Abstract
Zika virus (ZIKV) is an emerging mosquito-borne flavivirus, which caused an unprecedented epidemic in Latin America. Among all viral non-structural proteins in flavivirus, NS5 is the most highly conserved and has multiple crucial functions, including participating in viral RNA replication and suppressing host innate immunity. Although ZIKV NS5 prominently localizes in the nucleus during infection, its specific nuclear localization signal (NLS), and its role in viral replication and pathogenesis remain controversial. Here, we identified aa 11–90 and aa 370–406 regions that contain NLSs, which are critical for nuclear localization of ZIKV NS5. Further experiments demonstrated that nuclear localization of ZIKV NS5 predominantly participates in suppression of interferon regulatory factor 3 (IRF3)-mediated activation of type I IFN (IFN-I) transcription and inhibition of IFN-I downstream response independent of its effect on signal transducers and activators of transcription 2 (STAT2) degradation. These results suggest that subcellular localization of NS5 is important for its function on innate immune suppression, which provides new insight into ZIKV pathogenesis.
Keywords: Zika virus (ZIKV), NS5, nuclear localization signal (NLS), type I interferon (IFN-I), immune evasion
Introduction
Zika virus (ZIKV) is an important mosquito-transmitted pathogen belonging to the family Flaviviridae. In 1947, it was initially identified in Uganda and then drastically emerged in Asia and Latin America in the last t3 years [1, 2]. Recently, numerous reports have linked ZIKV infection to various neurological symptoms, such as Guillain–Barré syndrome and microcephaly, but the underlaying pathogenic mechanisms still need to be further elucidated [3, 4].
Similar to other flaviviruses, the single-stranded positive-sense RNA genome (~11 kb) of ZIKV encodes a polyprotein precursor (~3300 aa residues), which is subsequently cleaved into three structural (C, prM and E) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins by host and viral proteases [5, 6]. NS5 is the largest and the most conserved protein of flavivirus. It comprises two domains, an N-terminal methyltransferase (MTase) domain and an RNA-dependent RNA polymerase (RdRP) domain [7, 8]. During flavivirus infection, a replication complex constituted with NS5 and other non-structural proteins is assembled on the cytoplasmic side of the endoplasmic reticulum (ER) membrane with the help of host proteins [9–11].
Although viral replication primarily occurs in the cytoplasm, NS5 of flaviviruses, including ZIKV, Dengue virus (DENV) and yellow fever virus (YFV), unusually localizes in the nucleus with no clear role in viral replication and pathogenesis [12–14]. Previous research has demonstrated that DENV NS5 nuclear accumulation affects the production of virus and IL-8, and can also interact with host nuclear active spliceosomes to enhance its replication [15, 16]. It has been reported that proteins, which are larger than 45 kDa, require intrinsic targeting signals called nuclear localization signals (NLSs) to mediate its shuttle between the nucleus and cytoplasm [17]. In spite of the fact that ZIKV NS5 is closely related to DENV NS5, whose bipartite NLS is recognized by an importin α/β nuclear transporter within aa 320–368 and nuclear export signal (NES) is recognized by CRM1 within aa 369–405 [13, 15, 18], the NLSs of ZIKV NS5 and their role in viral replication and immune response remain poorly studied.
The type I interferon (IFN-I), as a vital component of the innate immune system, plays a crucial role in antiviral response in all vertebrates [19]. During flavivirus infection, NS5 has been reported as a key antagonist at different IFN-I signalling cascade stage by diverse strategies. For instance, Japanese encephalitis virus (JEV) NS5 inhibits IFN-I production by blocking the nuclear translocation of IRF-3 and NF-κB [20], whereas tick-borne encephalitis virus (TBEV) and West Nile virus (WNV) NS5 inhibit IFN-I signalling at ligand-receptor binding phase by suppressing surface expression of interferon alpha and beta receptor subunit 1 (IFNAR1) [21]. In the case of DENV and ZIKV infection, NS5 can degrade human STAT2 by a proteasome-dependent manner, thereby inhibiting IFN-I signalling [12, 22]. Because of its vital role, extensive research has focused on understanding the mechanisms of flavivirus NS5-mediated suppression of host immune system. However, it is still unclear whether the mechanisms are associated with the subcellular localization.
In this study, we show the aa 370–406, a conserved NLS in other flaviviruses, rather than the C-terminal 18 residues, which was previously demonstrated as a NLS in DENV NS5, is sufficient to lead the nuclear localization of ZIKV NS5. Furthermore, a new NLS-containing region aa 11–90 was identified in ZIKV NS5. Our results also reveal the relevance of NS5 subcellular localization with its function on modulating IFN-I response, which provides a new understanding on the mechanism of ZIKV immune evasion.
Methods
Cells lines, viruses and antibodies
The human embryonic kidney (HEK 293T) and Vero cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) supplemented with 100 U ml−1 penicillin, 100 g ml−1 streptomycin and 10 % fetal bovine serum (GIBCO). The C6/36 and HeLa cell lines were cultured in RPMI-1640 medium (Hyclone) supplemented with the same components as DMEM. C6/36 cells were grown at 28 °C in a 5 % CO2 incubator, while other kinds of cells were grown at 37 °C in a 5 % CO2 incubator.
The ZIKV SZ01 strain used in this study was kindly provided by Dr Bo Zhang (Wuhan Institute of Virology, Chinese Academy of Sciences) and propagated in C6/36 cells. Sendai virus (SeV) was kindly provided by Dr Ling Zhao (State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University).
Monoclonal mouse anti-ZIKV NS5 was generated in our lab. Antibodies against IRF3 (A11118, 1 : 1000), GAPDH (AC002,1 : 3000), Flag (AE005, 1 : 1000) and GFP (AE011, 1 : 1000) were purchased from ABclonal Technology. Antibody against STAT2 was from Cell Signaling Technology (72 604S, 1 : 1000).
