously by the intranasal and intraocular routes. Our study was planned to evaluate the pathogenicity and viral load, and peripheral immunogenicity in ferrets and hamsters inoculated by the intranasal route with a single low dose of the same SARS-CoV-2 clinical strain obtained after a single passage on cell culture. The infection of all inoculated hamsters and ferrets was confirmed in live animals and at PM.
In experimentally infected hamsters, the clinical signs are generally considered limited, except for the significant weight loss often reported. In our study, all hamsters tended to gain weight over time although at different rates and with lesser gain in inoculated hamsters. The weight loss or the lower weight gain could be due to the lack of appetite because of the loss of smell [18] and the clinical manifestations after virus replication (e.g. lethargy). Inoculated ferrets generally show even milder clinical signs of disease, independently of the challenge dose [8–11], without any weight loss, as reported in our study. The marked lethargy observed in ferrets in this study was also reported by others [8–11]. It is possible that transient hyperthermia was missed if shorter than 24 h (time interval between subcutaneous IPTT300 chip readings).
The kinetics, distribution and levels of RNA recovered in hamster tissues were similar to those reported in other studies [6, 7, 12], including with higher challenge doses, and generally higher than in ferrets, except in the nasal turbinates and large intestine where both species appeared to be on average infected at similar levels.
The pathogenicity of SARS-CoV-2 in the nasal cavity of the hamsters infected in this study involved respiratory and olfactory epithelium, including sustentacular cells [18]. In ferrets, a similar infection pattern has also been observed [9]. In this species, nasal washes provided non-invasive evidence of local infection with a consistently high level of viral RNA recovered from D2. The persistence of viral RNA over a long time-span increases the potential for detecting the infection, but should not be considered representative of the presence of infectious virus. The infectiousness of nasal washes was demonstrated in Vero6 cells on D2 and D4 only, and therefore over a short period of time and before the onset of clinical signs (lethargy) and the development of detectable serological responses. Such samples would be valuable to collect in hamsters in future experimental studies. In oral swabs, similar kinetics of viral RNA were observed in both species with a gradual decrease from D2 to complete absence on D14. The lack of positive samples in the retro-pharyngeal lymph nodes collected in ferrets suggests that SARS-CoV-2 did not drain through the oral mucosal, but targeted the nasal mucosa, infecting the trachea in both species, more severely in hamsters based on viral RNA recovery and histopathology findings, and finally the lungs, mostly in hamsters.
Both species developed a viral infection in the higher respiratory tract; however, hamsters developed a more severe infection in the lungs (measured by RNA levels and infectious virus isolation and histopathology) than ferrets. In ferrets, the presence of SARS-CoV-2 in the lungs was only detected in a few cells with the RNAscope ISH technique, and not related to the histopathological lesions. ISH was more sensitive than RNA extraction and cell culture in this study, probably detecting only small pieces of degraded viral RNA. This association in ferrets of mild infection without viral RNA is similar to what has been previously observed [8, 9]. The probe used for the S-gene in our ISH technique or the polyclonal antibody used by Kim et al. [8] will not detect replicating virus. A small amount of viral RNA in scattered cells can indicate just remnants of the inoculum. The presence of mild lesions in the lung from infected ferrets may indicate a mild inflammatory reaction within the airways due to damage to the respiratory epithelium. This discordance between ISH and RNA extraction may also originate from the dilution effect of the virus in tissues, the heterogeneous viral distribution (although the infection protocols did not target any specific part of the lung), or the inhibitory effect of blood contained in samples on RNA detection. One study [13] in hamsters highlighted that lesions were more severe in right lobes than in left lung lobes; in our study, samples from the left lung lobes were used for RNA extraction and if this difference between lung sides also occurs in ferrets, it may have induced an under-estimation of infection in this species. The right lung lobes submitted for histopathology presented signs of viral infection. The impact of a species difference (between ferrets and hamsters) in pulmonary pathogenicity at the inoculation dose used in these species is unclear. SARS-CoV-2 is detected in the lungs of the majority of human fatal cases [19–21] and ISARIC clinical investigations [22] reported shortness of breath or dyspnoea within the five most frequent symptoms, indicating common viral invasion of the lungs by SARS-CoV-2. However, lung viral analysis is not available in mild and moderate COVID-19 patients. The hamster model is a good proxy to study the invasion of the lungs by SARS-CoV-2 and may therefore bear some resemblance to more serious human cases, in spite of showing limited clinical signs such as weight loss.
SARS-CoV-2 RNA was also detected in our study in the intestinal tract of ferrets and hamsters, although at a lower level than in the upper respiratory tract. As in most human cases [23], no infectious virus was isolated from intestinal samples, while viral RNA was detected. The presence of SARS-CoV-2 in the brain has also been reported in other experimental studies [6, 10, 11], and rarely in human patients, although not systematically. In our study, viral RNA was detected in both species mostly in the olfactory bulb but also in the medulla oblongata. The dissemination to the olfactory bulb could be associated with loss of smell in hamsters [18] and potentially in ferrets. With regard to the impact of dissemination to the medulla oblongata, one hypothesis could be that its potential infection had an effect on respiratory regulation, but the present study was inconclusive. The impact of dissemination to the medulla oblongata is still unclear.
