Abstract
Pseudoviridae is a family of reverse-transcribing viruses with long terminal repeats (LTRs) belonging to the order Ortervirales. Pseudoviruses are commonly found integrated in the genomes of diverse plants, fungi and animals and are broadly known as Ty1/Copia LTR retrotransposons. Inside the cell, they form icosahedral virus particles, but unlike most other viruses, do not have an extracellular phase. This is a summary of the ICTV Report on the family Pseudoviridae, which is available at ictv.global/report/pseudoviridae.
Keywords: Pseudoviridae, ICTV Report, taxonomy
Virion
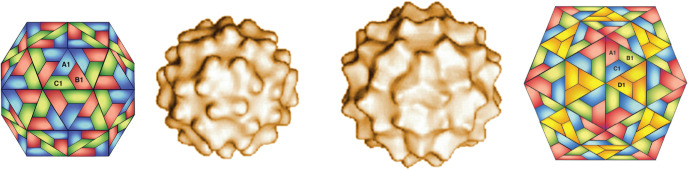
Pseudoviruses form intracellular, somewhat irregularly shaped virus-like particles (VLPs) 30–40 nm in diameter, which do not display infectivity and remain intracellular (Table 1). VLPs are formed by self-assembly of proteins encoded by the gag gene, namely the capsid (CP) and nucleocapsid (NC) proteins, which are homologous to the equivalent proteins of retroviruses and other members of the order Ortervirales [1]. Expression of truncated Gag protein variants yields icosahedral VLPs of different diameters but with a mean radius of 20 nm built on the T=3 or T=4 lattice (Fig. 1) [2, 3].
Table 1.
Characteristics of members of the family Pseudoviridae
|
Example: |
Saccharomyces cerevisiae Ty1 virus (M18706), species Saccharomyces cerevisiae Ty1 virus, genus Pseudovirus |
|---|---|
|
Virion |
Virions are icosahedral (T=3 or 4) and might be enveloped |
|
Genome |
Two identical copies of linear single-stranded RNA |
|
Replication |
Replication by reverse transcription primed with a host-encoded tRNA |
|
Translation |
Genomic RNA is translated into one or more polyproteins |
|
Host range |
Fungi, plants and animals |
|
Taxonomy |
Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Ortervirales, family Pseudoviridae; the genera Pseudovirus, Hemivirus and Sirevirus include >30 species |
Fig. 1.
Saccharomyces cerevisiae Ty1 virus particles formed from truncated capsid protein (aa 1–381). The surface structures of two forms (T=3, left; T=4, right) of around 30–40 nm determined by cryo-electron microscopy, are flanked by the corresponding schematic models. (Courtesy of H. Saibil, adapted from [3] with permission from American Society for Microbiology.)
Genome
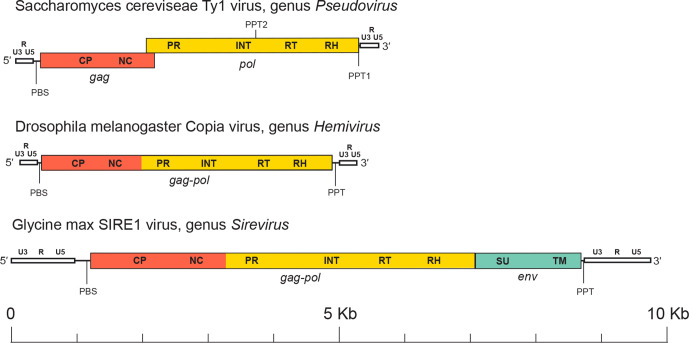
The genome of pseudovirids ranges from 4 kb to >9 kb and has an internal region flanked by two identical non-coding sequences called long terminal repeats (LTRs) (Fig. 2). LTRs are variable in size and contain three regions, named U3-R-U5 in analogy to retroviral LTRs. U3 contains promoters, R is repeated on each end of the transcript, and U5 constitutes the first portion of the reverse-transcribed genome. The internal region is delimited by two short motifs: the primer binding site (PBS), which is located downstream of the 5′-LTR and is usually complementary to the initiator tRNAMet, and the polypurine tract (PPT), which is upstream of the 3′-LTR. The internal region may contain one (gag-pol), two (gag and pol) or three (gag, pol and env) ORFs. The Gag polyprotein includes domains for the CP and NC proteins, while Pol includes domains for the protease (PR), integrase (INT) and reverse transcriptase-ribonuclease H (RT-RH). Members of the genus Sirevirus carry a third ORF downstream of gag-pol encoding a putative envelope protein [4].