Plasmid construction
Flag-tagged ZIKV NS5 and mutant constructs: To construct plasmid-encoding Flag-tagged wild-type NS5 (NS5-WT), the ZIKV NS5 gene was amplified by PCR from ZIKV SZ01 cDNA clone (GenBank accession number: KU866423.2) with the forward primer 5′- ATGGTACCGCCACCATGGGGGGTGGAACAGGAGAGAC-3′ and the reverse primer 5′- AACTCGAGTTACTTATCGTCGTCATCCTTGTAATCCAGCACTCCA-3′. The Flag-tag was fused to the C-terminal of ZIKV NS5 by adding its sequence to the reverse primer. Then the PCR fragment was digested with KpnI and XhoI and cloned into pre-cut pcDNA4.0 vector (Invitrogen) by using DNA ligation Kit (Takara). All ZIKV NS5 mutants mentioned in the article were generated based on NS5-WT by using overlap extension PCR. Briefly, NS5-WT construct was used as template for overlap extension PCR, and the PCR products were digested and cloned into pcDNA4.0 as described above.
Enhanced green fluorescent protein (EGFP)-tagged ZIKV NS5 truncated constructs: NS5-WT construct was used as template for PCR. The truncated NS5 fragments including aa 1–320, aa 1–406, aa 321–903 and aa 407–903 were digested with XhoI and KpnI and cloned into pre-cut pEGFP-C1 vector (Clontech).
EGFP-GST fluorescence reporter constructs: to create EGFP-GST vector, the glutathione S-transferase (GST) gene was amplified by PCR from pGEX-6P-1 plasmid (GE Healthcare) with the forward primer 5′- CCCAAGCTTCGATGTCCCCTATACTAGGTTATTGG-3′ and the reverse primer 5′- CGGGGTACCATCCGATTTTGGAGGATGGT-3′, and then the PCR product was digested with HindIII and KpnI and cloned into pre-cut pEGFP-C1 vector as described above. Gene fragments of 2×SV40 NLS (peptide sequence: PKKKRKVGPKKKRKVG), and C-terminal 18 amino acids of ZIKV NS5 (peptide sequence: LSTQVRYLGEEGSTPGVL) and DENV2 NS5 (peptide sequence: MPSMKRFRREEEEAGVLW) with KpnI and BamHI sticky ends were synthesized by Sangon Biotech, and then those fragments were cloned into a pre-cut EGFP-GST vector. The ZIKV NS5 fragments that were inserted into EGFP-GST were amplified by PCR using NS5-WT as a template. The PCR products were digested with KpnI and BamHI and cloned into the pre-cut EGFP-GST vector.
Dual-luciferase reporter plasmids: dual-luciferase reporter plasmids including p125-Luc for IFN-β, pIRF3-Luc for IRF3-responsive IFN-β, pNF-κB-Luc for NF-κB-responsive IFN-β, and pISRE-Luc were stored in our lab.
Immunofluorescence analysis
HeLa cells were seeded in 35 mm glass-bottom dishes and transfected with the specified plasmids. At an indicated time post-transfection, cells were fixed with pre-cold methanol for 10 mins at −20 °C. Following fixation, cells were washed three times by PBS and were blocked in PBS containing 1 % BSA for 1 h at room temperature (RT). Thereafter, cells were incubated with relevant primary antibody for 2 h at RT. After three washes with PBS, cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse or Alexa Fluor 555-conjugated goat anti-rabbit secondary antibody (Invitrogen, 1 : 1000) for 45 min. Then cells were washed and incubated with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen, 1 : 10000) for another 10 min at RT. Finally, cells were washed and observed using confocal laser scanning microscope (CLSM, Zeiss LSM 880 upright confocal microscope).
Dual-luciferase reporter assay
HEK 293 T cells were seeded in 48-well plates and co-transfected with a 500 ng empty vector, NS5-WT or NS5-E, 25 ng Renilla luciferase plasmid phRL-TK and 125 ng reporter plasmid p125-Luc, pIRF3-Luc, pNF-κB-Luc or pISRE-Luc. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. At 24 h post-transfection, cells were stimulated with 100 HAU ml−1 SeV for 16 h or 1000 IU ml−1 IFN-β for 45 min. Cells were harvested and lysed for measuring firefly and Renilla luciferase activities by Dual-Luciferase Reporter Assay System (Promega).
RNA extraction and quantitative real-time PCR
Total RNA in treated cells was extracted using TRIzol Reagent (Invitrogen), and 1 µg RNA was used to synthesize cDNA using a first-strand cDNA synthesis kit (TOYOBO). Quantitative real-time PCR was performed using a 7500 Real-Time PCR System (Applied Biosystems) and SYBR Green PCR Master Mix (TOYOBO). Data were normalized to the level of β-actin expression in each sample. The primer pairs used were as follows: β-actin: forward 5′-AGCGGGAAATCGTGCGTGAC-3′,reverse 5′-GGAAGGAAGGCTGGAAGAGTG-3′; IFN-β:forward 5′-TGCTCTGGCACAACAGGTAG-3′, reverse 5′-AGCCTCCCATTCAATTGCCA-3′.
Immunoprecipitation and Western blotting
HEK293T cells were transfected with the plasmids indicated in the figures. At 36 h post-transfection, cell extracts were prepared using RIPA buffer (Sigma) containing protease inhibitor cocktail (Roche). The cell lysate was incubated with the indicated antibodies at 4 °C overnight. Protein A+G agarose beads (25 µl; Beyotime) were added and incubated for another 3 h. The agarose beads were subsequently washed three times with wash buffer (0.05 M Tris-HCl with 0.15 M NaCl). The bound proteins were eluted by boiling in SDS-PAGE loading buffer for 10 min and used for Western blotting with the indicated antibodies.
For Western blotting, total cellular lysates were prepared as described above. Protein concentration was determined by using a BCA (bicinchoninic acid) protein assay kit (Thermo Scientific) and boiled in SDS-PAGE loading buffer at 95 °C for 10 min. Equivalent amounts of protein samples were separated by SDS-PAGE and electroblotted onto a polyvinylidene fluoride membrane (Roche) using a Mini Trans-Blot Cell (Bio-Rad). The membrane was blocked at RT for 2 h in PBS containing 3 % BSA, and then incubated the membrane with the indicated primary antibody overnight at 4 °C. After washing three times with TBS-Tween [50 mM Tris-HCl, 150 mM NaCl and 0.1 % (v/v) Tween 20, pH 7.4], the membrane was incubated with horseradish peroxidase-labelled goat anti-mouse or goat anti-rabbit IgG at RT for 45 min. Finally, the membranes were visualized with a chemiluminescence system (Bio-Rad) after three times of wash.