Antibody kinetics in ferrets and hamsters were comparable with those seen in patients with a seroconversion between D7 and D14 [23], and those seen in experimental models using hamsters and ferrets [6, 8–11]. In our study, the increase in antibody levels occurred faster in hamsters than in ferrets, from no detectable level at D4 to consistently high levels between D7 and D14 in hamsters and between D7 and D10 in ferrets. These observations suggest that in these species, the exposure to a low challenge viral dose inoculated by the intranasal route induced an early, significant and potentially protective humoral response. For both species, the increase of antibody levels slightly preceded or was concomitant with the disappearance of infectious virus (viral clearance), but further investigations would be required to conclude on the protective effect conferred by antibodies.
According to previous studies [9], reducing the challenge dose beyond the one used in our study impacts the reproducibility of infection success (only one out of six ferrets inoculated with 5×102 p.f.u. became infected in experiments by Ryan et al. [9]).To our knowledge, no study has performed inoculation with a lower dose than 103 p.f.u. in hamster. The lowest dose for which there is reproducibility of infection of all animals is still unknown.
A higher susceptibility in males has been suggested in humans [24–27]. No difference in the severity of the disease was measurable between sexes in our study. The low number of ferrets used in our study could be an explanation for this. Other factors not present in the male ferrets of our study could also explain the lack of increased severity: for instance, in more severe human cases, sex is often associated with comorbidities or an old age [19, 24].
COVID-19 is less frequently reported in children than in adults and the elderly [28]. Two studies in hamsters [12, 13] reported more severe lesions and clinical signs in older animals. The oldest ferrets used in experimental studies were 20 months old [8–11], so no conclusion can be drawn on the effect of age in this species. Some studies suggest that thymus-produced thymosin may have a protective effect [29, 30]. The youngest animals included in our study still presented a detectable active thymus at PM but its activity and influence on the disease pattern was not explored.
Conclusions
Hamsters and ferrets are valuable animal models to study COVID-19. Hamsters are easy to handle and maintain. Ferrets are more expensive than hamsters and require larger housing space, but a change in their generally active behaviour is a relatively easy clinical sign to detect in comparison with hamsters. The marked transient fatigue reported in the infected ferrets at days 7 and 8 was also reported by others [8, 9, 11] and likewise is a characteristic of the human COVID-19 condition that could be monitored in preventive or therapeutic experimental studies extending to that time-point. Infected hamsters showed remarkable replication of virus in the lung. In our study, the low challenge dose induced a lung infection very consistent within hamster groups and without severe weight loss seen with higher challenge doses; our protocol is therefore more in line with the 3Rs. Pulmonary infection of ferrets with SARS-CoV-2 requires higher infectious doses per animal when using the intratracheal route [8], and is not systematic [11]. A higher intranasal infection dose may therefore be required for smaller intragroup variability in future studies. Compared with tissue analysis at PM, longitudinal studies based on the collection (under general anaesthesia) of clinical samples such as oral and rectal swabs and nasal washes have the advantage of following the same animal over time, and therefore have the potential of using fewer animals. Both species provided samples with infectious virus up to D10, of relevance for excretion and potential for transmission. Under the conditions of our study, the model that seems most relevant is the hamster because of the systematic lung infection and its ease of supply and maintenance.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
We thank Euroferret for providing animals very rapidly in times of COVID-19 and severe confinement measures imposed in France. From Anses LRFSN, Jonathan Rieder, Mélanie Badré-Biarnais, Youssef Arnaout, Jean-Marc Boucher and Vanessa Bastid, for their investment in virological and serological analyses; Valère Brogat, Michel Munier and Nicolas Penel for animal care; Franca Rizzo for biosecurity and logistical support; Astrid Vabret for granting access to SARS-CoV2 strains isolated in the University Hospital of Caen, Normandy, France; Thomas Labadie from London School of Hygiene and Tropical Medicine, UK, for technical advice on viral titration of SARS-CoV2 UCN19 stocks; and Latifa Lakhdar, head of the Plateforme d’expérimentation animale, Anses - Laboratoire de Lyon.
Author contributions
Conceptualization; E.M.L., P.M., S.L. and A.S., methodology: F.B., M.W., E.P.M., S.M., S.La and P.M., validation: P.M., E.M.L., C.R., A.S. and S.L., formal analysis: J.S., M.A.G., J.V., C.P., S.M., S.La, E.P.M., C.R., and M.W., writing—original draft preparation: E.M.L. and P.M., writing—review and editing, all; figure: S.La; supervision: P.M. and E.M.L. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of the data; in the writing of the manuscript, or in the decision to publish the results.
Ethical statement
These experiments were approved by the Anses/ENVA/UPEC ethics committee and the French Ministry of Research (Apafis no. 24818-2020032710416319).