Fig. 2.
Pseudovirid genome organization. LTRs are white and show labels for the U3, R and U5 regions. Other labels are: capsid (CP); integrase (INT); long terminal repeat (LTR); nucleocapsid (NC); polypurine tract (PPT); primer binding site (PBS); protease (PR); reverse transcriptase (RT); ribonuclease H (RH); surface (SU); transmembrane (TM).
Replication
The replication of pseudovirids resembles that of retroviruses and occurs via reverse transcription in the VLP. The cellular tRNA molecule, typically initiator tRNAMet, which is packaged into the VLP, anneals to the viral RNA genome at the PBS complementary to the 3′-end of that tRNA and is used by RT as a primer to start reverse transcription. Once the full-length proviral cDNA is synthetized, it is imported into the nucleus, where it is integrated into a chromosomal target site by INT. The integrated form (equivalent to the retroviral provirus) is transcribed by the host RNA polymerase II to generate a new viral RNA molecule, which is translated to produce the Gag and Pol polyproteins and is packaged into the VLPs to reinitiate the replication cycle.
Taxonomy
Current taxonomy: ictv.global/taxonomy. The family Pseudoviridae belongs to the order Ortervirales [5] and includes the genera Pseudovirus, Hemivirus and Sirevirus. Current classification is based on host tropism, gene content and the length of the tail of the tRNA used as a primer to initiate reverse transcription. Hemiviruses differ from members of the genus Pseudovirus in that they use only a short segment of the tRNA. By contrast, sireviruses are restricted to plants and mostly encode a protein equivalent to retroviral Env downstream of the gag-pol gene [4]. Further updates and revisions in the genus demarcation of pseudovirids are expected to be based on phylogenetic criteria as described in the full ICTV Report (see Resources).
Resources
Full ICTV Report on the family Pseudoviridae: ictv.global/report/pseudoviridae.
Gypsy Database (GyDB) devoted to viruses and mobile genetic elements: http://gydb.org.
Funding information
B.S. was supported by the pre-doctoral research fellowship from Industrial Doctorates of MINECO (Grant 659 DI-17-09134). Production of this Profile, the ICTV Report, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Andrew J. Davison, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, Donald B. Smith, Richard J. Orton and Balázs Harrach.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CP, capsid; INT, integrase; LTR, long terminal repeat; NC, nucleocapsid; PBS, primer binding site; PPT, polypurine tract; PR, protease; RH, ribonuclease H; RT, reverse transcriptase; VLP, virus-like particles.
References
- 1.Krupovic M, Koonin EV. Homologous capsid proteins testify to the common ancestry of retroviruses, caulimoviruses, pseudoviruses, and metaviruses. J Virol. 2017;91:e00210-17. doi: 10.1128/JVI.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns NR, Saibil HR, White NS, Pardon JF, Timmins PA, et al. Symmetry, flexibility and permeability in the structure of yeast retrotransposon virus-like particles. EMBO J. 1992;11:1155–1164. doi: 10.1002/j.1460-2075.1992.tb05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer KJ, Tichelaar W, Myers N, Burns NR, Butcher SJ, et al. Cryo-electron microscopy structure of yeast Ty retrotransposon virus-like particles. J Virol. 1997;71:6863–6868. doi: 10.1128/JVI.71.9.6863-6868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson-Burch BD, Wright DA, Laten HM, Voytas DF. Retroviruses in plants? Trends Genet. 2000;16:151–152. doi: 10.1016/S0168-9525(00)01981-8. [DOI] [PubMed] [Google Scholar]
- 5.Krupovic M, Blomberg J, Coffin JM, Dasgupta I, Fan H, et al. Ortervirales: new virus order unifying five families of reverse-transcribing viruses. J Virol. 2018;92:e00515-18. doi: 10.1128/JVI.00515-18. [DOI] [PMC free article] [PubMed] [Google Scholar]