Statistical analysis
All results are presented as the mean ± sem. Statistical significance was determined by Student’s t-test, and a P-value<0.05 was considered to be statistically significant. Analyses were conducted using the PRISM v7.0 software program (GraphPad Software).
Results
Both aa 1-320 and aa 321-903 of ZIKV NS5 accumulate in the cell nucleus
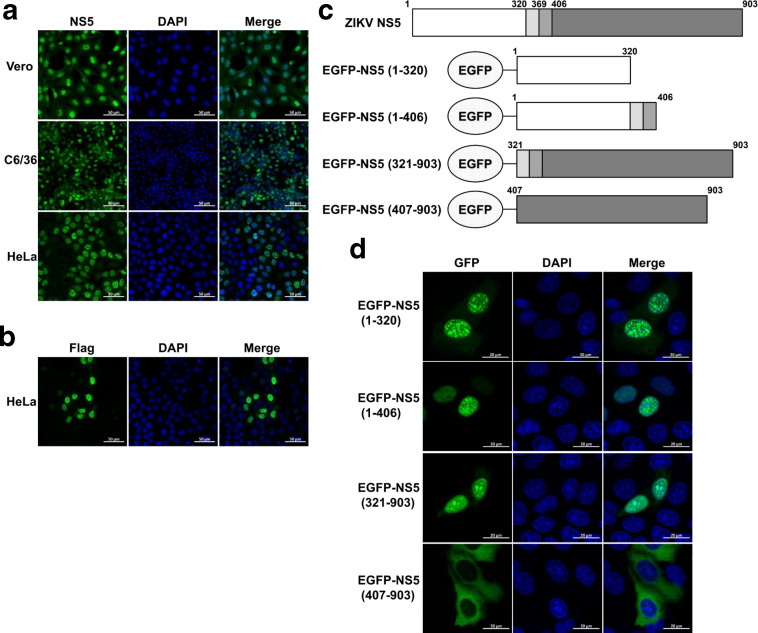
To examine the subcellular localization of ZIKV NS5, different host cells including HeLa, Vero and C6/36 were infected with ZIKV SZ01 strain. At 48 h post-infection, ZIKV NS5 was detected by immunofluorescence assay. As shown in Fig. 1a, ZIKV NS5 localized in the nucleus and formed punctuated NS5 nuclear bodies in all types of cells during infection, suggesting the prominent nuclear localization of ZIKV NS5 is independent of cell type. The subcellular localization of ZIKV NS5 was also assessed in HeLa cells transfected with plasmid encoding Flag-tagged NS5-WT, and the same localization pattern was observed (Fig. 1b).
Fig. 1.
Both aa 1–320 and aa 321–903 of ZIKV NS5 accumulate in the cell nucleus. Vero, C6/36 and HeLa cells were infected with ZIKV at a m.o.i. of 1. At 24 h post-infection, cells were subjected to immunofluorescence staining for ZIKV NS5 (a). HeLa cells were seeded in 35 mm glass-bottom dishes and transfected with 1 µg NS5-WT plasmids. At 24 h post-transfection, anti-Flag antibody was used for immunostaining (b). Diagram of EGFP-tagged constructs (c). HeLa cells were seeded in 35 mm glass-bottom dishes and transfected with indicated EGFP-tagged NS5 constructs. At 24 h post-transfection, anti-GFP antibody was used for immunostaining (d). Images were captured by Zeiss LSM 880 CLSM by 63×oil immersion lens. The images used are representative of two independent experiments.
To further explore which region in ZIKV NS5 contributes to its nuclear localization, plasmids encoding EGFP fused gene segments of residues 1–320, 1–406, 321–903 or 407–903 were constructed (Fig. 1c), given that the residues 320–405 are known as a conserved NLS in flavivirus [6, 23]. HeLa cells were transfected with these constructs individually, and the fluorescence distribution was observed at 24 h post-transfection by using a CLSM. As expected, we found that EGFP-NS5 (1–406) and EGFP-NS5 (321–903), both of which contain aa 321–406, strongly accumulated in the nucleus, while EGFP-NS5 (407–903) showed predominantly cytoplasmic localization (Fig. 1d), suggesting that aa 321–406 may contain the NLS of ZIKV NS5. Interestingly, we found that EGFP-NS5 (1–320) also accumulated in the nucleus, implying a possible new functional NLS within this region.
Both aa 11-90 and aa 370-406 of ZIKV NS5 contain NLSs
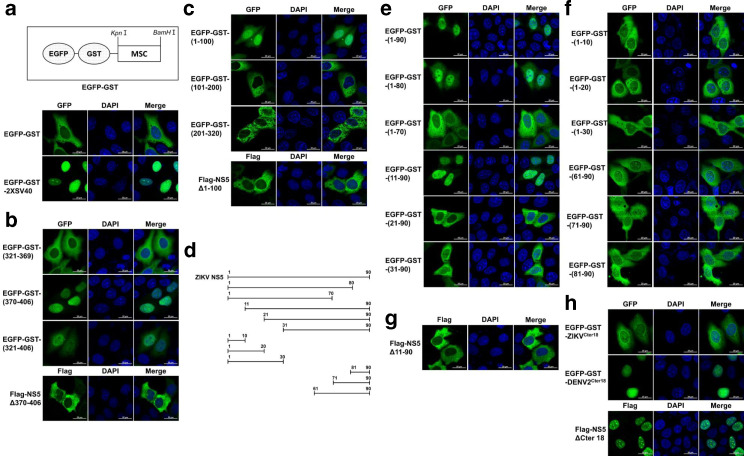
To further determine the sequences contributing to the ZIKV NS5 nuclear localization, a fluorescence reporter plasmid was generated by fusing GST to the C-terminal of EGFP (EGFP-GST) (Fig. 2a). The molecular weight of EGFP-GST fusion protein (~53 kDa) exceeds the exclusion limit of nuclear pore complex (NPC), so the nuclear direction ability of any predicted NLS can be validated by inserting it into this reporter plasmid [24, 25]. The NLS of canonical SV40 large T antigen was cloned into EGFP-GST plasmid to verify the feasibility of this model [26]. As shown in Fig. 2a, the EGFP-GST-2×SV40 fusion protein prominently localized in the nucleus, while the EGFP-GST mostly accumulated in the cytoplasm. Since aa 321–406 of ZIKV NS5 is highly conservative to aa 320–405 of DENV NS5, which contains two NLSs including bNLS (aa 320–368) and aNLS (aa 369–405) [13, 15, 18], we next explored if the corresponding regions of ZIKV NS5 could confer the NLS ability. To this end, aa 321–369, aa 370–406 and aa 321–406 of ZIKV NS5 were fused with EGFP-GST, respectively, and the plasmids were transfected into HeLa cells, followed by observation of fluorescence distribution at 24 h post-transfection. We found that EGFP-GST-(370-406) strongly accumulated in the nucleus, while EGFP-GST-(321-369) mainly accumulated in the cytoplasm, and EGFP-GST-(321-406) showed comparatively balanced distribution between the nucleus and the cytoplasm (Fig. 2b), suggesting aa 370–406 of ZIKV NS5 is a potential NLS. To further confirm the role of aa 370–406 on nuclear transportation of ZIKV NS5, HeLa cells were transfected with Flag-tagged ZIKV NS5 mutant lacking of aa 370–406 (Flag-NS5Δ370–406). As expected, cytoplasmic accumulation of Flag-NS5Δ370–406 was observed, which demonstrates that residues 370–406 are responsible for nuclear localization of ZIKV NS5 (Fig. 2b).