Footnotes
Abbreviations: CMC, carboxymethylcellulose; COVID- 19, Coronavirus Disease 2019; DMEM, Dulbecco's modified Eagle medium; D1 to D14, Day 1 to Day 14; FBS, Foetal bovine serum; ISH, in situ hybridization; MERS- CoV, Middle East Respiratory Syndrome coronavirus; OD, Optical density; p.f.u., plaque- forming unit; PM, post-mortem; RNA, ribonucleic acid; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TCID50, Fifty- percent tissue culture infectious dose; UTM, universal transport medium; UTM, universal transport medium.
Two supplementary tables are available with the online version of this article.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C, Preiser W, Günther S, Schmitz H, Doerr HW. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman CM, Frieman MB. Emergence of the middle East respiratory syndrome coronavirus. PLoS Pathog. 2013;9:e1003595. doi: 10.1371/journal.ppat.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JF-W, Zhang AJ, Yuan S, Poon VK-M, Chan CC-S, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sia SF, Yan L-M, Chin AWH, Fung K, Choy K-T, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020 doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y-I, Kim S-G, Kim S-M, Kim E-H, Park S-J, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan KA, Bewley KR, Fotheringham SA, Brown P, Hall Y. Dose-dependent response to infection with SARS-CoV-2 in the ferret model: evidence of protection to re-challenge [Internet] Microbiology. 2020 [Google Scholar]
- 10.Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J. Experimental transmission studies of SARS-CoV-2 in fruit bats, ferrets, pigs and chickens. SSRN Electronic Journal. 2020;28 doi: 10.2139/ssrn.3578792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Wen Z, Zhong G, Yang H, Wang C, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterrieder N, Bertzbach LD, Dietert K, Abdelgawad A, Vladimirova D, et al. Age-Dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Commission Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes; European Commission
- 15.Journal Officiel de la République Française Décret n 2013-118 Du 1er Février 2013 Relatif La Protection des Animaux Utilisés Des Fins Scientifiques. Paris, France: 2013. p. 2199. p. [Google Scholar]
- 16.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chrun T, Lacôte S, Urien C, Jouneau L, Barc C, et al. A Rift Valley fever virus GN ectodomain-based DNA vaccine induces a partial protection not improved by APC targeting. NPJ Vaccines. 2018;3:14. doi: 10.1038/s41541-018-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryche B, St Albin A, Murri S, Lacôte S, Pulido C, et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav Immun. 2020;89:579. doi: 10.1016/j.bbi.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edler C, Schröder AS, Aepfelbacher M, Fitzek A, Heinemann A, Heinrich F, et al. Dying with SARS-CoV-2 infection—an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int J Legal Med. 2020;134:1275–1284. doi: 10.1007/s00414-020-02317-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer I-M, Padera RF, Solomon IH, Kanjilal S, Hammer MM, et al. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol. 2020 doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martines RB, Ritter JM, Matkovic E, Gary J, Bollweg BC, et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerging Infect Dis. 21 mai. 2020;26 doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ISARIC (International Severe Acute Respiratory and Emerging Infections Consortium. ISARIC (International Severe Acute Respiratory and Emerging Infections Consortium Covid-19 report: 19 May 2010 [Internet]. Disponible sur: https://media.tghn.org/medialibrary/2020/05/ISARIC_Data_Platform_COVID-19_Report_19MAY20.pdf
- 23.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 24.Petrilli CM, Jones SA, Yang J, Rajagopalan H, O’Donnell L, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;2:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikpouraghdam M, Jalali Farahani A, Alishiri G, Heydari S, Ebrahimnia M, et al. Epidemiological characteristics of coronavirus disease 2019 (COVID-19) patients in Iran: a single center study. J Clin Virol. 2020;127:104378. doi: 10.1016/j.jcv.2020.104378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long L, Zeng X, Zhang X, Xiao W, Guo E, et al. Short-Term outcomes of COVID-19 and risk factors for progression. Eur Respir J. 2020;55:2000990. doi: 10.1183/13993003.00990-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H, Lie Y, Prinzen FW. Surveillance of COVID-19 in the general population using an online questionnaire: report from 18,161 Respondents in China. JMIR Public Health Surveill. 2020;6:e18576. doi: 10.2196/18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.She J, Liu L, Liu W. COVID‐19 epidemic: disease characteristics in children. J Med Virol. 2020;92:747–754. doi: 10.1002/jmv.25807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rehman S, Majeed T, Azam Ansari M, Ali U, Sabit H, et al. Current scenario of COVID-19 in pediatric age group and physiology of immune and thymus response. Saudi J Biol Sci. 2020;15 mai doi: 10.1016/j.sjbs.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Pang Y, Hu Z, Wu M, Wang C, et al. Thymosin alpha 1 (Tα1) reduces the mortality of severe COVID-19 by restoration of lymphocytopenia and reversion of exhausted T cells. Clin Infect Dis. 2020;22 mai doi: 10.1093/cid/ciaa630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.