Fig. 2.
Both aa 11–90 and aa 370–406 of ZIKV NS5 contain NLSs. Schematic illustration of EGFP-GST vector (a, upper panel). HeLa cells were transfected with reporter vector EGFP-GST, and the fluorescence distribution was observed 24 h later by using CLSM. The gene sequence of 2×SV40NLS was cloned into EGFP-GST and the recombinant plasmid was transfected into HeLa cells for 24 h to act as a positive control (a, lower panel). aa 370–406 and aa 1–320 of ZIKV NS5 were divided into several fragments and inserted into EGFP-GST. HeLa cells were transfected with these plasmids or indicated Flag-tagged NS5 truncated mutants. Anti-GFP or anti-Flag antibody was used for immunostaining at 24 h post-transfection (b and c). Schematic illustration of truncated fragments of ZIKV NS5 (d). HeLa cells were transfected to express the indicated gene fragments of ZIKV NS5 fused EGFP-GST or Flag-tagged NS5 truncated mutants. Anti-GFP or anti-Flag antibody was used for immunostaining at 24 h post-transfection (e–h). Images were captured by Zeiss LSM 880 upright confocal microscope by 63×oil immersion lens. The images used are representative of two independent experiments. MSC, multiple cloning sites.
Considering the notable finding that EGFP-NS5-(1-320) also accumulated in the nucleus, we next explored the potential NLS within aa 1–320. aa 1–320 of ZIKV NS5 was divided into three fragments (aa 1–100, aa 101–200 and aa 201–320), which were then inserted into EGFP-GST reporter plasmid. HeLa cells were transfected with these plasmids, and the fluorescence distribution was observed by CLSM at 24 h post-transfection. As shown in Fig. 2c, only EGFP-GST-(1-100) predominantly expressed in the nucleus, implying a possible NLS located within aa 1–100. cNLS Mapper was subsequently used to predict NLS within aa 1–100 [27]. However, no typical monopartite or bipartite NLS was identified, indicating a possibility of non-classical NLS in this region. Then the fluorescence distribution of chimeric EGFP-GST proteins fused with different C-terminal or N-terminal truncations of aa 1–100 was analysed (Fig. 2d). Only EGFP-GST-(1-90) and EGFP-GST-(11-90) showed the comparable quantity of nuclear accumulation with EGFP-GST-(1-100), while EGFP-GST-(1-70) and EGFP-GST-(21-90) were observed predominantly in cytoplasm, which suggests that aa 11–20 and aa 71–90 are essential for subcellular localization of EGFP-GST-(1-100) (Fig. 2e). To investigate whether aa 11–20 and aa 71–90 contain the sequence elements responsible for nuclear localization, residues 1–10, 1–20, 1–30, 61–90, 71–90 and 81–90 were fused with EGFP-GST individually. However, none of them could mediate nuclear transportation of EGFP-GST (Fig. 2f), indicating that no full-length NLS was included in these regions, and multiple stretches of amino acids in aa 11–90 may be responsible for nuclear localization. The critical role of aa 11–90 on nuclear localization of ZIKV NS5 was further confirmed by cytoplasmic accumulation of ZIKV NS5 mutant lacking of aa 11–90 (Flag-NS5Δ11–90) (Fig. 2g). Taken together, these results demonstrate that both aa 370–406 and aa 11–90 can determine the nuclear localization of ZIKV NS5.
A recent study has identified a motif within the C-terminal 18 amino acids (Cter18) of NS5 of DENV2 and possibly other flaviviruses as a key determinant for nuclear localization of NS5 [28]. To verify whether this region could also regulate the subcellular localization of ZIKV NS5, HeLa cells were transfected with the truncated mutant of ZIKV NS5 that lacks Cter18 (Flag-NS5ΔCter18). Inconsistent with findings of DENV2, Flag-NS5ΔCter18 of ZIKV showed a similar subcellular distribution with full-length NS5 (Fig. 2h). In addition, the Cter18 of ZIKV or DENV2 NS5 was integrated with EGP-GST. As shown in Fig. 2e, the chimeric protein EGFP-GST-DENV2Cter18 strikingly accumulated in the nucleus, while EGFP-GST-ZIKVCter18 mainly distributed in the cytoplasm. These findings demonstrated that Cter18 of ZIKV NS5 are not essential for nuclear localization of ZIKV NS5.
Residues K390R391P392R393 of ZIKV NS5 are essential for interaction with importin α
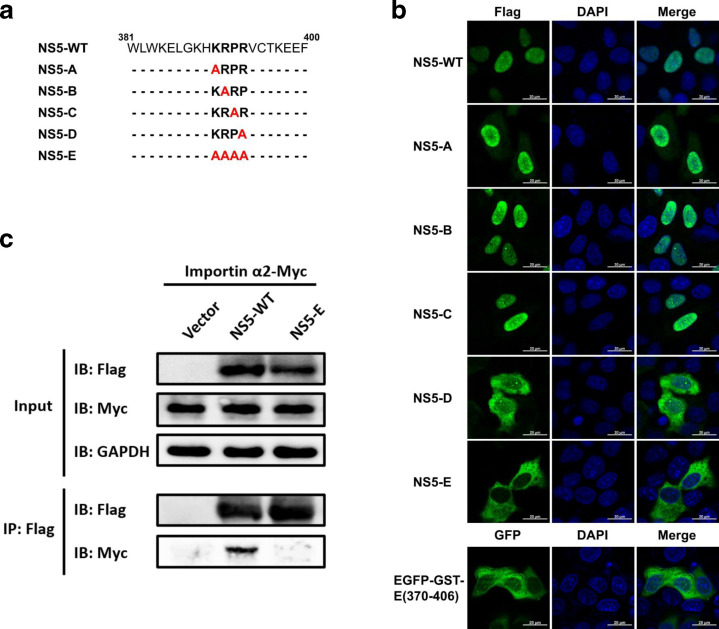
As reported previously, a conservative motif KRPR in NS5 of flaviviruses is crucial for interaction with importins [20], we construct a series of ZIKV NS5 mutants by introducing alanine substitutions in this motif, and their subcellular distribution was analysed (Fig. 3a). Construct NS5-E containing alanine substitutions of all residues K390R391P392R393 is the only mutant whose nuclear localization was completely impaired (Fig. 3b). And when these mutations were introduced into EGFP-GST-(370-406), its fluorescence distribution relocated to the cytoplasm (Fig. 3b), suggesting these residues are critical for nuclear localization of ZIKV NS5. To further examine whether this motif in ZIKV NS5 is responsible for interaction with importins, co-immunoprecipitation assay was performed with cells co-transfected with plasmid encoding NS5-WT or NS5-E and importin α2, which has been reported to direct the nuclear transport of ZIKV NS5 [29]. The result showed that importin α2 was co-immunoprecipitated with NS5-WT but not NS5-E, suggesting K390R391P392R393 as key residues involved in interaction of ZIKV NS5 with importin α.
Fig. 3.
Residues K390R391P392R393 of ZIKV NS5 are essential for interaction with importin α. Schematic illustration of Flag-tagged ZIKV NS5 mutant constructs (a). HeLa cells were transfected to express NS-WT and indicated mutants derivatives. At 24 h post-transfection, anti-Flag antibody was used for immunostaining and images were captured by Zeiss LSM 880 CLSM by 63×oil immersion lens (b). HEK 293 T cells were co-transfected with NS5-WT, NS5-E or vector plasmid and the importin α2-Myc. At 36 h post-transfection, immunoprecipitation assay was performed with whole-cell lysates by using anti-Flag antibody. Immunoprecipitates and input were analysed by Western blotting.
ZIKV NS5 nuclear localization contributes to the suppression of IFN-I production
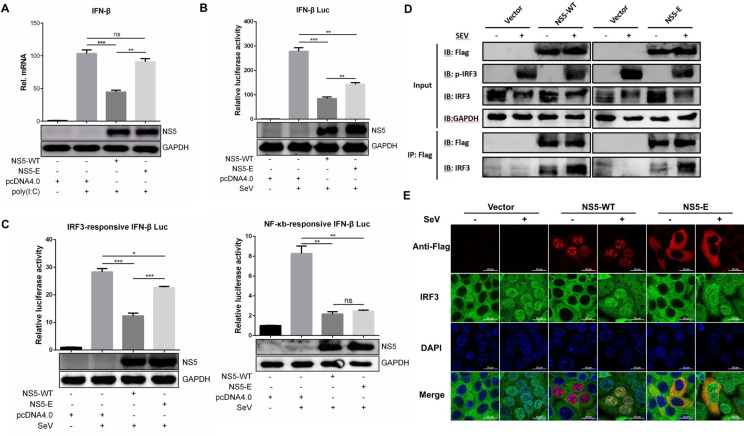
It has been reported that ZIKV NS5 could inhibit IFN-I production through interacting with IRF3 [12, 22]. To investigate whether this ability is associated with its subcellular localization, HeLa cells were transfected with plasmid encoding NS5-WT or NS5-E followed by poly(I:C) stimulation, and the induction of IFN-β was measured by qRT-PCR. As shown in Fig. 4a, NS5-WT significantly suppressed the expression of IFN-β, but NS5-E showed no obvious effect. Consistently, IFN-β promoter activity determined by luciferase reporter system was also inhibited by NS5-WT but not NS5-E (Fig. 4b). These results suggest that nuclear localization of ZIKV NS5 contributes to its inhibitory ability on IFN-I transcription.
Fig. 4.
ZIKV NS5 nuclear localization contributes to the suppression of IFN-I production. HeLa cells were seeded in 12-well plate and transfected with empty vector, NS5-WT or NS5-E, respectively. At 24 h post-transfection, cells were left untreated or transfected with 5 µg poly(I:C) and incubated for another 12 h. Cells were harvested for RNA extraction and subjected to detect mRNA level of IFN-β by qRT-PCR. Western blotting was performed to examine the expression of ZIKV NS5 protein (a). HEK 293 T cells were co-transfected with empty vector or indicated ZIKV NS5 plasmid, phRL-TK and p125-Luc (b), pIRF3-Luc or pNF-κB-Luc (c). After 24 h, cells were left untreated or treated with 100 HAU ml−1 SeV and luciferase activities were measured 12 h later. HEK 293 T cells were transfected with vector, NS5-WT or NS5-E plasmid. After 24 h, cells were left untreated or treated with 100 ml−1 SeV and Flag-tag immunoprecipitation assay was performed 12 h later (d). HeLa cells were transfected with vector, NS5-WT or NS5-E plasmid. At 24 h post-transfection, cells were left untreated or treated with 100 HAU ml−1 SeV. Anti-Flag and anti-IRF3 antibody were used for immunostaining 8 h later (e). Data are shown as means±sem of at least three independent experiments, with the error bars representing the standard deviations. ns, no significance; *, P<0.05; **, P<0.01; ***, P<0.001.
It has been well known that activated transcription factors such as IRF3 and NF-κB can translocate to the nucleus and induce IFN-I expression [30]. To elucidate whether nuclear localization of ZIKV NS5 plays a role in inhibiting the transcriptional activity of IRF3 or NF-κB. The IRF-3-responsive or NF-κB-responsive IFN-β promoter luciferase assay was performed with cells expressing NS5-WT or NS5-E. Compared with NS5-WT, NS5-E showed a similar inhibitory effect on NF-κB activation while an alleviated capability on IRF3 inhibition (Fig. 4c), suggesting that mutations on NLS of ZIKV NS5 interfere with its function on restraining IRF3 activation. Given the fact that ZIKV NS5 can interact with IRF3 [31], it is important to clarify whether the mutations would compromise their interaction or affected the nucleus-cytoplasmic shuttling of IRF3. To this end, co-immunoprecipitation and immunofluorescence assays were performed with cells expressing NS5-WT or NS5-E. As shown in Fig. 4d, both NS5-WT and NS5-E could interact with endogenous IRF3 either under the stimulation with SeV or not. The immunofluorescence assay revealed a strong co-localization of NS5-WT and IRF3 in cell nucleus after SeV stimulation (Fig. 4e). Interestingly, in the case of NS5-E, a cytoplasmic co-localization with IRF3 was observed in unstimulated cells without any impact on nuclear translocation of IRF3 after stimulation, suggesting that mutations on NS5 did not impair the interaction with IRF3. These findings indicate the possibility that nuclear interaction of NS5 and IRF3 is required for inhibition of IRF3-mediated IFN-I transcription.
ZIKV NS5 nuclear localization is associated with inhibition of IFN-I response
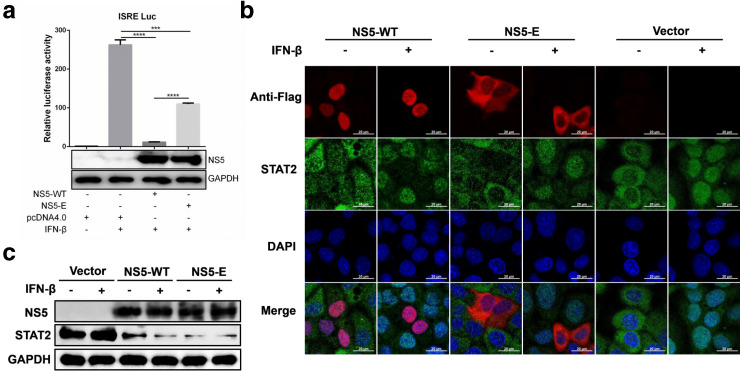
ZIKV NS5 has also been known as an antagonist of IFN-I signalling by degrading STAT2 [12, 22]. To investigate if this function of ZIKV NS5 is dependent on its subcellular localization, the activity of IFN-stimulated response element (ISRE) promoter was determined in cells transfected with plasmid encoding NS5-WT or NS5-E. As shown in Fig. 5a, although both NS5-WT and NS5-E significantly inhibited the activation of ISRE promoter, NS5-E showed relatively weaker inhibitory effect, indicating that ZIKV NS5 nuclear localization is partially involved in inhibition of IFN-I downstream signalling. To further determine whether NS5 nuclear localization contributes to STAT2 degradation, HeLa cells were transfected with plasmid expressing with NS5 WT or NS5-E, followed by IFN-β stimulation. The results showed that both NS5-WT and NS5-E degraded endogenous STAT2 (Fig. 5b, c), suggesting that ZIKV NS5 mediated degradation of STAT2 is independent on its nuclear accumulation. Taken together, these data demonstrate that ZIKV NS5 nuclear localization participates in inhibition of IFN-I response through a STAT2 degradation-independent mechanism.
Fig. 5.
ZIKV NS5 nuclear localization is associated with inhibition of IFN-I response. HEK 293 T cells were co-transfected with empty vector or indicated ZIKV NS5 plasmid, phRL-TK and pISRE-Luc. At 24 h post-transfection, cells were stimulated with 1000 IU ml−1 recombinant human IFN-β. After 45 min, luciferase activities were measured and the expression level of ZIKV NS5 was examined by Western blotting (a). HeLa cells were seeded in 12-well plate and transfected with 1 µg empty vector or indicated ZIKV NS5 plasmid. At 24 h post-transfection, cells were left untreated or treated with 1000 IU ml−1 recombinant human IFN-β for 45 min. Then the cells were fixed and subjected to immunofluorescence assay (b). Western blotting was performed to examine the expression level of ZIKV NS5 protein and the degradation of STAT2 (c). Data are shown as means±sem of at least three independent experiments, with the error bars representing the standard deviations. ***, P<0.001; ****, P<0.0001.
Discussion
Among flavivirus non-structural proteins, NS5 is most highly conserved and involved in many vital processes, including viral replication and innate immune suppression [9, 12, 20, 32]. Previous studies have revealed that ZIKV NS5 localizes to the nucleus through an importin α/β-dependent nuclear-transport mechanism [29]. However, the NLSs and the role of NS5 in the nucleus are still undefined. Here, our results showed that both aa 11–90 and aa 370–406 of ZIKV NS5 are responsible for the nuclear localization of ZIKV NS5, and K390R391P392R393 are critical residues for interaction of NS5 with importin α. Furthermore, we found that NS5 nuclear localization is associated with its inhibitory effect on IFN-I production and response.
Among all flaviviruses, DENV2 NS5 has well-characterized NLS. A typical bipartite NLS in aa 369–405 was identified as an indispensable element responsible for nuclear translocation and interaction with NS3 [13, 33, 34]. In this study, our results showed that aa 370–406 of ZIKV NS5, a highly conserved sequence with aa 369–405 of DENV and other flaviviruses (Fig. S1a, available in the online version of this article), is also sufficient to direct the protein into the nucleus, demonstrating a functional NLS within aa 370–406 of ZIKV NS5. In addition, our results showed that alanine substitutions of K390R391P392R393 completely abolish the nuclear accumulation of ZIKV NS5 and its interaction with importin α2, suggesting that K390R391P392R393 are key residues involved in importin α2-mediated NS5 nuclear transport. This finding is supported by our previous report, which demonstrated that the interaction of JEV NS5 with importins relies on K391R392P393R394 motif [20]. To further verify this conclusion, alanine substitutions of this motif were introduced within ZIKV infectious clone. However, no infectious viral particle could be rescued after mutation, suggesting that mutation of aa 390–393 in ZIKV NS5 is lethal to virus replication. This result supports that nuclear localization of ZIKV NS5 is critical for viral propagation. Considering the important role of NS5 on viral genomic replication, the lethality may also be caused by the impaired RdRp function of ZIKV NS5. A recent study also showed that introduction of NS5 NLS mutations into WNV infectious clone abolished nucleocytoplasmic shuttling of NS5 and resulted in lethality [35]. Besides aa 369–405, the Cter18 of DENV NS5 was also identified as a key determinant for differential subcellular localization between NS5 of DENV 1 and 2 [28]. Inconsistently, our results revealed that Cter18 of ZIKV NS5 is not required for its nuclear localization. This might be due to the low similarity of the motif KRFR, which was considered as a class 2 NLS responsible for the nuclear localization of DENV2 NS5 (Fig. S1b) [28, 36].
In addition to aa 370–406, we found aa 1–100 also has the ability to direct subcellular localization of reporter protein, suggesting a potential NLS within this region. Unfortunately, no NLSs specific to the importin α/β pathway was predicted by cNLS Mapper within this region, implying a non-classical NLS in aa 1–100. By inserting truncated mutants of aa 1–100 into fluorescence reporter plasmid, we found that both aa 11–20 and aa 70–90 are essential for its nuclear localization, suggesting there may be multiple stretches of amino acids within this region responsible for directing nuclear localization. Therefore, it is difficult to determine the key residues within aa 11–90 that contribute to NS5 subcellular localization by using site-directed mutagenesis. It is interesting to notice that NLS of aa 11–90 or aa 370–406 is sufficient to direct nuclear translocation of reporter protein (Fig. 2b, e), but both are required for nuclear localization of ZIKV NS5. Deletion of either aa 370–406 or aa 11–90 in NS5 altered its subcellular localization (Fig. 2b, g). This may be associated with the protein structure of NS5. However, the underlaying mechanism need to be further explored.
As already known that ZIKV NS5 could inhibit IFN-I production and downstream signalling [12, 22, 37]. It is interesting to understand whether this function is associated with subcellular localization of NS5. Our study demonstrated for the first time that nuclear accumulation of ZIKV NS5 contributes to its function on blocking of IRF3 activation and IFN-I production. Compared to NS5-WT, the NS5 mutant with alanine substitutions at K390R391P392R393 accumulated in the cytoplasmic and significantly inhibit IFN-β production and IRF3 activation. Moreover, the mutations did not affect the interaction between IRF3 and NS5 as well as the nuclear transportation of IRF3. These results raise the possibility that only ZIKV NS5, which interacts with IRF3 in the nucleus, could inhibit IFN-I production.
Our findings also reveal that nuclear localization of ZIKV NS5 contributes to its function on inhibiting the ISRE activation without affecting STAT2 degradation. Given that STAT2, STAT1 and IRF9 form a complex, which translocates into the nucleus and activate the transcription of ISGs [38]. It is incomprehensive to evaluate the activation of IFN-I downstream signalling simply by measuring STAT2 degradation. Other signal molecules, such as Janus kinases (Jak1 and Tyk2), STAT1 and IRF9, have also been known as antagonistic targets of viruses. For instance, DENV can inhibit IFN signalling through preventing the phosphorylation of Tyk2 [39]; WNV replication can inhibit the phosphorylation of both Jak1 and Tyk2 [40]; hepatitis C virus (HCV) impairs IFN signalling by suppressing the expression of Jak1 and the phosphorylation of STAT1 [41]; porcine bocavirus negatively regulates IFN signalling by targeting the DNA-binding domain of IRF9 [42]. These results imply a potential STAT2-independent mechanism involved in ZIKV NS5-mediate inhibition of IFN-I response.
Taken together, our study defined two regions of ZIKV NS5 (aa 11–90 and aa 370–406) are responsible for nuclear localization. In addition, we found that the nuclear localization of ZIKV NS5 is involved in its inhibitory effect on type I IFN production and response. These findings may provide new insight into the mechanisms for innate immune evasion and pathogenesis of ZIKV.
Supplementary Data
Funding information
Funding: This work was supported by National Key Research and Development Program of China (2016YFD0500407), National Natural Science Foundation of China (31825025, 31572517, 31972721), Fundamental Research Funds for the Central Universities (2662018QD025), and Natural Science Foundation of Hubei Province (2019CFA010).
Acknowledgements
We thank Dr Bo Zhang from Wuhan Institute of Virology, CAS for providing the ZIKV SZ01 strain.
Author contributions
J.Y. and S.B.C. conceived and designed the experiments. Z.K.Z., M.Y.T., W.H. and Z.J.F., performed the experiments. Z.K.Z. analysed the data. Z.K.Z. and J.Y. wrote the manuscript and prepared the figures. J.Y. and S.B.C. checked and finalized the manuscript. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: Cter18, C-terminal 18 amino acids; DAPI, 4',6-Diamidino-2-Phenylindole; DENV, dengue virus; EGFP, enhanced green fluorescent protein; GST, Glutathione S-transferase; HCV, Hepatitis C virus; IFNAR1, interferon alpha and beta receptor subunit 1; IFN-I, type I interferon; IRF3, interferon regulatory factor 3; JEV, Japanese encephalitis virus; MTase, methyltransferase; NES, nuclear export signal; NLS, nuclear localization signal; RdRP, RNA-dependent RNA polymerase; RT, room temperature; SeV, Sendai virus; STAT2, signal transducers and activators of transcription 2; TBEV, tick-borne encephalitis virus; WNV, West Nile virus; YFV, yellow fever virus; ZIKV, Zika virus.
One supplementary figure is available with the online version of this article.
References
- 1.Dick GWA, Kitchen SF, Haddow AJ. Zika virus. I. isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 2.Faria NR, Azevedo RdSdS, Kraemer MUG, Souza R, Cunha MS, et al. Zika virus in the Americas: early epidemiological and genetic findings. Science. 2016;352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 4.Cao-Lormeau VM, Blake A, Mons S, Lastère S, Roche C, et al. Guillain-Barré syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. The Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Z, Chan JFW, Tee KM, Choi GKY, Lau SKP, et al. Comparative genomic analysis of pre-epidemic and epidemic Zika virus strains for virological factors potentially associated with the rapidly expanding epidemic. Emerg Microbes Infect. 2016;5:1–12.:e22. doi: 10.1038/emi.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B, Yi G, Du F, Chuang YC, Vaughan RC, et al. Structure and function of the Zika virus full-length NS5 protein. Nat Commun. 2017;8:14762. doi: 10.1038/ncomms14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B, Tan XF, Thurmond S, Zhang ZM, Lin A, et al. The structure of Zika virus NS5 reveals a conserved domain conformation. Nat Commun. 2017;8:14763. doi: 10.1038/ncomms14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klema V, Padmanabhan R, Choi K. Flaviviral Replication Complex: Coordination between RNA Synthesis and 5’-RNA Capping. Viruses. 2015;7:4640–4656. doi: 10.3390/v7082837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye J, Chen Z, Zhang B, Miao H, Zohaib A, et al. Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PLoS One. 2013;8:e75188. doi: 10.1371/journal.pone.0075188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Ye J, Ashraf U, Li Y, Wei S, et al. MicroRNA-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J Virol. 2016;90:3722–3734. doi: 10.1128/JVI.03242-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, et al. Nuclear localization of dengue virus nonstructural protein 5 through its importin α/β-Recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8:795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 14.Buckley A, Gaidamovich S, Turchinskaya A, Gould EA. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J Gen Virol. 1992;73:1125–1130. doi: 10.1099/0022-1317-73-5-1125. [DOI] [PubMed] [Google Scholar]
- 15.Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. Crm1-Mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J Biol Chem. 2009;284:15589–15597. doi: 10.1074/jbc.M808271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, et al. The dengue virus NS5 protein Intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog. 2016;12:e1005841. doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, et al. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forwood JK, Brooks A, Briggs LJ, Xiao CY, Jans DA, et al. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem Biophys Res Commun. 1999;257:731–737. doi: 10.1006/bbrc.1999.0370. [DOI] [PubMed] [Google Scholar]
- 19.Versteeg GA, García-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye J, Chen Z, Li Y, Zhao Z, He W, et al. Japanese encephalitis virus NS5 inhibits type I interferon (IFN) production by blocking the nuclear translocation of IFN regulatory factor 3 and NF-κB. J Virol. 2017;91 doi: 10.1128/JVI.00039-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lubick KJ, Robertson SJ, McNally KL, Freedman BA, Rasmussen AL, et al. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, et al. Zika virus inhibits type‐I interferon production and downstream signaling. EMBO Rep. 2016;17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertzog J, Dias Junior AG, Rigby RE, Donald CL, Mayer A, et al. Infection with a Brazilian isolate of Zika virus generates RIG‐I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur J Immunol. 2018;48:1120–1136. doi: 10.1002/eji.201847483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maertens G, Cherepanov P, Debyser Z, Engelborghs Y, Engelman A. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. Journal of Biological Chemistry. 2004;279:33421–33429. doi: 10.1074/jbc.M404700200. [DOI] [PubMed] [Google Scholar]
- 25.Woodward CL, Wang Y, Dixon WJ, Htun H, Chow SA. Subcellular localization of feline immunodeficiency virus integrase and mapping of its karyophilic determinant. J Virol. 2003;77:4516–4527. doi: 10.1128/JVI.77.8.4516-4527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 27.Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, et al. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Yang SNY, Smith K, Forwood JK, Jans DA. Nuclear import inhibitor N -(4-hydroxyphenyl) retinamide targets Zika virus (ZIKV) nonstructural protein 5 to inhibit ZIKV infection. Biochem Biophys Res Commun. 2017;493:1555–1559. doi: 10.1016/j.bbrc.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Pichlmair A. Reis E Sousa C. innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Xia H, Luo H, Shan C, Muruato AE, Nunes BTD, et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun. 2018;9:414. doi: 10.1038/s41467-017-02816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison J, Laurent-Rolle M, Maestre AM, Rajsbaum R, Pisanelli G, et al. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontes MRM, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J Biol Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- 33.Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, et al. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem. 2002;277:36399–36407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Denman AJ, Russo A, Wagstaff KM, White PA, Jans DA, et al. Nucleocytoplasmic shuttling of the West Nile virus RNA-dependent RNA polymerase NS5 is critical to infection. Cell Microbiol. 2018;20:e12848. doi: 10.1111/cmi.12848. [DOI] [PubMed] [Google Scholar]
- 35.Tay MYF, Smith K, Ng IHW, Chan KWK, Zhao Y, et al. The C-terminal 18 amino acid region of dengue virus NS5 regulates its subcellular localization and contains a conserved arginine residue essential for infectious virus production. PLoS Pathog. 2016;12:e1005886. doi: 10.1371/journal.ppat.1005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowen JR, Quicke KM, Maddur MS, O’Neal JT, McDonald CE, et al. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog. 2017;13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LJ H, Hung LF, Weng CY, WL W, Chou P, et al. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J Immuno. 2005;174:8163–8172. doi: 10.4049/jimmunol.174.12.8163. [DOI] [PubMed] [Google Scholar]
- 40.Guo JT, Hayashi J, Seeger C. West Nile virus inhibits the signal transduction pathway of alpha interferon. J Virol. 2005;79:1343–1350. doi: 10.1128/JVI.79.3.1343-1350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee A, Di Bisceglie AM, Ray RB. Hepatitis C virus-mediated enhancement of microRNA miR-373 impairs the JAK/STAT signaling pathway. J Virol. 2015;89:3356–3365. doi: 10.1128/JVI.03085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Fang L, Wang D, Cai K, Zhang H, et al. Porcine bocavirus NP1 negatively regulates interferon signaling pathway by targeting the DNA-binding domain of IRF9. Virology. 2015;485:414–421. doi: 10.1016/j.virol.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.